Abstract
Aims/hypothesis
Glucose-dependent insulinotropic polypeptide (GIP) is an incretin hormone with anti-apoptotic effects on the pancreatic beta cell. The aim of this study was to generate transgenic mice with fluorescently labelled GIP-secreting K cells and to use these to investigate pathways by which K cells detect nutrients.
Methods
Transgenic mice were generated in which the GIP promoter drives the expression of the yellow fluorescent protein Venus. Fluorescent cells were purified by flow cytometry and analysed by quantitative RT-PCR. GIP secretion was assayed in primary cultures of small intestine.
Results
Expression of Venus in transgenic mice was restricted to K cells, as assessed by immunofluorescence and measurements of the Gip mRNA and GIP protein contents of purified cells. K cells expressed high levels of mRNA for Kir6.2 (also known as Kcnj11), Sur1 (also known as Abcc8), 1 Sglt1 (also known as Slc5a1), and of the G-protein-coupled lipid receptors Gpr40 (also known as Ffar1), Gpr119 and Gpr120. In primary cultures, GIP release was stimulated by glucose, glutamine and linoleic acid, and potentiated by forskolin plus 3-isobutyl-1-methylxanthine (IBMX), but was unaffected by the artificial sweetener sucralose. Secretion was half-maximal at 0.6 mmol/l glucose and partially mimicked by α-methylglucopyranoside, suggesting the involvement of SGLT1. Tolbutamide triggered secretion under basal conditions, whereas diazoxide suppressed responses in forskolin/IBMX.
Conclusions/interpretation
These transgenic mouse and primary culture techniques provide novel opportunities to interrogate the mechanisms of GIP secretion. Glucose-triggered GIP secretion was SGLT1-dependent and modulated by KATP channel activity but not determined by sweet taste receptors. Synergistic stimulation by elevated cAMP and glucose suggests that targeting appropriate G-protein-coupled receptors may provide opportunities to modulate GIP release in vivo.
Keywords: cAMP, GIP, Incretin, K cells, SGLT1
Glucose-dependent insulinotropic polypeptide (GIP) is an intestinal hormone that promotes insulin release and coordinates the fate of dietary fat. Together with glucagon-like peptide 1 (GLP-1), it is responsible for around 50% of the normal release of insulin following glucose ingestion, a phenomenon termed the incretin effect [1, 2]. In addition, GIP has been reported to stimulate fat deposition directly in adipocytes and has been postulated to link overnutrition to obesity [3-5].
Whilst the GLP-1 axis has proved a fruitful target for new glucose-lowering drugs, with both GLP-1 mimetics and inhibitors of GLP-1 degradation available for the treatment of type 2 diabetes [6], it is currently uncertain whether GIP receptor agonists will prove to be similarly beneficial, as pharmacological doses of GLP-1, but not GIP, have been shown to increase insulin release in this patient group [7]. The difference may be due, in part, to receptor desensitisation, which is rapid for GIP receptors and enhanced by hyperglycaemia [8]. Nevertheless, GIP, like GLP-1, has beneficial proliferative and anti-apoptotic effects on pancreatic islet cells [9], and dipeptidyl peptidase IV-resistant GIP analogues have been shown to improve glucose homeostasis in normal, obese and diabetic rodents [10, 11].
By contrast, there is increasing evidence that GIP plays a role in fat metabolism and that inhibiting the GIP axis may be of therapeutic value in obesity [12]. Thus, GIP-receptor knockout mice, whilst mildly glucose-intolerant [13], were protected against obesity in high-fat-fed and ob/ob models [14], and chronic interruption of GIP signalling by GIP receptor antagonists or K cell ablation was shown to have beneficial effects on weight gain, insulin sensitivity and glucose tolerance in various rodent models of obesity [15-17]. As antagonising GIP results in an acute reduction in glucose-triggered insulin secretion, as expected [18, 19], it is currently unclear whether its longer-term beneficial effects are due to the reduced circulating insulin levels or to the loss of extrapancreatic actions of GIP [20].
GIP secretion is triggered by the ingestion of carbohydrate or fat, elevated levels being detectable in the plasma within 10-20 min [21, 22]. As the GIP-containing K cells are located in the duodenal and jejunal epithelia and have apical surfaces opening into the gut lumen [23, 24], they are believed to sense luminal nutrients directly. Studies on isolated perfused rodent intestine have suggested that carbohydrate detection by K cells involves the sodium-dependent sugar uptake pathway, as the potency of different monosaccharides to trigger GIP release was found to match the properties of the intestinal Na+-coupled glucose transporters (e.g. sodium-dependent glucose transporter 1, SGLT1) [25, 26]. The finding that the SGLT inhibitor phloridzin impaired glucose-stimulated GIP secretion in rodent intestine [25] supported this view and further indicated that the mere presence of sugars in the gut lumen is not sufficient to trigger GIP secretion [27].
At the cellular and molecular levels very little is known about how K cells respond to glucose and other stimuli, largely because there is a lack of validated cell models for studying GIP release in vitro. GIP-releasing cell lines have been developed by subcloning STC-1 cells, but the STC-1 cell line was developed originally as a model of secretin secretion and also produces a wide range of other enteroendocrine peptides, including GLP-1 and cholecystokinin [28]. Although potentially useful for the identification of pathways in enteroendocrine cells in general, it is less clear whether results from STC-1 cells accurately mirror the behaviour of native K cells. Nevertheless, as identifying and culturing primary K cells has been fraught with difficulties, the use of such cell lines have so far provided the only reproducible method to study GIP secretion in vitro.
Although K cells are traditionally considered to be a distinct subset of enteroendocrine cells, there are reports in the literature of a subpopulation of K cells which stain for GLP-1 as well as GIP [29]. It is therefore perhaps not too surprising that the monosaccharide sensitivity of GIP secretion found in perfused intestinal preparations is similar to that reported for GLP-1 secretion, suggesting that similar mechanisms might underlie the glucose-dependent release of these two hormones. We previously reported that sugar-triggered GLP-1 secretion in the cell line GLUTag is determined by the activity of SGLTs, which can excite cells as a result of the coupled uptake of one or two Na+ ions for each molecule of glucose transported [30]. This generates a small inward current that is sufficient to depolarise the cells, triggering the firing of action potentials and Ca2+ entry through L-type voltage-gated Ca2+ channels. In addition, GLUTag cells express functional ATP-sensitive K+ (KATP) channels, the closure of which by glucose metabolism or sulfonylureas contributes to enhanced cell excitability and GLP-1 secretion [31]. By contrast, studies of GIP secretion using an STC-1 subclone (GIP/ins) suggested that nutrient sensing arises from the metabolic inhibition of AMP-dependent kinase-related kinases [32], and that native K cells do not express KATP channels [33]. It has also been proposed recently that glucose sensing by K and L cells may involve sweet taste receptor pathways, and that these enteroendocrine cells thereby resemble sweet-responsive lingual chemosensory cells [34, 35].
To enable the identification and characterisation of primary GIP-secreting K cells, we generated transgenic mice expressing a yellow fluorescent protein (Venus) under the control of the GIP promoter (GIP-Venus mice). Fluorescent intestinal cells from these mice, purified by flow cytometry, were used to identify the glucose-sensing machinery expressed in native K cells by quantitative RT-PCR and to evaluate the expression of certain lipid-sensitive G-protein-coupled receptors that have been postulated to play a role in fat detection by enteroendocrine cells. We also established primary cultures of adult mouse small intestine and used this model to investigate the functional properties of primary cultured K cells.
Methods
Generation of transgenic mice
Bacterial artificial chromosome (BAC) constructs containing the sequence of Venus driven by the rat GIP promoter were made by the RedEt technique [36] (see Electronic supplementary material [ESM]). Mice were bred in-house and animal procedures were approved by the local ethics committee and conformed to UK Home Office regulations.
