Abstract
The balance between stability and dynamics for active enzymes can be somewhat quantified by studies of intein splicing and cleaving reactions. Inteins catalyze the ligation of flanking host exteins while excising themselves. The potential for applications led to engineering of a mini-intein splicing domain, where the homing endonuclease domain of the Mycobacterium tuberculosis RecA (Mtu recA) intein was removed. The remaining domains were linked by several short peptides, but splicing activity in all was substantially lower than the full-length intein. Native splicing activity was restored in some cases by a V67L mutation. Using computations and experiments, we examine the impact of this mutation on the stability and conformational dynamics of the mini-intein splicing domain. Molecular dynamics simulations were used to delineate the factors that determine the active state, including the V67L mini-intein mutant, and peptide linker. We found that (1) the V67L mutation lowers the global fluctuations in all modeled mini-inteins, stabilizing the mini-intein constructs; (2) the connecting linker length affects intein dynamics; and (3) the flexibilities of the linker and intein core are higher in the active structure. We have observed that the interaction of the linker region and a turn region around residues 35-41 provides the pathway for the allostery interaction. Our experiments reveal that intein catalysis is characterized by non-linear Arrhenius plot, confirming the significant contribution of protein conformational dynamics to intein function. We conclude that while the V67L mutation stabilizes the global structure, cooperative dynamics of all intein regions appear more important for intein function than high stability. Our studies suggest that effectively quenching the conformational dynamics of an intein through engineered allosteric interactions could deactivate intein splicing or cleaving.
Keywords: Mini-intein, protein splicing, enzyme dynamics, allostery, conformational dynamics, non-linear Arrhenius plot
1. Introduction
Inteins are self-splicing protein domains that are found as insertions in a variety of host proteins, dividing their host protein into two segments, referred to as the N-extein and the C-extein. The self-catalyzed splicing function performed by inteins results in the excision of the intein and the ligation of the N- and C-exteins to form a mature and active host protein (Figure 1A) [1]. Inteins belong to the hedgehog intein superfamily of autoprocessing domains [2]. Due to the post-translational editing function, inteins have been used as powerful proteins in protein engineering, labeling, purification and control of protein function [3-10]. Inteins can also be used in drug design, as they are capable of undergoing selective activation of a protein, drug, or drug encapsulation in a viral coat [11]. A majority of the inteins found to date contain two independent domains – the homing endonuclease domain and the splicing domain [12]. In these bipartite inteins, the endonuclease domain is inserted into the splicing domain, dividing it into N- and C-terminal protein splicing domain segments [13-15]. Endonuclease activity is not required for protein splicing, and inteins can be engineered without the endonuclease domain (named minimal inteins, or mini-inteins) (Figure 1B) [16-23]. In general, while the removal of the endonuclease domain does not abolish the functionality of the splicing domain, in most cases the splicing activity is greatly diminished. It is commonly believed that the mini-inteins’ stability is disrupted by the deletion of the endonuclease domain. For example, a point mutation (V67L) could stabilize the protein’s core and restore the splicing activity of several Mycobacterium tuberculosis (Mtu) recA mini-inteins to that of the full-length Mtu intein [18]. This hypothesis is supported by observations that the V67L mutation slows hydrogen-exchange rates globally, indicating a shift to more stable conformations, and reduction in ensemble distribution, leading to splicing activation [24]. This conformational stabilization mechanism is consistent with temperature sensitive inteins, which have been optimized to be active at lower permissive temperatures, ranging from 18 degrees to 30 degrees, and are inactive at higher non-permissive temperatures [25, 26].
Figure 1.
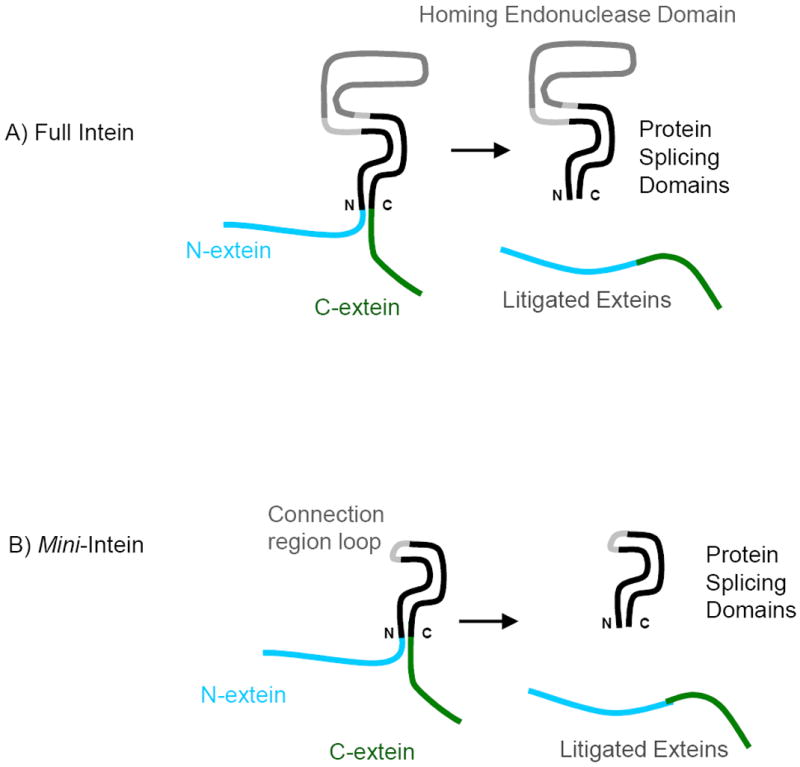
An illustration of the function of the full and mini-inteins. (A) Full intein has a large homing endonuclease domain (grey) and protein splicing domains (in black). The N- and Cexteins can be linked to form the ligated exteins. (B) In the mini-intein, the homing endonuclease domain is replaced by a short peptide (connection loop region).
Experimental data further indicate that the length of the peptide joining the two mini-intein segments, which replaces the removed endonuclease domain, can also affect intein activity (Figure 2) [18]. The Mtu recA mini-intein with longer loops tends to be more active, and that shortening the connection region beyond a certain point will inactivate the mini-intein. In one example of an inactive mini-intein, the endonuclease domain and part of the splicing domain (residues 96-402, 96Δ402) were removed. Other factors, such as temperature and pH, affect intein activity as well. The Mtu recA mini-intein has a pH optimum of approximately 6.2 and increased activity up to 37°C, above which the intein becomes unstable [23, 27]. Wu and his colleagues suggested that inteins tend to carry out their splicing activity faster at a lower pH [28]. Lower pH might change the protonation state of the active site histidine, which directly affects intein catalysis. However, the V67L mutation and the linker loop are distant from the active site (Figure 3), indicating a possible allosteric regulation mechanism.
Figure 2.
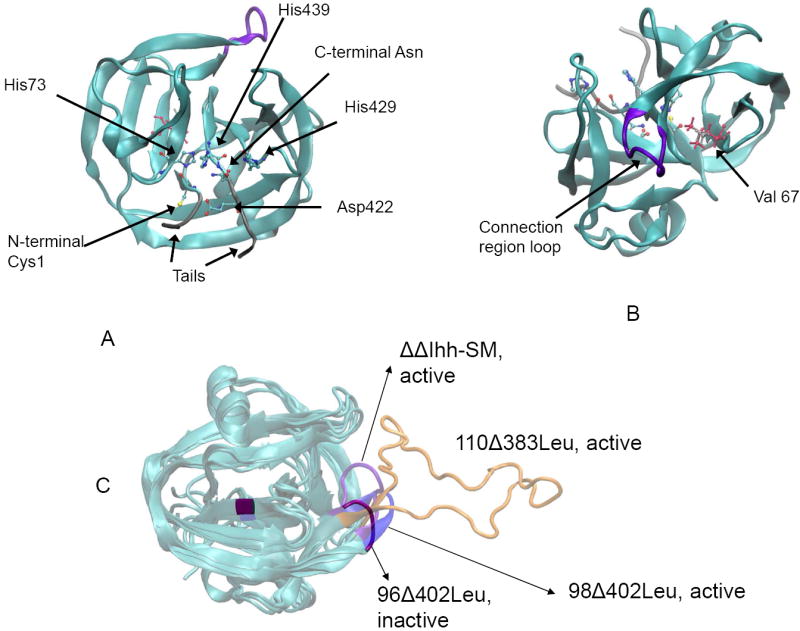
Intein activity can be regulated allostericaly by the V67L mutation and by changing loop length. (A) Conserved residues in proximity to the active site of the intein (built from PDB code 2L8L). (B) The endonuclease loop region (purple) and residue 67 are distant from the active site. V67 is shown in silver and V67L in pink (built from PDB code 2IMZ). (C) A comparison of the four structures being considered with different loop lengths (built from PDB code 2IMZ). Distinct colors indicate each structure. The large orange loop is for the active 110Δ383 mini-intein.
