Abstract
RecQ-like helicases are a highly conserved protein family that functions during DNA repair and, when mutated in humans, is associated with cancer and/or premature aging syndromes. The budding yeast RecQ-like helicase Sgs1 has important functions in double-strand break (DSB) repair of exogenously induced breaks, as well as those that arise endogenously, for example during DNA replication. To further investigate Sgs1’s regulation, we analyzed the subcellular localization of a fluorescent fusion of Sgs1 upon DNA damage. Consistent with a role in DSB repair, Sgs1 recruitment into nuclear foci in asynchronous cultures increases after ionizing radiation (IR) and after exposure to the alkylating agent methyl methanesulfonate (MMS). Yet, despite the importance of Sgs1 in replicative damage repair and in contrast to its elevated protein levels during S-phase, we find that the number of Sgs1 foci decreases upon nucleotide pool depletion by hydroxyurea (HU) treatment and that this negative regulation depends on the intra S-phase checkpoint kinase Mec1. Importantly, we identify the SUMO-targeted ubiquitin ligase (STUbL) complex Slx5-Slx8 as a negative regulator of Sgs1 foci, both spontaneously and upon replicative damage. Slx5-Slx8 regulation of Sgs1 foci is likely conserved in eukaryotes, since expression of the mammalian Slx5-Slx8 functional homologue, RNF4, restores Sgs1 focus number in slx8 cells and furthermore, knockdown of RNF4 leads to more BLM foci in U-2 OS cells. Our results point to a model where RecQ-like helicase subcellular localization is regulated by STUbLs in response to DNA damage, presumably to prevent illegitimate recombination events.
Keywords: Sgs1, BLM, STUbL, Slx5-Slx8, RNF4
1. Introduction
The genome is constantly exposed to DNA damage induced by environmental stress or endogenously through cellular metabolism. Preserving genome stability through accurate DNA repair is a crucial cellular function, where genome instability is associated with cancer predisposition and aging in humans [1]. One family of highly conserved proteins that is critical for error-free repair of DNA damage is the homologs of the bacterial RecQ DNA helicase [1, 2]. While bacteria express one helicase, RecQ, five genes have been identified in humans- BLM, WRN, RECQ1, RECQ4, and RECQ5 [1, 2]. Importantly, mutations in three of the human genes (BLM, WRN, and RECQ4) lead to severe heritable diseases, namely Bloom, Werner, and Rothmund-Thomson syndromes, respectively. Although they are distinct diseases, in common to all three syndromes are genetic instability and an increased cancer predisposition [1]. S. cerevisiae encodes two RecQ-like helicases, Sgs1 and Hrq1. Hrq1, most similar to metazoan RECQ4, was recently identified to be a member of the RecQ family [3, 4] and is also involved in the maintenance of genome stability [5]. The more characterized Sgs1 is considered most homologous to mammalian BLM [6-8], and functions in multiple processes that require unwinding of double-stranded DNA, such as DSB repair by homologous recombination (HR), telomere maintenance, and replication [1, 2].
Replication stress activates the intra S-phase checkpoint to prevent late origin firing, HR, and premature entry into mitosis, as well as inducing the expression of specialized proteins. In budding yeast, stalled replication forks with increased amounts of exposed single stranded DNA activate the Mec1 (mammalian ATR) dependent pathway of the checkpoint, ultimately promoting replication fork stabilization and DNA repair. DSBs that occur during S-phase on the other hand activate the Tel1 (mammalian ATM) mediated checkpoint pathway [Reviewed in [9-12]]. Consequently, mutants of checkpoint pathways accumulate aberrant replication intermediates [13-15]. Both Sgs1 and BLM mutant cells are hyper-sensitive to agents that interfere with replication, such as hydroxyurea (HU) [16, 17], and the respective proteins are found at stalled replication forks, as well as unperturbed forks in the case of Sgs1 [16, 17]. Sgs1 is required to effectively stabilize polymerases ε and α at stalled replication forks and may influence the stability of the entire replication complex [17-19]. One way to regulate RecQ-like helicases upon DNA damage is through their subcellular localization. For example, BLM is normally localized in PML bodies, but upon replicative damage BLM is SUMOylated and subsequently re-localized into nuclear DNA damage foci [20-22]. Although the yeast Sgs1 protein can form a nuclear focus [19, 23], it remains unknown if changes in subcellular localization of Sgs1 foci occur upon DNA damage.
Many genome-wide genetic screens have been performed to identify genes or pathways that functionally interact with Sgs1 [24-27]. A plasmid based synthetic lethality screen conducted in the Brill lab identified six genes whose deletions are not viable in an SGS1 null background, which they then termed SLX, synthetic lethal gene X [28]. Among the genes identified were both members of the SUMO-targeted ubiquitin ligase (STUbL) complex Slx5-Slx8. Consistent with the initial screen, disruption of either of these genes in an sgs1Δ cell results in synthetic lethality, however the basis of this genetic interaction is poorly understood. Slx5 and Slx8 form a hetero-dimeric ubiquitin E3 ligase complex, which recognizes and ubiquitinates substrates that have already been modified with SUMO [29-31]. Furthermore, Slx5 and Slx8 promote accurate replication [31, 32], where slx5 and slx8 mutants are highly sensitive to HU and genetically instable [28, 32-34]. A conserved function of STUbLs in maintenance of genome stability is underlined by the fact that depletion of the mammalian homolog RNF4 [35] causes increased sensitivity to DNA damage that requires HR for repair [31, 36, 37] and interferes with the telomeric DNA damage response [38].
To investigate the role of Sgs1 during DNA repair we analyzed a fluorescent fusion of endogenous Sgs1 and monitored its assembly into nuclear foci. Interestingly, after replication fork stalling by treatment with HU, the percentage of cells with an Sgs1 focus is significantly reduced despite up-regulation of overall Sgs1 protein levels. This repression or disassembly of Sgs1 foci depends on the checkpoint kinase Mec1. Consistently, an sgs1 allele, sgs1-D664Δ, that accumulates DNA replication intermediates upon DNA damage, but is proficient in HR [39], is defective in Sgs1 focus regulation, where Sgs1-D664Δ foci are no longer repressed upon replicative damage. Using this sgs1-D664Δ allele, we screened for factors needed for Sgs1’s role in DNA replication and identified a strong negative genetic interaction with deletions of the SUMO-dependent ubiquitin ligase (STUbL) complex, slx5 and slx8. Strikingly, wild-type Sgs1 focus regulation is impaired upon SLX8 disruption both spontaneously and when replication forks are stalled. Importantly, the increased focus number of Sgs1 in slx8Δ is rescued by expression of the mammalian STUbL RNF4, but not a catalytic site mutant. Finally, we find that similar to yeast, the human STUbL RNF4 also negatively regulates BLM focus number in human U-2 OS cells. These results indicate that the role of STUbLs in regulation of RecQ-like helicase nuclear foci is likely conserved. Our results point to a model in which Sgs1 foci are regulated by the Slx5-Slx8 complex, presumably to prevent illegitimate recombination as well as to allow replication fork restart.
2. Material and methods
2.1. Strains, plasmids, and media
Strains and plasmids used are listed in Table S1 and S2 of the supplemental information. With exception of the Y2H strains PJ69-4a and alpha [40], all strains are isogenic to W303 [41]. Media and plates were prepared according to standard protocols, yet with twice the amount of leucine [42]. Yeast transformation and epitope tagging was carried out as described [43, 44]. The YCplac33-SLX8 and pGAD-TOP3 plasmids were constructed using standard molecular cloning techniques by amplifying the ORF plus 500bp of promoter and 200bp of terminator (SLX8) or the ORF (TOP3) from purified yeast genomic DNA. SLX8 was inserted into the yeast shuttle vector YCplac33 [45] using the restriction endonucleases BamHI/ SalI and TOP3 into pGAD-C1 [40] using BamHI/ SalI.
2.2. Microscopy
Yeast cultures were grown at room temperature (RT) in SC with adenine (100 mg/ml) and if applicable treated with 100 mM hydroxyurea (Sigma-Aldrich) or 0.033% MMS (Sigma-Aldrich) for 2 or 2.5 hours, respectively. Prior to microscopy 1 ml of yeast culture was harvested (1500 g, 2 min, RT), mixed with an equal volume of liquid 1.4% agarose (in SC medium), and mounted on slides. Images were acquired using a Nikon TiE inverted live cell system with a 100× oil immersion objective (1.45 NA), a Photometrics HQ2 camera, and motorized Prior Z-stage. Stacks of 11 0.3-μm sections were captured using the following exposure times: differential interference contrast (DIC) (60 ms), YFP–Sgs1 and YFP-Sgs1-D664Δ (2×2 binning, 2 sec), Mcm2-RFP (2×2 binning, 1 sec), Rad52-CFP (1 sec/ 2×2 binning 500 msec). Nikon Elements software (Nikon Instruments) was used for acquisition and analysis. Images were exported from Nikon Elements as 8-bit RGB TIF files and enhanced for contrast and brightness using Photoshop (Adobe Systems Incorporated). All images depicted in one panel were adjusted in identical fashion. For analysis of Sgs1 focus formation, all 11 z-slices of each image (3 μm in total) were analyzed manually for an YFP-Sgs1 focus. To be counted as a focus, the signal had to be visible in at least 2-3 consecutive planes. Single z-slices are depicted in the figures.
For mammalian cell immunofluorescence U-2 OS cells were grown directly on cover slips according to standard procedures. Cells were fixed with 4% Formaldehyde for 30 min, permeabilized with 0.2% Triton X-100 (2 × 5 min), and washed with twice with 0.5% Tween20. BSA (New England Biolabs) was used for blocking (1 h at room temperature) and primary antibody incubation over night at 4°C. Cells were washed with TBS-T (TBS/ 0.2% Tween20) and incubated with secondary antibody for 2 h at room temperature. DNA was stained with DAPI (1 μg/ml) and cover slips mounted using Vectashield (Vector Labs). Image acquisition and analysis as described for yeast using Nikon Elements software (Nikon Instruments) and Photoshop (Adobe Systems Incorporated), using 7 (BLM) or 11 (BML/PML) z-steps of 0.5μm each. DIC (60 ms), Cy3 (BLM: 30/70 ms), GFP (PML/γH2AX: 30/70ms). Depicted images are maximum intensity projections of all planes. All images shown in one panel were processed in an identical fashion. Co-localization was assessed by visual analysis of BLM foci previously marked in the z-projections for co-localization with PML in multiple z-stack planes.
2.3. Western blots
Yeast whole-cell lysates were prepared by TCA precipitation as described [43]. Mammalian lysates were prepared by incubation of cells with RIPA buffer (150 mM NaCl, 50 mM Tris-HCl pH 7.2, 1% Triton X-100, 0.5 Na-deoxycholate, 0.1% SDS, 1mM EDTA, complete protease inhibitor cocktail (Roche), PhosSTOP phosphatase inhibitors (Roche), 2 mM PMSF, 20 mM NEM, 10 mM NaF) and addition of Benzonase (Novagen Millipore). Films of Western blot exposures were scanned and adjusted for contrast and brightness using Photoshop (Adobe Systems Incorporated).
2.4. Antibodies and dilutions
The following antibodies were used at the indicated dilutions: Myc (mouse monoclonal, Santa Cruz Biotechnology) 1:500 (WB); Clb2 (rabbit polyclonal, Santa Cruz Biotechnology) 1:2000 (WB); BLM (rabbit polyclonal, Bethyl Laboratories) 1:500 (IF); PML (mouse monoclonal, Santa Cruz Biotechnology) 1:500 (IF); γH2AX pS139 (mouse monoclonal, EMD Millipore) 1:500 (IF); RNF4 (rabbit polyclonal, gift of J. Palvimo) 1:1000 (WB); γ-tubulin (mouse monoclonal, Sigma Aldrich) 1:5000 (WB); Shp1 (rabbit polyclonal, gift of A. Buchberger) 1:10000 (WB); Anti-Rabbit Cy3 (Jackson Immunoresearch) 1:500 (IF); Anti-Mouse Alexis-488 (Invitrogen Molecular Probes) 1:500 (IF); Anti-Mouse HRP (GE Healthcare/ Jackson Immunoresearch); Anti-Rabbit HRP (GE Healthcare/ Jackson Immunoresearch).
