Abstract
Herpes zoster, commonly referred to as shingles, is caused by the varicella zoster virus (VZV). VZV initially manifests as chicken pox, most commonly in childhood, can remain asymptomatically latent in nerve tissues for many years and often re-emerges as shingles. Although reactivation may be related to immune suppression, aging and female sex, most inter-individual variability in re-emergence risk has not been explained to date. We performed a genome-wide association analyses in 22 981 participants (2280 shingles cases) from the electronic Medical Records and Genomics Network. Using Cox survival and logistic regression, we identified a genomic region in the combined and European ancestry groups that has an age of onset effect reaching genome-wide significance (P>1.0 × 10−8). This region tags the non-coding gene HCP5 (HLA Complex P5) in the major histocompatibility complex. This gene is an endogenous retrovirus and likely influences viral activity through regulatory functions. Variants in this genetic region are known to be associated with delay in development of AIDS in people infected by HIV. Our study provides further suggestion that this region may have a critical role in viral suppression and could potentially harbor a clinically actionable variant for the shingles vaccine.
Introduction
Herpes zoster is a significant, growing disease with a particular burden to the aging US population. There are more than a million cases of herpes zoster in the United States each year, with an annual rate of 3–4 cases per 1000 person, with the incidence increasing.1,2 Up to 3% of patients with herpes zoster require hospitalization.1 The risk is higher for women, persons of European ancestry compared with African ancestry, and persons with a family history of herpes zoster.1,3 Annual US costs of incident herpes zoster infections are $1.1 billion.4
Clinically, herpes zoster presents as painful, usually unilateral, vesicular skin infection that follows a dermatomal distribution. Of those affected with herpes zoster and depending on age, 10–50% will be left with chronic postherpetic neuralgia.1 In addition, herpes zoster has recently been shown to be a risk factor for cerebrovascular disease and myocardial infarction in a retrospective cohort study 106 601 herpes zoster cases and 213 202 control in the United Kingdom.5
Although a vaccine is now available, it is only 50% effective and currently is under-utilized. The herpes zoster vaccine is recommended for people aged ⩾60 years, by the Centers for Disease Control's Advisory Committee on Immunization Practices, as two-thirds of herpes zoster cases occur in people who are >60 years. The vaccine is Food and Drug Administration (FDA) approved for patients aged >50 years who have not experienced the disease.
Human herpes viruses, including the varicella zoster virus (VZV), are likely to have a long evolutionary history and coevolved with Homo sapiens.6 Thus host defenses have likely co-evolved with herpes viruses. Viral infections are generally recognized only when they cause symptomatic disease as viral activity within a host often does not result in any clinical symptoms.6 The major risk factor for clinical symptoms of herpes zoster is increasing age.1,6 T-cell immunity decreases with advancing age, which correlates with increased risk of clinical symptoms of herpes zoster.1 Genetic polymorphisms of immune system genes, in particular, may impact viral latency. Understanding what regulates the level of viral productivity, and the resulting immune response, may lead to clinical strategies to prevent or treat clinical disease.6
We performed a joint and genetic ancestry-stratified genome-wide association analyses to identify common genetic variants associated with herpes zoster diagnoses extracted from electronic medical records (EMRs) in adult participants from the electronic Medical Records and Genomics (eMERGE) Network. The Network is a National Human Genome Research Institute-funded consortium engaged in the development of methods and best practices for using the EMRs as tools for genomic research.7,8 The Network comprises a multi-ethnic cohort of roughly 57 000 individuals linked to EMRs for phenotype mining from nine participating sites (seven adult; two pediatric) in the United States.7
Results
Demographics by case–control status
We provide the demographic measures of the eMERGE cohort by case–control status and overall (Table 1). Out of the 22 981 participants eligible for these analyses, 2280 participants (~10%) were categorized as herpes zoster cases. The breakdown of participants by site is outlined. Geisinger Health System, Group Health Research Institute (GHRI)/University of Washington, Mayo Clinic and Marshfield Clinic cohorts were primarily comprised of participants of European ancestry and were enriched for older participants. Both the Northwestern University and Vanderbilt University cohorts tended to include younger participants and more patients of African ancestry compared to the other eMERGE sites. The Icahn School of Medicine Mount Sinai sample was also younger and has higher representation of Hispanic participants, in addition to representation of African and European ancestry. As expected, females were over-represented in cases (61%) as compared with controls (55%). Cases had a lower median body mass index (BMI; 27.9 kg m−2) than controls (28.8 kg m−2). Censored age, defined as onset age for cases and last observed BMI age for controls, was well matched. We also delineate the breakdown of genetically determined ancestry by case–control status (Table 1). There were a higher proportion of European ancestry participants in the case group (88%) as compared with the controls (79%), as expected. Conversely, there are fewer African ancestry participants in the case group (8%) compared with the controls (16%). Both the case and control groups comprise 2% Hispanic. There is a slight case–control difference for history of herpes simplex in case versus control (5% versus 2%, respectively) and chemotherapy >1 year prior to censored age (7% in cases vs 6% in controls) in this sample.
