Abstract
Tamoxifen is the standard-of-care treatment for estrogen receptor-positive premenopausal breast cancer. We examined tamoxifen metabolism via blood metabolite concentrations and germline variations of CYP3A5, CYP2C9, CYP2C19 and CYP2D6 in 587 premenopausal patients (Asians, Middle Eastern Arabs, Caucasian-UK; median age 39 years) and clinical outcome in 306 patients. N-desmethyltamoxifen (DM-Tam)/(Z)-endoxifen and CYP2D6 phenotype significantly correlated across ethnicities (R2: 53%, P<10−77). CYP2C19 and CYP2C9 correlated with norendoxifen and (Z)-4-hydroxytamoxifen concentrations, respectively (P<0.001). DM-Tam was influenced by body mass index (P<0.001). Improved distant relapse-free survival (DRFS) was associated with decreasing DM-Tam/(Z)-endoxifen (P=0.036) and increasing CYP2D6 activity score (hazard ratio (HR)=0.62; 95% confidence interval (CI), 0.43–0.91; P=0.013). Low (<14 nM) compared with high (>35 nM) endoxifen concentrations were associated with shorter DRFS (univariate P=0.03; multivariate HR=1.94; 95% CI, 1.04–4.14; P=0.064). Our data indicate that endoxifen formation in premenopausal women depends on CYP2D6 irrespective of ethnicity. Low endoxifen concentration/formation and decreased CYP2D6 activity predict shorter DRFS.
Introduction
The standard-of-care for estrogen receptor (ER)-positive breast cancer patients who are functionally premenopausal is a 5-year treatment with the selective ER modulator tamoxifen1 as aromatase inhibitors are of limited use due to the strong hypothalamic–pituitary control of estrogen levels.2 Tamoxifen for 5 years substantially lowers the yearly relapse rates and mortality in primary breast cancer.3 Data from the Adjuvant Tamoxifen, Longer Against Shorter trial and the Adjuvant Tamoxifen Treatment offers more trial indicate further benefit by maintaining tamoxifen treatment for 10 years with 25% mortality reduction relative to 5 years.4, 5 Despite this success, about 50% of patients do not benefit from tamoxifen and frequent adverse drug reactions (ADR) including hot flashes, vasomotor and gynecologic symptoms as well as depression and diminished sexual functioning prevent particularly young women from staying on the drug.6, 7 Toward a personalized strategy of premenopausal breast cancer treatment, the possible association between serum concentration of the active tamoxifen metabolite endoxifen and the occurrence of side effects has been addressed;7, 8 yet, the potential of such an approach regarding treatment outcome in young women is unclear.
Tamoxifen, which acts at the ER, requires conversion into active metabolites (Z)-4-hydroxytamoxifen and (Z)-endoxifen that have up to 100-fold higher ER affinity than the parent drug.9, 10, 11 The cytochrome P450 enzyme CYP2D6 has a major role in the formation of endoxifen12 in postmenopausal women.13, 14, 15 CYP2D6 is highly polymorphic with more than 100 genetic variants (http://www.imm.ki.se/cypalleles/cyp2d6.htm) contributing to the high interindividual variability of enzyme activity. Impaired metabolism by CYP2D6 can be accurately predicted by loss- and reduced-function alleles resulting in the poor metabolizer (PM) and intermediate metabolizer (IM) phenotypes. Likewise, functional and duplicated CYP2D6 alleles correlate with extensive (EM) and ultra rapid (UM) metabolizer phenotypes, respectively.16 There are significant interethnic differences of CYP2D6 allele frequencies across geographic regions and populations leading to a shift of metabolizer phenotype prevalence with higher frequencies of IMs in Asians and UMs in Arabic/North African countries as compared with populations of European descent.17
Clinical outcome of adjuvant tamoxifen in postmenopausal patients is influenced by their CYP2D6 metabolizer phenotype which can be predicted by genetic testing18, 19, 20 using germline rather than tumor DNA.21, 22 However, the current debate on the validity of the postmenopausal data and the lack of CYP2D6 association with clinical outcome in some of these studies21, 22, 23, 24, 25, 26, 27, 28 point to the need of combined pharmacokinetic and pharmacogenetic analyses particularly in the case of testing the hypothesis in another patient group, i.e. premenopausal patients. Recently, lower endoxifen concentrations were shown to be associated with poor clinical outcome in a mixed cohort of pre- and postmenopausal patients.14 Notably, the in vitro pharmacological modeling of endoxifen concentrations for the treatment of ER positive breast cancer showed the essential requirement of endoxifen to block breast cancer cell growth in the presence of high estrogen concentrations equivalent to premenopausal patients.29, 30 Therefore, it is reasonable to hypothesize that variable endoxifen formation contributes to tamoxifen efficacy in premenopausal patients. Here, we present combined pharmacokinetic and pharmacogenetic analyses in purely premenopausal breast cancer patient cohorts of different ethnic origin to evaluate (i) the factors that influence active tamoxifen metabolite concentrations with a particular emphasis on CYP2D6 and (ii) whether tamoxifen metabolite concentrations and/or genetic variants of drug-metabolizing enzymes (DME) are suitable biomarkers for the prediction of clinical outcome.
Patients and methods
Patients and study design
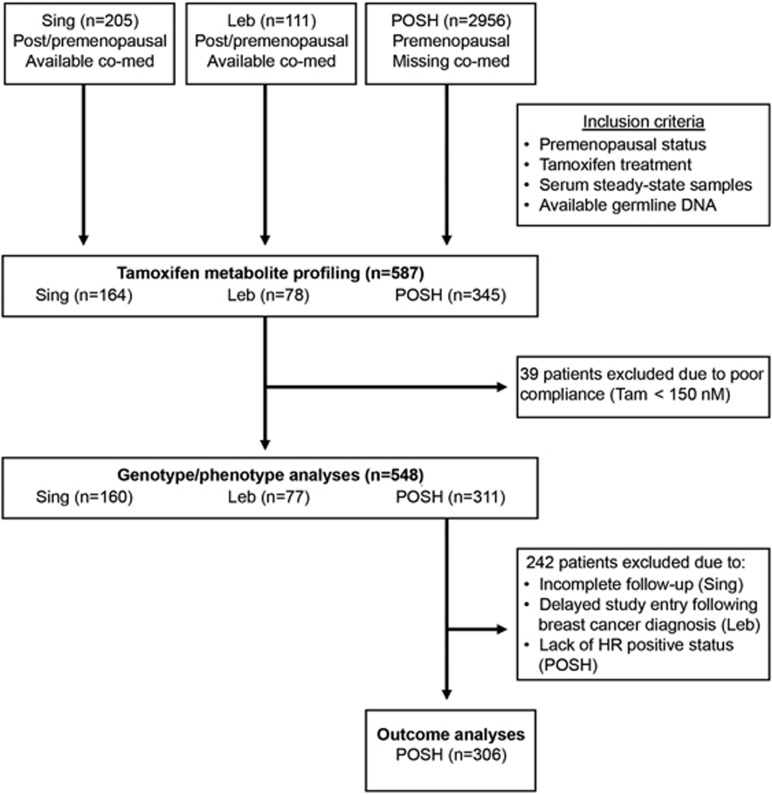
Three ethnic groups of prospectively recruited hormone receptor-positive premenopausal breast cancer patients with adjuvant tamoxifen treatment were investigated (Figure 1). Of these, 164 Asian patients in part previously described31 (136 Chinese, 14 Malays and 14 Indians) have been provided by the Division of Medical Sciences, Humphrey Oei Institute of Cancer Research, Singapore. Another 78 consecutively recruited patients (2009–2011) in part previously described32 have been provided by the Hematology-Oncology Division, Internal Medicine, American University of Beirut, Lebanon (Leb). Furthermore, 345 patients were selected from the Prospective study of Outcomes in Sporadic versus Hereditary breast cancer (POSH) cohort, an observational cohort study comprised mainly of Caucasian patients (2956 patients recruited between 2001 and 2007) by the Cancer Sciences Academic Unit and University of Southampton Clinical Trials Unit, University of Southampton, UK.33, 34 Selection of patients for our current study was based on the availability of tamoxifen steady-state serum and germline DNA. Patients taking CYP2D6 inhibitors were excluded from the Singapore cohort and comprised only seven patients who received weak CYP2D6 inhibitors, venlafaxine, escitalopram or clomipramine in the Lebanon cohort. For the POSH cohort, data on co-medication was not available. In total, 587 premenopausal patients were investigated for the quantitation of tamoxifen metabolites and genotyping. Figure 1 shows the patient inclusion scheme to explain the underlying rationale for the survival analyses, which was performed in the POSH cohort but not in the Singapore and Lebanon cohorts due to incomplete follow-up and delayed study entry of their patients, respectively (Figure 1).