Intestinal epithelial cell isolation for flow cytometry
Venus-expressing transgenic mice aged 2–6 months were killed by cervical dislocation and the gut was collected into ice-cold Leibovitz-15 (L-15) medium (PAA Laboratories, Yeovil, UK). The upper small intestine (incorporating 10-15 cm of gut, immediately distal to the pylorus), was opened, rinsed in L-15 and chopped into 1-2 mm pieces. Tissue was digested twice for 30 min with 1 mg/ml collagenase-XI in calcium-free Hanks’ balanced salt solution at 37°C. Cell suspensions were filtered through 70 μm nylon strainers (BD Biosciences, Bedford, MA, USA) and centrifuged at 300 g for 5 min, and pellets were resuspended in L-15 supplemented with 10% vol/vol fetal bovine serum (FBS).
Flow cytometry
A Cytomation MoFlo cell sorter (488 nm excitation; Beckman Coulter, High Wycombe, UK) was used to separate populations of >95% pure Venus-positive or - negative cells. Settings were as follows: 90 μm nozzle, 38-40 pounds per square inch, 58-60 kHz, plates charged at 3200 V. Single cells were selected by their side and forward scatter and pulse width. Venus-positive cells were selected by their fluorescence at 530 and 580 nm, and Venus-negative cells by a gate that excluded Venus fluorescence. Cells were sorted into RNAlater (Ambion, Austin, TX, USA) or lysis buffer for protein extraction (see below).
RNA extraction and quantitative RT-PCR
Total RNA from FACS-sorted cells was isolated using a microscale RNA isolation kit (Ambion) according to the manufacturer’s instructions. RNA was reverse-transcribed according to standard protocols. Quantitative RT-PCR was performed with a 7900 HT Fast Real-Time PCR system (Applied Biosystems, Foster City, CA, USA). The PCR reaction mix consisted of first-strand cDNA template, primer pairs (see ESM Table 2), 6-carboxyfluorescein/quencher probes (Bioresearch Technologies, Novato, CA, USA and Applied Biosystems) and PCR Master Mix (Applied Biosystems). Expression was compared with that of β-actin (Actb) measured on the same sample in parallel, giving a CT difference (ΔCT) for β-actin minus the test gene. Reactions in which the test gene was undetectable were assigned a CT value of 40, and where mRNA was detectable in fewer than two samples, this is marked in Table 1.
Table 1.
Gene expression in Kpos and Kneg cells measured by quantitative RT-PCR
| Gene | Kpos cells | Kneg cells | p value (Kpos vs Kneg) |
|---|---|---|---|
| Gip | +8.0±0.6 | −4.0±1.0 | 0.00013 |
| Gcg | −4.4±0.6 | −7.5±1.2 | 0.022 |
| Pyy | +0.1±1.0 | −9.5±1.2 | 0.00055 |
| Kir6.2 | −1.5±0.4 | −9.1±1.8 | 0.0054 |
| Surl | −0.1±0.1 | −9.0±0.7 | 0.00023 |
| Gck | −3.2±0.3 | −11.9±1.7 | 0.0074 |
| Sgltl | −1.2±0.3 | −1.4±0.3 | 0.75 |
| Glutl | −6.8±0.9 | −7.1±0.2 | 0.78 |
| Glut2 | −5.6±0.2 | −3.7±0.3 | 0.0080 |
| Glut3 | −12.9±0.4 (<) | −11.5±0.9 | − |
| Glut5 | −2.9±0.2 | −4.0±0.2 | 0.026 |
| TaslRl | −11.0±1.5 | −15.3±1.3 | 0.10 |
| Tas1R2 | −13.3±0.3 (<) | −14.0±0.6 (<) | − |
| TaslR3 | −8.2±1.9 | −9.6±0.5 | 0.52 |
| Gnat3 | −15.9±1.0 (<) | −15.7±1.3 (<) | − |
| Gpr40 | −1.7±0.3 | −8.3±1.3 | 0.0071 |
| Gprl20 | −3.2±0.5 | −9.5±0.7 | 0.0020 |
| Gprll9 | −4.7±0.8 | −13.1±1.1 | 0.00086 |
Data are CT differences (ΔCT), i.e. the CT for the gene of interest minus the CT for β-actin measured in parallel in the same sample (±SEM)
CTs were allocated a value of 40 if the gene was not detected
(<) marks data where fewer than two samples gave a detectable reading for the test gene, indicating that the expression level was less than the value indicated
Significance was compared by Student’s t test
Immunohistochemistry
Freshly isolated mouse duodenum/jejunum was fixed in 4% wt/vol para-formaldehyde for 48 h, cryoprotected in 20% wt/vol sucrose for 48 h and embedded in OCT (optimum cutting temperature compound) prior to sectioning. Duodenal/jejunal sections (7 μm) were first permeabilised with 0.1% vol/vol Triton X-100, blocked with 10% vol/vol goat serum in PBS, and incubated with a GIP antibody (1:250; Millipore, Billerica, MA, USA) overnight at 4°C. For SGLT1 staining of tissue sections, an antigen retrieval step was required. Briefly, sections were heated at 125°C for 3 min in trisodium citrate buffer (10 mmol/l, pH 6), allowed to cool, rinsed with PBS and blocked as described above. Subsequently, sections were incubated with an SGLT1 antibody (1:100) [37] for 3 h at room temperature. Because of the loss of Venus fluorescence as a consequence of antigen retrieval, a fluorescein isothiocyanate-conjugated GFP antibody (1:100; Abcam, Cambridge, UK) was also added for 3 h to label the K cells. Bound SGLT1 and GIP antibodies were detected using an Alexa 633-conjugated secondary antibody (1:500; Invitrogen, Paisley, UK). Tissue samples stained with the secondary antibody alone served as a control. All presented images were captured using a Zeiss LSM 510 Meta confocal microscope (Carl Zeiss, Welwyn Garden City, UK) with a 63×/NA 1.4 objective, at a resolution of 1024 × 1024 pixels and an optical slice thickness of 1.5 μm.
Primary crypt culture
Intestinal tissue from the top half of the small intestine, collected as above, was digested with 0.4 mg/ml collagenase XI. Digested tissue was centrifuged at 300 g and resuspended in Dulbecco’s modified Eagle medium (25 mmol/l glucose) supplemented with 10% vol/vol FBS, 2 mmol/l l-glutamine, penicillin and streptomycin. This tissue suspension was plated on Matrigel (BD Biosciences, Bedford, MA, USA) - coated 24-well plates and incubated at 37°C under 5% CO2.
GIP secretion
Secretion studies were performed 24-36 h after plating, using primary cultures of small intestine tissue from either GIP-Venus or control mice. As preliminary experiments showed that expression of the fluorescent transgene in K cells did not affect GIP secretion in vitro, transgenic and control mice were thereafter used interchangeably. Cultures were washed thoroughly and incubated in the presence or absence of test reagents in bath solution (see below) containing 0.1% wt/vol fatty acid-free BSA for 2-4 h at 37°C. The medium was collected and centrifuged to remove any contaminating cells. The plated cells were then treated with lysis buffer containing: 50 mmol/l Tris–HCl, 150 mmol/l NaCl, 1% vol/vol Igepal-CA 630, 0.5% wt/vol deoxycholic acid and one tablet of complete EDTA-free protease inhibitor cocktail (Roche, Basel, Switzerland). Plates were freeze–thawed and mechanically disrupted to extract intracellular GIP. GIP was assayed in the supernatant fraction and cell extracts using an enzyme-linked immunosorbent assay (GIP Total ELISA Kit; Millipore). GIP secretion in each well was expressed as a fraction of the total GIP measured in that well, and was normalised, as indicated, to the basal secretion measured in parallel on the same day.
Solutions
The standard bath solution contained (mmol/l): 4.5 KCl, 138 NaCl, 4.2 NaHCO3, 1.2 NaH2PO4, 2.6 CaCl2, 1.2 MgCl2 and 10 HEPES (pH 7.4, NaOH). Oleoylethanolamide (OEA), forskolin (fsk) and IBMX were prepared as 1000× stock solutions in DMSO, phloridzin as a 1000× stock in ethanol, tolbutamide as a 100 mmol/l stock solution in NaOH, and diazoxide as a 68 mmol/l stock solution in NaOH. Other agents were dissolved directly in bath solution. All chemicals were supplied by Sigma Aldrich (Poole, UK) unless otherwise stated.