Table 1. Summary statistics of demographic data and phenotypes by eMERGE participating site and combined.
| Case | Control | Overall | |
|---|---|---|---|
| N | 2280 | 20 701 | 22 981 |
| Site | |||
| Geisinger | 10% (222) | 13% (2696) | 13% (2918) |
| Group Health/UW | 25% (572) | 12% (2475) | 13% (3047) |
| Mayo Clinic | 13% (291) | 22% (4523) | 21% (4814) |
| Marshfield | 32% (720) | 12% (2543) | 14% (3263) |
| Mt Sinai | 7% (157) | 15% (3095) | 14% (3252) |
| Northwestern | 6% (127) | 10% (1974) | 9% (2101) |
| Vanderbilt | 8% (191) | 16% (3395) | 16% (3586) |
| Sex (female) | 61% (1 387) | 55% (11 443) | 56% (12 830) |
| Median BMI (kg m−2) | 24.7, 27.9, 31.7 | 25.2, 28.8, 33.6 | 25.1, 28.7, 33.4 |
| Censored age | 57, 67, 77 | 56, 66, 77 | 56, 66, 77 |
| Ancestry | |||
| African | 8% (173) | 16% (3312) | 15% (3485) |
| European | 88% (2 016) | 79% (16 407) | 80% (18 423) |
| Hispanic | 2% (41) | 2% (580) | 2% (621) |
| H. simplex (yes) | 5% (103) | 2% (377) | 2% (480) |
| Chemotherapy (yes) | 7% (164) | 6% (1213) | 6% (1377) |
Abbreviations: BMI, body mass index; eMERGE, electronic Medical Records and Genomics. The three numbers for BMI and age represent quartiles of the distributions (25th, 50th and 75th).
Genome-wide association study
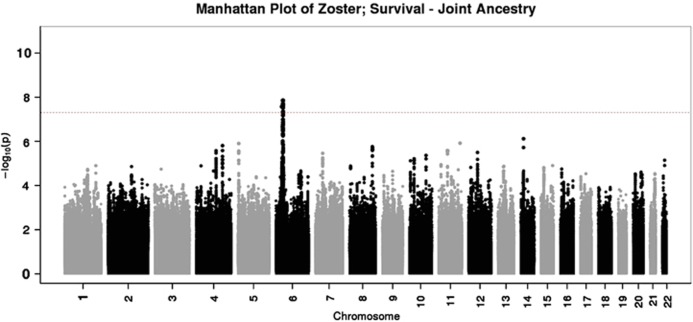
We provide the association results for herpes zoster association analyses stratified by genetically determined ancestry (Table 2). This included Cox regression analyses in the joint and European ancestry groups, as well as logistic regression analyses. Both the Cox and logistic regression models suggested a strong association on chromosome 6 in the human leukocyte antigen (HLA) region, specifically tagging HLA Complex P5 (HCP5) and upstream HLA-B in the beta block of the class 1 region (Figures 1 and 2, respectively).
Table 2. Summary of effects of loci that reached genome-wide significance for the joint (n=22 981, survival=25 986) and European ancestry (n=18 423, survival=22 679) analyses.
| Chr | SNP | A | BP | Joint MAF | Cox Joint | Cox European | Logistic Joint | Logistic European | Gene | Function |
|---|---|---|---|---|---|---|---|---|---|---|
| P-value | P-value | P-value (MAF) | P-value (MAF) | |||||||
| HR (95% CI) | HR (95% CI) | OR (95% CI) | OR (95% CI) | |||||||
| 6 | rs114045064 | C | 31332239 | 0.18 | 2.75 × 10−7 | 5.06 × 10−9 | 6.70 × 10−6 (17.51) | 2.24 × 10−8 (17.14) | HLA-B | Upstream |
| 0.81 (0.75–0.88) | 0.77 (0.71–0.85) | 0.82 (0.75–0.89) | 0.78 (0.71–0.85) | |||||||
| 6 | rs112660930 | T | 31332078 | 0.18 | 3.01 × 10−7 | 5.26 × 10−9 | 7.69 × 10−6 (17.57) | 2.56 × 10−7 (17.16) | HLA-B | Upstream |