2.5. Spore size analysis
As described in [46] by analyzing the size of individual spore colonies from tetrads using the Dissection Reader software developed by John Dittmar (Rothstein Lab) for use with ImageJ. The Dissection Reader script can be downloaded (http://www.rothsteinlab.com/tools/apps/dissection_reader). An average of 5-9 spores was analyzed for each genotype, and standard deviations were calculated. Wild-type spores were set to one, and the mutants were analyzed relative to wild-type growth.
2.6. Yeast-2-hybrid
The yeast strain PJ-69a [40] was transformed with the indicated plasmids and plated on medium lacking tryptophan and leucine. To test for an interaction, cultures of the co-transformants were grown in selective medium and a volume corresponding to 0.5 OD600 was plated on the respective plates. Plates were incubated at 30°C, imaged after 3-5 days by scanning, and processed using Photoshop (Adobe Systems Incorporated).
2.7. Cell cycle synchronization
alpha factor/ nocodazole arrest for microscopy
bar1Δ strains were grown as indicated for microscopy and arrested for three hours at room temperature in SC medium with adenine containing 2 μM alpha factor (gift of A. Buchberger) or 15 μg/ml nocodazole (Assay Designs, Fisher Scientific).
alpha factor arrest/ release
Overnight cultures of wild-type and mutant strains were diluted to an OD600 of 0.1 in 50 ml YPD. The cultures were then grown at room temperature for approximately four hours until reaching an OD600 of 0.3 - 0.35. 10 μM alpha factor (gift of A. Buchberger) in DMSO were added, and the cells were allowed to arrest for three hours at 25°C. Directly before addition of alpha factor, a control sample from the asynchronous culture was collected, and the pellet was frozen in liquid nitrogen. The cultures were then washed two times with equal volumes of YPD and resuspended to a final OD600 of approximately 0.5 in YPD. The released cultures were allowed to continue growing and an equal amount of cells was collected at each time-point, pelleted, and frozen in liquid nitrogen. Whole cell lysates of each time-point were analyzed by Western blot.
2.8. Cycloheximide (CHX) chase
CHX chase experiments were performed essentially as described [47]. Briefly, yeast strains were grown to logarithmic phase in YPD at room temperature. Translation was stopped by addition of cycloheximide (Sigma-Aldrich) to a final concentration of 0.5 mg/ml and cultures shifted to 30°C. Equal cell numbers were harvested for each time-point and TCA protein extracts prepared as described [43]. Sgs1-9myc was detected by Western blot against myc and quantification of scanned films carried out using NIH ImageJ software [48].
2.9. Cell culture and siRNA transfection
U-2 OS cells (ATCC) were cultured in DMEM/ 10% FBS at 5% CO2. siRNA was transfected using RNAimax (Invitrogen) according to the manufacturer’s protocol 48 h prior to analysis. siRNA pools (Thermo Fisher, Dharmacon) were used for both the non-targeting control (siGENOME non-targeting siRNA Pool #1; D-001206-12-05) and RNF4 (siGENOME Human RNF4 siRNA SMARTpool; M-006557-03-0005).
2.10. Real-Time PCR
U-2 OS cells were grown and treated as for immunofluorescence but trypsinized and harvested. 8*10^4 cells were resuspended in PBS, lysed and reverse transcription was performed using the Cells-to-CT kit (Applied Biosystems). Comparative CT experiments were performed according to the manufacturer’s protocol using TaqMan Fast Universal No AmpErase UNG PCR Master Mix and TaqMan gene expression assays for RNF4 and actin (all Applied Biosystems).
3. Results
3.1. Sgs1 forms nuclear foci that are negatively regulated by replication fork stalling
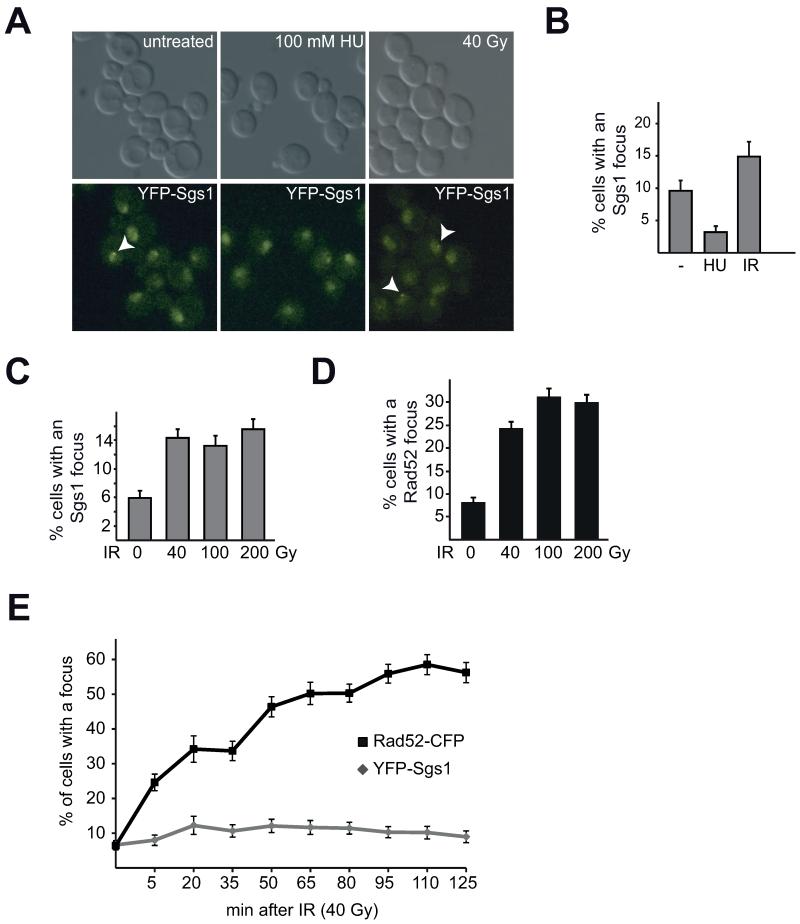
To analyze the role of the budding yeast RecQ-like helicase Sgs1, we created an N-terminal yellow fluorescent protein fusion at the endogenous SGS1 locus (YFP-Sgs1) and monitored YFP-Sgs1 expressing cells for formation of spontaneous nuclear foci. Consistent with previous reports [19, 23] live-cell fluorescent imaging reveals that Sgs1 forms nuclear foci in approximately 6-10% of untreated asynchronous cells depending upon the genetic background (Figure 1A, B). To further characterize the nature of nuclear Sgs1 foci, we analyzed Sgs1 focus formation upon different DNA damaging conditions. To do so, we first incubated YFP-Sgs1 expressing cultures with hydroxyurea (HU) for two hours, which should primarily stall but not collapse replication forks [49, 50]. Surprisingly, replication fork stalling significantly reduces the number of cells with an Sgs1 focus from approximately 10% to 3% (Figure 1A, B; p< 0.01). To determine if Sgs1 focus repression generally occurs after DNA damage, we then used 40 Gy γ-irradiation (IR) to induce multiple DSBs (approximately 4-7 DSBs per cell [51]). In striking contrast to treatment with HU, irradiation leads to an increase in the number of cells with an Sgs1 focus (Figure 1A, B; p < 0.01). Importantly, unlike Rad52 foci, increasing the γ-irradiation dose, and therefore the number of DSBs per cell, does not result in an increase in the percentage of cells with an Sgs1 focus (Figure 1C, D). While Sgs1 foci form within 20 minutes after 40 Gy γ-irradiation (6.7% untreated to 12.3% after 20 min, p < 0.05), they do not significantly change over time, which is in contrast to Rad52 foci, which increase over the entire time-course (Figure 1E). Since we analyzed an asynchronous cell population, these results suggest that Sgs1 foci may not form during every cell cycle stage.
Fig. 1. Sgs1 forms nuclear foci that are regulated by hydroxyurea (HU).
A) Asynchronous cultures of YFP-Sgs1 were treated as indicated (100 mM HU for 2 hours or 40 Gy IR and analyzed after 30 min) and analyzed by live-cell microscopy. Representative z-plane images for YFP-Sgs1 are shown, each arrow indicates a focus. B) Quantification of A). Graph depicts the average percentage of cells with a focus and standard error of the mean (SEM) for each condition (n>100 per condition). C) Sgs1 foci do not increase with increasing amounts of IR. Analysis of a strain expressing YFP-Sgs1 and Rad52-CFP without and 10- 15 min after irradiation with the indicated doses. Graph depicts percentage of cells with an YFP-Sgs1 focus and SEM [n(untreated)= 536, n(40 Gy)= 843, n(100 Gy)= 567, n(200 Gy)= 718 cells; p>0.1 for irradiated samples]. D) Rad52 focus number increases with increasing IR doses. Analysis of C) for Rad52 foci (p<0.025 for irradiated samples). E) Sgs1 foci form early after irradiation and do not increase over time. Graph depicts the percentage of cells with an YFP-Sgs1 or Rad52-CFP focus at the indicated time-points with the SEM [n>150 cells per time-point].
3.2. Sgs1 foci are cell cycle regulated
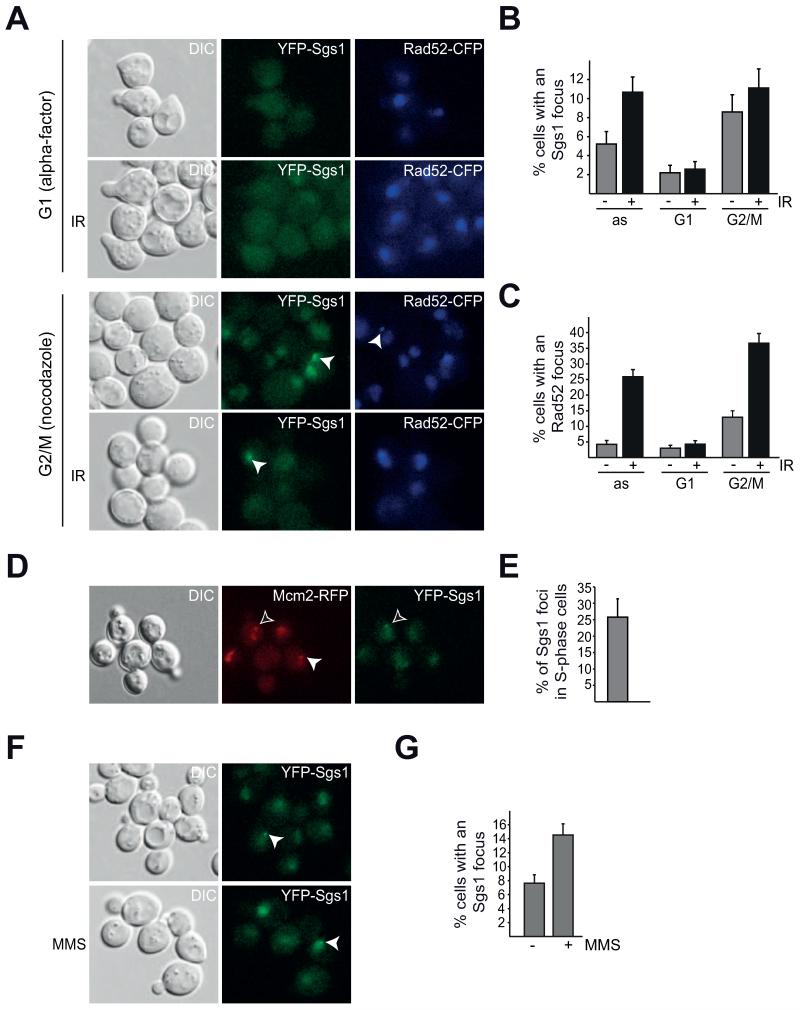
To address the possibility that YFP-Sgs1 foci are cell cycle regulated, we arrested cells expressing YFP-Sgs1 and Rad52-CFP in G1 using the mating pheromone alpha factor and in G2/M by addition of the spindle poison nocodazole. In addition to determining the percentage of untreated cells with an Sgs1 or Rad52 focus, we also analyzed the effect of irradiation at each cell cycle stage (Figure 2A, B, C). Interestingly, we find that the YFP-Sgs1 signal in G1-arrested cells is weak overall, with no strong detectable nuclear staining (Figure 2A). Furthermore, the percentage of cells with an Sgs1 focus is significantly decreased in G1 and does not increase following IR treatment (Figure 2B, p > 0.1). These results are consistent with the notion that HR is repressed during G1, since a homologous template is unavailable for repair [52]. Accordingly, Rad52 foci are also repressed during a G1 arrest and similarly do not respond to irradiation (Figure 2A, C; p > 0.1). Upon nocodazole treatment, Sgs1 foci do not significantly increase after irradiation during G2/M (Figure 2B, p > 0.1), while Rad52 foci do (Figure 2C, p < 0.005). These results suggest that the majority of Sgs1 focus regulation likely occurs during S/G2 and that the increase in Sgs1 foci observed upon IR treatment of asynchronous cultures (Figure 1A, B) may be a response to DSB induction as well as an effect of an altered cell cycle profile after irradiation.