Data analysis
For expression data, mean, standard error and significance calculations were performed on the raw ΔCT data. Statistical comparisons were performed by Student’s two- or one-sample t test or regression analysis, as indicated, with a threshold for significance of p=0.05.
Results
Generation and characterisation of transgenic mice
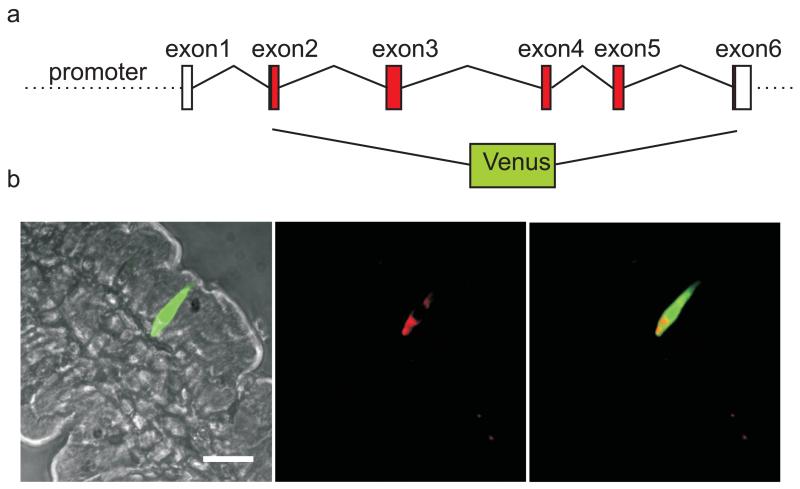
Transgenic mice were created using a construct based on a BAC containing ~200 kb around the rat Gip locus, in which the Gip coding sequence was replaced by that of Venus (Fig. 1a). We generated two founder strains that faithfully transmitted the transgene to their offspring, each containing a single copy number, as assessed by quantitative PCR. The GIP-Venus mice had yellow fluorescent cells in their gut epithelium, readily visible in living tissue specimens by epifluorescence microscopy. The highest density of fluorescent cells occurred in the duodenum and jejunum, consistent with the previously reported localisation of K cells [38]. Immunofluorescence staining with a GIP antibody confirmed that expression of the transgene was K cell-specific (Fig. 1b), and as no difference was observed between the two strains they were used interchangeably for the remainder of the study.
Fig. 1.
Generation of transgenic mice and confirmation of cell-specific Venus expression. a The BAC construct for making transgenic mice was made by cloning Venus into the coding region of Gip (for further details see ESM). b Colocalisation of direct Venus fluorescence (green) with GIP immunofluorescence (red) in the small intestine. Scale bar, 20 μm
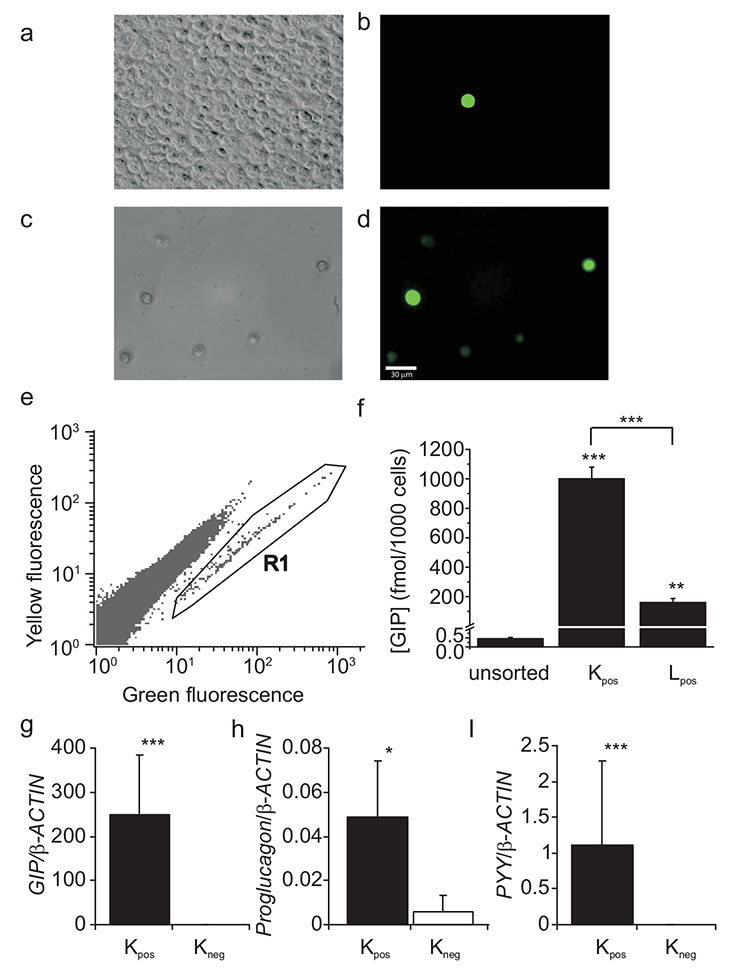
By flow cytometry, we collected populations of up to 20,000 Venus-positive K cells (Kpos) at a purity of ~95%, as assessed by direct epifluorescence microscopy (Fig. 2a–c), as well as control (Venus-negative) cells (Kneg). Kpos cells contained 4000-fold more G ip message and 2000-fold more GIP protein than Kneg cells, confirming that the fluorescent marker was correctly targeted to the K cell population (Fig. 2d, e and Table 1). Levels of Gip mRNA and GIP protein were eightfold and sixfold higher, respectively, in Kpos cells than in fluorescently tagged GLP-1-producing L cells from the small intestine [39].
Fig. 2.
Collection and analysis of Venus-positive and control cells from the intestine. a, b Single-cell enzymatic digest of the upper small intestine from a GIP-Venus mouse, before (a) and after (b) FACS sorting. Differential interference contrast (DIC) images are shown on the left, with the corresponding fluorescence at 488 nm excitation on the right. After FACS sorting, >95% of the collected cells were Venus-positive. Scale bar, 30 μm. c K cells were collected from the upper small intestine by FACS sorting, using gates on pulse width and side and forward scatter to select single cells, and on yellow (580 nm) and green (530 nm) fluorescence to select either Venus-positive (R1) or Venus-negative cells. The excitation wavelength was 488 nm. d Histogram showing GIP protein content in unsorted, Kpos and Lpos cells from the upper small intestine. GIP content was analysed after cell extraction by ELISA. Lpos cells were FACS-sorted in a similar way to Kpos cells, using a transgenic mouse expressing Venus under the control of the proglucagon promoter [39]. Each bar represents three or four samples from different mice. **p<0.01, ***p<0.001 (Student’s t test) e, f, g Histograms showing relative gene expression of Gip, Gcg and Pyy in Kpos (black columns) and Kneg cells (white columns) from the upper small intestine. Expression was analysed by quantitative RT-PCR and compared with that of β-actin in the same sample. Three to six samples from separate mice were analysed for each bar. Data are presented as geometric mean, and the error bar was calculated from the log(base 2) data. Significance differences between Kpos and Kneg cells were found using Student’s t test; *p<0.05, ***p<0.001.
Kpos cells expressed ninefold higher levels of proglucagon (GCG) and 780-fold higher levels of mRNA for peptide YY (Pyy) than Kneg cells (Fig 2f and g). By comparison, however, small intestinal L cells had 90-fold more Gcg mRNA (p=0.0007) and >60-fold more GLP-1 protein than Kpos cells, but similar levels of Pyy mRNA (data not shown).