| 0.81 (0.75–0.88) | 0.77 (0.71–0.85) | 0.82 (0.75–0.89) | 0.78 (0.71–0.86) | |||||||
| 6 | rs116062713 | C | 31433566 | 0.08 | 5.22 × 10−8 | 1.04 × 10−7 | 1.75 × 10−6 (7.82) | 5.52 × 10−6 (9.07) | HCP5 | Upstream |
| 0.72 (0.65–0.82) | 0.73 (0.64–0.82) | 0.73 (0.65–0.83) | 0.74 (0.65–0.84) | |||||||
| 6 | rs114864815 | T | 31428987 | 0.08 | 4.54 × 10−8 | 6.94 × 10−8 | 1.19 × 10−6 (7.69) | 3.68 × 10−6 (9.14) | HCP5 | 3′ UTR |
| 0.73 (0.65–0.82) | 0.73 (0.64–0.82) | 0.73 (0.65–0.83) | 0.74 (0.65–0.84) |
| SNP | Logistic African | Logistic Hispanic |
HapMap pilot 1 low coverage panel MAF |
|||
|---|---|---|---|---|---|---|
| P-value (MAF) OR (95% CI) | P-value (MAF) OR (95% CI) | I2Heterogeneity index | CEU | YRI | CHB+JPT | |
| rs114045064 | 1.87 × 10−1 (0.18) | 8.26 × 10−1 (0.26) | 79.74 | 0.23 | 0.15 | 0.03 |
| 1.20 (0.92–1.57) | 1.06 (0.63–1.78) | |||||
| rs112660930 | 1.77 × 10−1 (0.18) | 8.33 × 10−1 (0.26) | 79.99 | 0.23 | 0.13 | NA |
| 1.20 (0.92–1.58) | 1.06 (0.63–1.77) | |||||
| rs116062713 | NA (0.02) | NA (0.04) | NA | 0.16 | NA | NA |
| NA | NA (NA) | |||||
| rs114864815 | NA (0.01) | NA (0.04) | NA | 0.16 | NA | NA |
| NA (NA) | NA (NA) | |||||
Abbreviations: BP, base pair; CI, confidence interval; HCP5, HLA Complex P5; HR, hazards ratio; MAF, minor allele frequency; NA, not available; OR, odds ratio; SNP, single-nucleotide polymorphism; UTR, untranslated region. The African ancestry and Hispanic analyses included 3460 and 584 participants, respectively. Actual numbers for the analyses may be different due to missing phenotype and/or covariate data. The BP positions are defined from the GRCh37/hg19 build. Both rs116062713 and rs114864815 did not meet the MAF threshold for the African ancestry and Hispanic analyses. The HapMap abbreviations are defined as follows: (1) CEPH (Utah residents with ancestry from northern and western Europe) (abbreviation: CEU); (2) Yoruba in Ibadan, Nigeria (abbreviation: YRI); and (3) Han Chinese in Beijing, China (abbreviation: CHB); and Japanese in Tokyo, Japan (abbreviation: JPT). For HapMap allele frequencies, each SNP was merged into the corresponding variant: (1) rs114045064 was merged into rs2596551; (2) rs112660930 was merged into rs2596550; and (3) rs116062713 was merged into rs75640364; and rs114864815 was merged into rs77349273.
Figure 1.
Manhattan plot of P-values generated using Cox regression analysis in the joint ancestry sample.
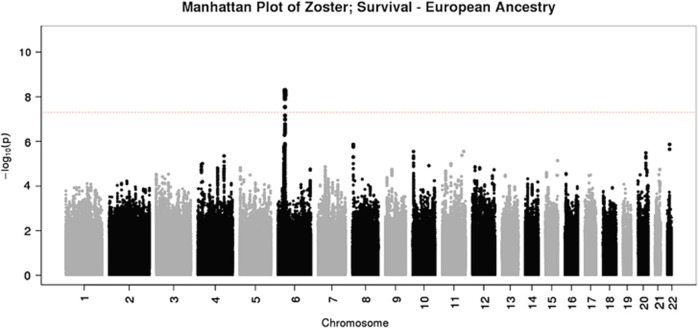
Figure 2.
Manhattan plot of P-values generated using Cox regression analysis in the European ancestry sample.