Figure 1.
Study flow diagram of premenopausal study. co-med, co-medication; HR, hormone receptor; Leb, Lebanon; POSH, Prospective study of Outcomes in Sporadic versus Hereditary breast cancer; Sing, Singapore; Tam, tamoxifen.
Steady-state blood samples of patients treated with tamoxifen (20 mg per day) were collected on-site within the first year of treatment and immediately stored at −20 °C. Study approvals were obtained from the National Cancer Centre Ethics Review Committee (Singapore), American University of Beirut Institutional Review Board (Lebanon) and South and West MultiCentre Research Ethics Committee (MREC 00/6/69; POSH). All patients provided informed consent.
Measurement of tamoxifen and metabolites
Tamoxifen and its metabolites DM-Tam, (Z)-4-hydroxytamoxifen ((Z)-4-OH-Tam), (Z)-endoxifen, N-desmethyltamoxifen (DM-Tam), N,N-Didesmethyltamoxifen (DDM-Tam) and (Z)-norendoxifen ((Z)-4-OH-DDM-Tam; Supplementary Figure 1) were measured by liquid chromatography tandem mass spectrometry in the multiple reaction monitoring mode on a 6460 triple quadrupole mass spectrometer (Agilent Technologies, Waldbronn, Germany) as described previously.13 All analyses refer to the active (Z)-isomers of the tamoxifen metabolites which were separated from their respective inactive (E)-isomers.35
DNA isolation and genotyping
DNA samples were genotyped for CYP2D6 alleles associated with null (PM: *3, *4, *5 and *6) and reduced (IM: *9, *10 and *41) CYP2D6 function by matrix-assisted, laser desorption/ionization, mass spectrometry.18, 31 CYP2D6 gene duplication (UM) was determined via TaqMan Copy Number Assay (Applied Biosystems, Foster City, CA, USA). Absence of variant alleles or gene duplications was assigned to normal CYP2D6 function (EM). Individuals with ambiguous genotypes were verified by AmpliChip P450 assay (Roche Molecular Diagnostics, Mannheim, Germany), which revealed additional CYP2D6 alleles *14, *15 and *17. For genotype interpretation, we applied a CYP2D6 activity score36 to predict PM, IM, EM and UM phenotypes: PM/PM (0), PM/IM (0.5), IM/IM (0.75), PM/EM (1), IM/EM (1.5), EM/EM (2) and EM/UM (3). In survival analyses, CYP2D6 activity scores were classified into three phenotype classes: PM (0), hetEM/IM (0.5–1.5) and EM and UM (2 to 3). Genetic testing was also performed for CYP2C9 (*2, *3), CYP2C19 (*2,*3,*17) and CYP3A5*3, known to be involved in tamoxifen metabolism using TaqMan or Sequenom mass array genotyping.13
Study aims, power calculation and statistical analyses
Primary aim of this study was to determine factors that influence active tamoxifen metabolite concentrations with special emphasis on CYP2D6-catalyzed metabolite endoxifen. Thus, we performed a power calculation assuming a 50% decrease or increase of square root transformed endoxifen or log-transformed metabolic ratio (MR) DM-Tam/endoxifen per one unit of CYP2D6 activity score as a relevant gene-dose effect.13, 37 On the basis of the s.d. of the plasma concentrations in the three cohorts (excluding poorly/non-compliant patients), we computed that the sample size in the smallest cohort (n=77, Leb) would already provide 92.9% power in the analyses. Secondary aim was to test whether tamoxifen metabolite concentrations and/or genetic variants of DME can reliably predict clinical outcome in premenopausal patients. As there is no such data published in a purely premenopausal clinical setting, we consider this an exploratory investigation.
SPSS (version 20, Chicago, IL) and R-3.01 including libraries coin-1.0–22, MASS-7.3–27, mfp-1.4.9, party-1.0–8 and survival-2.37–4 (www.r.project.org) were used for statistical analyses. Clinical characteristics and DME genotypes were compared between study cohorts by Kruskal–Wallis and χ2-tests. Tamoxifen concentrations across all patients displayed two splits operationally used to define non-compliance (⩽40 nM) and poor compliance (40–150 nM). Frequencies of CYP2D6 phenotypes between these groups were compared by χ2-tests. All following analyses excluded poorly/non-compliant patients.
Correlations of square root transformed endoxifen concentrations or log-transformed MR DM-Tam/endoxifen and CYP2D6 activity score were examined by linear models with stratification for ethnicity.
Survival analyses were strictly confined to the POSH cohort because of delayed study entry following breast cancer diagnosis and incomplete follow-up in the Lebanon and Singapore cohorts, respectively. The endpoint was distant relapse-free survival (DRFS) defined as the time from diagnosis to the earliest occurrence of distant metastasis or death from any cause. First, we tested which clinical characteristics, tamoxifen metabolites or CYP genotypes were associated with DRFS using univariate Cox regression and general asymptotic independence tests. Step-wise model selection (R-library mfp) revealed nodal status and chemotherapy use to be included as covariates in subsequent multivariate Cox regression. Conditional inference trees (P-value cutoff 0.15) and receiver operating characteristic analyses on DRFS were applied to identify cutoffs for MR DM-Tam/endoxifen and endoxifen concentration. For endoxifen, there was neither a split nor a significant association in multivariate Cox regression, hence we explored non-monotonic effects by dividing the POSH population into quarters. Associations between DRFS and classified endoxifen concentrations, MR DM-Tam/endoxifen or CYP2D6 phenotypes were investigated by Kaplan–Meier analyses and multivariate Cox regression adjusted for nodal status and chemotherapy use. All Cox models were stratified for ethnicity. All statistical tests were two-sided and statistical significance was defined as P<0.05.