Figure 3.
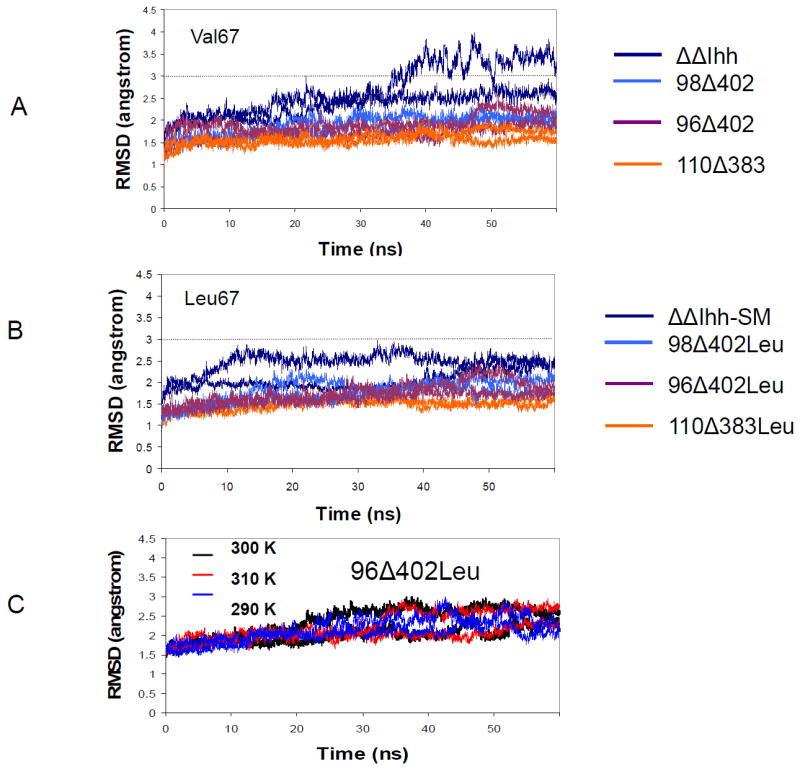
RMSD trajectories from molecular dynamics simulations indicate V67L mutation stabilizes intein. However 96Δ402Leu intein does not show large structural variations in the temperature range from 290 K to 310 K. Residues in the loop regions (97-401) are not included in the RMSD calculations. (A) native V67 inteins (B) mutant V67L inteins. (C) RMSD of the 96Δ402Leu model as a function of temperature.
Protein dynamics and conformational changes often underlie allosteric regulation [29-33], and conformational dynamics are indispensable to enzyme function [34]. The splicing function of intein consists of three catalytic steps (N–X acyl shift; trans-esterification; and Asn cyclization), and requires large conformational changes. A rigid conformation may prevent the sampling of active conformations which are important for protein splicing [35, 36]. The optimization of intein function needs a delicate balance of protein structure, stability and conformational dynamics, explaining why mutations observed in evolved inteins are difficult to rationalize [37]. It is important to understand the structural and dynamical aspects of inteins for intein engineering and the improvement of intein-based technologies[38].
In this study, we explore intein stability and dynamics using a combination of molecular dynamics simulations with experimental measurements of the temperature dependence of intein cleaving kinetics. Extensive molecular dynamics simulations were designed to investigate the regulation of intein stability and dynamics by the V67L mutation and the linker region. For both active and inactive inteins, we monitor the global structural changes as well as the conserved residues across most inteins [20, 22, 23, 39] (Figure 2A). These residues are in proximity to the active site of the intein, and have been shown to be important for normal intein function. Our study demonstrates that protein dynamics, rather than stability, differentiates between active and inactive inteins. The active structure has higher flexibility in both the connection region and the intein core, while the inactive structure has a lower flexibility in these regions. Finally, our experimental study revealed that intein catalysis is characterized by a non-linear Arrhenius plot, confirming the significant contribution of protein conformational dynamics to intein function.
MATERIALS AND METHODS
1. Protein structure selection
We simulated eight types of inteins. The nuclear magnetic resonance (NMR) structure of Mtu recA mini-intein (PDB ID 2L8L) [40], was chosen to be the starting point of active intein (see Figure 2A). N- and C- terminal ‘tails’ were added to simulate the presence of the N- and C-exteins, with the respective sequences GEGHG and CSPPF. The structure is referred to as ‘ΔΔIhh-SM’. To simulate the effect of linker loop length, we started with the x-ray structure of the splicing domain of the Mtu RecA ΔI-SM mini-intein (PDB ID: 2IMZ) [22]. This structure failed to resolve the linker peptide, so we added the missing residues as a random loop. This structure is designated as 110Δ383, and contains residues 1-110 and 383-440 of the native intein along with the added tails (Figure 2C). Further loop deletions result in the final two structures, 98Δ402 and 96Δ402, which are named similarly to 110Δ383 above. These four structures were modeled with the V67L stabilizing mutation (ΔΔIhh-SM, 110Δ383Leu, 98Δ402Leu, and 96Δ402Leu), and with the wild-type valine residue (ΔΔIhh, 110Δ383, 98Δ402, and 96Δ402) as well (see Table 1 for a summary of the structural data) [18, 22]. All eight structures have known experimental activities from published data. These experimental data indicate which intein will be active and which will be inactive in vitro; and consequently, the in silico simulations of these eight models could provide insight into what controls the activity of inteins and how.
Table 1.
Intein systems simulated in this study
| Structure | Residue 67 | Number of Residuesa | Sequence at connection region | Activity |
|---|---|---|---|---|
| ΔΔIhh | Val | 149 | (94) -AVRDVETGE – LRT - (407) | Poor |
| 110Δ383 | Val | 178 | (94) -AQPRRFDGFGDSAPIPARVQALADALDDKFLHDMLA – EELRT - (407) | Poor |
| 98Δ402 | Val | 147 | (94) -AQPRR – EELRT - (407) | Inactive |
| 96Δ402 | Val | 145 | (94) -AQP – EELRT - (407) | Inactive |
| ΔΔIhh-SM | Leu | 149 | (94) -VRDVETGE – LRT - (407) | Active |
| 110Δ383-Leu | Leu | 178 | (94) -AQPRRFDGFGDSAPIPARVQALADALDDKFLHDMLA – EELRT - (407) | Active |
| 98Δ402-Leu | Leu | 147 | (94) -AQPRR – EELRT - (407) | Active |
| 96Δ402-Leu | Leu | 145 | (94) -AQP – EELRT - (407) | Inactiveb |
N- and C- terminal ‘tails’ were added to simulate the presence of the N- and C-exteins, with the respective sequences GEGHG and CSPPF
The 96-L structure will become active at 20°C.
2. Molecular Dynamics Simulations
The simulated molecules were solvated in a TIP3P water box [41, 42] with a margin of at least 15 Å from any point on the edge of the water box. Sodium and chloride ions were added to the system up to a concentration of 0.10 M. MD simulations were performed using the NAMD package [43] and the CHARMM27 force field [44] with a constant pressure of 1 atm and a temperature of 300K. The exceptions to these base conditions are four 96Δ402 runs, two of which at 290K and two at 310K. The time step was 2fs with a SHAKE constraint on all bonds with hydrogen atoms [45]. Long-range electrostatic interactions were calculated with the Particle Mesh Ewald method [46]. Each model was simulated twice, with different starting conditions for each trajectory. The first MD runs were performed after 8000 steps of minimization and two 50 ps heating (250K and 280K) and 100 ps equilibration runs at the simulation temperatures (290K, 300K, and 310K). The second set of MD simulations was performed after 3000 steps of minimization, 30 ps heating at 250°K, 50 ps heating at 280K, and 200 ps equilibration runs at the simulation temperatures (290K, 300K, and 310K). The CHARMM program [44, 47] was used to analyze trajectories. The correlation matrix was obtained by computing the covariances of the spatial residue displacements of 60 ns MD trajectories for selected pairs of residues. The RMSD (root mean squared deviation) and RMSF (root mean square fluctuations) analysis of the dynamic trajectories was obtained using starting crystal structures as the reference.
3. Experimental intein cleaving rate constants
The protein construct pET/CBD-I-MBP was used for the experimental determination of intein cleaving rate constants. Here, CBD refers to the chitin-binding domain, I refers to the ΔI-CM (Cleavage Mutant) intein[23], and MBP refers to the E. coli maltose binding protein. This construct was chosen due to its high solubility and ease of expression and purification.