Fig. 2. Sgs1 foci are cell cycle regulated.
A) Sgs1 focus formation is cell cycle dependent. B) Quantification of Sgs1 foci in A. C) Quantification of Rad52 foci in A. D) Spontaneous Sgs1 foci are present during S-phase. A YFP-Sgs1 Mcm2-RFP strain was analyzed as in Fig.1), the filled arrows in the middle and right panel mark S-phase nuclei, and YFP-Sgs1 foci, respectively. The black arrow points to an early S-phase cell with an YFP-Sgs1 focus. E) Percentage of YFP-Sgs1 cells with a Sgs1 focus that are also in early S-phase, marked by nuclear Mcm2-RFP puncta [n(foci total)=62] in D) depicted with SEM. F) Sgs1 focus formation is induced after exposure of a YFP-Sgs1 Rad52-CFP to 0.033% MMS for 2.5 hours. G) Quantification of F) depicted with SEM [n(untreated)= 516, n(MMS)= 502; p<0.005]
However, we find that Sgs1 foci are repressed upon HU treatment (Figure 1B). Since HU depletes cells of dNTP pools resulting in an S-phase arrest [53], we speculated that the reduction of Sgs1 foci observed upon HU treatment may be due to replication fork stalling and not to an inability of Sgs1 to form foci during S-phase. To examine if YFP-Sgs1 forms foci during S-phase, we identified S-phase cells by live-cell imaging of a strain expressing a fluorescently tagged variant of the essential replication protein Mcm2. This strain, where Mcm2-RFP is the only source of Mcm2, is not slow growing nor sensitive to HU or methyl methanesulfonate (MMS) (Supplemental Figure S1), indicating that the fusion protein is likely functional. Mcm2-RFP localizes to the nucleus and forms small nuclear puncta only during S-phase (Figure 2D). Of the YFP-Sgs1 foci that form, approximately 25% of them are in early S-phase cells, as marked by nuclear punctate Mcm2-RFP (Figure 2D, E), thus demonstrating that spontaneous Sgs1 foci form during S-phase. Therefore, repression of Sgs1 focus number during HU treatment is likely specific to replication fork stalling.
To further support our hypothesis that Sgs1 focus repression during HU is due to replication fork stalling and not a response to all types of replicative stress, we analyzed Sgs1 focus formation after addition of the alkylating agent MMS (Figure 2F), which induces DNA damage that blocks replication fork progression and must be bypassed or repaired to avoid fork collapse and DSB formation [54]. Interestingly, we observe a significant increase in cells with an Sgs1 focus (Figure 2F, G, p < 0.005), which is similar to IR treatment (Figure 1). These results indicate that repression of Sgs1 foci following HU is specific for nucleotide pool depletion and not to S-phase.
3.3. Sgs1 focus repression during replication fork stalling requires the intra S-phase checkpoint kinase Mec1
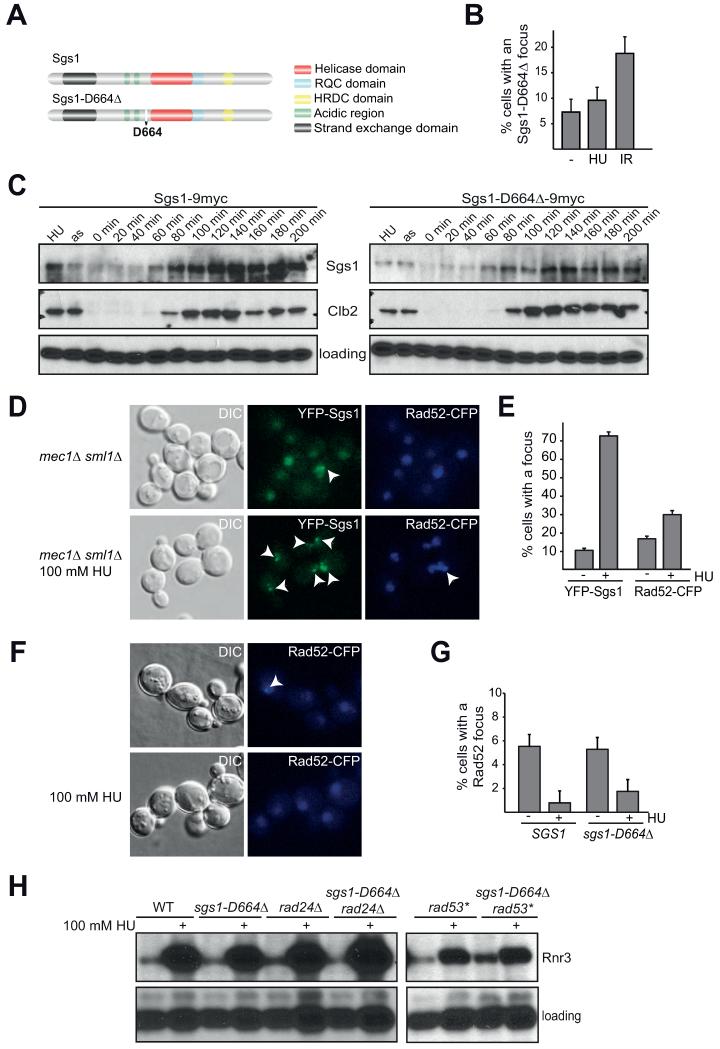
Our analysis of YFP-Sgs1 suggests that regulation of Sgs1 focus formation is tightly linked to DNA replication and the repair of replicative damage. We previously characterized the separation-of-function allele sgs1-D664Δ, which is specifically defective in replication, but not HR [39, 46]. Thus we used this allele to further analyze the function and regulation of Sgs1 nuclear foci. sgs1-D664Δ is a complete deletion of Asp 664, located between the acidic regions and the helicase domain (Figure 3A). Since upon replicative damage, sgs1-D664Δ cells accumulate replication intermediates as assayed by two-dimensional gel electrophoresis (X-structures) [39], we asked whether regulation of YFP-Sgs1-D664Δ focus formation is altered. Intriguingly, Sgs1-D664Δ foci are not regulated upon HU treatment, but remain approximately constant without a statistically significant change (7.3% vs. 9.6%, p > 0.1; Figure 3B). In contrast to HU, the number of cells with an Sgs1-D664Δ focus increases similarly to a wild-type strain after γ-irradiation (Figure 3B; p < 0.01), suggesting that the regulation of Sgs1 foci upon HU treatment differs from focus induction in response to IR. Additionally, analysis of the sgs1-D664Δ allele suggests that repression of Sgs1 foci may be required for its role in preventing HR at illegitimate substrates such as a stalled replication fork. In this scenario, premature HR could cause the accumulation of aberrant replication intermediates, which is consistent with the increased X-structures observed in the sgs1-D664Δ allele [39].
Fig. 3. The intra S-phase checkpoint is required for Sgs1 focus regulation.
A) Schematic model of Sgs1 illustrating the position of D664, which is deleted in sgs1-D664Δ. B) Sgs1-D664Δ foci are not repressed by replication fork stalling. Asynchronous YFP-Sgs1-D664Δ were treated as indicated (100 mM HU for 2 hours or 40 Gy IR and analyzed after 30 min) and analyzed by live-cell microscopy as in Fig.1A). Graph depicts the percentage of cells with a focus with SEM. C) Sgs1 protein levels fluctuate during the cell cycle. Sgs1-9myc and Sgs1-D664Δ-9myc strains were arrested with α-factor, released, and samples taken every 20 min. The first two lanes are protein extracted from HU arrested and asynchronous cells. Western blot against Sgs1/Sgs1-D664Δ-9myc (myc), Clb2, Shp1 (loading). D) Sgs1 focus repression upon HU treatment requires Mec1. YFP-Sgs1 Rad52-CFP mec1Δ sml1Δ bar1Δ strains were analyzed without treatment or after incubation with 100 mM HU for 2 hours. Representative z-planes are shown. E) Quantification of the percentage of cells with one or more Sgs1 or Rad52 foci in D) with SEM [n(untreated) = 716, n(HU)= 438 cells; p (untreated to HU)< 0.005 for Sgs1 and Rad52]. F) Rad52 foci are repressed in sgs1-D664Δ upon treatment with HU. Cultures of Rad52-CFP and sgs1-D664Δ Rad52-CFP were directly analyzed or treated with 100 mM HU for 2 hours and analyzed by live-cell microscopy. G) Quantification of the percentage of cells with Rad52-CFP foci in F) with the SEM plotted [n (−/+HU)= 271/254 (SGS1), n (−/+HU)= 265/220 (sgs1-D664Δ)]; p < 0.005 (WT untreated to HU), p < 0.05 (sgs1-D664Δ untreated to HU); p > 0.1 (WT to sgs1-D664Δ, both conditions)]. H) Analysis of Rnr3 expression in different genetic backgrounds. Cultures of the indicated genotypes were grown to log phase and divided into the untreated control or treated with 100 mM HU for 2 hours. Whole cell lysates were analyzed by Western blot against HA (Rnr3-3HA) and a loading control (Shp1). rad53* is the checkpoint-deficient mutant rad53-K227A used as a positive control.
One possibility is that the reduced Sgs1 foci observed after HU treatment could be the result of a reduction in overall Sgs1 protein levels. To test this, we released G1-arrested 9myc-tagged Sgs1 or Sgs1-D664Δ cells into medium containing 100 mM HU and analyzed Sgs1 protein levels by Western blot. Strikingly, Sgs1 levels were significantly higher after HU treatment than in an asynchronous (as) culture (Figure 3C; compare the first two lanes in the Sgs1-9myc blot), thus indicating that the reduction of Sgs1 foci by HU is not merely due to a decrease in overall protein levels. Importantly, this finding suggests that an active mechanism to repress focus formation or to disassemble foci when replication forks are stalled likely exists.
Sgs1 protein levels fluctuate during the cell cycle and peak during S-phase [17, 19]. To determine if Sgs1-D664Δ protein levels are mis-regulated during the cell cycle, and thereby possibly cause focus mis-regulation, we synchronized 9myc tagged Sgs1 or Sgs1-D664Δ expressing strains in G1 with alpha factor and monitored protein levels of Sgs1 by Western blot after alpha factor release. Sgs1 protein levels are indeed cell cycle regulated (Figure 3C) and even though Sgs1-D664Δ protein levels are consistently lower, they do fluctuate similar to wild-type Sgs1, indicating that cell cycle regulation of its protein levels is likely not the cause of the phenotypic differences of sgs1-D664Δ cells.