Results
Patient characteristics and cytochrome P450 genotypes
The median age of diagnosis was 39.1 years (range 22–59 years) (Table 1). Follow-up, age, body mass index (BMI), ethnicity, tumor size and proportion of patients treated with chemotherapy differed among cohorts. Genotypes were successfully obtained from 583 patients for CYP2D6 (99%) and in 97–99% for CYP3A5, CYP2C9 and CYP2C19. Genotypes met Hardy–Weinberg Equilibrium with few exceptions of minor deviations (Table 2). Notably, UM frequency in the POSH cohort was lower than expected38 likely due to sample size.
Table 1. Clinical characteristics of premenopausal patients.
|
Singapore (n=164) |
Lebanon (n=78) |
POSH (n=345) |
Overall (n=587) |
P-valuea | |||||
|---|---|---|---|---|---|---|---|---|---|
| No. of patients | % | No. of patients | % | No. of patients | % | No. of patients | % | ||
| Follow-up, years | <0.001 | ||||||||
| Median | 4.0 | 4.1 | 6.4 | 5.5 | |||||
| Range | 1.3–14.2 | 1.9–8.5 | 1.2–11.6 | 1.2–14.2 | |||||
| Age at diagnosis, years | <0.001 | ||||||||
| Median | 47.0 | 43.0 | 37.5 | 39.1 | |||||
| Range | 30.0–59.0 | 24.0–51.0 | 22.0–41.0 | 22.0–59.0 | |||||
| BMI | <0.001 | ||||||||
| ⩽30 | 153 | 93.3 | 62 | 79.5 | 272 | 78.8 | 487 | 82.9 | |
| >30 | 10 | 6.1 | 16 | 20.5 | 65 | 18.8 | 91 | 15.5 | |
| Unknown | 1 | 0.6 | 8 | 2.3 | 9 | 1.5 | |||
| Ethnicity | <0.001 | ||||||||
| Caucasian | 320 | 92.8 | 320 | 54.5 | |||||
| Asian | 164 | 100 | 5 | 1.4 | 169 | 28.8 | |||
| African | 15 | 4.3 | 15 | 2.6 | |||||
| Middle Eastern | 78 | 100 | 78 | 13.3 | |||||
| Unknown | 5 | 1.4 | 5 | 0.9 | |||||
| Tumor size (cm) | 0.004 | ||||||||
| ⩽2 | 95 | 57.9 | 30 | 38.5 | 174 | 50.4 | 299 | 50.9 | |
| 2–5 | 58 | 35.4 | 35 | 44.9 | 142 | 41.2 | 235 | 40.0 | |
| >5 | 11 | 6.7 | 13 | 16.7 | 20 | 5.8 | 44 | 7.5 | |
| Unknown | 9 | 2.6 | 9 | 1.5 | |||||
| Nodal status | 0.08 | ||||||||
| Negative | 92 | 56.1 | 40 | 51.3 | 159 | 46.1 | 291 | 49.6 | |
| Positive | 69 | 42.1 | 38 | 48.7 | 184 | 53.3 | 291 | 49.6 | |
| Unknown | 3 | 1.8 | 2 | 0.6 | 5 | 0.9 | |||
| Grading | 0.21 | ||||||||
| G1 | 14 | 17.9 | 38 | 11.0 | 52 | 8.9 | |||
| G2 | 35 | 44.9 | 178 | 51.6 | 213 | 36.3 | |||
| G3 | 27 | 34.6 | 125 | 36.2 | 152 | 25.9 | |||
| Unknown | 164 | 100 | 2 | 2.6 | 4 | 1.2 | 170 | 29.0 | |
| Hormone receptor status | 0.472 | ||||||||
| (ER or PR) Positive | 164 | 100 | 78 | 100 | 340 | 98.6 | 582 | 99.1 | |
| (ER and PR) Negative | 3 | 0.9 | 3 | 0.5 | |||||
| Unknown | 2 | 0.6 | 2 | 0.3 | |||||
| HER2 status | 0.252 | ||||||||
| Positive | 43 | 26.2 | 18 | 23.1 | 66 | 19.1 | 127 | 21.6 | |
| Negative | 112 | 68.3 | 60 | 76.9 | 158 | 45.8 | 330 | 56.2 | |
| Unknown | 9 | 5.5 | 121 | 35.1 | 130 | 22.1 | |||
| Chemotherapy | 0.001 | ||||||||
| Yes | 143 | 87.2 | 69 | 88.5 | 261 | 75.7 | 473 | 80.6 | |
| No | 21 | 12.8 | 9 | 11.5 | 84 | 24.3 | 114 | 19.4 | |
| CYP2D6 phenotypes | <0.001 | ||||||||
| PM/PM | 4 | 5.1 | 30 | 8.7 | 34 | 5.8 | |||
| hetEM/IM | 129 | 78.7 | 37 | 47.4 | 205 | 59.4 | 371 | 63.2 | |
| EM/UM | 33 | 20.1 | 37 | 47.4 | 110 | 31.9 | 180 | 30.7 | |
| Unknown | 2 | 1.2 | 2 | 0.3 | |||||
Abbreviations: BMI, body mass index; EM, extensive metabolizer; ER, estrogen receptor; HER2, human epidermal growth factor receptor 2; het, heterozygous; PM, poor metabolizer; POSH, Prospective study of Outcomes in Sporadic versus Hereditary breast cancer; PR, progesterone receptor; IM, intermediate metabolizer; UM, ultra rapid metabolizer.
Refers to test for differences between the three cohorts.
Table 2. Observed frequencies of DME gene variants.