The CBD-I-MBP fusion protein was expressed in E. coli strain BLR (Novagen) by inoculating a 2ml overnight culture of Luria Broth (LB) supplemented with 100 ug/ml ampicillin with a single, isolated colony from an LB-Amp plate. The inoculum culture was incubated with shaking at 37°C, 250 RPM, for 16-20 hours. The following morning, 1ml of this culture was used to inoculate an expression culture consisting of 200 ml of Terrific Broth (TB) supplemented with 100 μg/ml ampicillin in a 1 liter baffled flask. The expression culture was incubated with shaking at 37°C, 150 RPM, for ~2-3 hours, until the absorbance at 600nm wavelength OD600 of the culture reached ~0.4-0.8. The culture was then cooled to 20°C in a shaking water bath for 30 minutes. After the culture had been cooled, protein overexpression was induced by addition of IPTG to a final concentration of 0.5 mM. The culture was then incubated at 16°C, 150 RPM, for 20-24 hours. After expression, the cells were harvested by centrifugation at 4°C and resuspended in 10 ml ice cold column equilibration buffer (40 mM phosphate buffer, pH 8.0, 500 mM NaCl, 2mM EDTA). The cells were then lysed by sonication and clarified by centrifugation at 4°C. The clarified lysate was diluted 1:5 into column equilibration buffer and loaded into a 50-ml Superloop attached to an AKTA Purifier 10 chromatography system. Protein purification was performed using a 2 ml bed volume of chitin affinity resin (NEB), packed into a water-jacketed XK-16/20 chromatography column (GE). The water jacket allowed for precise control of the temperature of the column. The CBD-I-MBP fusion protein was purified from the host cell proteins by loading the diluted lysate onto the chitin column and washing for several column volumes with column equilibration buffer.
In order to determine intein rate constants, the column was brought to the appropriate temperature using an attached water bath. Intein self-cleavage was induced by shifting the pH from 8.0 to 6.2 in a step change. The flow rate was reduced to between 0.010 and 0.025 ml/min to allow intein self-cleavage to occur in flow-through mode. The release kinetics of MBP was tracked by monitoring the absorbance at 280 nm (A280). The MBP release kinetics could then be fit using a first-order exponential decay model, as previously described [27], to determine the intein cleaving rate constant, k (hr-1). The portion of the curve at pH 6.2 was used for the fit, to ensure that the rate constant was determined at a constant pH for each temperature. This process was repeated between 2-4 times at each temperature tested from 25.5 to 37°C.
RESULTS
1. The global stability of intein does not fully correlate with intein activity
The point mutation V67L is capable of restoring the Mtu RecA mini-intein splicing activity to normal levels after the intein has been disrupted by the removal of the endonuclease domain. We first examine the change in global stability seen in the intein with residue V67 versus V67L by comparing the RMSD values of the inteins (Figure 3). The residues in the loop regions (97-401) are not included in the RMSD calculations to allow comparison of core domain residues only. Previously, an NMR study [24] showed that the V67L mutation stabilizes the ΔΔIhh-CM structure, which differs in one position (Gly 422) from the ΔΔIhh-SM studied here (Asp 422). Consistently, we found that the RMSD for the ΔΔIhh-SM (V67L, blue line, Figure 3B) is smaller than ΔΔIhh (V67, blue line, Figure 3A). The (still small) drop in the RMSD value between the two ΔΔIhh structures, indicates that the V67L mutation has a global stabilizing effect which could promote stability that in turn increases intein activity, in agreement with earlier experimental data [18].
Overall, all three V67L mini-inteins (110Δ383Leu, 98Δ402Leu, and 96Δ402Leu) have smaller RMSDs than ΔΔIhh-SM, indicating that the loop sequence in the ΔΔIhh-SM (VRDVETG) can also affect the global stability of intein. Even though the 110Δ383-Leu structure is the most stable and most active mini-intein, one cannot conclude that intein activity is a direct consequence of protein stability. Unlike the stabilization effect of the V67L mutation on the ΔΔIhh inteins, the V67L mutation has a negligible effect on the other mini-inteins (Figures 3A and 3B), with only about a 0.10 Å difference in RMSD for the 110Δ383, 98Δ402, and 96Δ402 models (compared with 110Δ383-Leu, 98Δ402-Leu, and 96Δ402-Leu, respectively).
The temperature effect also does not correlate with intein stability. Hiraga and his colleagues showed that the 96Δ402 mini-intein is active at a temperature of 20°C (293K) only when the V67L mutation is present [18]. The intein will be inactive at higher temperatures, making its use in mammalian cells implausible. We simulated the 96Δ402Leu structure at three temperatures: 310K, 300K, and 290K. RMSD calculations on these trajectories show that increasing the temperatures to 310K results in a negligible decrease in the RMSD value from the average value of the 300K run. Decreasing the simulation temperature to 290K, results in a marginally larger decrease in RMSD. Overall, both of these drops are insignificant and show that the global stability of the 96Δ402Leu intein is unlikely to be affected by a change in the temperature range studied here (Figure 3C).
2. The active 110Δ383Leu intein has higher dynamics in core region
An insight into the question of whether an intein with a dynamic or a stable core would be the most active might be obtained from the RMSF values of the residues. We plotted out the RMSF value for all eight structures in Figure 4 (Figure 4A for the V67 inteins and Figure 4B for the V67L inteins). RMSF analysis shows that the most active V67L intein form, 110Δ383Leu, has a very dynamic structure overall. The connection region has the highest fluctuation, which is expected, as the connection loop is largely unrestrained, allowing it to move rapidly. Interestingly, the structure also has a highly dynamic core region (containing residues 55 to 155). This trend continues to be seen even when residue V67 is present. On the other hand, the inactive 96Δ402 models have the most stable core region, in both the V67 and V67L (96Δ402Leu) structures. Both the ΔΔIhh-SM and 98Δ402Leu structures tend to exhibit RMSF values between those of the 110Δ383Leu and 96Δ402Leu models. The results indicated that the shortening of the connection region loop past a certain point (as exemplified in the 96Δ402 model) will quench the dynamics of the intein molecule and inactivate its enzymatic activity. Lengthening the linker loop allows the intein some flexibility, which is necessary for the splicing activity.
Figure 4.
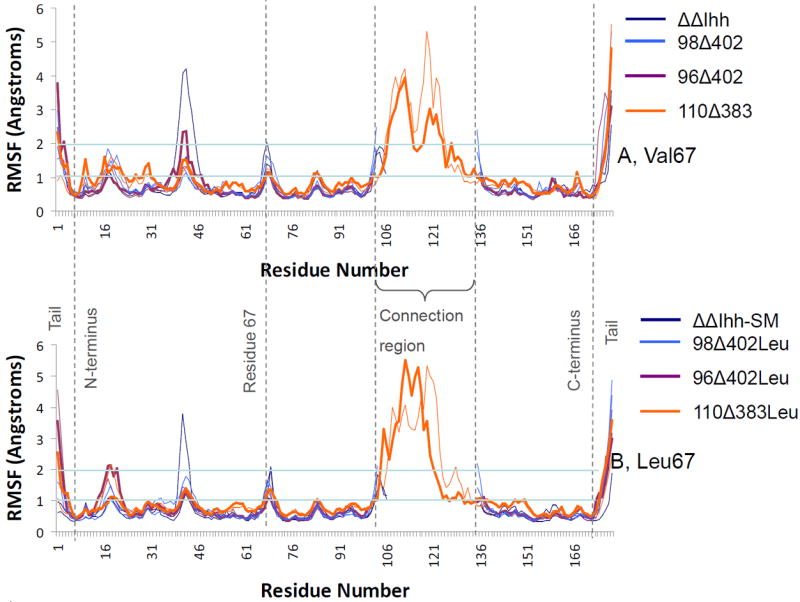
Root Mean Squared Fluctuations (RMSF) of the backbone Cα atoms for each amino acid in the simulated inteins reveal that the active 110Δ383Leu intein has higher dynamics in core region. (A) V67 inteins; (B) V67L inteins. The lines for 110Δ383 and 96Δ402 inteins have been bolded for effect.
A number of other trends are also shown in the RMSF graphs. The V67 inteins have very dynamic peripheral regions (especially around residues 10 to 27), whilst the V67L inteins show less marked dynamics in these regions. The stabilization of this region appears to be an effect of the presence of the V67L mutation. There are also large spikes in the RMSF values in the tail regions of the intein. This is to be expected, as these regions are unconstrained and free to move. Another trend in both graphs reflects local dynamic effects in the region around residue 67, which is also to be expected. Previous work by Du et al. has shown that extensive chemical shift perturbation occurs in the region around V67L [24]. A third notable trend is the two peaks around residues 19 and 41 in Figure 4, which appear to correlate with the length of the linker loop. The ΔΔIhh-SM, 98Δ402Leu and 96Δ402Leu inteins all tend to spike in RMSF around these two regions, whilst the 110Δ383Leu does not. Given the sudden increase in flexibility in these regions, it is possible that the movement of this region is restricted when a long connecting region is present. When the connection region loop is as long as it is in the 110Δ383 structure it occupies the space that residues around 19 and 41 move into when the longer loop is not present. We conclude that changing the linker loop length will affect the flexibility for the regions around residues 19 and 41.