Expression of glucose-sensing machinery
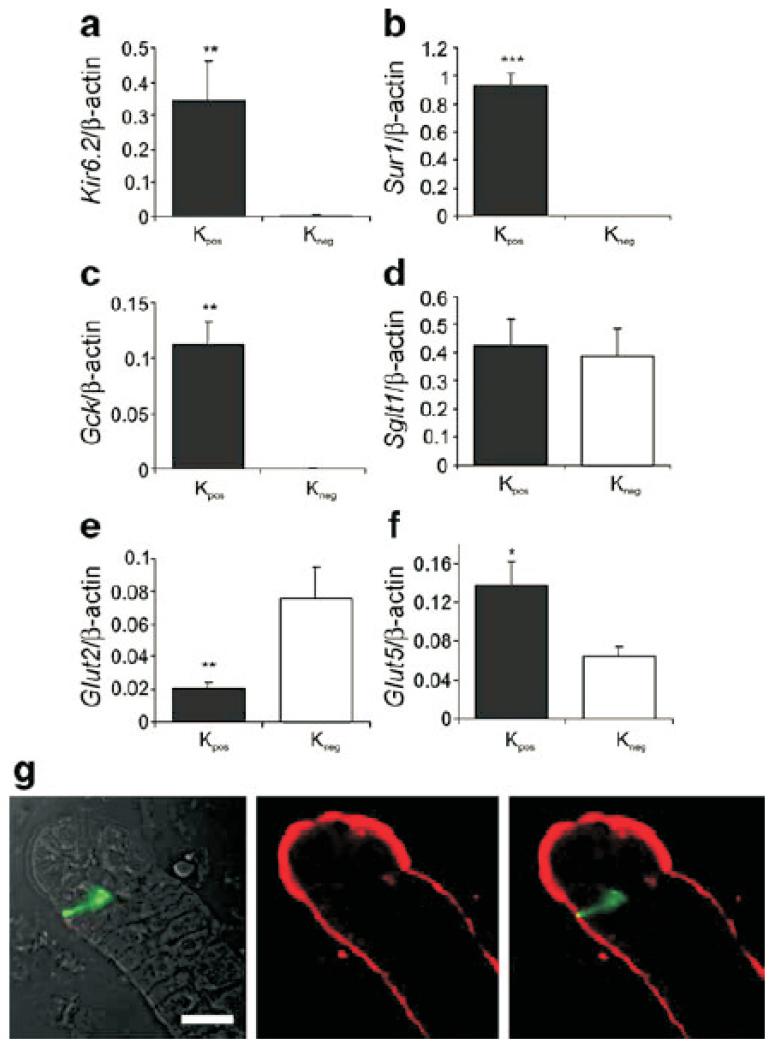
To identify the glucose-sensing machinery expressed in K cells, we compared the expression in Kpos and Kneg cells of a range of candidate glucose-sensing components, including KATP channel subunits (Kir6.2 [also known as Kcnj11], Sur1 [also known as Abcc8]), glucokinase (Gck) and Sglt1 (also known as Slc5a1), facilitative glucose transporters (Glut1, −2, −3 and −5 [also known as Slc2a1, −2, −3 and −5]), taste receptor subunits (Tas1r1, −2 and −3) and α-gustducin (Gnat3). Gene expression levels of Kir6.2, Sur1 and Gck were higher in Kpos than Kneg cells by 190-fold, 470-fold and 420-fold, respectively (Fig. 3a and Table 1). In comparison with expression levels measured in pancreatic beta cells (F. Gribble, unpublished data), Kpos cells had similar levels of Kir6.2 and Gck expression and only twofold lower expression of Sur1. Both Kpos and Kneg cells exhibited high expression of Sglt1, and the protein was localised to the apical surface by immunofluorescence staining (Fig. 3b) Expression of facilitative glucose transporters in Kpos cells was detected in the relative order Glut5>Glut2>Glut1>Glut3, and whereas Glut1, −3 and −5 were expressed at similar levels in Kpos compared with Kneg cells, Glut2 expression was fourfold lower in the Kpos than Kneg cells (p=0.008). Of the sweet taste receptor subunits (made up of Tas1r2 and Tas1r3), Tas1r3 expression was detectable at similar levels in both Kpos and Kneg cells (Table 1), but Tas1r2 and Gnat3 expression levels were below the detection limit in Kpos cells.
Fig. 3.
Expression of candidate glucose-sensing machinery in K cells. a–f Histograms showing relative gene expression of KATP channel subunits (Kir6.2, Sur1), Gck, Sglt1, Glut2 and Glut5 in Kpos (black columns) and Kneg cells (white columns) from the upper small intestine. Expression was analysed by quantitative RT-PCR and compared with that of β-actin in the same sample. Each column represents three to six samples from separate mice. Data are presented as geometric mean, and the error bars were calculated from the log(base 2) data. Significance comparisons between Kpos and Kneg cells were calculated by Student’s t test; *p<0.05, **p<0.01, ***p<0.001. g Immunostaining for Venus (green) and SGLT1 (red) in the duodenum/jejunum, showing the apical localisation of SGLT1 on the villus. Because of the thickness of the optical slice (1.5 μm), only the apical tip of the K cell can be clearly seen. Scale bar, 20 μm
Nutrient-stimulated GIP secretion in vitro
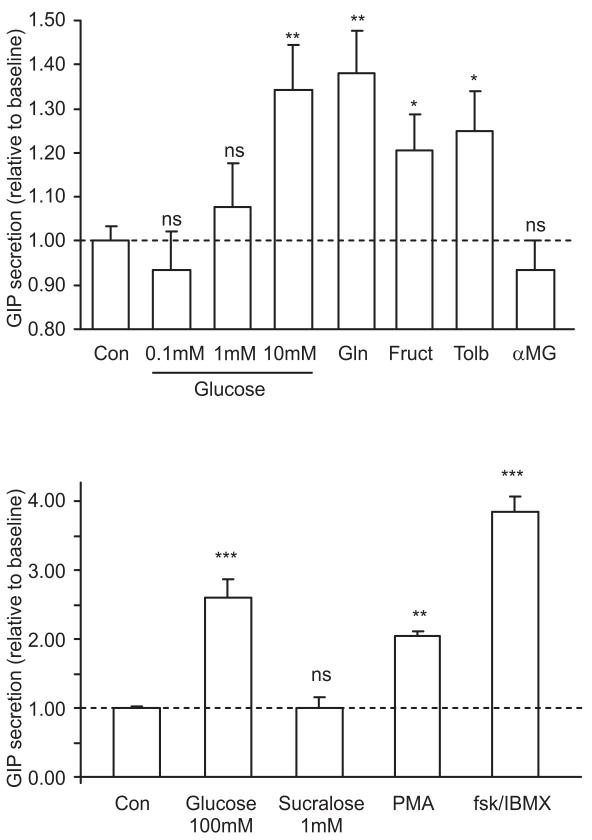
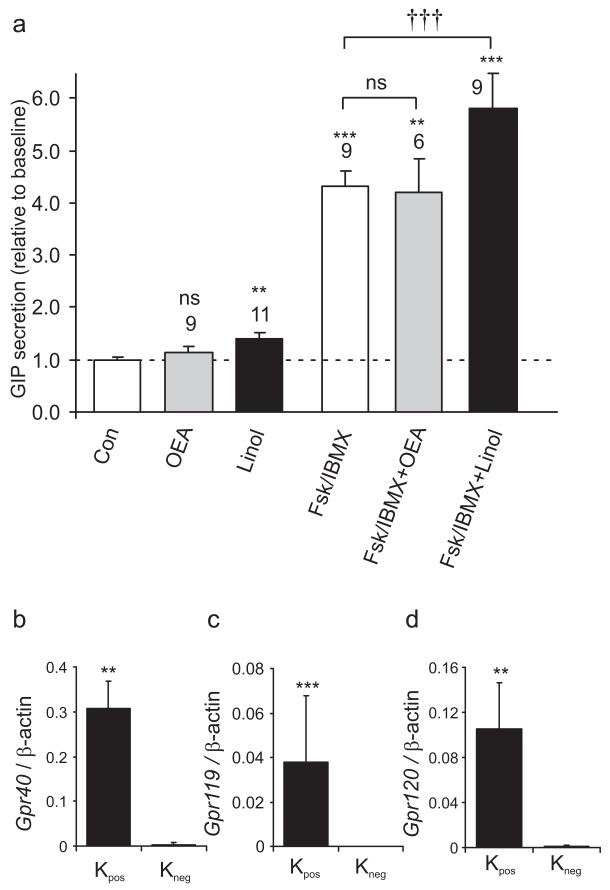
We next investigated the nutrient sensitivity of GIP secretion in vitro, using primary cultures of adult mouse small intestine. As shown in Fig. 4a, GIP secretion was stimulated 1.34-fold by 10 mmol/l glucose and 1.38-fold by 10 mmol/l glutamine, an amino acid that potently stimulates GLP-1 release from GLUTag cells [40]. Consistent with the finding that K cells express KATP channels, we also observed a 1.25-fold stimulation of secretion by 500 μmol/l tolbutamide. GIP release was stimulated 1.21-fold by 10 mmol/l fructose and 2.6-fold by 100 mmol/l glucose. At such high concentrations, glucose may have non-specific effects, perhaps related to its osmolarity. To test whether this effect of high glucose might alternatively be attributable to the activation of sweet taste receptors, we examined the response to the artificial sweetener sucralose (1 mmol/l). However, consistent with the undetectable levels of Tas1r2 and Gnat3 by quantitative RT-PCR, sucralose did not stimulate GIP release (Fig. 4b). GIP secretion was also increased 1.4-fold by linoleic acid (100 μmol/l), 2.0-fold by the protein kinase C activator phorbol 12-myristate 13-acetate (PMA; 1 μmol/l) and 3.8-fold by a combination of fsk plus IBMX (fsk/IBMX; 10 μmol/l of each), but was unaffected by OEA (10 μmol/l), a low-affinity agonist of G-protein-coupled receptor (GPR)119 (Figs 4b and 5a).