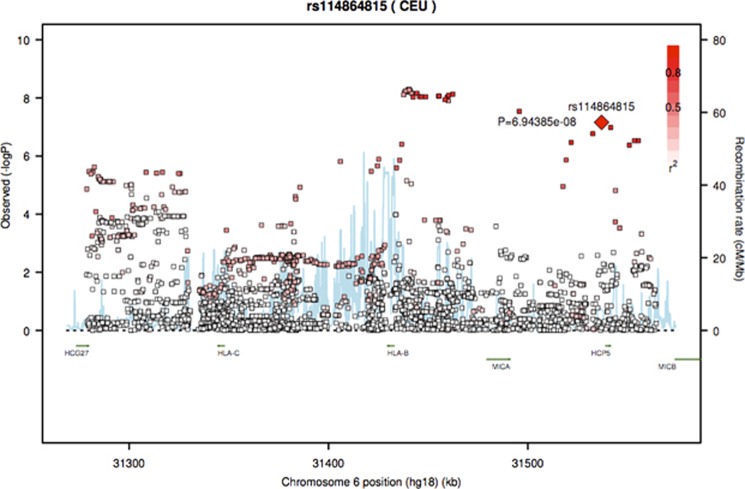
The variants driving the association in the European ancestry group and tagging HCP5 were rs116062713 (P=1.04 × 10−7) and rs114864815 (P=6.94 × 10−8) located upstream and at the 5′ untranslated region of HCP5, respectively. In each case, the minor allele frequency ((MAF)=0.08) conferred protection, with hazard ratios (HR)=0.73 (95% confidence interval (CI): 0.64–0.82). In the joint ancestry survival analyses, these variants nearly reached genome-wide significance (P=5.22 × 10−8 and 4.54 × 10−8), with HR~0.72 (95% CI: 0.65–0.82). With logistic regression analyses, both the joint and European ancestry analyses yielded associations in this region, with P~4.00x10−6 and odds ratio (OR)~0.74 (95% CI: ~0.65–0.83). Manhattan plots and corresponding QQ plots are provided (Supplementary Figures S1–S3, respectively). We illustrate the regional linkage disequilibrium (LD) plot using annotated P-values from the Cox regression analysis in the European ancestry sample (Figure 3). LD, with respect to rs114864815, was generated using the same European ancestry sample and is annotated with shades of red. The background recombination rate illustrated in blue was generated from the 1000 Genomes Pilot 1 data. There is a region in high LD with the HCP5 variants upstream of HLA-B that yielded the most significant association results in the European ancestry survival analyses (Figure 3). The most significant single-nucleotide polymorphisms (SNPs) (rs114045064 and rs112660930) reached genome-wide significance (P=5.06 × 10−9 and 5.26 × 10−9), with HR=0.77 (95% CI: 0.71–0.85). These SNPs are tagging a repeated untranslated region region of the retrogene of dihydrofolate reductase (DHFR) gene. With respect to logistic regression analyses, both the joint and European ancestry analyses yielded associations in this region, with P~1.00 × 10−7 and OR~0.80 (95% CI:~0.73–0.87). None of these variants reached genome-wide significance in the African ancestry and Hispanic groups (Table 2).
Figure 3.
Regional LD plot using the SNP Annotation and Proxy Search software.25
We provide a dot plot illustrating median age of zoster onset for case subjects of genetically determined European ancestry by the SNPs associated with herpes zoster, as compared with the overall median of 67.96 (Supplementary Figure S4). Each SNP in the HCP5 region is categorized as having 0, versus 1 or 2 copies of the minor allele. For each of these SNPs, having one or two copies of the minor allele was associated with a later median age of herpes zoster onset, as compared with the major allele.
PheWAS (phenotype-wide association study)
We have included the top PheWAS associations (P<0.01) for two SNPs associated with zoster (rs114864815 and rs114045064) with respective International Classification of Diseases (ICD)-9 code, the description of the phenotype, ß, OR, standard error, P-value, the number of cases and controls and MAF (Supplementary Table S1). The list was enriched in two general areas, including inflammatory and inflammatory disorders or infections, and cancers of the skin and mucosal areas. All of the association results are illustrated in a PheWAS association plot (Supplementary Figure S5). Phenotypes are grouped along the x axis by categorization within the PheWAS code hierarchy.9,10
The strongest association in the PheWAS was herpes zoster with P=1.54 × 10−5 and 1.57 × 10−7 and OR=0.73 and 0.75 with rs114864815 and rs114045064, respectively. There were also many inflammatory disorders or infections among the top associations with the same variants: (1) Acute pancreatitis (P=2.24 × 10−3 and 1.69 × 10−2); (2) Acute tonsillitis (P=1.51 × 10−3 and 4.48 × 10−3); (3) Cerebral atherosclerosis (P=2.10 × 10−3 and 5.29 × 10−3); (4) Vaginitis and vulvovaginitis (rs114045064, P=2.70 × 10−3); (5) Impetigo (rs114045064, P=2.99 × 10−3); (6) Peptic ulcer (rs114045064, P=3.63 × 10−3); (7) Meningitis (P=3.87x10−3 and 6.22x10−3); (8) Acute bronchitis and bronchiolitis (P=6.12x10−3 and 4.23 × 10−3); (9) Lichen planus (rs114045064; P=4.80 × 10−3); (10) Acute pharyngitis (rs114045064; P=6.39 × 10−3); and (11) Dyshidrosis (rs114045064; P=9.51 × 10−3).
There were was also an enrichment of cancers of the skin and mucosal areas among the top associations with rs114864815 and rs114045064, respectively: (1) Malignant neoplasm of oral cavity and pharynx (rs114864815; P=3.78 × 10−3); (2) Cancer of nasal cavities (rs114045064; P=3.82 × 10−3); (3) Carcinoma in situ of skin (rs114864815; P=4.12 × 10−3); and (4) Non-melanoma skin cancer (P=5.38 × 10−3 and 6.34 × 10−3). Although not meeting the P<0.01 threshold, an association with herpes simplex was suggestive (rs114864815, rs114045064; P=8.31 × 10−2 and 1.69 × 10−2). Distributions of P-values by PheWAS category based on the associations for both rs114864815 and rs114045064 are illustrated using box-and-whisker plots (Supplementary Figure S6).