3. Shorted loop decreases the relative motion between the N and C splicing domain segments
To further investigate the cooperative fluctuation of the individual residues, we computed the covariance matrix of the residue displacements for three of the V67L structures, 110Δ383Leu, 98Δ402Leu, and 96Δ402Leu (Figures 5A, B, and C respectively). The covariance matrix takes into account the direction of movement of each residue in each snapshot during the last nanosecond of each trajectory. We chose to focus on these three models in order to look for any patterns that may give us hints as to why the latter structure, 96Δ402Leu is inactive at this simulation temperature whilst the other two are both active.
Figure 5.
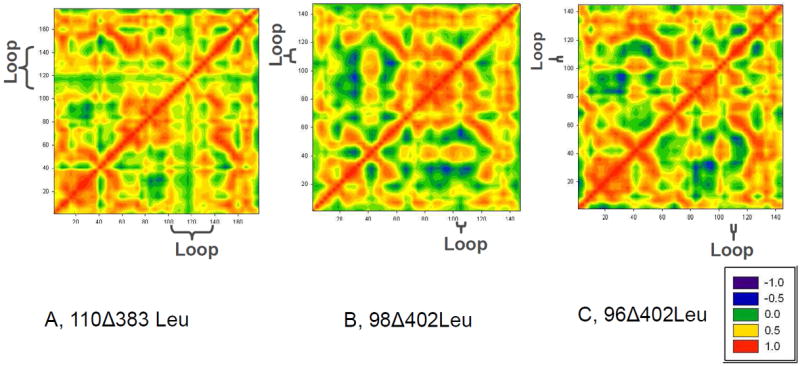
Covariance Matrices of backbone Cα atom movement indicate that the different regions in intein are more rigidly connected with a shorter loop. Covariance Matrices in the last nanosecond of the MD simulations were plotted. Red color indicates that two atoms move in same direction, blue a movement in opposite directions, and green color that the atoms move randomly with respect to each other. A) 110Δ383Leu; B) 98Δ402Leu; C) 96Δ402Leu. All are deletion mutants. A and B are active mutants; C is an inactive mutant.
The matrices present a few interesting trends. With a long loop around the 120-140 region, the covariance matrix of the 110Δ383Leu structure has marked blocks (Figure 5A), which are divided around residues 40 and 120. The line at residue 120 occurs in the middle of the connection region loop, while the line at residue 40 has been shown previously in Figure 4 to correlate directly with the length of the connection region loop. This lattice-like shape is weaker in either the 98Δ402Leu or the 96Δ402Leu mini-inteins, indicating that the different regions in intein are now more rigidly connected. The relative motion between N- and C- splicing domain segments is reflected in the lower right (or upper left) corner of the covariance matrix. As can be seen in Figure 5, the red/orange region around residue 40 becomes continuous in the 96Δ402Leu structure, indicating decreased relative motion between the N- and C- segments. Since intein splicing requires precise orientation of the two splicing domain segments, increased rigidity between the two segments could inhibit intein splicing.
The likely interaction pathway of the long loop region with the N- and C- segments is through the interactions with turn region of 35-41 (40-46 in figure 4). As indicated in figure 6, the turn region (colored green) and the loop region (colored yellow) has directly contact through Leu40. A nearby arginine Arg43 form salt bridges with two nearby glutamate residues (labeled E126 and E127 for ΔΔIhh-SM and E159 and E160 for 110Δ383Leu), which are close to the Csegment. Examination of the loop contact and the salt bridge distances revealed clear correlation with intein activities. We observed the sidechain of Arg98 forms hydrogen bond with the carbonyl oxygen of Leu40. The hydrogen bonding is stronger for active ΔΔIhh-SM than for inactive ΔΔIhh (V67) (Figure 6D). Similar results were observed for 98Δ402 mini-intein. However, the Arg98 is deleted in 96Δ402 mini-intein, where the loop region lost contact with the turn region completely. In the 110Δ383 intein, Asp126 in the loop forms salt bridge directly with the Arg43 (Figure 6C). As indicated in Figure 6E, the active 110Δ383Leu intein forms stronger salt bridge than the less active 110Δ383 V67 intein. When examining the salt bridges between Arg43 with nearby glutamate residues, we found that the active inteins have stronger salt bridge between Arg43-Glu127 (Arg43-Glu160 for 110Δ383), while inactive inteins prefer Arg-Glu126 (Arg43-Glu159 for 110Δ383) interactions (Figure 7). This trend is most strongly reflected in inactive ΔΔIhh (V67) and active ΔΔIhh-SM (Figures 7A and 7E).
Figure 6.
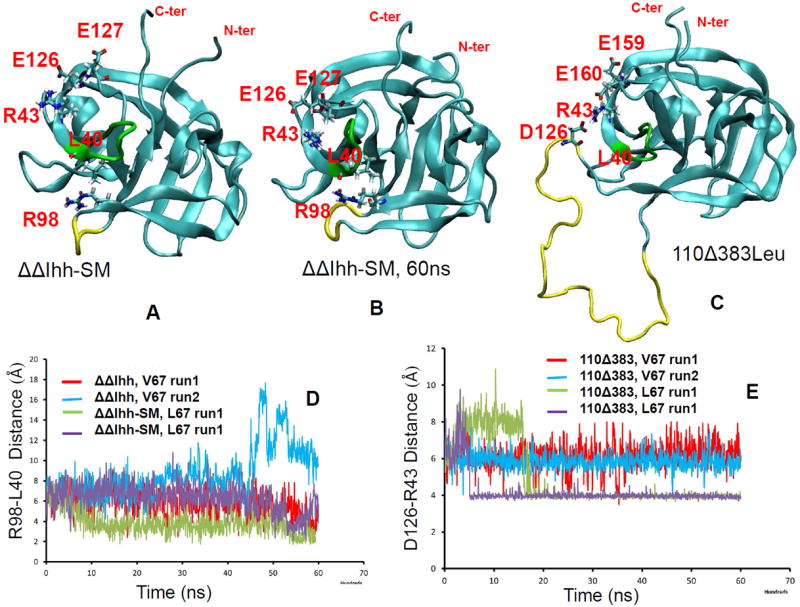
MD simulations revealed interaction pathway connecting linker region to active site of intein. The turn region (colored green) and the loop region (colored yellow) has directly contact through Leu40. A nearby arginine Arg43 form salt bridges with two nearby glutamate residues (labeled E126 and E127 for ΔΔIhh-SM and E159 and E160 for 110Δ383Leu), which are close to the C- segment. (A) the starting structure the intein used in MD simulations (built from PDB code 2L8L), where R98 and L40 has limited sidechain contact and R43 forms salt bridge with E126. (B) snapshot from MD simulation at 60 ns. R98 sidechain form hydrogen bond with backbone oxygen of L40. (C) D126 in linker forms salt bridge with R43 and provided strong interaction with the turn region (colored green). (D) HN(R98)…O(L40) distance trajectories from MD simulations of ΔΔIhh-SM intein. (E) N(R43)…Cγ(D126) distance trajectories from MD simulations of110Δ383 inteins.
Figure 7.
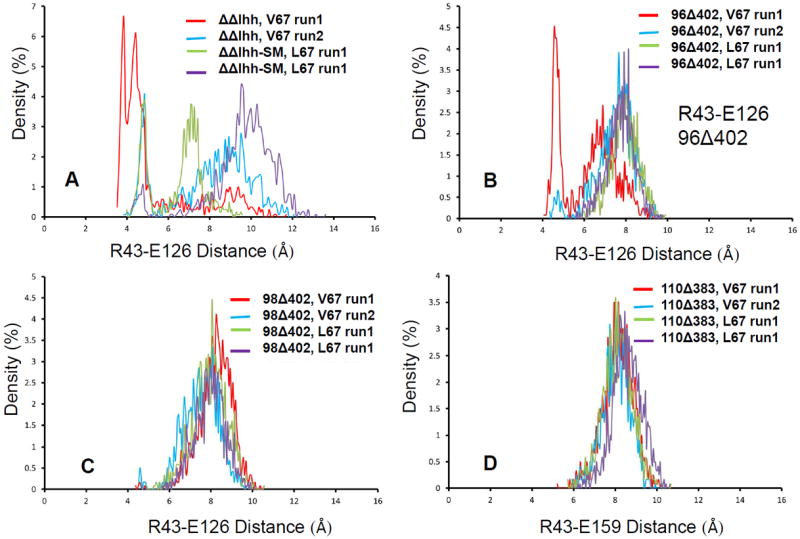
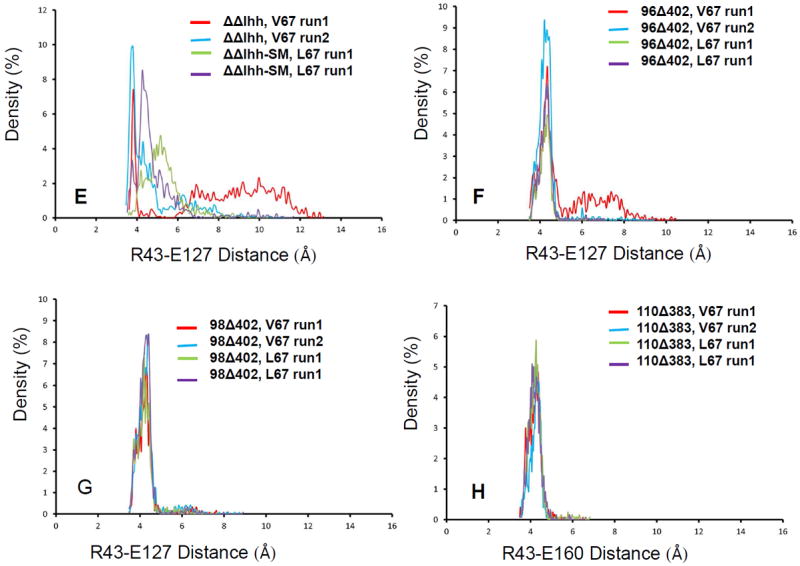
MD simulations revealed interaction reveal active and inactive inteins have different preference of salt bridges. The figures plot salt bridge distance distributions for of Cz (R43)…Cδ(E126) in ΔΔIhh-SM intein (A), 96Δ402 intein (B), and 98Δ402 intein (C). (D) is the distance distributions for of Cz (R43)…Cδ(E159) for 110Δ383 intein. (E-G) are (R43)…Cδ(E127) in ΔΔIhh-SM intein (A), 96Δ402 intein (B), and 98Δ402 intein, respectively. (H) is the distance distributions for of Cz (R43)…Cδ(E160) for 110Δ383 intein.