One critical mechanism to ensure proper replication is the intra S-phase or replication checkpoint [9-12]. Treatment of cells with HU causes replication fork stalling [53] and checkpoint activation [15, 55]. Mec1 is a central kinase in the intra S-phase checkpoint that responds to different types of DNA damage, including replication stress. Upon replicative damage Mec1 activates the downstream checkpoint kinases Chk1 and Rad53 (reviewed in [10]). Additionally, a role for Mec1 in stabilizing the polymerase at a stalled replication fork has been proposed [10, 17]. Thus, the intra S-phase checkpoint could be responsible for repression of Sgs1 foci. To address this possibility, we analyzed Sgs1 focus formation upon HU treatment in the checkpoint mutant background mec1Δ sml1Δ (Figure 3D, E). In contrast to a wild-type cell, where Sgs1 foci are repressed upon HU treatment (Figure 1B), Sgs1 foci strikingly increase to approximately 70% in mec1Δ sml1Δ cells when HU treated (Figure 3E). Furthermore we observe multiple Sgs1 foci per cell in the checkpoint mutant (Figure 3D). Therefore, Sgs1 focus regulation during HU depends on the intra S-phase checkpoint kinase Mec1.
Since we find that repression of Sgs1 foci depends on a functional intra S-phase checkpoint and Sgs1-D664Δ foci are not repressed by HU treatment, we next analyzed checkpoint signaling in sgs1-D664Δ. To do this, we first monitored Rad52 focus repression (Figure 3F, G), indicative of repressed HR [56, 57], and then subsequently we analyzed the expression of a downstream checkpoint-target gene, RNR3 (Figure 3H). Activation of the intra S-phase checkpoint effector kinase Rad53 in budding yeast can occur through two parallel pathways: via Sgs1 or Rad24 [18, 19]. Activated Rad53 phosphorylates multiple downstream targets leading to a cell cycle arrest, inhibition of late origin firing, inhibition of HR, and the elevated expression of downstream genes such as RNR3 [9]. Additionally, Rad52 foci are repressed upon intra S-phase checkpoint activation [56]. To test if checkpoint activation is functional in sgs1-D664Δ, we analyzed Rad52 focus formation in SGS1 and sgs1-D664Δ cells treated with HU [Figure 3F, G; p < 0.005 (WT) and p < 0.05 (sgs1-D664Δ)], and find that Rad52 foci are similarly repressed in both strain backgrounds (Figure 3F, G; p > 0.1). Since a checkpoint defect in sgs1 is reported to be only clearly visible upon additional deletion of RAD24 [58], we analyzed the double mutant of sgs1-D664Δ rad24Δ for Rnr3 expression (Figure 3H). Similar to the single mutant sgs1-D664Δ, checkpoint activation, as determined by the strong increase in Rnr3 protein levels, is functional in sgs1-D664Δ rad24Δ. In contrast the positive control rad53-K227A (rad53*, Figure 3H) is partially defective in checkpoint activation. In summary, these results indicate that the intra S-phase checkpoint of sgs1-D664Δ is largely functional. Furthermore, our results demonstrate that Sgs1 focus regulation depends on the checkpoint kinase Mec1 and occurs independently of or in parallel to Rad52 focus regulation.
3.4. The STUbL complex Slx5-Slx8 genetically interacts with Sgs1
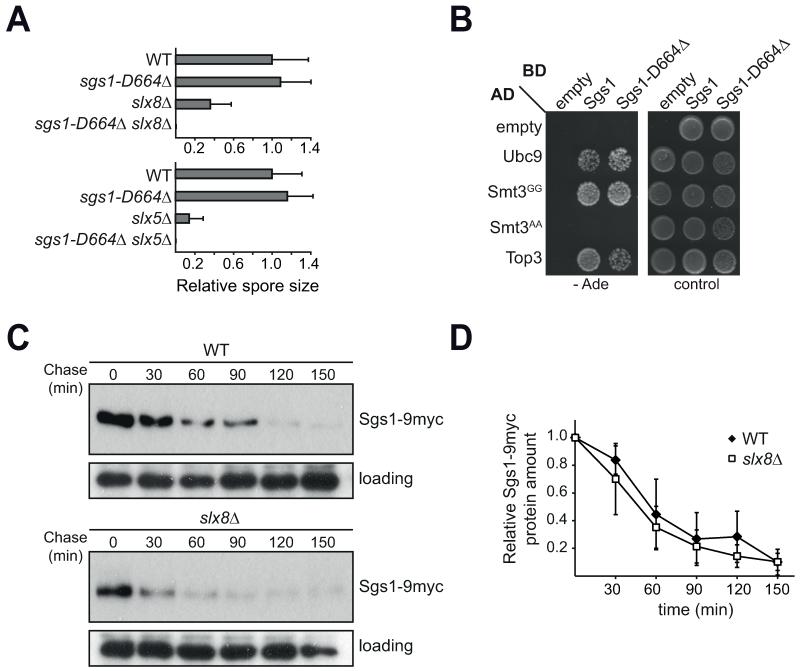
To identify genes that regulate Sgs1 in vivo, we selectively screened known negative genetic interactors of sgs1 for a synthetic sickness or lethal phenotype with sgs1-D664Δ. Only very few genetic interactions of sgs1Δ were fully phenocopied by sgs1-D664Δ, most striking were deletions of either of the two components of the yeast SUMO-targeted ubiquitin ligase (STUbL) complex Slx5-Slx8 (Figure 4A). This suggests that Sgs1 is involved in a SUMO and ubiquitin regulated process. To address the possibility of a physical interaction between Sgs1 and components of the SUMO/ubiquitin pathway, we analyzed Sgs1 and Sgs1-D664Δ in directed Y2H assays with Ubc9, Smt3GG, Smt3AA, and Top3 as a positive control [59] (Figure 4B). Indeed, both Sgs1 as well as the mutant Sgs1-D664Δ strongly interact with the SUMO E2 Ubc9 and the yeast wild-type SUMO Smt3. Interestingly, for both Sgs1 and Sgs1-D664Δ the interaction with SUMO is abolished when a non-conjugatable Smt3AA mutant is used (Figure 4B). This indicates that SUMO conjugation is required for its interaction with Sgs1 in the Y2H assays, either directly through modification of Sgs1 itself or by selective binding of a SUMOylated substrate to Sgs1. Given the central role SUMO plays in DNA repair [60], we wanted to determine if other HR proteins interact with SUMO. Therefore, we analyzed the interaction of several HR factors with Smt3 by Y2H (Supplemental Figure S2). While the positive control Rad52 showed an interaction with Smt3, other HR factors, including Rad55 and Rad59, do not (Supplemental Figure S2). Therefore, not all HR proteins interact with Smt3, suggesting that there is likely specificity for the interaction of Sgs1 with Smt3 and Ubc9 (Figure 4B).
Fig. 4. Sgs1 interacts with the SUMO and ubiquitin systems.
A) Spore size analysis of progeny of sgs1-D66Δ crossed with slx5Δ and slx8Δ. Graph shows relative colony size normalized to wild-type (WT), 5-9 individual spores of each genotype were quantified. B) Y2H of Sgs1 and Sgs1-D664Δ. The denoted proteins fused to either the activation (AD) or binding (BD) domain were expressed in the reporter strain PJ69-4a. Growth on the control plate (SC-Leu-Trp) shows transformation of both plasmids, while growth on the stringent selection plate (SC-Ade) indicates a strong interaction of the two fusion proteins. C) Sgs1-9myc protein stability does not depend on Slx8. Cycloheximide (CHX) chase of Sgs1-9myc in WT and slx8Δ. Whole-cell lysates of each time-point were analyzed by Western blot against Sgs1 (myc) and Shp1 (loading). D) Quantification of B), the Western blot signal of each time-point was normalized to the 0 min value using ImageJ. The mean of three independent experiments with standard deviation is plotted.
Sgs1 is SUMOylated [61] at lysine 621 in vivo and in vitro [62], making it a possible direct substrate of STUbLs. Furthermore, Sgs1 was suggested as a potential ubiquitination substrate in a large-scale screen for yeast proteins that exhibit a large difference in predicted and experimental molecular weight [63]. Ubiquitination of a substrate by STUbLs frequently leads to its degradation [64]. To test if Sgs1 is targeted to the 26S proteasome by the Slx5-Slx8 complex, we monitored protein stability of Sgs1-9myc in wild-type and slx8Δ cells (Figure 4C, D). Sgs1-9myc is indeed degraded over time; however, this degradation does not depend on the presence of Slx8 in untreated asynchronous cultures (Figure 4C, D). While we observe less overall Sgs1 protein in slx8Δ (Figure 4C), most likely due to an altered cell cycle profile, our results of the CHX chase show that SLX8 deletion does not impair degradation of the majority of Sgs1 in untreated asynchronous cultures (Figure 4D).
3.5. Regulation of Sgs1 foci requires the Slx5-Slx8 complex
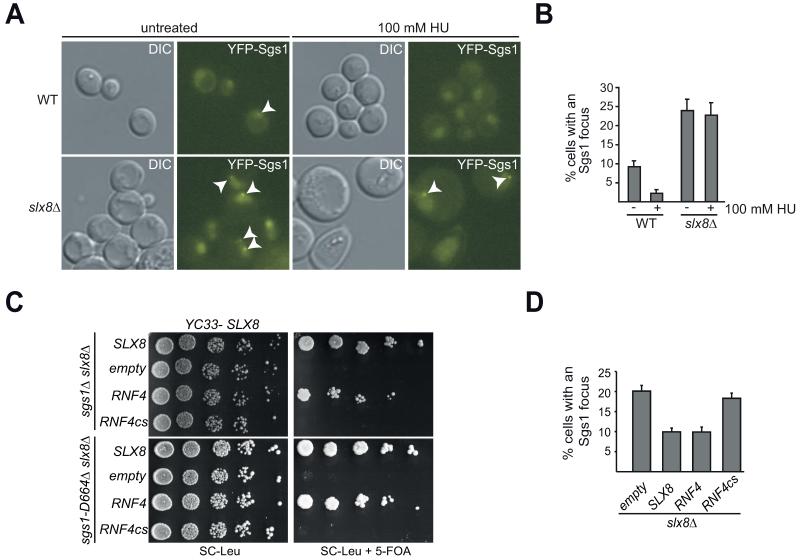
Since the Slx5-Slx8 complex does not regulate the majority of Sgs1 protein levels, we hypothesized that the STUbL complex may affect Sgs1 function by alternative mechanisms, such as the regulation of its recruitment into nuclear foci (Figure 5A, B). It was previously demonstrated that SLX8 disruption leads to an increase in the number of Rad52 foci [65], which is likely linked to the hyper-recombination [32] and increased gross chromosomal rearrangements [33] observed in slx8Δ cells. Consistent with the increase in HR, deletion of SLX8 also results in more cells with a nuclear Sgs1 focus. Importantly, similar to the sgs1-D664Δ background, these Sgs1 foci are neither repressed nor disassembled following HU treatment (Figure 5A, B).
Fig. 5. Sgs1 foci are regulated by the STUbL complex Slx5-Slx8.
A) Sgs1 focus regulation depends on Slx8. Microscopy as in Fig. 1A of YFP-Sgs1 and slx8Δ YFP-Sgs1 without or with 100 mM HU. B) Quantification of A) with SEM plotted. Graph depicts the percentage of cells with an YFP-Sgs1 focus [n (−/+HU)= 423/321 (WT), n (−/+HU)= 213/172 (slx8Δ)]. C) Rescue of synthetic lethality shown in Fig. 3A by the mammalian STUbL RNF4. Serial dilutions of sgs1Δ slx8Δ and sgs1-D664Δ slx8Δ transformed with a rescuing URA3-marked SLX8 plasmid (YC33-SLX8) and the indicated RNF4 plasmids (pRS415 based). Counter-selection against YC33-SLX8 on 5-FOA reveals RNF4 but not RNF4cs rescue of synthetic lethality. D) The mammalian STUbL RNF4 rescues Sgs1 focus number in slx8Δ. Microscopy as in A) of slx8Δ strains transformed with the indicated plasmids. Percentage of cells with an YFP-Sgs1 focus with SEM [n=781 (pRS415), n=1107 (SLX8), n=566 (RNF4), n=885 (RNF4cs)].