|
Genotype frequency |
Minor allele frequency |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Allele | Variant | rs number | Function | Genotypes | Sing (n=164) | Leb (n=78) | POSH (n=345) | Total (n=587) | Sing | Leb | POSH | Total |
| CYP2D6 | ||||||||||||
| *3 | 2549delA | rs35742686 | Abrogated | A/A | 1.00 | 1.00 | 0.97 | 0.98 | 0 | 0 | 0.02† | 0.01 |
| A/del | 0 | 0 | 0.03 | 0.02 | ||||||||
| del/del | 0 | 0 | 0.003 | 0.002 | ||||||||
| *4 | g1846 G>A | rs3892097 | Abrogated | G/G | 0.99 | 0.76 | 0.63 | 0.75 | 0.003 | 0.141 | 0.21 | 0.15 |
| G/A | 0.01 | 0.21 | 0.31 | 0.21 | ||||||||
| A/A | 0 | 0.04 | 0.06 | 0.04 | ||||||||
| *5 | Chromosomal deletion | Abrogated | wt/wt | 0.87 | 0.94 | 0.93 | 0.91 | 0.067 | 0.032 | 0.035 | 0.04 | |
| wt/del | 0.13 | 0.06 | 0.07 | 0.09 | ||||||||
| del/del | 0 | 0 | 0 | 0 | ||||||||
| *6 | 1707delT | rs5030655 | Abrogated | T/T | 1.00 | 0.97 | 0.97 | 0.98 | 0 | 0.013 | 0.02 | 0.01 |
| T/del | 0.00 | 0.03 | 0.03 | 0.02 | ||||||||
| del/del | 0 | 0 | 0 | 0 | ||||||||
| *9 | 2615_2617delAAG | rs5030656 | Reduced | AAG/AAG | 1.00 | 1.00 | 0.94 | 0.97 | 0 | 0 | 0.03† | 0.02 |
| AAG/del | 0 | 0 | 0.05 | 0.03 | ||||||||
| del/del | 0 | 0 | 0.01 | 0.003 | ||||||||
| *10 | g100 C>T | rs1065852 | Reduced | C/C | 0.31 | 0.99 | 0.97 | 0.79 | 0.488π | 0.006 | 0.02 | 0.15 |
| C/T | 0.40 | 0.01 | 0.03 | 0.13 | ||||||||
| T/T | 0.29 | 0 | 0 | 0.08 | ||||||||
| *41 | g2988 G>A | rs28371725 | Reduced | G/G | 0.93 | 0.72 | 0.79 | 0.82 | 0.037 | 0.154 | 0.11 | 0.1 |
| G/A | 0.06 | 0.26 | 0.19 | 0.16 | ||||||||
| A/A | 0.01 | 0.03 | 0.01 | 0.01 | ||||||||
| *XN | Duplication | Increased | 2 copies | 0.99 | 0.87 | 0.99 | 0.98 | 0.007‡ | 0.128‡ | 0.003‡ | 0.02‡ | |
| >2 copies | 0.01 | 0.13 | 0.002 | 0.02 | ||||||||
| CYP2C19 | ||||||||||||
| *2 | 681 G>A | rs4244285 | Abrogated | G/G | 0.48 | 0.79 | 0.75 | 0.67 | 0.326 | 0.116 | 0.145 | 0.19 |
| G/A | 0.40 | 0.18 | 0.23 | 0.27 | ||||||||
| A/A | 0.13 | 0.03 | 0.02 | 0.05 | ||||||||
| *3 | 636 G>A | rs4986893 | Abrogated | G/G | 0.90 | 0.90 | 0.052 | nd | nd | 0.05 | ||
| G/A | 0.09 | 0.09 | ||||||||||
| A/A | 0.01 | 0.01 | ||||||||||
| *17 | −806 C>T | rs12248560 | Increased | C/C | 0.92 | 0.62 | 0.67 | 0.74 | 0.04 | 0.216 | 0.2# | 0.15 |
| C/T | 0.08 | 0.32 | 0.26 | 0.22 | ||||||||
| T/T | 0.00 | 0.05 | 0.07 | 0.04 | ||||||||
| CYP2C9 | ||||||||||||
| *2 | c430 C>T | rs1799853 | Reduced | C/C | 1.00 | 0.83 | 0.75 | 0.83 | 0 | 0.087 | 0.14 | 0.09 |
| C/T | 0.00 | 0.17 | 0.23 | 0.15 | ||||||||
| T/T | 0.00 | 0.00 | 0.02 | 0.01 | ||||||||
| *3 | c1075 A>C | rs1057910 | Reduced | A/A | 0.91 | 0.88 | 0.85 | 0.87 | 0.046 | 0.06 | 0.08 | 0.07 |
| A/C | 0.08 | 0.12 | 0.14 | 0.13 | ||||||||
| C/C | 0.01 | 0.00 | 0.003 | 0.002 | ||||||||
| CYP3A5 | ||||||||||||
| *3 | 6986 A>G | rs776746 | Markedly reduced | A/A | 0.10 | 0.01 | 0.02 | 0.05 | 0.72 | 0.91 | 0.91Φ | 0.85 |
| A/G | 0.36 | 0.15 | 0.14 | 0.21 | ||||||||
| G/G | 0.54 | 0.83 | 0.84 | 0.75 | ||||||||
Abbreviations: c, cDNA; del, deletion; DME, drug-metabolizing enzymes; g, gDNA; Leb, Lebanon; nd, not determined; POSH, Prospective study of Outcomes in Sporadic versus Hereditary breast cancer; Sing, Singapore; w, wildtype; wt, wildtype referring to major, that is, functional alleles.
Note-Nucleotide positions refer to numbering according to the ATG start codon. CYP2D6*14, *15 and *17 alleles were detected by AmpliChip quality control only and were not included in the table.
*XN, refers to duplication of EM alleles resulting in UM phenotype.
‡Refers to prevalence of UM phenotype based on multiple copies of functional alleles.
Genotype frequencies that deviate from HWE have the following P values: †P=0.02; πP=0.045; #P=0.006; ΦP=0.003.
Tamoxifen metabolite profiling, compliance and CYP2D6 activity
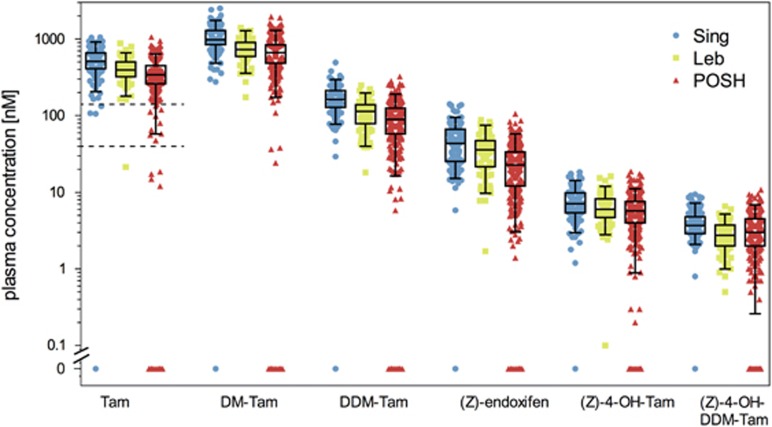
There was a strong interindividual variability for tamoxifen and the five metabolites in each ethnic group (Figure 2). According to two tamoxifen concentration splits, we defined non-compliant (⩽40 nM), poorly compliant (40–150 nM) and compliant (>150 nM) patients (Figure 2, dashed lines). Twenty-four POSH patients were non-compliant (7%) in their first-year serum sample, 10 patients were poorly compliant (2.9%). Of the other two cohorts, two patients were below 40 nM, whereas three patients were below 150 nM. There were no differences in CYP2D6 phenotype frequencies between poorly/non-compliant patients and the remaining patients, therefore subsequent genotype-phenotype correlations were not biased by excluding poorly/non-compliant patients (data not shown).
Figure 2.
Metabolic profiling for tamoxifen (Tam) and five measured metabolites, N-desmethyltamoxifen (DM-Tam), N,N-didesmethyltamoxifen (DDM-Tam), (Z)-endoxifen, 4-hydroxytamoxifen [(Z)-4-OH-DDM-Tam] and norendoxifen [(Z)-4-OH-DDMT-Tam] in study cohorts from Singapore (Sing, N=164), Lebanon (Leb, N=78) and Prospective study of Outcomes in Sporadic versus Hereditary breast cancer (POSH, N=345). Metabolite concentrations are presented as boxplots with whiskers defined by the 5th and 95th percentiles and extreme values outside the whiskers. The two dashed lines for Tam delineate putative non-compliant (⩽40 nM) and poorly compliant (40–150 nM) patients as defined from Tam plasma concentrations. Patients with Tam concentrations <150 nM were excluded from further analyses.