4. Shortened loops disturb the balance between Asp 422 and the N and C Terminals
Several conserved residues have been determined experimentally and computationally to be critical for intein function [20, 22, 23, 39]. Among these are the first residue Cys or Ser, the last Asn, the penultimate His439, the upstream His73, and Asp422. The dynamics of these residues in the active state could affect the ability of the intein to splice or cleave. We examined nine distances between experimentally validated critical residues, looking for a noticeable change between active and inactive intein models. Table 2 lists the distances that have been considered. The distances are averaged over the last 30 ns of simulation. These distances are classified into two categories: those known to be active (ΔΔIhh-SM, 110Δ383Leu, 98Δ402Leu, 96Δ402Leu at 290K), and those known to be inactive (V67 inteins and the 96Δ402Leu intein at 300K and 310K). Examining the differences between the active and inactive category revealed two important distances – the distance between the C-terminal intein residue (Asn440) and Asp422, and the distance between the intein N-terminus and Asp422 (Figure 8, Table 3). For the active 110Δ383Leu, the dynamic structure allows the adjustment of the relative distances between Cys1 - Asp422 and Asn440 - Asp422. Eventually, Asp422 locates with equal distances to Cys1 and Asn440 (Figure 8A). However, the inactivated 96Δ402 intein does not have sufficient flexibility, remaining stuck at unequal distances between Cys1 - Asp422 and Asn440 - Asp422. As a result, Asp422 tends to be closer to the C-terminus of the mini-inteins in inactive structures, with a notably shorter distance in Figure 8B.
Table 2.
List of active site atomic distances monitored in MD simulations
| Distance Number | Residue Name | Atom name | Residue Number | Residue Name | Atom name | Residue Number |
|---|---|---|---|---|---|---|
| 1 | GLY | CA, Cα carbon | -1 * | ASN | CA, Cα carbon | 440 |
| 2 | CYS | SG, Sulfur | 1 | ASP | OD1, sidechain carboxy oxygen | 422 |
| 3 | CYS | SG, Sulfur | 1 | ASP | OD2, sidechain carboxy oxygen | 422 |
| 4 | ASN | ND2, sidechain nitrogen | 440 | ASP | OD1, sidechain carboxy oxygen | 422 |
| 5 | ASN | ND2, sidechain nitrogen | 440 | ASP | OD2, sidechain carboxy oxygen | 422 |
| 6 | CYS | SG, Sulfur | 1 | HIS | ND1, sidechain nitrogen | 73 |
| 7 | CYS | SG, Sulfur | 1 | HIS | NE2, sidechain nitrogen | 73 |
| 8 | ASN | ND2, sidechain nitrogen | 440 | HIS | ND1, sidechain nitrogen | 73 |
| 9 | ASN | ND2, sidechain nitrogen | 440 | HIS | NE2, sidechain nitrogen | 73 |
The numbering of intein starts from Cys1, The amino acids in N-terminal extein, which will be cleaved from intein and ligated with C-terminal extein, is numbered negatively from Gly -1.
Figure 8.
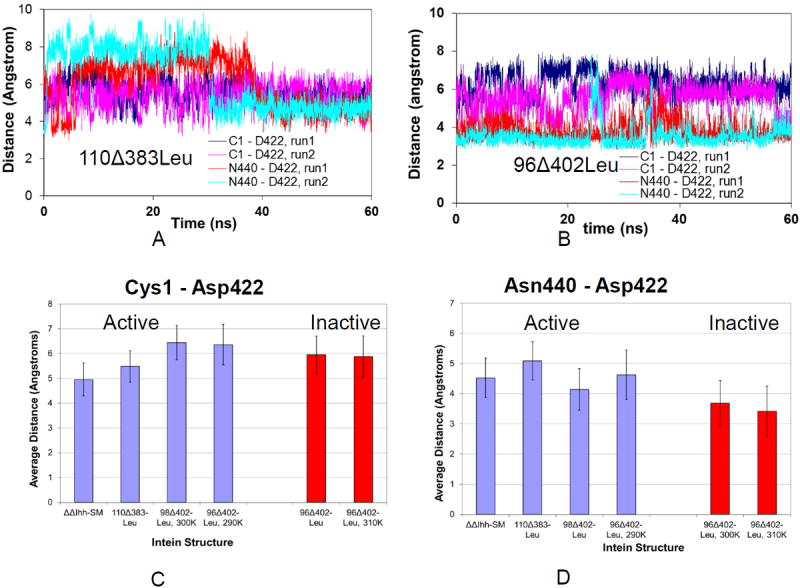
Molecular dynamics trajectories and a histogram illustrating that the distances of Asp422 from Cys1 and Asn440 are different in active and inactive inteins. (A) Asp422 in active intein 110Δ383Leu has similar distances from Cys1 and Asn440. (B) Asp422 of inactive intein 96Δ402Leu has different distances from Cys1 and Asn440. The average distances of Cys1-Asp 422 in different inteins are shown in (C) and that of Cys1-Asn440 in (D). On the X-axis of (C) and (D) these distances are displayed for the different constructs which were simulated.
Table 3.
Active inteins have smaller distance differences between Cys1-Asp422 and Asn440-Asp422.
| Run1 | Run2 | |||||
|---|---|---|---|---|---|---|
| C1 – D422, Å | N440 – D422, Å | Difference, Å | C1 – D422, Å | N440 – D422, Å | Difference, Å | |
| ΔΔIhh-SM | 4.09±0.5 | 3.84±0.93 | 0.25 | 5.82±0.82 | 5.20±1.1 | 0.62 |
| ΔΔIhh | 4.99±1.45 | 3.76±0.69 | 1.23 | 7.10±0.50 | 4.51±0.63 | 2.58 |
| 110Δ383Leu | 5.45±0.55 | 5.45±2.22 | 0.0 | 5.51±0.72 | 4.73±0.59 | 0.78 |
| 110Δ383 | 4.76±1.04 | 3.60±0.75 | 1.27 | 4.93±5.92 | -5.92±0.26 | 0.42 |
| 98Δ402Leu | 6.5±0.6 | 3.21±0.44 | 3.29 | 6.37±0.77 | 5.06±1.13 | 1.31 |
| 98Δ402 | 5.52±1.01 | 4.58±0.83 | 0.94 | 6.24±0.86 | 4.94±1.11 | 1.30 |
| 96Δ402Leu | 6.24±0.62 | 3.82±0.75 | 2.41 | 5.67±0.88 | 3.55±0.57 | 2.12 |
| 96Δ402 | 5.68±0.93 | 4.53±1.11 | 1.15 | 5.90±1.06 | 3.39±0.57 | 2.50 |
| 96Δ402Leu, 310K | 5.48±1.03 | 3.37±0.70 | 2.12 | 6.28±0.63 | 3.46±0.65 | 2.81 |
| 96Δ402Leu, 290K | 6.28±0.83 | 4.01±0.62 | 2.28 | 6.44±0.8 | 5.24±0.97 | 1.20 |
Table 3 lists the distances between Cys1 - Asp422 and Asn440 - Asp422 for all structures simulated in this study. As can be seen in Table 3, both active ΔΔIhh-SM and 110Δ383Leu structures have similar distances between Cys1-Asp422 and Asn440-Asp422. The ranges of the distance differences between Cys1-Asp422 and Asn440-Asp422 are 0.25 to 0.62 Å and 0.0 to 0.78 Å for ΔΔIhh-SM and 110Δ383Leu, respectively. With the shortened loops in the 98Δ402Leu and 96Δ402Leu structures, the corresponding distance differences increased to the range of 1.31 - 3.29 Å and 2.12 – 2.41 Å respectively. It is interesting to note that at the high 310 K temperature, the distance difference for the 96Δ402Leu structure increased to 2.12 – 2.81 Å, while the distance difference decreased to 1.20 – 2.28 Å at the lower temperature of 290 K. The decreased distance differences between Cys1-Asp422 and Asn440-Asp422 at 290 K suggest why 96Δ402Leu could be active at that temperature.