Discussion
We were able to ascertain a robust herpes zoster phenotype from EMR data using an electronic phenotyping algorithm. We found strong evidence for protective variants in HCP5 among participants of the European ancestry. Our results suggest that genomic variation in the HLA region may be associated with resistance to herpes zoster. The major histocompatibility complex in humans, or HLA system, is an excellent candidate region for assessing such polymorphisms with its large number of immune-related genes. The same variants implicated here for herpes zoster risk have been shown to have a role in the progression of host HIV infection.11
Given the fact that HCP5 is non-coding and is related to human endogenous retroviruses, we hypothesize that this region could be serving a host-driven regulatory function for herpes zoster. Herpes viruses are large (100–200 nm) and contain double-stranded DNA, which also contain a large number of non-coding RNAs that serve various regulatory functions.6 Associations in non-coding regions such as HCP5 support the notion that genomic variation of regulatory functions can be associated with risk of viral reactivation.
Consistent with previous literature,1,3 we found associations between female sex, European ancestry and older ages with herpes zoster. Evaluation of age of onset effects suggests that these variants influence age of onset of herpes zoster, which we have demonstrated in the case participants of European ancestry (Supplementary Figure S4).
There are several limitations to this study. Our sample sizes of participants with herpes zoster for both African ancestry (n=173) and Hispanic (n=41) are too small for ancestry-specific genomic association studies. We also do not have information on whether controls had initial exposure to VZV. The chickenpox vaccine became available in the United States in 1995. Thus the vast majority of our controls would not have been vaccinated for chickenpox. More than 95% of the US adults experienced VZV (usually as children) before the FDA's licensure of the varicella vaccine.12 Controls who never had primary infection with VZV are not at risk of reactivation in the form of herpes zoster and could reduce our power but should not lead to false-positive genetic associations. However, we also cannot distinguish between a protection from a primary VZV infection, extended VZV latency and protection from re-emergence. Any or all of these factors would lead to reduced risk of a clinical diagnosis of herpes zoster. We do not know that the age of shingles diagnosis for cases was their first episode of herpes zoster, and some controls may have had herpes zoster that was not captured in the electronic health record; however, both of these occurrences would typically bias effect size towards the null. Finally, these results have not yet been replicated, although this region's role in the progression of host HIV infection is in strong support of viral control. Future evaluation should test the validity of this association.
The PheWAS results were encouraging for multiple reasons. We were able to reproduce our original herpes zoster association results using only ICD-9 codes to identify cases. Both the PheWAS and the primary logistic/Cox regression phenotypes were dichotomous phenotypes and relied on ICD-9 codes to define cases and controls. However, there are differences in the analyses, with the PheWAS being less detailed. The primary analysis phenotype definitions included consideration of ICD-9 miscoding (Methods) and other possible confounding effects, such as zoster vaccination, chemotherapy and prior infections. Participants also had to be ⩾40 years and have continual health system enrollment prior to diagnosis. The PheWAS model adjusted for decade of birth, sex, eMERGE study site and the first three principal components (eigenvectors) from a combined ancestry principal component analyses (PCA). For the logistic/Cox regression models, sex, log10 median BMI, study site and eigenvectors 1 and 2 both for joint and genetic ancestry-stratified groups were utilized as covariates. The results also highlighted HLA genotype associations with an enrichment of inflammatory and inflammatory disorders or infections, thus supporting this region's vital role in immune response. Some of these HLA associations, such as the acute pancreatitis, have been demonstrated before.13 We were also able to demonstrate a suggestive association of SNPs associated with zoster with herpes simplex. This is plausible, because VZV and herpes simplex are ancestrally related.14 We did remove participants with a history of herpes simplex for the European survival analyses, and the results did not change (data not shown). The top PheWAS associations were also enriched for phenotypes with cancers of the skin and mucosal areas, which may also have an infectious link.
In conclusion, we identified HCP5 SNPs that predict risk of herpes zoster diagnosis and age of onset of the disease. Participants with rs114864815 are at less risk and are affected at later ages. We also identified variants that predict risk of herpes zoster near the retrogene DHFR (upstream HLA-B). Inhibitors of DHFR have been shown to potentiate the antiviral effects of acyclovir on herpes viruses.15 It is unclear whether variation in one or both of these gene regions are causal with onset of herpes zoster or is a surrogate in strong LD with the true causal variant. Further evaluation of this region will elucidate its function and its association with the susceptibility of herpes zoster. This region is also an excellent candidate for cerebrovascular disease and early onset myocardial infarction as demonstrated in the retrospective cohort study in the United Kingdom where herpes zoster was shown to be a risk factor.5 The mining of EMRs for phenotypes such as herpes zoster are providing unique opportunities for discovery that normally would not be possible with traditional genotype association studies. This field is relatively young, and in many cases, like these results, all available data are utilized and replication is not possible. As the field matures, and more bio-repositories linked to EMRs come on line, replication data will be available.