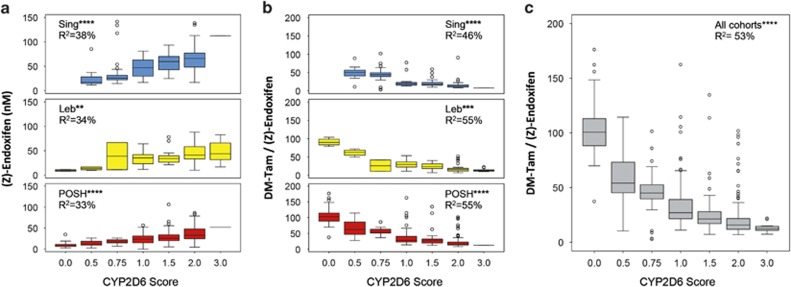
The compliant patients from each cohort are shown according to genotype-predicted CYP2D6 activity scores (Table 3). There was a strong gene-dose effect for an association between the CYP2D6 activity score and endoxifen concentrations (P<10−40) in all ethnic cohorts (Figure 3a). The contribution of CYP2D6 to the interindividual variability of endoxifen formation via DM-Tam, as deduced from MR DM-Tam/(Z)-endoxifen, was 46% (Singapore, P<10−17), 55% (Lebanon, P<10−9) and 55% (POSH, P<10−46), respectively (Figure 3b). Altogether, 53% of the interindividual variability of endoxifen formation from DM-Tam was attributed to CYP2D6 (Figure 3c; R2=0.53; P<10−77).
Table 3. CYP2D6 activity scores of compliant patients.
|
Singapore (n=160) |
Lebanon (n=77) |
POSH (n=311) |
Overall (n=548) |
|||||
|---|---|---|---|---|---|---|---|---|
| CYP2D6 activity scores | No. of patients | % | No. of patients | % | No. of patients | % | No. of patients | % |
| 0.0 | 0 | 4 | 5.2 | 25 | 8.0 | 29 | 5.3 | |
| 0.5 | 12 | 7.5 | 4 | 5.2 | 28 | 9.0 | 44 | 8.0 |
| 0.75 | 52 | 32.5 | 2 | 2.6 | 7 | 2.25 | 61 | 11.1 |
| 1.0 | 13 | 8.1 | 15 | 19.5 | 95 | 30.5 | 123 | 22.4 |
| 1.5 | 48 | 30.0 | 15 | 19.5 | 53 | 17.0 | 116 | 21.2 |
| 2.0 | 32 | 20.0 | 27 | 35.1 | 102 | 32.8 | 161 | 29.4 |
| 3.0 | 1 | 0.6 | 10 | 13.0 | 1 | 0.3 | 12 | 2.2 |
| Unknown | 2 | 1.2 | 2 | 0.4 | ||||
Abbreviation: POSH, Prospective study of Outcomes in Sporadic versus Hereditary breast cancer.
Figure 3.
Steady-state plasma concentrations of endoxifen and metabolic ratio DM-Tam/(Z)-endoxifen in premenopausal breast cancer patients according to genotype-based CYP2D6 activity score: (a) Endoxifen concentrations in Singapore (upper panel, N=160, R2=0.38; P<10−12), Lebanon (middle panel, N=77, R2=0.34; P<10−4) and POSH (lower panel, N=306, R2=0.33; P<10−21) cohorts. (b) Metabolic ratios of DMT/(Z)-endoxifen in Singapore (upper panel, R2=0.46; P<10−17), Lebanon (middle panel, R2=0.55; P<10−9) and POSH (lower panel, R2=0.55; P<10−46) cohorts. (c) Metabolic ratio DM-Tam/(Z)-endoxifen across all cohorts (N=548, R2=0.53; P<10−77). Data are presented as boxplots with whiskers defined by 5th and 95th percentiles and extreme values outside the whiskers. DM-Tam, N-desmethyltamoxifen. **P<10−3; ***P<10−5; ****P<10−10. POSH, Prospective study of Outcomes in Sporadic versus Hereditary breast cancer.
Other factors influencing tamoxifen metabolism
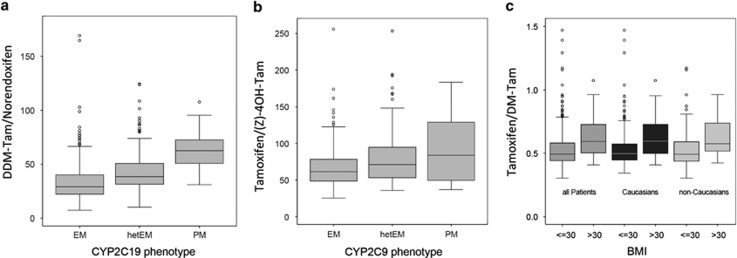
The formation of norendoxifen from DDM-Tam precursor and (Z)-4-OH-Tam converted from tamoxifen was decreased in patient carriers of CYP2C19*2 and/or *3 (Figure 4a, P<0.001) and CYP2C9*2 and/or*3 alleles (Figure 4b), P<0.001, respectively. Lower levels of DM-Tam were observed in patients with a BMI higher than 30, both in Caucasians and non-Caucasians (Figure 4c, P<0.001). Notably, DM-Tam formation did not depend on the CYP3A5*3 reduced-function allele. Multivariate regression analyses across all cohorts showed that the combined genetic (CYP2C9, CYP2C19, CYP3A5) and non-genetic factors (age, BMI) contributed to only 2.8% of DM-Tam/endoxifen ratio as compared with 53% by CYP2D6.
Figure 4.
Impact of CYP2C19, CYP2C9 and body mass index on tamoxifen metabolite ratios: (a) Metabolic ratio (MR) DDM-Tam/norendoxifen according to the loss-of-function alleles CYP2C19*2/*3 predicting EM, hetEM (heterozygous *2 or *3) and PM (homozygous *2 or *3). (b) MR tamoxifen/(Z)-4-OH-TAM according to CYP2C9 *2/*3 reduced activity alleles defining hetEM (heterozygous *2 or *3) and PM (homozygous *2 or *3) versus EM with normal activity (absence of *2 or *3). (c) MR tamoxifen/DM-Tam stratified by BMI (⩽30 or >30) in all patients, Caucasians and non-Caucasians. Data are presented as boxplots with whiskers defined by 5th and 95th percentiles and extreme values outside the whiskers. BMI, body mass index; DDM-Tam, DiDesmethyltamoxifen; DM-Tam, Desmethyltamoxifen; EM, extensive metabolizer; hetEM, heterozygous EM; PM, poor metabolizer; (Z)-4-OH-TAM, (Z)-4-hydroxytamoxifen.