To determine if other mutants that exhibit more Rad52 foci display a similar increase in Sgs1 foci, we deleted the DSB repair gene RAD59. While rad59Δ cells display a ten-fold increase in Rad52 foci ([65] and data not shown), there is no significant change in the percentage of cells with an YFP-Sgs1 focus (Supplemental Figure S3). This result does not exclude the possibility that deletion of other DNA repair factors may also alter Sgs1 focus regulation. However, our findings indicate that Slx5-Slx8 likely regulate a distinct sub-pathway of HR which involves Sgs1 foci.
The vertebrate homolog of the Slx5-Slx8 complex, RNF4, performs similar functions in maintaining genome integrity [31, 36, 37]. We analyzed the ability of RNF4 and a catalytically inactive mutant, RNF4cs ([66]; RNF4-C136S, C139S, C177S, C180S), to rescue the synthetic lethality of sgs1Δ slx8Δ and sgs1-D664Δ slx8Δ by using a shuffle-strain approach. As expected [66, 67] expression of mammalian RNF4 under control of a yeast promoter rescues the growth defect of sgs1Δ slx8Δ, while a catalytic site mutant does not (Figure 5C). We find this is also true for sgs1-D664Δ slx8Δ demonstrating that the requirement of the catalytic activity of the STUbL is fully phenocopied by sgs1-D664Δ (Figure 5C). To determine if RNF4 may be performing a similar function with regard to Sgs1 focus formation, we examined Sgs1 foci in slx8Δ cells where mammalian RNF4 is expressed. Importantly, expression of the mammalian STUbL RNF4 restores the overall Sgs1 focus number in slx8Δ cells to wild-type levels (Figure 5D). In contrast, the catalytic site mutant RNF4cs does not rescue YFP-Sgs1 focus number in slx8Δ cells (Figure 5D). This strongly suggests that the E3 ubiquitin ligase activity of the STUbL is required to properly regulate Sgs1 foci. To determine if SUMOylation of Sgs1 is similarly required for focus regulation we analyzed YFP-sgs1-K621R (Supplemental Figure S4), a strain mutated in the major SUMOylation site of Sgs1 [61, 62]. In untreated cultures, focus formation of this mutant is indistinguishable from wild-type Sgs1 (Supplemental Figure S4, percentage of cell with a focus: 10.6% for both Sgs1 and Sgs1-K621R; p > 0.1). Additionally, similar to wild-type Sgs1, the double sgs1-K621R slx8Δ mutant is viable and exhibits a strong increase in YFP-Sgs1-K621R foci (22.5%; p < 0.005). This is consistent with the initial characterization of this sgs1 mutant [62], where the authors showed that the sgs1-K621R allele has a primary defect in telomere maintenance and not in replication or recombination.
3.6. The mammalian STUbL RNF4 affects BLM focus number
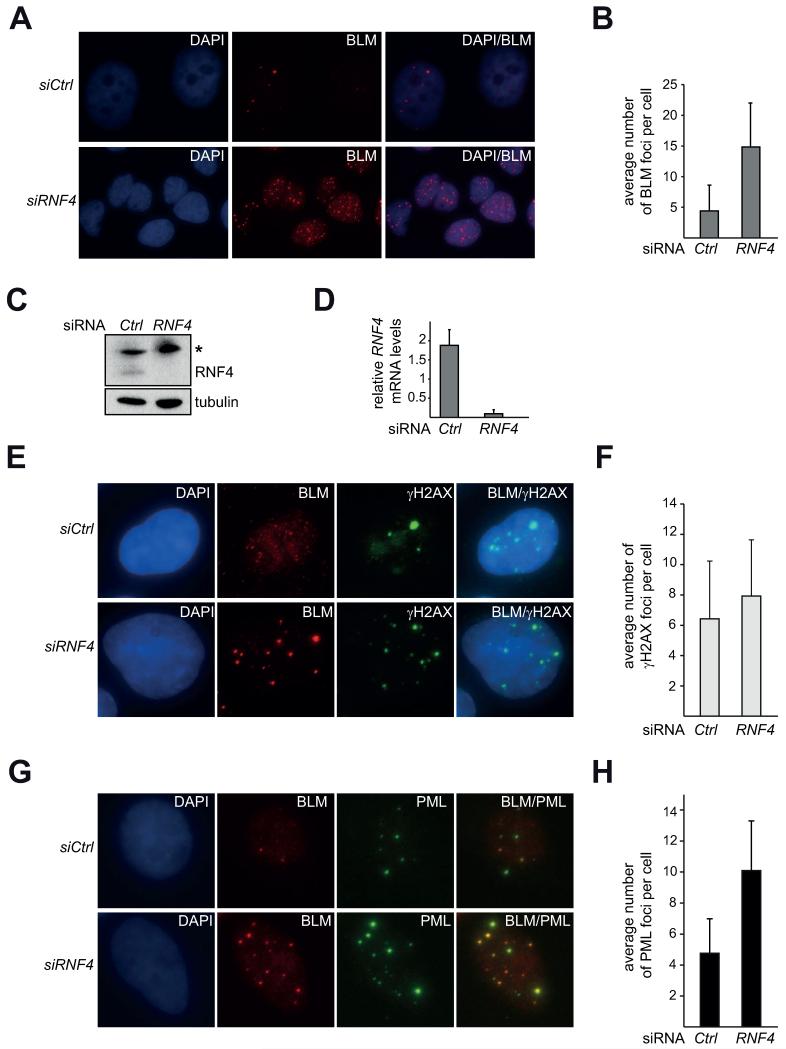
Since we find that expression of mammalian RNF4 rescues Sgs1 nuclear focus regulation when the yeast SLX8 is absent (Figure 5D), we hypothesized that RNF4 might similarly regulate human BLM foci. We focused on the human RecQ helicase homolog BLM for our analysis, as the domain architectures of Sgs1 and BLM proteins are very similar and both primarily function as a complex with their respective Top3 and Rmi1 homologues [1]. Using siRNA, we depleted RNF4 from the human U-2 OS cell line (Figure 6C, D) and strikingly observe that the average number of BLM foci per cell increases by approximately 3.5 fold, when compared to a non-specific siRNA control (Figure 6A, B). These findings suggest that the regulation of RecQ-like helicase foci by STUbLs is likely conserved and that BLM/ Sgs1 focus numbers increase in the absence of the STUbL RNF4/ Slx5-Slx8.
Fig. 6. BLM focus regulation is affected by RNF4 knockdown.
A) Knockdown of RNF4 increases the average number of BLM foci per cell. Z-projections of BLM immunofluorescence (IF) in U-2 OS cells with DAPI stain 48 h after transfection with non-targeting (siCtrl) or siRNA against RNF4 (siRNF4). B) Quantification of A). BLM foci were counted in DAPI/BLM z-projection overlays. Average number of foci per cell with standard deviation [n(cells)=121 (siCtrl), n=120 (siRNF4)]. C), D) RNF4 knockdown efficiency. C) Lysates of cells in A) were analyzed by Western blot against RNF4. γtubulin serves as a loading control. (*) denotes an unspecific cross-reactive band. D) qPCR for RNF4 mRNA-levels. Representative experiment of triplicates normalized to the untreated control. Comparative CT RNF4: actin. E) Immunofluorescence against γH2AX and BLM in U-2 OS cells 48 h after transfection with non-targeting (siCtrl) or siRNA against RNF4 (siRNF4). Representative maximum intensity z-projections are shown. F) Quantification of E). Graph depicts the average number of γH2AX foci upon treatment with non-targeting siRNA (Ctrl) or siRNA against RNF4 (RNF4) with standard deviations plotted [n(cells)= 68 (Ctrl); n(cells)= 59 (RNF4)].G) Immunofluorescence against PML and BLM as in E). H) Quantification of PML-NBs in G) [n(cells)= 101 (Ctrl); n(cells)= 98 (RNF4)].
Since RNF4 functions in DNA repair [36-38], we next wanted to test the possibility that the RNF4 knockdown leads to a severe activation of the DNA damage response. Thus we analyzed cells for changes in γH2AX foci, which strongly increase upon DNA damage such as γ-irradiation. We find that the average number of γH2AX foci per cell is slightly increased in RNF4 knockdown cells, but not to an extent comparable to DNA damaging treatment (Figure 6E, F). This is consistent with the previously reported finding that an RNF4 knockdown only slightly affects the percentage of γH2AX positive untreated cells [37]. Therefore these results suggest that the increase of BLM foci observed when RNF is depleted is likely not solely due to an increased DNA damage response.
BLM [68], as well as other DNA repair proteins such as the MRN complex, RAD51 and RPA [69] co-localize with the promyelocytic protein (PML) at a distinct nuclear structure called PML nuclear body (NB) [70, 71]. Interestingly, PML is modified by SUMO [72] and RNF4 is required for the degradation of SUMOylated PML in vivo [73, 74]. Cellular regulation of PML-NBs is crucial, as PML-NBs are mis-regulated in leukemia and absent in some solid tumors [70]. Furthermore, PML −/− cells display an elevated number of sister chromatid exchanges (SCEs) [68], a characteristic phenotype of Bloom syndrome cells [75], strongly suggesting a physiological importance of BLM recruitment to PML-NBs. We hypothesized that the association of BLM with PML-NBs is an important contributor to genome stability that is dependent upon RNF4. We therefore investigated if RNF4 knockdown causes 1) an alteration in PML-NB number and 2) affects BLM localization to PML-NBs. To address this hypothesis, we performed immunofluorescence against PML and BLM in cells treated with control siRNA or siRNA against RNF4 and find that the number of PML bodies increases approximately two-fold upon RNF4 knockdown (Figure 6G, H). Despite the increase in BLM foci upon RNF4 knockdown, the majority of these foci co-localize with PML bodies (87% +/− 1.1%). Therefore, the increase in BLM foci in mammalian cells may be due to an altered PML body homeostasis upon RNF4 knockdown.
4. Discussion
In this study we provide insights into the cellular regulation of the RecQ-like helicase Sgs1 and identify STUbLs as a regulator of yeast and mammalian RecQ-like helicase recruitment into nuclear foci. Both budding yeast Sgs1 as well as the mammalian RecQ-like helicase BLM function to either promote or repress HR depending on the cell cycle stage during which the DNA damage occurs, as well as the type of damage incurred [1]. One way to regulate protein activity is through its localization, as well as its post-translational modification [22]. Recent studies show that the activity of the mammalian homolog BLM is regulated by different forms of post-translational modification, which in turn affect its localization and protein-protein interactions [20-22, 76]. In contrast, no mechanism of regulation by alteration of subcellular localization has been reported for the yeast RecQ-like helicase Sgs1. Here, we demonstrate that similar to BLM, the subcellular localization of Sgs1 is regulated in response to DNA damage. We find that Sgs1 forms cell cycle-regulated nuclear foci, which are significantly reduced if cells are treated with hydroxyurea (HU), even though overall Sgs1 protein levels are not decreased (Figure 1A, B; Figure 2C). The reduced number of Sgs1 foci upon HU treatment is specific to replication fork stalling and is not observed upon other types of DNA damage such as IR, MMS, or generally during S-phase (Figure 1, Figure 2). We also do not observe a clear co-localization of YFP-Sgs1 with Mcm2-RFP (Figure 2D), which is in line with our hypothesis that Sgs1 foci form predominantly upon DSB-inducing damage and not at unperturbed replicons. Furthermore, the finding that Sgs1-D664Δ foci are not regulated upon HU treatment (Figure 3B), indicates a potential importance of Sgs1 focus regulation during replication fork stalling, as sgs1-D664Δ cells specifically exhibit replication defects [39]. Importantly, we find the STUbL complex Slx5-Slx8 is a negative regulator of Sgs1 focus number. The effect of STUbLs on RecQ-like helicases is likely conserved, as expression of mammalian RNF4 in a yeast slx8Δ mutant restores Sgs1 focus number to wild-type levels. Intriguingly, this rescue depends on the catalytic activity of RNF4, indicating that ubiquitination is likely required for regulation of Sgs1.