Fig. 4.
Glucose-dependent GIP secretion from primary intestinal cultures. a Mixed intestinal cultures from the upper small intestine were incubated for 2–4 h in bath solution containing additions as indicated: control (Con), glucose (0.1, 1 or 10 mmol/l), glutamine (Gln, 10 mmol/l), fructose (Fruct, 10 mmol/l), tolbutamide (Tolb, 500 μmol/l) or αMG (10 mmol/l). Number of wells: glucose 0.1, 1, 10 mmol/l, 11, 13, 27, respectively; Gln, 12; Fruct, 9; Tolb, 19; αMG, 12. GIP was measured in the supernatant fraction and cell extracts, and is expressed relative to basal secretion measured in parallel on the same day (indicated by dashed line). Error bars represent 1 SE and significance is shown relative to baseline, analysed by single-factor t test; *p<0.05, **p<0.01. b Responsiveness of GIP secretion to a higher concentration of glucose (100 mmol/l) compared with sucralose (1 mmol/l), PMA (1 μmol/l) or fsk/IBMX (10 μmol/l of each). Number of wells: glucose, 13; sucralose, 5; PMA, 3; fsk/IBMX, 53. Experiments were carried out as in a. Error bars represent 1 SE, and significance is shown relative to baseline (shown by the dashed line) using a single-factor t test; **p<0.01, ***p<0.001
Fig. 5.
Responses to lipids and expression of candidate G-protein coupled receptors in K cells. a GIP responses to fatty acids. Mixed intestinal cultures from the upper small intestine were incubated in bath solution containing additions as indicated: control (Con), oleoylethanolamide (OEA 10 μmol/l), linoleic acid (Linol, 100 μmol/l), fsk/IBMX (10 μmol/l of each). Number of wells: OEA, 9; Linol 11; fsk/IBMX, 9; fsk/IBMX+OEA, 6; fsk/IBMX+Linol, 9. GIP was measured in the supernatant fraction and cell extracts, and is expressed relative to basal secretion measured in parallel on the same day (indicated by dashed line). Error bars represent 1 SE, and significance is shown relative to baseline, analysed by single-factor t test. **p<0.01, ***p<0.001. Significance of differences between different combinations that included fsk/IBMX was tested by regression analysis. †††p<0.001. Gene expression of Gpr40 (b), Gpr119 (c) and Gpr120 (d) in Kpos (black columns) and Kneg cells (white columns) from the upper small intestine relative to that of β-actin as measured by quantitative RT-PCR. Each column represents three or four samples. Data are presented as geometric mean, and the error bar was calculated from the log(base 2) data. Significances of comparisons between Kpos and Kneg cells were calculated by Student’s t test performed on the log(base 2) data. **p<0.01, ***p<0.001
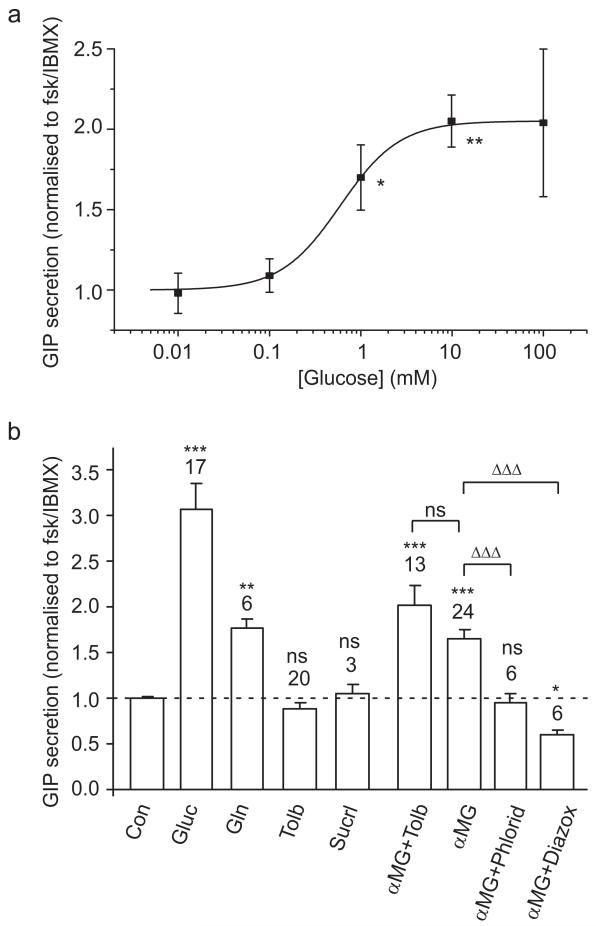
We next examined the effect of candidate secretagogues in the presence of fsk/IBMX (Fig. 6). Under this condition, glucose was a more potent stimulus, enhancing GIP secretion 3.1-fold at 10 mmol/l, compared to the 1.34-fold stimulation seen in the absence of fsk/IBMX. Secretion was also stimulated 1.8-fold by glutamine (10 mmol/l), 1.4-fold by linoleic acid (100 μmol/l), and again not by OEA or sucralose under these conditions (Figs 5a and 6b). Measurement of the dose–response relationship for glucose in the presence of fsk/IBMX (Fig 6a) revealed an ED50 of 0.6 mmol/l. Unlike the findings under basal conditions, in fsk/IBMX we observed a highly significant, 1.7-fold stimulation of GIP release by the non-metabolisable glucose analogue α-methyl glucopyranoside (αMG; 10 mmol/l) which was abolished by addition of phloridzin (5 μmol/l), a specific inhibitor of SGLTs (Fig. 6b). Diazoxide (340 μmol/l) inhibited αMG-triggered GIP secretion even below the baseline level observed in fsk/IBMX alone. Taken together with the finding that tolbutamide failed to trigger GIP secretion under these conditions, this suggests that fsk/IBMX acts at least in part via closure of KATP channels in K cells. The greater effect of glucose in comparison with αMG is not due to its combined action on both SGLT1- and KATP channel-dependent pathways, as the further addition of tolbutamide did not significantly enhance secretion compared with that triggered by αMG alone (Fig. 6b). An additional metabolic component of glucose is, however, suggested by the finding that, under depolarising conditions (fsk/IBMX, 30 mmol/l KCl and 340 μmol/l diazoxide), 10 mmol/l glucose triggered a further 1.3-fold increase in GIP secretion (p=0.016, n=6).
Fig. 6.