Methods
Selection and description of participants in eMERGE
We developed an algorithm to extract individuals with herpes zoster from the EMR while removing participants with significant comorbidities to assess genetic association. This algorithm was designed at the University of Washington and developed and implemented at the GHRI, both in Seattle, WA, USA. To enhance our covariate data, we augmented traditional EMR-based structured data (for example, diagnosis and procedure codes, labs and medication records) with medication information extracted from unstructured clinical notes (for example, progress notes and hospital discharge summaries) using Natural Language Processing.16, 17, 18 Natural Language Processing-derived information provides richer and more complete information on medication exposures as many medications are only referenced in clinical notes.
Briefly, cases were identified if participants were aged ⩾40 years with at least one diagnosis code for herpes zoster and at least 1 year of continuous health system enrollment prior to diagnosis. Suppression of the immune system, whether by disease or by immunosuppressant drugs, is a major risk factor for herpes zoster and was considered as an exclusion. Exclusionary criteria included any of the following: (1) Vaccination for herpes zoster within the last year for cases and ever for controls; (2) Two or more diagnosis codes for HIV; (3) Cancer of the blood or bone marrow up to 12 months prior to the index date; (4) Chemotherapy up to 12 months prior to the diagnosis date; (5) Steroid use 21 days prior to index date; and (6) History of transplant immunosuppression medications at any strength. Controls were selected if they were never vaccinated for VZV (except for survival analyses, see below), had no evidence of diagnosis codes for herpes zoster and they did not have an above listed exclusion.
As a standard best practice in eMERGE, manual chart validation was performed at two separate sites to ensure the phenotype data-mining algorithm performed appropriately. To ensure the herpes zoster EMR algorithm correctly classified participants, 25 cases and 25 controls were randomly selected at both GHRI and Vanderbilt University, respectively (n total=100). At GHRI, there was a positive predicted value of 100% for the cases and 96% for the controls. At Vanderbilt University, the positive predicted value was 96, and 100% for the cases and controls, respectively. eMERGE investigators have developed Phenotype KnowledgeBase (PheKB), a repository for phenotype creation, validation and execution of phenotype algorithms across the network and beyond.7,19 This tool was utilized for the herpes zoster EMR algorithm to facilitate development and revising of the phenotype. The algorithm and implementation data are available on PheKB (Web Resources).
Genotyping and imputation
The eMERGE Coordinating Center (CC) at the Pennsylvania State University (PSU) performed genotype imputations for the eMERGE Phase-II project, which includes all participants' samples from eMERGE-I and eMERGE-II, using the BEAGLE software package (Version 3.3.1, Seattle, WA, USA).20 Details of the genotyping platforms for the nine eMERGE sites (seven adult sites and two pediatric) can be found on the PSU eMERGE CC web site (Web Resources). The majority of samples were genotyped on the Human 660 Quad, with additional samples genotyped by the OmniExpress chip and others. Imputation was performed for all autosomes, with a reference panel selected from the 1000 Genomes Project (October 2012 release). For the analyses presented here, variants were included if the allelic R2⩾0.7, call rate was ⩾99% and MAF was >0.03. The allelic R2 is the accuracy of imputed genotypes in terms of the squared correlation between the allele dosage (number of minor alleles) of the most likely imputed genotype and the allele dosage of the true genotype.20 Further quality control/quality assurance measures are outlined in the imputation guide provided on the PSU eMERGE CC website (Web Resources).
Defining genetically determined ancestry
PCA were performed using the SNPRelate R statistical computing package.21 Prior to inclusion into the correlation matrix, postimputation variants were selected after LD pruning at r=0.5 and a MAF>0.03. For all 38 250 participants, genetically determined European ancestry was assigned to all participants with eigenvectors 1 and 2 values within 4 s.d. from the medians of eigenvectors 1 and 2 of self-identified European ancestry participants. For genetically determined African ancestry, we identified all participants with values within 2 s.d. from the medians of eigenvector 1 and 2 of self-identified African ancestry participants. For the Hispanic group, we identified all participants with values within 1 s.d. from the medians of eigenvector 1 and 2 of self-identified Hispanic participants.22 The ancestry classifications clustered with the genotyped HapMap reference samples as we have recently published.23 Summaries regarding genetically determined ancestry are listed (Table 1). To control for ancestry-specific admixture, PCA was performed within each respective ancestry group. This included 18 423 participants of European ancestry, 3485 of African ancestry and 621 Hispanic.