DNA damage during S-phase can cause a replication fork to either stall, stall and then subsequently collapse, or to directly collapse (Figure 7). Treatment of cells with HU depletes nucleotide pools and causes replication fork stalling, while at the same time activating the intra S-phase checkpoint. Upon checkpoint activation the stalled polymerase is stabilized by a mechanism partly dependent on Sgs1 [77] and allows replication fork re-start. In this situation, HR is repressed to prevent illegitimate recombination events (Figure 7). Our results show that Sgs1 foci are repressed during HU treatment (Figure 1A, B), even though Sgs1 protein levels are elevated in S-phase (Figure 3C), leading us to hypothesize that there is an active mechanism to prevent focus formation or to promote focus disassembly upon replication fork stalling. If recombination at a stalled fork is not properly inhibited, this leads to increased or aberrant HR, likely causing the accumulation of illegitimate replication intermediates such as X-structures [13, 61, 78]. Interestingly, we find that the YFP-Sgs1-D664Δ focus number is not decreased upon HU treatment, while the regulation of Sgs1-D664Δ foci in response to IR remains intact (Figure 3B). Previously we demonstrated that the sgs1-D664Δ mutant exhibits a defect in replication, which is marked by an increased accumulation of X-structures after MMS treatment [39]. Therefore, we propose a functional link between proper processing of stalled replication forks, likely inhibiting HR, and the repression of Sgs1 foci (Figure 7). Collectively these results point us to a model where HR is actively repressed at a stalled replication fork to prevent illegitimate recombination. In contrast, the HR machinery would be recruited when a fork collapses to enable repair and re-start. This model is further supported by our finding that Sgs1 foci increase after irradiation and MMS treatment, which both can cause DSBs that require HR for repair (Figure 7). Interestingly, replication stress can also lead to sister-chromatid bridging that causes the formation of different types of anaphase bridges [79]: chromatin bridges and ultra-fine bridges (UFBs). BLM localizes to UFBs and Bloom syndrome (BS) cells contain increased numbers of anaphase bridges [80]. Very recently similar structures were identified in budding yeast. Sgs1 localizes to UFBs and sgs1Δ mutants exhibit increased numbers of chromatin bridges [81]. Thus, given the defects of sgs1-D664Δ in the resolution of replication intermediates and our hypothesis that HR at stalled replication forks may not be properly repressed in this mutant, an increased incidence of chromatin bridges in anaphase is possible.
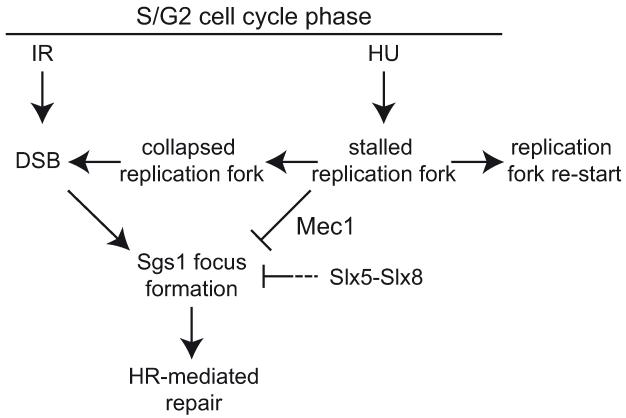
Fig. 7. Model of Sgs1 focus regulation with respect to the types of DNA damage incurred.
Treatment with hydroxyurea (HU) stalls replication forks, which can either re-start or collapse. During replication fork stalling, Sgs1 focus formation is repressed dependent on the intra S-phase checkpoint kinase Mec1. If replication forks collapse, or cells are irradiated (IR) or treated with methyl methanesulfonate (MMS), double-strand breaks (DSB) form. DSBs cause or promote the formation of Sgs1 foci and repair by homologous recombination (HR). The Slx5-Slx8 complex negatively affects Sgs1 focus formation either by inhibiting their formation or promoting their disassembly, perhaps to regulate illegitimate recombination events.
Furthermore, we find that Sgs1 foci are regulated upon intra S-phase checkpoint activation. Like Sgs1, formation of spontaneous Rad52 foci is inhibited during replication fork stalling by HU [56, 57, 82]. Inhibition of the central checkpoint kinases Mec1 and Tel1 restores Rad52 focus formation during an HU-induced S-phase arrest, indicating that the repression of Rad52 foci depends on intra S-phase checkpoint signaling, but not Rad53 activity [56]. Similarly, we demonstrate that deletion of MEC1 leads to a striking increase of Sgs1 foci upon HU-treatment. Furthermore, the morphology of these foci differs from a wild-type cell, as we frequently observe more than one Sgs1 focus per cell (Figure 3D, E). However, using the sgs1-D664Δ allele, we demonstrate that Rad52 focus repression and checkpoint activation are both functional, while Sgs1-D664Δ foci are mis-regulated (Figure 3B). It has been speculated that Rad52 focus inhibition is mediated through its phosphorylation by Mec1 or Chk1 [56] and it appears conceivable that a similar mechanism of Sgs1 focus regulation may exist. Recent work has indeed uncovered Sgs1 as a Mec1 substrate upon checkpoint activation [58]. Using an sgs1 mutant affecting a region proximal to D664, the authors demonstrate that Sgs1 phosphorylation is necessary for efficient Rad53 recruitment and activation [58]. As we find a strong induction of the downstream checkpoint target gene RNR3 through Dun1, which is itself activated by Rad53, we do not believe that Rad53 activity is significantly reduced in sgs1-D664Δ. Therefore, the defect in Sgs1-D664Δ focus regulation is likely not an effect of reduced Rad53 activity. However, we cannot exclude that an additional checkpoint-dependent phosphorylation event on Sgs1 is required for focus regulation and is absent in the sgs1-D664Δ mutant. This scenario appears plausible since BLM is modified by ATR upon replication fork stalling to allow fork re-start [83, 84].
Post-translational modification by SUMO and/or ubiquitin is important for regulation of many DSB repair proteins including BLM and Sgs1 [20-22, 62, 76, 85]. Interestingly, we find that upon deletion of SLX8, the number of Sgs1 foci significantly increases, both spontaneously and with damage (Figure 5A, B). Thus, Slx5-Slx8 is important for the regulation of Sgs1 foci in an unperturbed cell. This function is likely conserved in higher eukaryotes, as mammalian RNF4 rescues Sgs1 focus formation in yeast slx8Δ (Figure 5D). Importantly, our results provide strong evidence that ubiquitination is required for the regulation of Sgs1 foci, as the catalytic mutant of RNF4 is not able to rescue this phenotype in slx8Δ (Figure 5D). The STUbL complex Slx5-Slx8 could control Sgs1 focus formation by ubiquitination of a critical factor at a damaged replication fork, a hypothesis suggested by previous studies [34]. While it is tempting to speculate that Sgs1 may be the unknown critical substrate, we are currently technically unable to provide clear evidence that Sgs1 is itself modified by Slx5-Slx8. Despite the fact that we do weakly detect Sgs1 in a His-ubiquitin pull-down (data not shown), the modified pool only represents a very small protein amount at the detection limit of our experimental system, making it difficult to differentiate if a certain fraction of this modification depends on Slx5-Slx8. However, our results do provide evidence against Slx5-Slx8 targeting bulk amounts of Sgs1 to the 26S proteasome for degradation in untreated asynchronous cultures, as Sgs1 protein stability in a CHX chase is independent of Slx8 (Figure 4C). This does not fully exclude Sgs1 as a Slx5-Slx8 substrate, as ubiquitination and degradation of Sgs1 may only affect a small pool of the total cellular protein, i.e. at a damaged replication fork. Alternatively, ubiquitination by Slx5-Slx8 may not target Sgs1 to the proteasome, but rather modulate its localization or ability to interact with other proteins through addition of an alternative ubiquitin chain type such as lysine 63-linkage. An alternative hypothesis that is equally plausible is that replication stress, for example caused by elevated 2μ DNA levels in slx8Δ cells [32], may lead to an increased number of collapsed or severely damaged replication forks [32], which in turn are marked by an Sgs1 focus. In this scenario the mis-regulation of a critical Slx5-Slx8 substrate causes severe defects during replication in an Sgs1-independent manner. Sgs1 is then required to act on these structures and in doing so is recruited into a nuclear focus. Due to the pleiotropic phenotypes of both sgs1 as well as slx8 mutants it is currently difficult to differentiate between the two possibilities.
Importantly, the mammalian STUbL RNF4 has also been implicated in DSB repair as it accumulates at micro-irradiation-induced DSBs, dependent on the DNA repair proteins MDC1, NBS1, RNF8, 53BP1, and BRCA1 [36, 37]. Consistent with its recruitment to a damage site, RNF4 promotes DSB repair and its depletion sensitizes cells to IR and HU [36, 37]. RNF4 regulates the dynamics of multiple DNA damage associated proteins such as RPA1 and MDC1. Interestingly, it has been proposed that BLM may itself be an RNF4 substrate [37]. This hypothesis would be in line with our experimental data demonstrating that RNF4 affects BLM focus regulation. BLM is SUMOylated in vivo and a non-SUMOylate-able mutant of BLM does not localize to PML bodies, but rather induces and localizes to DNA damage foci [21]. Regulation of BLM function is critical to human health, as mutations in BLM cause Bloom syndrome [86], as well as potentially lead to cancer predisposition [87-89]. RNF4 itself is also a potential target for certain cancer treatments [90], making it even more important to fully understand the mechanistic details of how RNF4 regulates BLM.
In summary, we identify a novel regulatory mechanism of Sgs1/ BLM function through recruitment into nuclear foci and find the STUbL ligases Slx5-Slx8 and RNF4 to be regulators of RecQ-like helicase foci. This adds an additional level of regulation to BLM and its sub-nuclear localization.
Supplementary Material
Highlights.
-
-
S. cerevisiae RecQ-like helicase Sgs1 forms nuclear foci
-
-
HU treatment causes a reduction of Sgs1 foci dependent on the checkpoint kinase Mec1
-
-
Deletion of the STUbL Slx5-Slx8 alters Sgs1 focus regulation
-
-
Expression of the mammalian STUbL RNF4 rescues Sgs1 focus regulation in yeast slx8Δ
-
-
Focus numbers of the mammalian Sgs1 orthologue BLM are increased upon RNF4 knockdown
Acknowledgements
We thank R. Rothstein for strains, reagents, and critical reading of the manuscript. A. Buchberger for generously sharing materials and advice. We thank S. Jentsch, L. Symington, and D. Branzei for providing Y2H plasmids, S. Brill for supplying the SLX8 and RNF4 plasmids, J. Palvimo for the RNF4 antibody, and X. Zhao for the slx5Δ and slx8Δ strains. We thank the Center for Biologic Imaging at the University of Pittsburgh for help with microscopy, R. Sobol and E. Fouquerel for help with the qPCR analysis, and C. Wollny for cell culture advice.
Funding
This study was supported by National Institutes of Health (GM088413) and The Ellison Medical Foundation (AG-NS-0935-12) to K.A.B.
Abbreviations
- HU
hydroxyurea
- Y2H
yeast-2-hybrid
- DSB
double-strand break
- HR
homologous recombination
- STUbL
SUMO-targeted ubiquitin ligase
- SUMO
small ubiquitin-like modifier
- YFP
yellow fluorescent protein
- CFP
cyan fluorescent protein
- RFP
red fluorescent protein
- DIC
differential interference contrast
- DAPI
4′,6-diamidino-2-phenylindole
- MMS
methyl methanesulfonate
- SCE
sister chromatid exchange
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
None.