GIP secretion in the presence of elevated cAMP. a Mixed intestinal cultures from the upper small intestine were incubated for 2-4 h in bath solution containing fsk/IBMX (10 μmol/l of each) plus different glucose concentrations, as indicated. GIP was measured in the supernatant fraction and cell extracts, and is expressed relative to the secretion in wells containing fsk/IBMX measured in parallel on the same day. The data were fitted with a logistic equation [y=A+(1−A)/(1+(x/ED50)n)], with a maximal amplitude A=2.1, an ED50=0.6 mmol/l and slope factor n of 1.3. Error bars represent 1 SE, and significance is shown relative to fsk+IBMX, analysed by single-factor t test; *p<0.05, **p<0.01. b GIP responses to agonists in the presence of fsk/IBMX (10 μmol/l of each). Experiments were carried out as in a, with additions as indicated: control (Con), glucose (Gluc, 10 mmol/l), glutamine (Gln, 10 mmol/l), tolbutamide (Tolb, 500 μmol/l), sucralose (Sucrl, 1 mmol/l), αMG (10 mmol/l), phloridzin (Phlorid, 5 μmol/l), diazoxide (Diazox, 340 μmol/l). Number of wells: Gluc, 17; Gln, 6; Tolb, 20; Sucrl, 3; αMG+Tolb, 13; αMG, 24; αMG+Phlorid, 6; aMG+Diazox, 6. Secretion was normalised to that measured in fsk/IBMX alone, measured in parallel on the same day (indicated by the dashed line). Error bars represent 1 SE, and significance is shown relative to fsk/IBMX, analysed by a single-factor t test; *p<0.05, **p<0.01, ***p<0.001. Significance of differences between different combinations that included αMG were tested by Student’s t test; ns non significant, †††p<0.001
In view of the stimulation of GIP secretion by linoleic acid, PMA and elevated cAMP, we investigated whether K cells express candidate G-protein-coupled receptors that are coupled to the activation of these pathways. Recent reports suggest that GPR40 and 120 (Gq-coupled) as well as GPR119 (Gαs-coupled) may play a role in the regulation of enteroendocrine hormone release by long-chain fatty acids and OEA, respectively [41-43], raising the possibility that these and/or related receptors might provide novel targets for harnessing the endogenous release of incretin hormones. By quantitative RT-PCR, we detected expression of Gpr40 (also known as Ffar1), Gpr120 and Gpr119 in the Kpos cell population, and only very low levels in the corresponding Kneg cells (Fig. 5b,c and d and Table 1).
Discussion
To date, there has been relatively poor understanding of how K cells release GIP, largely because living enteroendocrine cells are difficult to identify, and there are no reliable models for studying GIP secretion in vitro. The transgenic mice we have generated, with fluorescently labelled K cells, enable the identification and purification of K cells from freshly isolated tissue. Our data establish that K cells express the potential glucose sensing machinery, Kir6.2, Sur1, Gck and Sglt1, as well as lipid-sensing receptors, Gpr40, Gpr119 and Gpr120. This correlates functionally with our findings that GIP secretion was stimulated by glucose, glutamine, fatty acids (linoleic acid), PMA and cAMP elevation in primary intestinal cultures of adult rodent intestine.
Immunofluorescence staining of fixed tissue and quantification of Gip mRNA and protein in FACS-purified Kpos cells confirmed that expression of the fluorescent protein in GIP-Venus mice is specific to the K cell population. In agreement with previous findings, we also detected gene expression of Gcg and Pyy in the K cell pool, suggesting some overlap with L cells. However, whereas Gip expression was six- to eightfold higher in K than L cells, proglucagon expression showed the reverse pattern, with 90-fold higher levels in L than in K cells. These findings are consistent with the results of immunofluorescence studies which showed that GIP and GLP-1 are colocalised in a subpopulation of small intestinal K and L cells [29].
Glucose (10 mmol/l) triggered GIP secretion 1.3-fold under basal conditions, but 3-fold in the presence of fsk/IBMX, indicating a synergistic interaction between these two stimulatory pathways. In fsk/IBMX, we could also detect a 1.7-fold stimulation of GIP release by αMG, indicating that secretion can be triggered by monosaccharides independently of glycolytic metabolism. The responsiveness of GIP secretion to 10 mmol/l αMG, taken together with the measured ED50 for glucose of 0.6 mmol/l, close to the Km for glucose and αMG of SGLT1 (~0.3 mmol/l [44]), suggests that SGLT1 plays a key role in glucose-triggered GIP release. This is consistent with previous data from whole intestinal preparations showing that GIP secretion was triggered by luminal substrates of SGLTs [25]. The submillimolar responsiveness of cultured K cells to glucose and lack of responsiveness to sucralose, together with low gene expression levels of sweet taste receptor subunits and Gnat3 in Kpos cells by quantitative RT-PCR, suggest that sweet taste receptors do not underlie the observed sugar-triggered GIP release. Although these findings are consistent with a recent report that orally administered sweeteners do not trigger GIP or GLP-1 release in rats [45], other recent studies have suggested that the sweet taste receptor pathway underlies glucose-triggered incretin hormone secretion [34, 35]. Further studies are required to evaluate whether the low expression levels of TAS1 receptor subunits play a role in GIP and GLP-1 secretion in vivo
The presence of functional KATP channels in K cells was confirmed in secretion studies by the findings that GIP release was triggered by tolbutamide under basal conditions and inhibited by diazoxide in the presence of fsk/IBMX. This was supported by the high expression levels of Kir6.2 and Sur1 mRNAs in purified K cells (>100-fold higher than in neighbouring Kneg cells) and is consistent with a previous observation that Kir6.2 was detectable in human K cells by immunostaining [46]. As tolbutamide responsiveness was lost in the presence of fsk/IBMX, yet under these conditions diazoxide inhibited secretion, our data suggest that elevated cAMP brings about the closure of KATP channels in K cells. cAMP-induced closure of KATP channels in K cells could also allow small αMG-associated transporter currents to become a sufficient trigger for GIP release. As the open KATP current would only be expected to inhibit the depolarising action of SGLT1 if these conductances occurred in the same cell, the finding that diazoxide inhibits the stimulatory action of αMG (in fsk/IBMX) also supports the idea that SGLT1 in the K cells themselves is involved in αMG-triggered GIP secretion.
The threefold greater secretory response to glucose than to αMG could be explained by an additional metabolic responsiveness to glucose, as glucose triggered a further elevation of GIP secretion under conditions designed to bypass any depolarising action of SGLT1. The ability of fructose and glutamine to stimulate GIP release under basal conditions also favours a role for metabolism in K cells. As fructose is not a substrate for SGLT1, but may enter K cells using the highly expressed facilitative glucose transporter GLUT5 [47], the response to this sugar cannot be explained by any known electrogenic uptake mechanism.
Elevation of cAMP by fsk/IBMX was a potent trigger of GIP release in our experiments, and was synergistic with the action of glucose. The effectiveness of cAMP elevation and PMA as GIP secretagogues suggests that harnessing pathways coupled to the activation of adenylate cyclase and/or protein kinase C might provide novel pathways to modulate GIP release in vivo. It has been suggested, although not definitively demonstrated, that lipids might physiologically modulate the secretion of GLP-1 and GIP through activation of the G-protein-coupled receptors GPR40, GPR119 and GPR120 [41, 42]. We detected a moderate but significant increase in GIP secretion in response to the potential GPR40 and GPR120 agonist linoleic acid at both basal and elevated cAMP concentrations, consistent with Gq-coupled signal transduction from these receptors. The GPR119 agonist AR231453 was also recently shown to increase secretion of GIP and GLP-1 in rodents in vivo, although the authors were unable to demonstrate expression of Gpr119 in K cells [42]. Our data show, however, that Gpr119 expression in K cells is enhanced 330-fold compared with the neighbouring cells, and is similar to that found in small intestinal and colonic L cells (data not shown). These new tools to measure the expression profile of adult K cells and the functional responsiveness of GIP secretion in primary culture offer unique future opportunities to interrogate the mechanisms underlying GIP release.