Val67 has different effects on the distance distributions for the ΔΔIhh and 110Δ383 than for the shortened loop inteins 98Δ402 and 96Δ402. For ΔΔIhh and 110Δ383, the differences are larger than in the Leu67 form, being 1.23 - 2.58 Å and 0.42 – 1.27 Å, respectively. The large change in the distance difference between Cys1-Asp422 and Asn440-Asp422 for the ΔΔIhh (Val 67) and ΔΔIhh-SM (Leu67) also corroborates its activity losses. However, the V67L transition for the 98Δ402 and 96Δ402 structures shows that Val67 may have smaller distance distributions than the Leu67 form, which is not consistent with the observed intein activity. Apparently, other factors also contribute to the regulation of intein splicing.
Nevertheless, our simulations have shown that the ability of a residue to be flexible within a balanced range of distances between Cys1-Asp422 and Asn440-Asp422 is crucial to the activity of an intein. It is known that Asp422 aids intein activity by bridging the N- and C-termini, interacting with both [22, 23]. We show that the Asp422 is more flexible in the active structures than in the inactive structures, as the active structures are able to attain a greater range of distance values, which supports the mechanism that Asp422 is crucial for interacting with both the N- and C-termini. Though the residue is in close proximity to both termini, it must still maintain a level of flexibility in order to correctly assume the conformations it requires to properly carry out the intein splicing activity. Overall, we can conclude from this that an active intein will tend to fluctuate and be more dynamic around the active site, especially around Asp422.
5. Intein catalysis is characterized by non-linear Arrhenius plot
In order to investigate the role of protein dynamics in intein catalysis, we examined its cleaving kinetics at different temperatures from 20.5 °C to 42 °C. As indicated in Figure 9, an Arrhenius plot of the transformed data clearly shows the non-linearity of the rate constant with respect to temperature. Specifically, two transition temperatures are observed, indicating a strong coupling between conformational dynamics and activation energy [48-54]. There are two transition points along the Arrhenius plot, with one around 28.3 °C and another around 34.8 °C. The decreasing slop around 28.3 °C indicates that activation energy decreases from 28.3 °C to 34.8 °C. Apparently, the Mtu RecA mini-intein has higher conformational dynamics at temperatures above 28.3 °C, leading to a decrease in activation energy. The most likely region that contributes to this transition is the loop region. The second point around 34.8 °C indicates that the overall stability of the Mtu RecA mini-intein is becoming strongly perturbed above this temperature, leading to the increase of activation energy. There is no indication of protein unfolding around the 34.8 °C transition temperature, indicating that the intein activity is perturbed by dynamics rather than global structural stabilities. Our simulations at 310K also indicated that intein maintains its global structural stability at 310K. Previously, it was found that chemical shift peak of H429 becomes stronger from 5 °C to 55 °C, indicating the higher structural dynamics from low temperatures to high temperature[24]. Therefore, it is possible that the balance needed for key catalytic residues to mediate C-termini interactions is lost when temperature goes too high.
Figure 9.
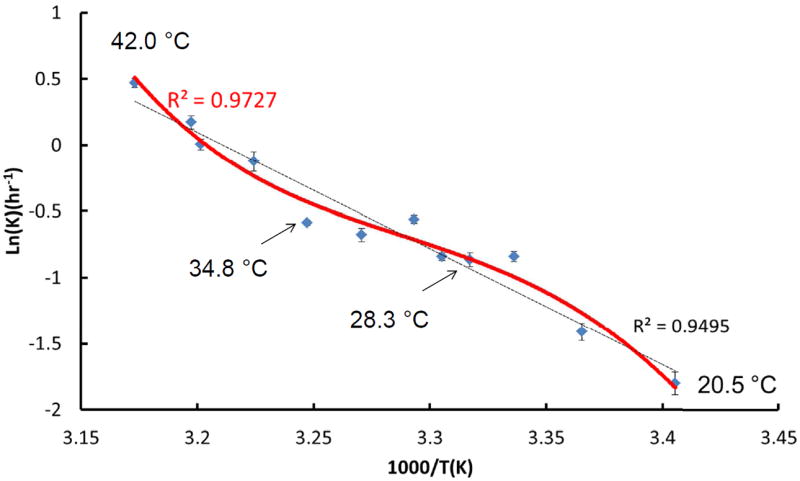
Temperature dependence of the catalytic constant in the form of an Arrhenius plot for the Mtu recA intein reveals a strong coupling between conformational dynamics and activation energy. The dashed line represents a linear fit of the experimental observation using all temperature data, while the solid line has the best fit of experimental observation.
DISCUSSION
A recent crystal structure of active intein core did not show substantial conformational change, probably due to the missing of C-extein residues [55]. The activation energy for intein cleavage was found to be significantly higher than those observed for typical enzyme-catalyzed reactions, and its strong temperature dependence suggest that large conformational changes are needed for the cleavage reaction to occur [27]. There are two possible underlying mechanisms for the high activation energy. First, quantum mechanical calculations [55, 56] have shown that C-terminal cleavage already has an activation barrier of 19 to 28 kcal/mol. Second, the energy needed for the large conformational dynamics may be coupled with the activation energy of the chemical bond cleavage. To elucidate the underlying conformational mechanism of intein activity, our study computationally modeled a number of inteins with known activities, and experimentally examined the temperature dependence of intein catalytic kinetics for one well-known example. We investigated the dynamics, stability, and possible structural changes due to changing conditions, and searched for possible reasons behind them. We further questioned whether an active intein should maintain a stable protein structure, and at the same time whether, as would be expected under such circumstances, an active intein would have dynamic correlations among the different regions to allow the intein to catalyze the splicing/cleaving activity at the active site.
The balance between stability and dynamics in any enzyme is difficult to quantify. The enzyme, in this case intein, must be sufficiently stable, but it also must be sufficiently dynamic so that it can perform its function. Our simulation results support the experimental indication that the V67L mutation stabilizes the intein (as shown by the increased global stability in the presence of the V67L mutation), and the proposition that this stabilization leads to higher intein activity. However, we found that the V67L mutation restores the active state dynamics of the Mtu RecA mini-intein after deletion of the endonuclease domain. We further investigated the roles that global stability and dynamics play in determining the difference between the active and inactive intein by analyzing inteins with different linker loop lengths and at different temperatures. We found that while shortening the linker loop between the splicing domain segments stabilizes the intein, shortening the loop too much will result in the intein structure becoming so rigid that it is unable to perform its splicing function. An intein with the V67L mutation is only functional if the linker loop where the endonuclease domain was removed is flexible enough to allow intein activity. The fact that intein activity can be modulated by linker sequence and length is consistent with general observations that linker regions can allosterically control protein functions [57]. We have observed that the interaction of the linker region and a turn region around residues 35-41 provided the pathway for this allostery interaction. Generally, the contact between the positively charged Arg98 and hydrophobic Leu40 is not favored. It is interesting to see that the sidechain-backbone interaction compensate the mismatch between the charged residue Arg98 and hydrophobic residue Leu40. Experimentally, it has been found that mutation of the Arg98 to more hydrophobic Cystein or Proline can improve inteins’ activity[19], probably with better hydrophobic interactions with Leu40. As a strong support to above suggestion, we found the active 110Δ383Leu intein forms stronger salt bridge than the less active 110Δ383 V67 intein (Figure 6E), and it is interesting to see how small perturbation of V67L mutation can lead to large allosteric effects of salt bridge between R43 and D126, which eventually effects overall dynamics around active site. The requirement of dynamics for intein function can also be reflected by the Asp422, which is one of the several conserved residues critical to splicing [22, 23]. Asp422 aids intein activity by facilitating reactions at both the N- and the C-termini. In order to do so, the residue must maintain a certain level of flexibility, which is required to assume the conformations needed for proper intein activity. We find that the distances between Asp422 and the N- and C-termini cover a wide range of values, while maintaining an equal distance only in the active inteins. Asp422 tends to fluctuate more in active inteins and that increased activity can be associated with increased fluctuations around the active site.