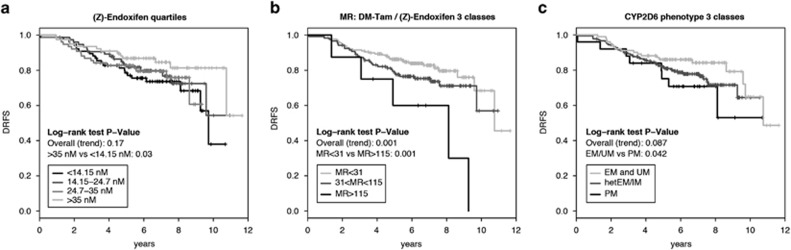
Tamoxifen metabolites and clinical outcome
POSH outcome analysis (N=306) was performed excluding hormone receptor-negative and poorly/non-compliant patients. There was no association between endoxifen concentrations and DRFS given the lack of a significant trend (Table 4, Figure 5a). However, following Madlensky et al.,14 when we classified patients into percentiles, we observed that patients with low endoxifen concentrations (<14.15 nM) had higher risk for distant relapse or death compared with those with high concentrations (>35 nM, P=0.03, Figure 5a), and showed a trend by multivariate Cox regression analysis of an increased hazard ratio (HR) of 1.94; 95% confidence interval (CI) 0.96–3.93; P=0.064 (Table 4). We next explored the MR DM-Tam/endoxifen and observed an increased HR with increasing MR (decreasing endoxifen formation rate) in multivariate Cox regression: HRper 1 unit=1.007; 95% CI 1.000–1.014; P=0.036 (Table 4). Because the per unit effect of this HR is rather small on a linear scale, we applied two cutoff values demonstrating worse (MR>115), moderate (MR 31–115) and better DRFS (MR<31) by Kaplan–Meier analysis (Log-rank P=0.001; Figure 5b). This was confirmed in multivariate Cox regression with an increased HR for patients with MR>115 (low endoxifen formation rate) compared with patients with MR<31 (high endoxifen formation rate): HR=3.82; 95% CI 1.47–9.89; P=0.006 (Table 4).
Table 4. Cox proportional hazard models of DRFS for (Z)-endoxifen plasma levels, MR DM-Tam/(Z)-endoxifen or CYP2D6 activity score/phenotype.
| Quantitative analyses | Categorical analyses | |||||||
|---|---|---|---|---|---|---|---|---|
| |
HR (per unit)a |
95% CI |
Wald P |
|
Factor |
HR |
95% CI |
Wald P |
| (Z)-Endoxifen (nM) | (Z)-Endoxifen (nM) | >35 | 1 (Reference) | |||||
| Unadjusted | 0.989 | 0.973–1.004 | 0.142 | 24.7–35 | 1.61 | 0.76–3.38 | 0.210 | |
| Adjusted | 0.992 | 0.977–1.007 | 0.291 | 14.15–24.7 | 1.45 | 0.69–3.06 | 0.329 | |
| <14.15 | 1.94 | 0.96–3.93 | 0.064 | |||||
| Nodal status | Negative | 1 (Reference) | ||||||
| Positive | 2.09 | 1.11–3.92 | 0.022 | |||||
| Chemotherapy | No | 1 (Reference) | 0.243 | |||||
| Yes | 1.65 | 0.71–3.83 | ||||||
| MR DM-Tam/(Z)-Endoxifen | MR DM-Tam/(Z)-Endoxifen | <31 | 1 (Reference) | |||||
| Unadjusted | 1.008 | 1.001–1.015 | 0.025 | 31–115 | 1.43 | 0.86–2.37 | 0.167 | |
| Adjusted | 1.007 | 1.000–1.014 | 0.036 | >115 | 3.82 | 1.47–9.89 | 0.006 | |
| Nodal status | Negative | 1 (Reference) | ||||||
| Positive | 2.09 | 1.12–3.89 | 0.021 | |||||
| Chemotherapy | No | 1 (Reference) | ||||||
| Yes | 1.62 | 0.70–3.74 | 0.260 | |||||
| CYP2D6 activity score | CYP2D6 phenotype class | EM/UM | 1 (Reference) | |||||
| Unadjusted | 0.611 | 0.425–0.88 | 0.008 | hetEM/IM | 1.55 | 0.88–2.72 | 0.132 | |
| Adjusted | 0.623 | 0.429–0.905 | 0.013 | PM | 1.98 | 0.82–4.79 | 0.128 | |
| Nodal status | Negative | 1 (Reference) | ||||||
| Positive | 2.15 | 1.14–4.04 | 0.018 | |||||
| Chemotherapy | No | 1 (Reference) | ||||||
| Yes | 1.60 | 0.69–3.73 | 0.277 | |||||
Abbreviations: CI, confidence interval; DRFS, distant recurrence-free survival; HR, hazard ratio; MR DM-Tam/(Z)-endoxifen, metabolic ratio N-desmethyltamoxifen/(Z)-endoxifen; TAM, tamoxifen.
Node status was binary coded with factor levels negative vs positive; chemotherapy was binary coded with levels no (no chemotherapy given) and yes (chemotherapy given); CYP2D6 and tamoxifen metabolite predictor variables were coded categorically. Models were corrected for nodal status and chemotherapy and stratified for ethnicity.
For univariate analysis, HR refers to an increase by one unit (1 nM) of steady-state endoxifen plasma concentration, increase of the dimensionless MR DM-Tam/(Z)-endoxifen ranging between 8 (highest) and 176 (lowest) endoxifen formation rates and an increase by one unit of CYP2D6 phenotype score ranging from 0 (PM) to 3 (UM).
Figure 5.
Kaplan–Meier analyses for an association between (Z)-endoxifen concentrations, metabolic ratio (MR) Desmethyltamoxifen (DM-Tam)/(Z)-endoxifen or CYP2D6 phenotype score and distant relapse-free survival (DRFS) in the POSH cohort. Kaplan–Meier analyses for DRFS and the three predictor variables classified into groups. (a) Steady-state endoxifen concentrations split into four equally-sized patient groups (<14.1, 14.1–24.7, 24.7–35 and >35 nM), (b) MR DM-Tam/(Z)-endoxifen classified by conditional inference trees into three splits (<31, 31–115 and >115), (c) CYP2D6 phenotypes grouped into EM (plus UM), hetEM and PM. Corresponding Mantel–Cox log-rank tests are stratified for ethnicity. EM, extensive metabolizer; hetEM/IM, heterozygous EM/IM; IM, intermediate metabolizer; PM, poor metabolizer; POSH, Prospective study of Outcomes in Sporadic versus Hereditary breast cancer; UM, ultra rapid metabolizer.
Importantly, none of the remaining metabolites including tamoxifen and DM-Tam showed an association between plasma concentrations and clinical outcome (Supplementary Figure 2).
CYP2D6 genotype and clinical outcome
Cox modeling revealed a significant linear association between CYP2D6 activity score and outcome (DRFS), both in univariate (P=0.008) and multivariate models (HRper 1 CYP2D6 score unit=0.62; 95% CI 0.43–0.91; P=0.013; Table 4). When grouping activity scores into the phenotype classes EM/UM, hetEM/IM and PM, Kaplan–Meier analysis indicated that PM patients have worse DRFS compared to EM subjects (Figure 5c; log-rank test P=0.042). When adjusted for nodal status and chemotherapy use, the association was no longer significant (HRPM vs EM=1.98; 95% CI 0.82–4.79; P=0.13; Table 4).