Supplementary Material
Acknowledgements
We would like to thank the Wellcome Trust, MRC, Lister Institute and St John’s College, Cambridge for funding. We are grateful to S. McCallum (Cambridge Institute for Medical Research), who advised on and performed the FACS sorting, A. Miyawaki (Riken Institute, Japan) for the gift of the Venus containing plasmid and GlaxoSmithKline for the gift of quantitative RT-PCR primers.
Abbreviations
- αMG
α-methyl glucopyranoside
- BAC
bacterial artificial chromosome
- fsk
forskolin
- GIP
glucose-dependent insulinotropic polypeptide
- GLP-1
glucagon-like peptide 1
- GPR40, 119, 120
G-protein-coupled receptor 40, 119, 120
- IBMX
3-isobutyl-1-methylxanthine
- KATP
ATP-sensitive potassium channel
- OEA
oleoylethanolamide
- PMA
phorbol 12-myristate 13-acetate
- SGLT1
sodium-dependent glucose transporter 1
Footnotes
Duality of interest
The authors declare that there is no duality of interest associated with this manuscript
References
- 1.Nauck MA, Bartels E, Orskov C, Ebert R, Creutzfeldt W. Additive insulinotropic effects of exogenous synthetic human gastric inhibitory polypeptide and glucagon-like peptide-1-(7-36) amide infused at near-physiological insulinotropic hormone and glucose concentrations. J Clin Endocrinol Metab. 1993;76:912–917. doi: 10.1210/jcem.76.4.8473405. [DOI] [PubMed] [Google Scholar]
- 2.Baggio LL, Drucker DJ. Biology of incretins: GLP-1 and GIP. Gastroenterology. 2007;132:2131–2157. doi: 10.1053/j.gastro.2007.03.054. [DOI] [PubMed] [Google Scholar]
- 3.Yip RGC, Wolfe MM. GIP biology and fat metabolism. Life Sci. 2000;66:91–103. doi: 10.1016/s0024-3205(99)00314-8. [DOI] [PubMed] [Google Scholar]
- 4.Getty-Kaushik L, Song DH, Boylan MO, Corkey BE, Wolfe MM. Glucose-dependent insulinotropic polypeptide modulates adipocyte lipolysis and reesterification. Obesity. 2006;14:1124–1131. doi: 10.1038/oby.2006.129. [DOI] [PubMed] [Google Scholar]
- 5.Kim SJ, Nian C, McIntosh CH. Activation of lipoprotein lipase by glucose-dependent insulinotropic polypeptide in adipocytes. A role for a protein kinase B, LKB1, and AMP-activated protein kinase cascade. J Biol Chem. 2007;282:8557–8567. doi: 10.1074/jbc.M609088200. [DOI] [PubMed] [Google Scholar]
- 6.Drucker DJ, Nauck MA. The incretin system: glucagon-like peptide-1 receptor agonists and dipeptidyl peptidase-4 inhibitors in type 2 diabetes. Lancet. 2006;368:1696–1705. doi: 10.1016/S0140-6736(06)69705-5. [DOI] [PubMed] [Google Scholar]
- 7.Nauck MA, Heimesaat MM, Orskov C, Holst JJ, Ebert R, Creutzfeldt W. Preserved incretin activity of glucagon-like peptide 1 [7-36 amide] but not of synthetic human gastric inhibitory polypeptide in patients with type-2 diabetes mellitus. J Clin Invest. 1993;91:301–307. doi: 10.1172/JCI116186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu G, Kaneto H, Laybutt DR, et al. Downregulation of GLP-1 and GIP receptor expression by hyperglycaemia – possible contribution to impaired incretin effects in diabetes. Diabetes. 2007;56:1551–1558. doi: 10.2337/db06-1033. [DOI] [PubMed] [Google Scholar]
- 9.Trumper A, Trumper K, Horsch D. Mechanisms of mitogenic and anti-apoptotic signaling by glucose-dependent insulinotropic polypeptide in beta(INS-1)-cells. J Endocrinol. 2002;174:233–246. doi: 10.1677/joe.0.1740233. [DOI] [PubMed] [Google Scholar]
- 10.Hinke SA, Gelling RW, Pederson RA, et al. Dipeptidyl peptidase IV-resistant [D-Ala(2)]glucose-dependent insulinotropic polypeptide (GIP) improves glucose tolerance in normal and obese diabetic rats. Diabetes. 2002;51:652–661. doi: 10.2337/diabetes.51.3.652. [DOI] [PubMed] [Google Scholar]
- 11.O’Harte FP, Mooney MH, Kelly CM, Flatt PR. Improved glycaemic control in obese diabetic ob/ob mice using N-terminally modified gastric inhibitory polypeptide. J Endocrinol. 2000;165:639–648. doi: 10.1677/joe.0.1650639. [DOI] [PubMed] [Google Scholar]
- 12.Kieffer TJ. GIP or not GIP? That is the question. Trends Pharmacol Sci. 2003;24:110–112. doi: 10.1016/S0165-6147(03)00031-2. [DOI] [PubMed] [Google Scholar]
- 13.Miyawaki K, Yamada Y, Yano H, et al. Glucose intolerance caused by a defect in the entero-insular axis: a study in gastric inhibitory polypeptide receptor knockout mice. Proc Natl Acad Sci U S A. 1999;96:14843–14847. doi: 10.1073/pnas.96.26.14843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miyawaki K, Yamada Y, Ban N, et al. Inhibition of gastric inhibitory polypeptide signaling prevents obesity. Nat Med. 2002;8:738–742. doi: 10.1038/nm727. [DOI] [PubMed] [Google Scholar]
- 15.Althage MC, Ford EL, Wang S, Tso P, Polonsky KS, Wice BM. Targeted ablation of GIP-producing cells in transgenic mice reduces obesity and insulin resistance induced by a high fat diet. J Biol Chem. 2008;283:18365–18376. doi: 10.1074/jbc.M710466200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Irwin N, McClean PL, O’Harte FP, Gault VA, Harriott P, Flatt PR. Early administration of the glucose-dependent insulinotropic polypeptide receptor antagonist (Pro(3))GIP prevents the development of diabetes and related metabolic abnormalities associated with genetically inherited obesity in ob/ob mice. Diabetologia. 2007;50:1532–1540. doi: 10.1007/s00125-007-0692-2. [DOI] [PubMed] [Google Scholar]
- 17.McClean PL, Irwin N, Cassidy RS, Holst JJ, Gault VA, Flatt PR. GIP receptor antagonism reverses obesity, insulin resistance, and associated metabolic disturbances induced in mice by prolonged consumption of high-fat diet. Am J Physiol Endocrinol Metab. 2007;293:E1746–E1755. doi: 10.1152/ajpendo.00460.2007. [DOI] [PubMed] [Google Scholar]
- 18.Ebert R, Creutzfeldt W. Influence of gastric inhibitory polypeptide antiserum on glucose-induced insulin secretion in rats. Endocrinology. 1982;111:1601–1606. doi: 10.1210/endo-111-5-1601. [DOI] [PubMed] [Google Scholar]
- 19.Tseng CC, Kieffer TJ, Jarboe LA, Usdin TB, Wolfe MM. Postprandial stimulation of insulin release by glucose-dependent insulinotropic polypeptide (GIP). Effect of a specific glucose-dependent insulinotropic polypeptide receptor antagonist in the rat. J Clin Invest. 1996;98:2440–2445. doi: 10.1172/JCI119060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McClean PL, Gault VA, Irwin N, McCluskey JT, Flatt PR. Daily administration of the GIP-R antagonist (Pro3)GIP in streptozotocin-induced diabetes suggests that insulin-dependent mechanisms are critical to anti-obesity-diabetes actions of (Pro3)GIP. Diabetes Obes Metab. 2008;10:336–342. doi: 10.1111/j.1463-1326.2007.00712.x. [DOI] [PubMed] [Google Scholar]