Association analyses
Our primary aim was to robustly identify variants associated with herpes zoster diagnosis. Because age is a major risk factor for shingles, we assessed survival using Cox proportional hazard analysis with end points of age at loss to follow-up, death or VZV vaccination for controls, versus age at first zoster episode for cases. When age of onset is variable and early onset may indicate an increased genetic susceptibility, Cox regression may have more statistical power than logistic regression.24 We used the coxph R function with the base survival package in R for Cox proportional-hazards regression analyses. We implemented this function using the R-plugin feature in PLINK.25 For variables with repeated measures considered for covariates (BMI and age at a given participant visit), median values were utilized in the association models.
We performed a joint analysis of all participants and genetic ancestry-stratified analyses of each ancestry group separately. For the joint analysis, we included covariates for sex, log10 median BMI, study site and eigenvectors 1 and 2. For the European ancestry analyses, eigenvectors 1 and 2 derived from the joint ancestry analysis were dropped from the model and replaced with eigenvectors 1 and 2 derived from the European-specific PCA analyses. SNP genotypes were coded as 0, 1 and 2 copies of the minor allele based on the most probable genotype (additive genotypic model). For completeness, logistic regression analyses of zoster case/control status as a dichotomous phenotype was performed in PLINK25 with the given covariates and genotypes.25
PheWAS
To determine pleiotropic effects of the SNPs associated with zoster (rs114864815, rs114045064), we assessed a range of associated clinical phenotypes (PheWAS) in 28 580 genotyped individuals of European ancestry from seven different eMERGE sites with EMR-linked DNA biobanks with Vanderbilt University.9,10
The PheWAS algorithm identified 1619 unique PheWAS ‘phenotypes' from 6 994 816 unique dates of interaction with the EMR. Per dichotomous phenotype, participants were grouped as a corresponding ‘case' using distinct ICD-9 billing codes from each participant's records. Under the presumption that acute diseases such as infections and infectious complications like zoster may not be billed multiple times, a ‘case' was a record that had a single, valid ICD-9 code that maps to PheWAS case group, and ‘controls' were assigned if the participant did not have any ICD-9 codes belonging to the exclusion code grouping corresponding for that case. Association tests were performed using logistic regression in R with the PheWAS package, adjusted for decade of birth, self-reported sex, study site and the first three principal components.9,10
Other plots
The regional LD plot was created using the SNP Annotation and Proxy Search software.26 The background recombination rate was generated from the 1000 Genomes Pilot 1 data, specifically the CEPH (Utah residents with ancestry from northern and western Europe) sample. The dot plot of median values by genotype was created using the summary function from the Hmisc R statistical computing package (Web Resources).
Web Resources
eMERGE web site: http://www.gwas.net
eMERGE Coordinating Center genotyping data: http://emerge.mc.vanderbilt.edu/genotyping-data-released
PheKB phenotype: http://phekb.org/phenotype/herpes-zoster
R package Hmisc: http://cran.r-project.org/web/packages/Hmisc/index.html
R package rms: http://cran.r-project.org/web/packages/rms/index.html
R package SNPRelate: https://github.com/zhengxwen/SNPRelate
R package PheWAS: https://knowledgemap.mc.vanderbilt.edu/research/content/phewas-r-package
Acknowledgments
We are grateful to all the participants of the eMERGE study. This study was supported by the following U01 grants from the National Human Genome Research Institute (NHGRI), a component of the National Institutes of Health (NIH), Bethesda, MD, USA: (1) U01HG006375 (Group Health/University of Washington); (2) U01HG006382 (Geisinger Health System); (3) U01HG006379 (Mayo Clinic); (4) U01HG006389 (Essentia Health and Marshfield Clinic Research Foundation); (5) U01HG006388 (Northwestern University); (6) HG004438 (Center for Inherited Disease Research, Johns Hopkins University); (7) HG004424 (Broad Institute of Harvard and MIT); (8) U01HG006378, U01HG006385, U01HG006385 (Vanderbilt University); (9) U01HG006380 (The Mt Sinai Hospital); (10) U01HG006828 (Cincinnati Children's Hospital Medical Center/Harvard); and (11) U01HG006830 (Children's Hospital of Philadelphia). Additional support was provided by a State of Washington Life Sciences Discovery Fund award to the Northwest Institute of Genetic Medicine (to GPJ).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Genes and Immunity website (http://www.nature.com/gene)
Supplementary Material
References
- Cohen JI. Herpes zoster. N Engl J Med. 2013;369:1766–1767. doi: 10.1056/NEJMc1310369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimland D, Moanna A. Increasing incidence of herpes zoster among veterans. Clin Infect Dis. 2010;50:1000–1005. doi: 10.1086/651078. [DOI] [PubMed] [Google Scholar]
- Hicks LD, Cook-Norris RH, Mendoza N, Madkan V, Arora A, Tyring SK. Family history as a risk factor for herpes zoster: a case-control study. Arch Dermatol. 2008;144:603–608. doi: 10.1001/archderm.144.5.603. [DOI] [PubMed] [Google Scholar]