References
- [1].Bernstein KA, Gangloff S, Rothstein R. The RecQ DNA helicases in DNA repair. Annu. Rev. Genet. 2010;44:393–417. doi: 10.1146/annurev-genet-102209-163602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Chu WK, Hickson ID. RecQ helicases: multifunctional genome caretakers. Nat. Rev. Cancer. 2009;9:644–654. doi: 10.1038/nrc2682. [DOI] [PubMed] [Google Scholar]
- [3].Barea F, Tessaro S, Bonatto D. In silico analyses of a new group of fungal and plant RecQ4-homologous proteins. Comput. Biol. Chem. 2008;32:349–358. doi: 10.1016/j.compbiolchem.2008.07.005. [DOI] [PubMed] [Google Scholar]
- [4].Kwon SH, Choi DH, Lee R, Bae SH. Saccharomyces cerevisiae Hrq1 requires a long 3′-tailed DNA substrate for helicase activity. Biochem. Biophys. Res. Commun. 2012;427:623–628. doi: 10.1016/j.bbrc.2012.09.109. [DOI] [PubMed] [Google Scholar]
- [5].Choi DH, Lee R, Kwon SH, Bae SH. Hrq1 functions independently of Sgs1 to preserve genome integrity in Saccharomyces cerevisiae. J. Microbiol. 2013;51:105–112. doi: 10.1007/s12275-013-3048-2. [DOI] [PubMed] [Google Scholar]
- [6].Ellis NA, Groden J, Ye TZ, Straughen J, Lennon DJ, Ciocci S, Proytcheva M, German J. The Bloom’s syndrome gene product is homologous to RecQ helicases. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- [7].Nordlund P, Reichard P. Ribonucleotide reductases. Annu. Rev. Biochem. 2006;75:681–706. doi: 10.1146/annurev.biochem.75.103004.142443. [DOI] [PubMed] [Google Scholar]
- [8].Kitamura E, Blow JJ, Tanaka TU. Live-cell imaging reveals replication of individual replicons in eukaryotic replication factories. Cell. 2006;125:1297–1308. doi: 10.1016/j.cell.2006.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Branzei D, Foiani M. Maintaining genome stability at the replication fork. Nat. Rev. Mol. Cell Biol. 2010;11:208–219. doi: 10.1038/nrm2852. [DOI] [PubMed] [Google Scholar]
- [10].Friedel AM, Pike BL, Gasser SM. ATR/Mec1: coordinating fork stability and repair. Curr. Opin. Cell Biol. 2009;21:237–244. doi: 10.1016/j.ceb.2009.01.017. [DOI] [PubMed] [Google Scholar]
- [11].Segurado M, Tercero JA. The S-phase checkpoint: targeting the replication fork. Biology of the cell / under the auspices of the European Cell Biology Organization. 2009;101:617–627. doi: 10.1042/BC20090053. [DOI] [PubMed] [Google Scholar]
- [12].Labib K, De Piccoli G. Surviving chromosome replication: the many roles of the S-phase checkpoint pathway. Philosophical transactions of the Royal Society of London. Series B, Biological sciences. 2011;366:3554–3561. doi: 10.1098/rstb.2011.0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Liberi G, Maffioletti G, Lucca C, Chiolo I, Baryshnikova A, Cotta-Ramusino C, Lopes M, Pellicioli A, Haber JE, Foiani M. Rad51-dependent DNA structures accumulate at damaged replication forks in sgs1 mutants defective in the yeast ortholog of BLM RecQ helicase. Genes Dev. 2005;19:339–350. doi: 10.1101/gad.322605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Sogo JM, Lopes M, Foiani M. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science. 2002;297:599–602. doi: 10.1126/science.1074023. [DOI] [PubMed] [Google Scholar]
- [15].Lopes M, Cotta-Ramusino C, Pellicioli A, Liberi G, Plevani P, Muzi-Falconi M, Newlon CS, Foiani M. The DNA replication checkpoint response stabilizes stalled replication forks. Nature. 2001;412:557–561. doi: 10.1038/35087613. [DOI] [PubMed] [Google Scholar]
- [16].Sengupta S, Linke SP, Pedeux R, Yang Q, Farnsworth J, Garfield SH, Valerie K, Shay JW, Ellis NA, Wasylyk B, Harris CC. BLM helicase-dependent transport of p53 to sites of stalled DNA replication forks modulates homologous recombination. EMBO J. 2003;22:1210–1222. doi: 10.1093/emboj/cdg114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Cobb JA, Bjergbaek L, Shimada K, Frei C, Gasser SM. DNA polymerase stabilization at stalled replication forks requires Mec1 and the RecQ helicase Sgs1. EMBO J. 2003;22:4325–4336. doi: 10.1093/emboj/cdg391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Bjergbaek L, Cobb JA, Tsai-Pflugfelder M, Gasser SM. Mechanistically distinct roles for Sgs1p in checkpoint activation and replication fork maintenance. EMBO J. 2005;24:405–417. doi: 10.1038/sj.emboj.7600511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Frei C, Gasser SM. The yeast Sgs1p helicase acts upstream of Rad53p in the DNA replication checkpoint and colocalizes with Rad53p in S-phase-specific foci. Genes Dev. 2000;14:81–96. [PMC free article] [PubMed] [Google Scholar]
- [20].Ouyang KJ, Woo LL, Zhu J, Huo D, Matunis MJ, Ellis NA. SUMO modification regulates BLM and RAD51 interaction at damaged replication forks. PLoS Biol. 2009;7:e1000252. doi: 10.1371/journal.pbio.1000252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Eladad S, Ye TZ, Hu P, Leversha M, Beresten S, Matunis MJ, Ellis NA. Intra-nuclear trafficking of the BLM helicase to DNA damage-induced foci is regulated by SUMO modification. Hum. Mol. Genet. 2005;14:1351–1365. doi: 10.1093/hmg/ddi145. [DOI] [PubMed] [Google Scholar]
- [22].Böhm S, Bernstein KA. The role of post-translational modifications in fine-tuning BLM helicase function during DNA repair. DNA Repair (Amst) 2014;22C:123–132. doi: 10.1016/j.dnarep.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Tkach JM, Yimit A, Lee AY, Riffle M, Costanzo M, Jaschob D, Hendry JA, Ou J, Moffat J, Boone C, Davis TN, Nislow C, Brown GW. Dissecting DNA damage response pathways by analysing protein localization and abundance changes during DNA replication stress. Nat. Cell Biol. 2012;14:966–976. doi: 10.1038/ncb2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Pan X, Ye P, Yuan DS, Wang X, Bader JS, Boeke JD. A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell. 2006;124:1069–1081. doi: 10.1016/j.cell.2005.12.036. [DOI] [PubMed] [Google Scholar]
- [25].Pan X, Yuan DS, Xiang D, Wang X, Sookhai-Mahadeo S, Bader JS, Hieter P, Spencer F, Boeke JD. A robust toolkit for functional profiling of the yeast genome. Mol. Cell. 2004;16:487–496. doi: 10.1016/j.molcel.2004.09.035. [DOI] [PubMed] [Google Scholar]
- [26].Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, Schuldiner M, Gebbia M, Recht J, Shales M, Ding H, Xu H, Han J, Ingvarsdottir K, Cheng B, Andrews B, Boone C, Berger SL, Hieter P, Zhang Z, Brown GW, Ingles CJ, Emili A, Allis CD, Toczyski DP, Weissman JS, Greenblatt JF, Krogan NJ. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–810. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- [27].Slater ML. Effect of reversible inhibition of deoxyribonucleic acid synthesis on the yeast cell cycle. J. Bacteriol. 1973;113:263–270. doi: 10.1128/jb.113.1.263-270.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Mullen JR, Kaliraman V, Ibrahim SS, Brill SJ. Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics. 2001;157:103–118. doi: 10.1093/genetics/157.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Ii T, Mullen JR, Slagle CE, Brill SJ. Stimulation of in vitro sumoylation by Slx5-Slx8: evidence for a functional interaction with the SUMO pathway. DNA Repair (Amst) 2007;6:1679–1691. doi: 10.1016/j.dnarep.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Ii T, Fung J, Mullen JR, Brill SJ. The yeast Slx5-Slx8 DNA integrity complex displays ubiquitin ligase activity. Cell Cycle. 2007;6:2800–2809. doi: 10.4161/cc.6.22.4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Prudden J, Pebernard S, Raffa G, Slavin DA, Perry JJ, Tainer JA, McGowan CH, Boddy MN. SUMO-targeted ubiquitin ligases in genome stability. EMBO J. 2007;26:4089–4101. doi: 10.1038/sj.emboj.7601838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Burgess RC, Rahman S, Lisby M, Rothstein R, Zhao X. The Slx5-Slx8 complex affects sumoylation of DNA repair proteins and negatively regulates recombination. Mol. Cell. Biol. 2007;27:6153–6162. doi: 10.1128/MCB.00787-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Zhang C, Roberts TM, Yang J, Desai R, Brown GW. Suppression of genomic instability by SLX5 and SLX8 in Saccharomyces cerevisiae. DNA Repair (Amst) 2006;5:336–346. doi: 10.1016/j.dnarep.2005.10.010. [DOI] [PubMed] [Google Scholar]
- [34].Nagai S, Dubrana K, Tsai-Pflugfelder M, Davidson MB, Roberts TM, Brown GW, Varela E, Hediger F, Gasser SM, Krogan NJ. Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science. 2008;322:597–602. doi: 10.1126/science.1162790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Hakli M, Karvonen U, Janne OA, Palvimo JJ. SUMO-1 promotes association of SNURF (RNF4) with PML nuclear bodies. Exp. Cell Res. 2005;304:224–233. doi: 10.1016/j.yexcr.2004.10.029. [DOI] [PubMed] [Google Scholar]
- [36].Yin Y, Seifert A, Chua JS, Maure JF, Golebiowski F, Hay RT. SUMO-targeted ubiquitin E3 ligase RNF4 is required for the response of human cells to DNA damage. Genes Dev. 2012;26:1196–1208. doi: 10.1101/gad.189274.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Galanty Y, Belotserkovskaya R, Coates J, Jackson SP. RNF4, a SUMO-targeted ubiquitin E3 ligase, promotes DNA double-strand break repair. Genes Dev. 2012;26:1179–1195. doi: 10.1101/gad.188284.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Groocock LM, Nie M, Prudden J, Moiani D, Wang T, Cheltsov A, Rambo RP, Arvai AS, Hitomi C, Tainer JA, Luger K, Perry JJ, Lazzerini-Denchi E, Boddy MN. RNF4 interacts with both small ubiquitin-like modifier and nucleosomes to promote the DNA damage response. EMBO Rep. 2014 doi: 10.1002/embr.201338369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Bernstein KA, Shor E, Sunjevaric I, Fumasoni M, Burgess RC, Foiani M, Branzei D, Rothstein R. Sgs1 function in the repair of DNA replication intermediates is separable from its role in homologous recombinational repair. EMBO J. 2009;28:915–925. doi: 10.1038/emboj.2009.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Thomas BJ, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- [42].Sherman F, Fink GR, Hicks JB. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1986. [Google Scholar]