We have observed non-linear Arrhenius plot of intein catalysis. A similar case was reported for 3-isopropylmalate dehydrogenase. The linker which is responsible for the hinge-bending domain motions in 3-isopropylmalate dehydrogenase controls its conformational dynamics and leads to an unusual dependence of the catalytic efficiency on temperature [53]. While the 3-isopropylmalate dehydrogenase has a simple curved Arrhenius plot, a Sigmoidal Arrhenius plot is observed for the dissociation constant of 3-isopropylmalate [53]. In general, it is believed that enzymes have evolved such that the lowest-energy states are the most active, thereby leading to low activation energies even as the temperature is increased [48, 51]. A convex Arrhenius plot indicates a decreasing activation energy with increasing temperature [51], as is observed for the Mtu RecA mini-intein in the 30.5 °C to 35 °C temperature range. In many cases, the lowest-energy states in enzyme catalysis can be conformationally flexible and are favorable at higher temperatures, leading to observed biphasic rate constants with changes in temperature [48-54]. For example, wild-type tetrameric thermophilic alcohol dehydrogenase from Bacillus stearothermophilus (ht-ADH) has small kink in its Arrhenius plot [54], indicating an obvious yet weak change in activation energy. Notably, a ht-W87A mutation in this enzyme can abolish this temperature dependent breakpoint [54]. Some enzymes like the E1 component of the Escherichia coli pyruvate dehydrogenase multienzyme complex (E1ec) [50] and Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) [52] have large temperature-dependent activation energy changes. This is due to the fact that that the protein conformational ensemble and dynamics modulate cooperativity in the rate-determining catalytic step. Interestingly, similar to the W87A mutation in ht-ADH, a D549A mutation in the dynamic loop of E1ec also abolishes its biphasic kinetic behavior [50].
Our experimental examination of the dependence of intein cleaving kinetics on temperature clearly reveals a strong coupling between conformational dynamics and activation energy. This dependence suggests the need for a balance between stability and dynamics in intein splicing. Our results provide further clarify temperature sensitive inteins [25, 26], and may also suggest potential methods to reversibly control inteins’ function for various applications. For example, it is still a challenge to tightly control intein function within mammalian cells, which has hindered the adoption of self-cleaving tag methods in these expression hosts. Although evolutionary and other more direct protein engineering approaches can be used to control intein function, our studies additionally suggest that effectively quenching the conformational dynamics of an intein through engineered allosteric interactions could deactivate intein splicing or cleaving as well.
Conformational dynamics are indispensable to enzyme function.
MD simulations revealed that loop length affects intein dynamics and the linker and core flexibility are higher in the active structure.
Catalysis is characterized by non-linear Arrhenius plot, confirming protein conformational dynamics importance to intein function.
Acknowledgments
This project has been funded in whole or in part with Federal funds from the National Cancer Institute, National Institutes of Health, under contract number HHSN261200800001E. M. Cronin thanks student intern program in Frederick National Lab. All simulations were performed using the high-performance computational facilities of the Biowulf PC/Linux cluster at the NIH, Bethesda, MD (http://biowulf.nih.gov) and ABCC at Frederick National Laboratory. D. Wood was supported by startup funds from the Ohio State University.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Paulus H. Protein splicing and related forms of protein autoprocessing. Annu Rev Biochem. 2000;69:447–496. doi: 10.1146/annurev.biochem.69.1.447. [DOI] [PubMed] [Google Scholar]
- 2.Perler FB. Protein splicing of inteins and hedgehog autoproteolysis: structure. function, and evolution, Cell. 1998;92:1–4. doi: 10.1016/s0092-8674(00)80892-2. [DOI] [PubMed] [Google Scholar]
- 3.Evans TC, Jr, Benner J, Xu MQ. The cyclization and polymerization of bacterially expressed proteins using modified self-splicing inteins. The Journal of biological chemistry. 1999;274:18359–18363. doi: 10.1074/jbc.274.26.18359. [DOI] [PubMed] [Google Scholar]
- 4.Kwon Y, Coleman MA, Camarero JA. Selective immobilization of proteins onto solid supports through split-intein-mediated protein trans-splicing. Angew Chem Int Ed Engl. 2006;45:1726–1729. doi: 10.1002/anie.200503475. [DOI] [PubMed] [Google Scholar]
- 5.Romanelli A, Shekhtman A, Cowburn D, Muir TW. Semisynthesis of a segmental isotopically labeled protein splicing precursor: NMR evidence for an unusual peptide bond at the N-extein-intein junction. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:6397–6402. doi: 10.1073/pnas.0306616101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seyedsayamdost MR, Yee CS, Reece SY, Nocera DG, Stubbe J. pH Rate profiles of FnY356-R2s (n = 2, 3, 4) in Escherichia coli ribonucleotide reductase: evidence that Y356 is a redox-active amino acid along the radical propagation pathway. Journal of the American Chemical Society. 2006;128:1562–1568. doi: 10.1021/ja055927j. [DOI] [PubMed] [Google Scholar]
- 7.Zuger S, Iwai H. Intein-based biosynthetic incorporation of unlabeled protein tags into isotopically labeled proteins for NMR studies. Nature biotechnology. 2005;23:736–740. doi: 10.1038/nbt1097. [DOI] [PubMed] [Google Scholar]
- 8.Ramirez M, Guan D, Ugaz V, Chen Z. Intein-triggered artificial protein hydrogels that support the immobilization of bioactive proteins. Journal of the American Chemical Society. 2013;135:5290–5293. doi: 10.1021/ja401075s. [DOI] [PubMed] [Google Scholar]
- 9.Selgrade DF, Lohmueller JJ, Lienert F, Silver PA. Protein scaffold-activated protein trans-splicing in mammalian cells. Journal of the American Chemical Society. 2013;135:7713–7719. doi: 10.1021/ja401689b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wood DW, Camarero JA. Intein Applications: From Protein Purification and Labeling to Metabolic Control Methods. The Journal of biological chemistry. 2014 doi: 10.1074/jbc.R114.552653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paulus H. Protein splicing inhibitors as a new class of antimycobacterial agents. Drugs of the Future. 2007;32:973–984. [Google Scholar]
- 12.Perler FB. InBase: the Intein Database. Nucleic Acids Res. 2002;30:383–384. doi: 10.1093/nar/30.1.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chevalier BS, Stoddard BL. Homing endonucleases: structural and functional insight into the catalysts of intron/intein mobility. Nucleic Acids Res. 2001;29:3757–3774. doi: 10.1093/nar/29.18.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gimble FS. Invasion of a multitude of genetic niches by mobile endonuclease genes. FEMS Microbiol Lett. 2000;185:99–107. doi: 10.1111/j.1574-6968.2000.tb09046.x. [DOI] [PubMed] [Google Scholar]
- 15.Liu XQ. Protein-splicing intein: Genetic mobility, origin, and evolution. Annu Rev Genet. 2000;34:61–76. doi: 10.1146/annurev.genet.34.1.61. [DOI] [PubMed] [Google Scholar]
- 16.Derbyshire V, Wood DW, Wu W, Dansereau JT, Dalgaard JZ, Belfort M. Genetic definition of a protein-splicing domain: functional mini-inteins support structure predictions and a model for intein evolution. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:11466–11471. doi: 10.1073/pnas.94.21.11466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Du Z, Liu Y, Zheng Y, McCallum S, Dansereau J, Derbyshire V, Belfort M, Belfort G, Van Roey P, Wang C. 1H, 13C, and 15N NMR assignments of an engineered intein based on Mycobacterium tuberculosis RecA. Biomol NMR Assign. 2008;2:111–113. doi: 10.1007/s12104-008-9098-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hiraga K, Derbyshire V, Dansereau JT, Van Roey P, Belfort M. Minimization and stabilization of the Mycobacterium tuberculosis recA intein. J Mol Biol. 2005;354:916–926. doi: 10.1016/j.jmb.2005.09.088. [DOI] [PubMed] [Google Scholar]
- 19.Hiraga K, Soga I, Dansereau JT, Pereira B, Derbyshire V, Du Z, Wang C, Van Roey P, Belfort G, Belfort M. Selection and structure of hyperactive inteins: peripheral changes relayed to the catalytic center. J Mol Biol. 2009;393:1106–1117. doi: 10.1016/j.jmb.2009.08.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shemella P, Pereira B, Zhang Y, Van Roey P, Belfort G, Garde S, Nayak SK. Mechanism for intein C-terminal cleavage: a proposal from quantum mechanical calculations. Biophysical journal. 2007;92:847–853. doi: 10.1529/biophysj.106.092049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shingledecker K, Jiang SQ, Paulus H. Molecular dissection of the Mycobacterium tuberculosis RecA intein: design of a minimal intein and of a trans-splicing system involving two intein fragments. Gene. 1998;207:187–195. doi: 10.1016/s0378-1119(97)00624-0. [DOI] [PubMed] [Google Scholar]