Discussion
This is the first study that investigated a purely premenopausal breast cancer population for interindividual variability of tamoxifen metabolism and its influence on clinical outcome. In contrast to postmenopausal patients,13, 18, 19, 20, 37 little is known regarding the clinical relevance of variable tamoxifen metabolism for drug response in premenopausal patients. Although postmenopausal patients may receive aromatase inhibitor treatment as an alternative to tamoxifen, a personalized approach in the premenopausal setting must confront the lack of available alternatives and address the dilemma of young women stopping tamoxifen prematurely due to ADRs. Emerging data report a link between steady-state endoxifen concentrations and ADRs;7, 8 however, the two retrospective studies reporting an association between endoxifen blood concentrations and clinical outcome do not reflect the premenopausal setting. Notably, Madlensky et al.14 provided the first clinical outcome data for endoxifen; however, their study lacked premenopausal subgroup analysis. Similarly, a small exploratory study performed with 48 oophorectomized women is not reminiscent of the premenopausal setting.39
Our comprehensive approach with premenopausal women accounts for potential determinants of tamoxifen response, including tamoxifen metabolites, CYP2D6 phenotypes and drug compliance. Notably, compliance is a serious issue as non-compliance pertains to up to 50% of patients by year five of the recommended tamoxifen treatment and has been associated with poor survival.40, 41, 42, 43 In our study, tamoxifen compliance assessment via blood concentrations was limited to the first year of treatment and the poor/non-compliance rate of 10% (POSH) was lower than expected.41, 44 Although first-year compliance may not be predictive for compliance for the full 5-year treatment duration, this has been accounted for by excluding all poorly/non-compliant patients from subsequent analyses. Regardless of the true compliance rate known to us we suggest, that outcome stratification based on CYP2D6 metabolism would be perhaps even better in a fully compliant population. We observed a clear CYP2D6 gene-dose effect for plasma endoxifen and endoxifen formation from DM-Tam. This strong effect was independent of age, BMI or non-CYP2D6 DME polymorphisms, which together contributed less than 3% of the observed variability in endoxifen formation. The CYP2D6 effect is similar to that observed in postmenopausal patients and holds true across ethnic groups (Asians, Europeans and Middle Eastern Arabs) independent of the differences in CYP2D6 allele frequencies.45 Our data clearly show that endoxifen formation follows the same principle in pre- and postmenopausal women driven by CYP2D6. Yet, its contribution to formation is estimated at approximately 53% suggesting that other factors contribute to the bioavailability of endoxifen. Additional findings of a correlation between impaired CYP2C9 activity and decreased 4-OH-Tam formation, as well as CYP2C19 loss-of-function and decreased formation of the anti-estrogen norendoxifen, corroborate previous in vitro observations and point to additional relevant metabolic pathways involved in the formation of antiestrogens.46, 47 Moreover, a potential non-genetic influence on tamoxifen metabolism by adipose tissue acting as sequestering ‘compartment' for lipophilic tamoxifen metabolites is supported by the observed association between BMI and decreased DM-Tam concentrations.
To understand the clinical relevance of tamoxifen metabolism in premenopausal patients, we conducted an outcome analysis (POSH) after excluding all patients with incomplete follow-up and/or delayed study entry (Singapore, Lebanon). The association of endoxifen concentrations with clinical outcome was inconclusive; yet, when we grouped endoxifen levels into distinct classes, patients with low concentrations (<14.1 nM) were at increased risk of distant relapse or death compared with patients with high endoxifen (>35 nM; Figure 5a). Importantly, the low endoxifen class contained 88% of all PMs (24 of 27 patients) thereby supporting the notion of a link between deficient CYP2D6 phenotype, reduced endoxifen concentration, and impaired clinical efficacy of tamoxifen. Our data are in line with those of Madlensky et al.14 who reported an distant recurrence rate for patients with low endoxifen concentrations (5.9 ng ml−1, 15.8 nM) in a patient cohort comprising pre- and postmenopausal patients. In contrast to endoxifen alone, using MR DM-Tam/endoxifen as a surrogate for CYP2D6 endoxifen formation demonstrated a strong association between high MR (low endoxifen concentrations) and an increased risk for recurrence or death, thereby substantiating the clinical relevance of the CYP2D6-mediated pathway for tamoxifen response. Together these data support the antagonistic potency of active metabolites at the ER, rather than the inhibition of ER by abundant tamoxifen and major metabolites through receptor saturation48, 49 in premenopausal patients. This is further substantiated by the lack of an outcome association with variable concentrations of tamoxifen and DM-Tam (Supplementary Figure 2). Notably, our in vivo data confirm the in vitro prediction by Maximov et al.29, 30 who demonstrated that endoxifen, rather than ER low-affinity tamoxifen metabolites, matters in blocking breast cancer cell replication in the presence of estrogen concentrations equivalent to premenopausal patients.
Our observation that CYP2D6 PM or low activity score patients had less favorable outcome compared with EM or high activity score patients supports the relevance of the CYP2D6 genotype for the prediction of tamoxifen outcome in premenopausal women, a hypothesis that has been originally postulated for the postmenopausal setting.18, 19, 20, 50 This association was considerably stronger when using activity scores as compared with classifying into three CYP2D6 phenotype groups (Table 4) supporting a gene-dose effect. Unlike in postmenopausal breast cancer where chemotherapy has been suggested to abrogate the CYP2D6 tamoxifen outcome effect,51 our data do not support this notion despite the large majority of POSH patients having received chemotherapy as it is the standard in the premenopausal setting. Combination chemotherapy produces chemical oophorectomy in the majority of premenopausal women over the age of 4052 resulting in lower circulating estradiol levels compared with premenopausal patients taking tamoxifen alone;53 however, the ovarian function of most POSH patients may have remained intact as inferred from their median age of 37.5 years. Another confounding factor in pharmacogenetic studies is the deviation from Hardy–Weinberg Equilibrium, which has been observed in the POSH cohort for CYP2D6*3 and *9, likely due to rare frequency or population admixture. Borderline deviation from Hardy–Weinberg Equilibrium was observed for CYP2D6*10 in the Singapore cohort, which could be attributed to the *36-*10 genotype (not analyzed)54 and/or population admixture (17% Malays/Indians vs 83% Chinese). Because genotyping was done from germline DNA, Hardy–Weinberg Equilibrium deviation is not considered a confounder in our outcome analysis.
A limitation of this study is its moderate sample size, given that more than 800 patients would be required to detect a CYP2D6-related clinical effect in postmenopausal women with sufficient power.22 This low power might explain the lack of significance with CYP2D6 phenotype in multivariate Cox analysis (P=0.13). However, breast cancer in young women is less frequent than in postmenopausal women, resulting in smaller available cohorts with simultaneous collection of serum, genomic DNA and detailed follow-up per patient. We therefore restricted our outcome analyses to the 306 compliant patients for whom, both pharmacogenetic and pharmacokinetic data were available. This design differs from a previous preliminary dataset with more patients but without control for compliance.55 It may be argued that this design may give rise to a selection bias; however, we believe that strict inclusion criteria including first-year compliance, serum availability for pharmacokinetic measurements and a reasonable follow-up time are integral to CYP2D6-related outcome analysis. Notwithstanding, our study presents the largest analyses of purely premenopausal patients for which pharmacokinetic and pharmacogenetic outcome associations have been described. No information on co-medication was available for the POSH cohort, a reason why we cannot comment on their possible influence on the results. In the Singapore and Lebanon cohorts, pharmacokinetic analyses were likely not influenced because no patient received moderate/strong CYP2D6 inhibitors. Collectively, our data for the association of tamoxifen outcome with endoxifen formation/concentrations in premenopausal patients must be considered exploratory therefore, its potential clinical relevance awaits confirmation by independent studies.