- [43].Knop M, Siegers K, Pereira G, Zachariae W, Winsor B, Nasmyth K, Schiebel E. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast. 1999;15:963–972. doi: 10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- [44].Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno-Borchart A, Doenges G, Schwob E, Schiebel E, Knop M. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- [45].Gietz RD, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- [46].Bernstein KA, Mimitou EP, Mihalevic MJ, Chen H, Sunjaveric I, Symington LS, Rothstein R. Resection Activity of the Sgs1 Helicase Alters the Affinity of DNA Ends for Homologous Recombination Proteins in Saccharomyces cerevisiae. Genetics. 2013:1241–1251. doi: 10.1534/genetics.113.157370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Böhm S, Lamberti G, Fernandez-Saiz V, Stapf C, Buchberger A. Cellular functions of Ufd2 and Ufd3 in proteasomal protein degradation depend on Cdc48 binding. Mol. Cell. Biol. 2011;31:1528–1539. doi: 10.1128/MCB.00962-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Petermann E, Orta ML, Issaeva N, Schultz N, Helleday T. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol. Cell. 2010;37:492–502. doi: 10.1016/j.molcel.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Slater ML. Effect of reversible inhibition of deoxyribonucleic acid synthesis on the yeast cell cycle. J Bacteriol. 1973;113:263–270. doi: 10.1128/jb.113.1.263-270.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Ma W, Resnick MA, Gordenin DA. Apn1 and Apn2 endonucleases prevent accumulation of repair-associated DNA breaks in budding yeast as revealed by direct chromosomal analysis. Nuc. Acids Res. 2008;36:1836–1846. doi: 10.1093/nar/gkm1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Mathiasen DP, Lisby M. Cell cycle regulation of homologous recombination in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 2014;38:172–184. doi: 10.1111/1574-6976.12066. [DOI] [PubMed] [Google Scholar]
- [53].Bianchi V, Pontis E, Reichard P. Changes of deoxyribonucleoside triphosphate pools induced by hydroxyurea and their relation to DNA synthesis. J. Biol. Chem. 1986;261:16037–16042. [PubMed] [Google Scholar]
- [54].Tercero JA, Diffley JF. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature. 2001;412:553–557. doi: 10.1038/35087607. [DOI] [PubMed] [Google Scholar]
- [55].Pellicioli A, Lucca C, Liberi G, Marini F, Lopes M, Plevani P, Romano A, Di Fiore PP, Foiani M. Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO J. 1999;18:6561–6572. doi: 10.1093/emboj/18.22.6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Barlow JH, Rothstein R. Rad52 recruitment is DNA replication independent and regulated by Cdc28 and the Mec1 kinase. EMBO J. 2009;28:1121–1130. doi: 10.1038/emboj.2009.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Alabert C, Bianco JN, Pasero P. Differential regulation of homologous recombination at DNA breaks and replication forks by the Mrc1 branch of the S-phase checkpoint. EMBO J. 2009;28:1131–1141. doi: 10.1038/emboj.2009.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Hegnauer AM, Hustedt N, Shimada K, Pike BL, Vogel M, Amsler P, Rubin SM, van Leeuwen F, Guenole A, van Attikum H, Thoma NH, Gasser SM. An N-terminal acidic region of Sgs1 interacts with Rpa70 and recruits Rad53 kinase to stalled forks. EMBO J. 2012;31:3768–3783. doi: 10.1038/emboj.2012.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Sollier J, Driscoll R, Castellucci F, Foiani M, Jackson SP, Branzei D. The Saccharomyces cerevisiae Esc2 and Smc5-6 proteins promote sister chromatid junction-mediated intra-S repair. Mol. Biol. Cell. 2009;20:1671–1682. doi: 10.1091/mbc.E08-08-0875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Bergink S, Jentsch S. Principles of ubiquitin and SUMO modifications in DNA repair. Nature. 2009;458:461–467. doi: 10.1038/nature07963. [DOI] [PubMed] [Google Scholar]
- [61].Branzei D, Sollier J, Liberi G, Zhao X, Maeda D, Seki M, Enomoto T, Ohta K, Foiani M. Ubc9- and mms21-mediated sumoylation counteracts recombinogenic events at damaged replication forks. Cell. 2006;127:509–522. doi: 10.1016/j.cell.2006.08.050. [DOI] [PubMed] [Google Scholar]
- [62].Lu CY, Tsai CH, Brill SJ, Teng SC. Sumoylation of the BLM ortholog, Sgs1, promotes telomere-telomere recombination in budding yeast. Nucleic Acids Res. 2009:488–498. doi: 10.1093/nar/gkp1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Seyfried NT, Xu P, Duong DM, Cheng D, Hanfelt J, Peng J. Systematic approach for validating the ubiquitinated proteome. Anal. Chem. 2008;80:4161–4169. doi: 10.1021/ac702516a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Sriramachandran AM, Dohmen RJ. SUMO-targeted ubiquitin ligases. Biochim. Biophys. Acta. 2013:75–85. doi: 10.1016/j.bbamcr.2013.08.022. [DOI] [PubMed] [Google Scholar]
- [65].Alvaro D, Lisby M, Rothstein R. Genome-Wide Analysis of Rad52 Foci Reveals Diverse Mechanisms Impacting Recombination. PLoS Genet. 2007;3:e228. doi: 10.1371/journal.pgen.0030228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Mullen JR, Das M, Brill SJ. Genetic evidence that polysumoylation bypasses the need for a SUMO-targeted Ub ligase. Genetics. 2011;187:73–87. doi: 10.1534/genetics.110.124347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Uzunova K, Gottsche K, Miteva M, Weisshaar SR, Glanemann C, Schnellhardt M, Niessen M, Scheel H, Hofmann K, Johnson ES, Praefcke GJ, Dohmen RJ. Ubiquitin-dependent proteolytic control of SUMO conjugates. J. Biol. Chem. 2007;282:34167–34175. doi: 10.1074/jbc.M706505200. [DOI] [PubMed] [Google Scholar]
- [68].Zhong S, Hu P, Ye T, Stan R, Ellis N, Pandolfi P. A role for PML and the nuclear body in genomic stability. Oncogene. 1999;18:7941–7947. doi: 10.1038/sj.onc.1203367. [DOI] [PubMed] [Google Scholar]
- [69].Bischof O, Kim SH, Irving J, Beresten S, Ellis NA, Campisi J. Regulation and localization of the Bloom syndrome protein in response to DNA damage. J. Cell Biol. 2001;153:367–380. doi: 10.1083/jcb.153.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JL, Toufighi K, Mostafavi S, Prinz J, St Onge RP, VanderSluis B, Makhnevych T, Vizeacoumar FJ, Alizadeh S, Bahr S, Brost RL, Chen Y, Cokol M, Deshpande R, Li Z, Lin ZY, Liang W, Marback M, Paw J, San Luis BJ, Shuteriqi E, Tong AH, van Dyk N, Wallace IM, Whitney JA, Weirauch MT, Zhong G, Zhu H, Houry WA, Brudno M, Ragibizadeh S, Papp B, Pal C, Roth FP, Giaever G, Nislow C, Troyanskaya OG, Bussey H, Bader GD, Gingras AC, Morris QD, Kim PM, Kaiser CA, Myers CL, Andrews BJ, Boone C. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Melnick A, Licht JD. Deconstructing a disease: RARalpha, its fusion partners, and their roles in the pathogenesis of acute promyelocytic leukemia. Blood. 1999;93:3167–3215. [PubMed] [Google Scholar]
- [72].Muller S, Matunis MJ, Dejean A. Conjugation with the ubiquitin-related modifier SUMO-1 regulates the partitioning of PML within the nucleus. EMBO J. 1998;17:61–70. doi: 10.1093/emboj/17.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Tatham MH, Geoffroy MC, Shen L, Plechanovova A, Hattersley N, Jaffray EG, Palvimo JJ, Hay RT. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat. Cell Biol. 2008;10:538–546. doi: 10.1038/ncb1716. [DOI] [PubMed] [Google Scholar]
- [74].Lallemand-Breitenbach V, Jeanne M, Benhenda S, Nasr R, Lei M, Peres L, Zhou J, Zhu J, Raught B, de The H. Arsenic degrades PML or PML-RARalpha through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat Cell Biol. 2008;10:547–555. doi: 10.1038/ncb1717. [DOI] [PubMed] [Google Scholar]
- [75].German J, Crippa LP, Bloom D. Bloom’s syndrome. III. Analysis of the chromosome aberration characteristic of this disorder. Chromosoma. 1974;48:361–366. doi: 10.1007/BF00290993. [DOI] [PubMed] [Google Scholar]
- [76].Tikoo S, Madhavan V, Hussain M, Miller ES, Arora P, Zlatanou A, Modi P, Townsend K, Stewart GS, Sengupta S. Ubiquitin-dependent recruitment of the Bloom syndrome helicase upon replication stress is required to suppress homologous recombination. EMBO J. 2013;32:1778–1792. doi: 10.1038/emboj.2013.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Cobb JA, Schleker T, Rojas V, Bjergbaek L, Tercero JA, Gasser SM. Replisome instability, fork collapse, and gross chromosomal rearrangements arise synergistically from Mec1 kinase and RecQ helicase mutations. Genes Dev. 2005;19:3055–3069. doi: 10.1101/gad.361805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Branzei D, Vanoli F, Foiani M. SUMOylation regulates Rad18-mediated template switch. Nature. 2008;456:915–920. doi: 10.1038/nature07587. [DOI] [PubMed] [Google Scholar]
- [79].Liu Y, Nielsen CF, Yao Q, Hickson ID. The origins and processing of ultra fine anaphase DNA bridges. Curr. Opin. Genet. Dev. 2014;26C:1–5. doi: 10.1016/j.gde.2014.03.003. [DOI] [PubMed] [Google Scholar]
- [80].Chan KL, North PS, Hickson ID. BLM is required for faithful chromosome segregation and its localization defines a class of ultrafine anaphase bridges. EMBO J. 2007;26:3397–3409. doi: 10.1038/sj.emboj.7601777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Germann SM, Schramke V, Pedersen RT, Gallina I, Eckert-Boulet N, Oestergaard VH, Lisby M. TopBP1/Dpb11 binds DNA anaphase bridges to prevent genome instability. J. Cell Biol. 2014;204:45–59. doi: 10.1083/jcb.201305157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Lisby M, Barlow JH, Burgess RC, Rothstein R. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- [83].Davies SL, North PS, Dart A, Lakin ND, Hickson ID. Phosphorylation of the Bloom’s syndrome helicase and its role in recovery from S-phase arrest. Mol. Cell. Biol. 2004;24:1279–1291. doi: 10.1128/MCB.24.3.1279-1291.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Davalos AR, Kaminker P, Hansen RK, Campisi J. ATR and ATM-dependent movement of BLM helicase during replication stress ensures optimal ATM activation and 53BP1 focus formation. Cell Cycle. 2004;3:1579–1586. doi: 10.4161/cc.3.12.1286. [DOI] [PubMed] [Google Scholar]
- [85].Ouyang KJ, Yagle MK, Matunis MJ, Ellis NA. BLM SUMOylation regulates ssDNA accumulation at stalled replication forks. Front. Genet. 2013;4:167. doi: 10.3389/fgene.2013.00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].German J, Sanz MM, Ciocci S, Ye TZ, Ellis NA. Syndrome-causing mutations of the BLM gene in persons in the Bloom’s Syndrome Registry. Hum. Mutat. 2007;28:743–753. doi: 10.1002/humu.20501. [DOI] [PubMed] [Google Scholar]
- [87].Goss KH, Risinger MA, Kordich JJ, Sanz MM, Straughen JE, Slovek LE, Capobianco AJ, German J, Boivin GP, Groden J. Enhanced tumor formation in mice heterozygous for Blm mutation. Science. 2002;297:2051–2053. doi: 10.1126/science.1074340. [DOI] [PubMed] [Google Scholar]
- [88].Gruber SB, Ellis NA, Scott KK, Almog R, Kolachana P, Bonner JD, Kirchhoff T, Tomsho LP, Nafa K, Pierce H, Low M, Satagopan J, Rennert H, Huang H, Greenson JK, Groden J, Rapaport B, Shia J, Johnson S, Gregersen PK, Harris CC, Boyd J, Rennert G, Offit K. BLM heterozygosity and the risk of colorectal cancer. Science. 2002;297:2013. doi: 10.1126/science.1074399. [DOI] [PubMed] [Google Scholar]
- [89].Thompson ER, Doyle MA, Ryland GL, Rowley SM, Choong DY, Tothill RW, Thorne H, Barnes DR, Li J, Ellul J, Philip GK, Antill YC, James PA, Trainer AH, Mitchell G, Campbell IG. Exome sequencing identifies rare deleterious mutations in DNA repair genes FANCC and BLM as potential breast cancer susceptibility alleles. PLoS Genet. 2012;8:e1002894. doi: 10.1371/journal.pgen.1002894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Saito M, Fujimitsu Y, Sasano T, Yoshikai Y, Ban-Ishihara R, Nariai Y, Urano T, Saitoh H. The SUMO-targeted ubiquitin ligase RNF4 localizes to etoposide-exposed mitotic chromosomes: Implication for a novel DNA damage response during mitosis. Biochem. Biophys. Res. Commun. 2014:83–88. doi: 10.1016/j.bbrc.2014.03.106. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.