- 21.Elliot RM, Morgan LM, Tredger JA, Deacon S, Wright J, Marks V. Glucagon-like peptide-1 (7-36)amide and glucose-dependent insulinotropic polypeptide secretion in response to nutrient ingestion in man: acute post-prandial and 24-h secretion patterns. J Endocrinol. 1993;138:159–166. doi: 10.1677/joe.0.1380159. [DOI] [PubMed] [Google Scholar]
- 22.Meier JJ, Nauck MA. Glucose-dependent insulinotropic polypeptide/gastric inhibitory polypeptide. Best Pract Res Clin Endocrinol Metab. 2004;18:587–606. doi: 10.1016/j.beem.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 23.Buffa R, Polak JM, Pearse AGE, Solcia E, Grimelius L, Capella C. Identification of the intestinal cell storing gastric inhibitory peptide. Histochemistry. 1975;43:249–255. doi: 10.1007/BF00499706. [DOI] [PubMed] [Google Scholar]
- 24.Buchan AM, Polak JM, Capella C, Solcia E, Pearse AG. Electronimmunocytochemical evidence for the K cell localization of gastric inhibitory polypeptide (GIP) in man. Histochemistry. 1978;56:37–44. doi: 10.1007/BF00492251. [DOI] [PubMed] [Google Scholar]
- 25.Sykes S, Morgan LM, English J, Marks V. Evidence for preferential stimulation of gastric inhibitory polypeptide secretion in the rat by actively transported carbohydrates and their analogues. J Endocrinol. 1980;85:201–207. doi: 10.1677/joe.0.0850201. [DOI] [PubMed] [Google Scholar]
- 26.Flatt PR, Kwasowski P, Bailey CJ. Stimulation of gastric inhibitory polypeptide release in ob/ob mice by oral administration of sugars and their analogues. J Nutr. 1989;119:1300–1303. doi: 10.1093/jn/119.9.1300. [DOI] [PubMed] [Google Scholar]
- 27.Fushiki T, Kojima A, Imoto T, Inoue K, Sugimoto E. An extract of Gymnema sylvestre leaves and purified gymnemic acid inhibits glucose-stimulated gastric inhibitory peptide secretion in rats. J Nutr. 1992;122:2367–2373. doi: 10.1093/jn/122.12.2367. [DOI] [PubMed] [Google Scholar]
- 28.Rindi G, Grant SG, Yiangou Y, et al. Development of neuroendocrine tumors in the gastrointestinal tract of transgenic mice. Heterogeneity of hormone expression. Am J Pathol. 1990;136:1349–1363. [PMC free article] [PubMed] [Google Scholar]
- 29.Mortensen K, Christensen LL, Holst JJ, Ørskov C. GLP-1 and GIP are colocalized in a subset of endocrine cells in the small intestine. Regul Pept. 2003;114:189–196. doi: 10.1016/s0167-0115(03)00125-3. [DOI] [PubMed] [Google Scholar]
- 30.Gribble FM, Williams L, Simpson AK, Reimann F. A novel glucose-sensing mechanism contributing to glucagon-like peptide-1 secretion from the GLUTag cell line. Diabetes. 2003;52:1147–1154. doi: 10.2337/diabetes.52.5.1147. [DOI] [PubMed] [Google Scholar]
- 31.Reimann F, Gribble FM. Glucose-sensing in glucagon-like peptide-1-secreting cells. Diabetes. 2002;51:2757–2763. doi: 10.2337/diabetes.51.9.2757. [DOI] [PubMed] [Google Scholar]
- 32.Li L, Wice BM. Bombesin and nutrients independently and additively regulate hormone release from GIP/Ins cells. Am J Physiol Endocrinol Metab. 2005;288:E208–E215. doi: 10.1152/ajpendo.00346.2004. [DOI] [PubMed] [Google Scholar]
- 33.Wang SY, Chi MM, Li L, Moley KH, Wice BM. Studies with GIP/Ins cells indicate secretion by gut K cells is KATP channel independent. Am J Physiol Endocrinol Metab. 2003;284:E988–E1000. doi: 10.1152/ajpendo.00398.2002. [DOI] [PubMed] [Google Scholar]
- 34.Jang HJ, Kokrashvili Z, Theodorakis MJ, et al. Gut-expressed gustducin and taste receptors regulate secretion of glucagon-like peptide-1. Proc Natl Acad Sci U S A. 2007;104:15069–15074. doi: 10.1073/pnas.0706890104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Margolskee RF, Dyer J, Kokrashvili Z, et al. T1R3 and gustducin in gut sense sugars to regulate expression of Na+-glucose cotransporter 1. Proc Natl Acad Sci U S A. 2007;104:15075–15080. doi: 10.1073/pnas.0706678104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, Muyrers JP, Testa G, Stewart AF. DNA cloning by homologous recombination in Escherichia coli. Nat Biotechnol. 2000;18:1314–1317. doi: 10.1038/82449. [DOI] [PubMed] [Google Scholar]
- 37.Mace OJ, Affleck J, Patel N, Kellett GL. Sweet taste receptors in rat small intestine stimulate glucose absorption through apical GLUT2. J Physiol. 2007;582:379–392. doi: 10.1113/jphysiol.2007.130906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Polak JM, Pearse AGE, Grimelius L, Marks V. Gastrointestinal apudosis in obese hyperglycaemic mice. Virchows Arch. B Cell Path. 1975;19:135–150. doi: 10.1007/BF02889363. [DOI] [PubMed] [Google Scholar]
- 39.Habib AM, Reimann F, Parker HE, Gribble FM. Expression of glucose-sensing machinery in intestinal GLP-1 secreting cells. Diabetologia. 2007;50(Suppl 1):S287. (Abstract) [Google Scholar]
- 40.Reimann F, Williams L, da Silva Xavier G, Rutter GA, Gribble FM. Glutamine potently stimulates glucagon-like peptide-1 secretion from GLUTag cells. Diabetologia. 2004;47:1592–1601. doi: 10.1007/s00125-004-1498-0. [DOI] [PubMed] [Google Scholar]
- 41.Hirasawa A, Tsumaya K, Awaji T, et al. Free fatty acids regulate gut incretin glucagon-like peptide-1 secretion through GPR120. Nat Med. 2005;11:90–94. doi: 10.1038/nm1168. [DOI] [PubMed] [Google Scholar]
- 42.Chu ZL, Carroll C, Alfonso J, et al. A role for intestinal endocrine cell-expressed g protein-coupled receptor 119 in glycemic control by enhancing glucagon-like peptide-1 and glucose-dependent insulinotropic peptide release. Endocrinology. 2008;149:2038–2047. doi: 10.1210/en.2007-0966. [DOI] [PubMed] [Google Scholar]
- 43.Overton HA, Babbs AJ, Doel SM, et al. Deorphanization of a G protein-coupled receptor for oleoylethanolamide and its use in the discovery of small-molecule hypophagic agents. Cell Metab. 2006;3:167–175. doi: 10.1016/j.cmet.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 44.Díez-Sampedro A, Lostao MP, Wright EM, Hirayama BA. Glycoside binding and translocation in Na(+)-dependent glucose cotransporters: comparison of SGLT1 and SGLT3. J Membr Biol. 2000;176:111–117. doi: 10.1007/s00232001081. [DOI] [PubMed] [Google Scholar]
- 45.Fujita Y, Speck M, Asadi A, Webb T, Kieffer TM. Incretin release from gut is enhanced by sugar but not by sweeteners in vivo. 68th Scientific Session of the American Diabetes Association in San Fransisco 2008: 1451-P (Abstract) 2008 http://professional.diabetes.org/Abstracts.
- 46.Nielsen LB, Ploug KB, Swift P, et al. Co-localisation of the Kir6.2/SUR1 channel complex with glucagon-like peptide-1 and glucose-dependent insulinotrophic polypeptide expression in human ileal cells and implications for glycaemic control in new onset type 1 diabetes. Eur J Endocrinol. 2007;156:663–671. doi: 10.1530/EJE-06-0756. [DOI] [PubMed] [Google Scholar]
- 47.Uldry M, Thorens B. The SLC2 family of facilitated hexose and polyol transporters. Pflugers Arch. 2004;447:480–489. doi: 10.1007/s00424-003-1085-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.