- 22.Van Roey P, Pereira B, Li Z, Hiraga K, Belfort M, Derbyshire V. Crystallographic and mutational studies of Mycobacterium tuberculosis recA mini-inteins suggest a pivotal role for a highly conserved aspartate residue. J Mol Biol. 2007;367:162–173. doi: 10.1016/j.jmb.2006.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wood DW, Wu W, Belfort G, Derbyshire V, Belfort M. A genetic system yields self-cleaving inteins for bioseparations. Nature biotechnology. 1999;17:889–892. doi: 10.1038/12879. [DOI] [PubMed] [Google Scholar]
- 24.Du Z, Liu Y, Ban D, Lopez MM, Belfort M, Wang C. Backbone dynamics and global effects of an activating mutation in minimized Mtu RecA inteins. J Mol Biol. 2010;400:755–767. doi: 10.1016/j.jmb.2010.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zeidler MP, Tan C, Bellaiche Y, Cherry S, Hader S, Gayko U, Perrimon N. Temperature-sensitive control of protein activity by conditionally splicing inteins. Nature biotechnology. 2004;22:871–876. doi: 10.1038/nbt979. [DOI] [PubMed] [Google Scholar]
- 26.Tan G, Chen M, Foote C, Tan C. Temperature-sensitive mutations made easy: generating conditional mutations by using temperature-sensitive inteins that function within different temperature ranges. Genetics. 2009;183:13–22. doi: 10.1534/genetics.109.104794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wood DW, Derbyshire V, Wu W, Chartrain M, Belfort M, Belfort G. Optimized single-step affinity purification with a self-cleaving intein applied to human acidic fibroblast growth factor. Biotechnology progress. 2000;16:1055–1063. doi: 10.1021/bp0000858. [DOI] [PubMed] [Google Scholar]
- 28.Wu W, Wood DW, Belfort G, Derbyshire V, Belfort M. Intein-mediated purification of cytotoxic endonuclease I-TevI by insertional inactivation and pH-controllable splicing. Nucleic Acids Res. 2002;30:4864–4871. doi: 10.1093/nar/gkf621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shkumatov AV, Chinnathambi S, Mandelkow E, Svergun DI. Structural memory of natively unfolded tau protein detected by small-angle X-ray scattering. Proteins. 2011;79:2122–2131. doi: 10.1002/prot.23033. [DOI] [PubMed] [Google Scholar]
- 30.Gunasekaran K, Ma B, Nussinov R. Is allostery an intrinsic property of all dynamic proteins? Proteins. 2004;57:433–443. doi: 10.1002/prot.20232. [DOI] [PubMed] [Google Scholar]
- 31.Tsai CJ, del Sol A, Nussinov R. Allostery: absence of a change in shape does not imply that allostery is not at play. Journal of molecular biology. 2008;378:1–11. doi: 10.1016/j.jmb.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ngu TT, Lee JA, Rushton MK, Stillman MJ. Arsenic metalation of seaweed Fucus vesiculosus metallothionein: the importance of the interdomain linker in metallothionein. Biochemistry. 2009;48:8806–8816. doi: 10.1021/bi9007462. [DOI] [PubMed] [Google Scholar]
- 33.Tsai CJ, Del Sol A, Nussinov R. Protein allostery, signal transmission and dynamics: a classification scheme of allosteric mechanisms. Mol Biosyst. 2009;5:207–216. doi: 10.1039/b819720b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma B, Nussinov R. Enzyme dynamics point to stepwise conformational selection in catalysis. Curr Opin Chem Biol. 2010;14:652–659. doi: 10.1016/j.cbpa.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Du Z, Liu J, Albracht CD, Hsu A, Chen W, Marieni MD, Colelli KM, Williams JE, Reitter JN, Mills KV, Wang C. Structural and mutational studies of a hyperthermophilic intein from DNA polymerase II of Pyrococcus abyssi. The Journal of biological chemistry. 2011;286:38638–38648. doi: 10.1074/jbc.M111.290569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frutos S, Goger M, Giovani B, Cowburn D, Muir TW. Branched intermediate formation stimulates peptide bond cleavage in protein splicing. Nature chemical biology. 2010;6:527–533. doi: 10.1038/nchembio.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Volkmann G, Mootz HD. Recent progress in intein research: from mechanism to directed evolution and applications. Cellular and molecular life sciences : CMLS. 2013;70:1185–1206. doi: 10.1007/s00018-012-1120-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eryilmaz E, Shah N, Muir T, Cowburn D. Structural and Dynamical Features of Inteins and Implications on Protein Splicing. The Journal of biological chemistry. 2014 doi: 10.1074/jbc.R113.540302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Du Z, Shemella PT, Liu Y, McCallum SA, Pereira B, Nayak SK, Belfort G, Belfort M, Wang C. Highly conserved histidine plays a dual catalytic role in protein splicing: a pKa shift mechanism. Journal of the American Chemical Society. 2009;131:11581–11589. doi: 10.1021/ja904318w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Du Z, Zheng Y, Patterson M, Liu Y, Wang C. pK(a) coupling at the intein active site: implications for the coordination mechanism of protein splicing with a conserved aspartate. Journal of the American Chemical Society. 2011;133:10275–10282. doi: 10.1021/ja203209f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jorgensen JCWL, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79 [Google Scholar]
- 42.M MW, Jorgensen WL. A five-site model for liquid water and the reproduction of the density anomaly by rigid, nonpolarizable potential functions. J Chem Phys. 2000;112:8910–8922. [Google Scholar]
- 43.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Jr, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, III, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. Journal of Physical Chemistry B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 45.Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical-Integration of Cartesian Equations of Motion of a System with Constraints -Molecular-Dynamics of N-Alkanes. Journal of Computational Physics. 1977;23:327–341. [Google Scholar]
- 46.Darden T, York D, Pedersen L. Particle Mesh Ewald -an N.Log(N) Method for Ewald Sums in Large Systems. Journal of Chemical Physics. 1993;98:10089–10092. [Google Scholar]
- 47.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. J Comput Chem. 1983;4:187–217. [Google Scholar]
- 48.Nagel ZD, Dong M, Bahnson BJ, Klinman JP. Impaired protein conformational landscapes as revealed in anomalous Arrhenius prefactors. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:10520–10525. doi: 10.1073/pnas.1104989108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zoldak G, Sut’ak R, Antalik M, Sprinzl M, Sedlak E. Role of conformational flexibility for enzymatic activity in NADH oxidase from Thermus thermophilus. European journal of biochemistry / FEBS. 2003;270:4887–4897. doi: 10.1046/j.1432-1033.2003.03889.x. [DOI] [PubMed] [Google Scholar]
- 50.Kale S, Jordan F. Conformational ensemble modulates cooperativity in the rate-determining catalytic step in the E1 component of the Escherichia coli pyruvate dehydrogenase multienzyme complex. The Journal of biological chemistry. 2009;284:33122–33129. doi: 10.1074/jbc.M109.065508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Truhlar D, Kohen A. Convex Arrhenius plots and their interpretation. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:848–851. doi: 10.1073/pnas.98.3.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hajdu I, Bothe C, Szilagyi A, Kardos J, Gal P, Zavodszky P. Adjustment of conformational flexibility of glyceraldehyde-3-phosphate dehydrogenase as a means of thermal adaptation and allosteric regulation. European biophysics journal : EBJ. 2008;37:1139–1144. doi: 10.1007/s00249-008-0332-x. [DOI] [PubMed] [Google Scholar]
- 53.Hajdu I, Szilagyi A, Kardos J, Zavodszky P. A link between hinge-bending domain motions and the temperature dependence of catalysis in 3-isopropylmalate dehydrogenase. Biophysical journal. 2009;96:5003–5012. doi: 10.1016/j.bpj.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nagel ZD, Cun S, Klinman JP. Identification of a long-range protein network that modulates active site dynamics in extremophilic alcohol dehydrogenases. The Journal of biological chemistry. 2013;288:14087–14097. doi: 10.1074/jbc.M113.453951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mujika JI, Lopez X, Mulholland AJ. Mechanism of C-terminal intein cleavage in protein splicing from QM/MM molecular dynamics simulations. Organic & biomolecular chemistry. 2012;10:1207–1218. doi: 10.1039/c1ob06444d. [DOI] [PubMed] [Google Scholar]
- 56.Shemella PT, Topilina NI, Soga I, Pereira B, Belfort G, Belfort M, Nayak SK. Electronic structure of neighboring extein residue modulates intein C-terminal cleavage activity. Biophysical journal. 2011;100:2217–2225. doi: 10.1016/j.bpj.2011.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ma B, Tsai CJ, Haliloglu T, Nussinov R. Dynamic allostery: linkers are not merely flexible. Structure. 2011;19:907–917. doi: 10.1016/j.str.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]