- Yawn BP, Itzler RF, Wollan PC, Pellissier JM, Sy LS, Saddier P. Health care utilization and cost burden of herpes zoster in a community population. Mayo Clin Proc. 2009;84:787–794. doi: 10.4065/84.9.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuer J, Pacou M, Gauthier A, Brown MM. Herpes zoster as a risk factor for stroke and TIA: a retrospective cohort study in the UK. Neurology. 2014;82:206–212. doi: 10.1212/WNL.0000000000000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinde B. Herpesviruses: latency and reactivation—viral strategies and host response. J Oral Microbiol. 2013;5 doi: 10.3402/jom.v5i0.22766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman O, Kuivaniemi H, Tromp G, Faucett WA, Li R, Manolio TA, et al. The Electronic Medical Records and Genomics (eMERGE) Network: past, present, and future. Genet Med. 2013;15:761–771. doi: 10.1038/gim.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty CA, Chisholm RL, Chute CG, Kullo IJ, Jarvik GP, Larson EB, et al. The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics. 2011;4:13. doi: 10.1186/1755-8794-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny JC, Bastarache L, Ritchie MD, Carroll RJ, Zink R, Mosley JD, et al. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat Biotechnol. 2013;31:1102–1110. doi: 10.1038/nbt.2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny JC, Ritchie MD, Basford MA, Pulley JM, Bastarache L, Brown-Gentry K, et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics. 2010;26:1205–1210. doi: 10.1093/bioinformatics/btq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellay J, Ge D, Shianna KV, Colombo S, Ledergerber B, Cirulli ET, et al. Common genetic variation and the control of HIV-1 in humans. PLoS Genet. 2009;5:e1000791. doi: 10.1371/journal.pgen.1000791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman GS, King PG. Review of the United States universal varicella vaccination program: herpes zoster incidence rates, cost-effectiveness, and vaccine efficacy based primarily on the Antelope Valley Varicella Active Surveillance Project data. Vaccine. 2013;31:1680–1694. doi: 10.1016/j.vaccine.2012.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawa S, Ota M, Yoshizawa K, Horiuchi A, Hamano H, Ochi Y, et al. HLA DRB10405-DQB10401 haplotype is associated with autoimmune pancreatitis in the Japanese population. Gastroenterology. 2002;122:1264–1269. doi: 10.1053/gast.2002.33022. [DOI] [PubMed] [Google Scholar]
- Grose C. Pangaea and the Out-of-Africa Model of Varicella-Zoster Virus Evolution and Phylogeography. J Virol. 2012;86:9558–9565. doi: 10.1128/JVI.00357-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prichard MN, Prichard LE, Shipman C., Jr Inhibitors of thymidylate synthase and dihydrofolate reductase potentiate the antiviral effect of acyclovir. Antiviral Res. 1993;20:249–259. doi: 10.1016/0166-3542(93)90024-d. [DOI] [PubMed] [Google Scholar]
- Nadkarni PM, Ohno-Machado L, Chapman WW. Natural language processing: an introduction. JAMIA. 2011;18:544–551. doi: 10.1136/amiajnl-2011-000464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Stenner SP, Doan S, Johnson KB, Waitman LR, Denny JC. MedEx: a medication information extraction system for clinical narratives. JAMIA. 2010;17:19–24. doi: 10.1197/jamia.M3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzuner O, Solti I, Cadag E. Extracting medication information from clinical text. JAMIA. 2010;17:514–518. doi: 10.1136/jamia.2010.003947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton KM, Peissig PL, Kho AN, Bielinski SJ, Berg RL, Choudhary V, et al. Validation of electronic medical record-based phenotyping algorithms: results and lessons learned from the eMERGE network. JAMIA. 2013;20:e147–e154. doi: 10.1136/amiajnl-2012-000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning BL, Browning SR. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet. 2009;84:210–223. doi: 10.1016/j.ajhg.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Levine D, Shen J, Gogarten SM, Laurie C, Weir BS. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics. 2012;28:3326–3328. doi: 10.1093/bioinformatics/bts606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manichaikul A, Palmas W, Rodriguez CJ, Peralta CA, Divers J, Guo X, et al. Population structure of Hispanics in the United States: the multi-ethnic study of atherosclerosis. PLoS Genet. 2012;8:e1002640. doi: 10.1371/journal.pgen.1002640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosslin DR, McDavid A, Weston N, Nelson SC, Zheng X, Hart E, et al. Genetic variants associated with the white blood cell count in 13,923 subjects in the eMERGE Network. Hum Genet. 2012;131:639–652. doi: 10.1007/s00439-011-1103-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Net JB, Janssens AC, Eijkemans MJ, Kastelein JJ, Sijbrands EJ, Steyerberg EW. Cox proportional hazards models have more statistical power than logistic regression models in cross-sectional genetic association studies. Eur J Hum Genet. 2008;16:1111–1116. doi: 10.1038/ejhg.2008.59. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O'Donnell CJ, de Bakker PI. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24:2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.