Our data support the notion that tamoxifen efficacy in premenopausal breast cancer patients is influenced by CYP2D6-mediated metabolism. If replicated, therapeutic drug level monitoring at steady-state could identify patients with high endoxifen levels or low DM-Tam/endoxifen ratio expected to have a lower risk of recurrence and who should therefore be encouraged to adhere to tamoxifen. On the basis of current research and developments toward improving therapeutic levels of endoxifen, patients with impaired/deficient CYP2D6 and suboptimal therapeutic endoxifen concentrations could be considered for the following: (i) increase of the standard tamoxifen dose as previously suggested for postmenopausal women,15, 56, 57 (ii) standard tamoxifen dose supplemented with endoxifen, a possibility currently explored by in silico modeling of CYP2D6 phenotype guided dosing schemes,58 (iii) treatment with (Z)-endoxifen hydrochloride of which first-in-man studies have already been published59 and further clinical trials are underway (www.clinicaltrials.gov: NCT01327781, NCT01273168) and (iv) aromatase inhibitor combined with ovarian suppression,60 as currently investigated in the Tamoxifen and EXemestane (TEXT) and Suppression of Ovarian Function Trial (SOFT) trials.61
Acknowledgments
We thank the patients, physicians, nurses and data managers for their participation in this study. This work was supported by the Robert Bosch Foundation, Stuttgart, Germany, Deutsche Forschungsgemeinschaft (DFG, SCHR 1323/2-1 and MU 1727/2-1), Germany and IZEPHA (Grant 21-0-0), Germany, The German Cancer Consortium (DKTK), American University of Beirut Medical Practice Plan (AUBMPP), and National Medical Research Council Grant NMRC/1159/2008 and NMRCG13161. PS is a fellow of the FP7 EU Marie Curie Initial Training Network FightingDrugFailure (GA238132).
Author contributions
HB, TM, PS, WS and MS designed the research. BC, RD, BE, DE, BG, SG, MZH, JSLL, TM, RCHN, PS, ES, WS, AT, NSW, YSY and NKZ performed the research. HB, BC, BE, DE, ME, TM, PS, WS, MS, SW and NKZ analyzed the data. BC, DE and AT contributed study materials or patients. WS, MS and HB led the study. qDE established the POSH study cohort. BC established the Singapore study cohort. NZK and AT established the Lebanon study cohort as co-principle investigators. BC, RD, BE, DE, SG, MZH, JSLL, RCHN, AT, NSW, YSY and NKZ collected samples and data. PS and WS performed the genotyping. BG and TM performed the drug metabolite analytics. PS, WS, TM and SW performed the statistical analysis. HB, BC, DE, TM, PS, WS, MS, SW and NKZ wrote the manuscript. All other authors contributed primarily to the patient ascertainment and/or data, and sample collection and preparation. All authors reviewed the final manuscript.
HB and MS report scientific collaborations with Roche Molecular Diagnostics and Siemens Healthcare Diagnostics Products in the previous 3 years. The remaining authors declared no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the The Pharmacogenomics Journal website (http://www.nature.com/tpj)
Supplementary Material
References
- Gnant M, Harbeck N, Thomssen C. St. Gallen 2011: summary of the consensus discussion. Breast Care (Basel) 2011;6:136–141. doi: 10.1159/000328054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winer EP, Hudis C, Burstein HJ, Wolff AC, Pritchard KI, Ingle JN, et al. American Society of Clinical Oncology technology assessment on the use of aromatase inhibitors as adjuvant therapy for postmenopausal women with hormone receptor-positive breast cancer: status report 2004. J Clin Oncol. 2005;23:619–629. doi: 10.1200/JCO.2005.09.121. [DOI] [PubMed] [Google Scholar]
- Early Breast Cancer Trialists' Collaborative Group Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet. 2005;365:1687–1717. doi: 10.1016/S0140-6736(05)66544-0. [DOI] [PubMed] [Google Scholar]
- Davies C, Pan H, Godwin J, Gray R, Arriagada R, Raina V, et al. Long-term effects of continuing adjuvant tamoxifen to 10 years versus stopping at 5 years after diagnosis of oestrogen receptor-positive breast cancer: ATLAS, a randomised trial. Lancet. 2013;381:805–816. doi: 10.1016/S0140-6736(12)61963-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray R, Rea D, Handley K, Bowden SJ, Perry P, Earl HM, et al. aTTom: long-term effects of continuing adjuvant tamoxifen to 10 years versus stopping at 5 years in 6,953 women with early breast cancer J Clin Oncol 2013316s (supplement abstract 5). [Google Scholar]
- Bowles EJA, Buist DSM, Chubak J, Yu O, Johnson J, Chestnut J, et al. Endocrine therapy initiation from 2001 to 2008 varies by age at breast cancer diagnosis and tumor size. J Oncol Pract. 2012;8:113–120. doi: 10.1200/JOP.2011.000417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorizio W, Wu AHB, Beattie MS, Rugo H, Tchu S, Kerlikowske K, et al. Clinical and biomarker predictors of side effects from tamoxifen. Breast Cancer Res Treat. 2012;132:1107–1118. doi: 10.1007/s10549-011-1893-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruddy KJ, Desantis SD, Gelman RS, Wu AHB, Punglia RS, Mayer EL, et al. Personalized medicine in breast cancer: tamoxifen, endoxifen, and CYP2D6 in clinical practice. Breast Cancer Res Treat. 2013;141:421–427. doi: 10.1007/s10549-013-2700-1. [DOI] [PubMed] [Google Scholar]
- Johnson MD, Zuo H, Lee K-H, Trebley JP, Rae JM, Weatherman RV, et al. Pharmacological characterization of 4-hydroxy-N-desmethyl tamoxifen, a novel active metabolite of tamoxifen. Breast Cancer Res Treat. 2004;85:151–159. doi: 10.1023/B:BREA.0000025406.31193.e8. [DOI] [PubMed] [Google Scholar]
- Lim YC, Desta Z, Flockhart DA, Skaar TC. Endoxifen (4-hydroxy-N-desmethyl-tamoxifen) has anti-estrogenic effects in breast cancer cells with potency similar to 4-hydroxy-tamoxifen. Cancer Chemother Pharmacol. 2005;55:471–478. doi: 10.1007/s00280-004-0926-7. [DOI] [PubMed] [Google Scholar]
- Brauch H, Mürdter TE, Eichelbaum M, Schwab M. Pharmacogenomics of tamoxifen therapy. Clin Chem. 2009;55:1770–1782. doi: 10.1373/clinchem.2008.121756. [DOI] [PubMed] [Google Scholar]
- Desta Z, Ward BA, Soukhova NV, Flockhart DA. Comprehensive evaluation of tamoxifen sequential biotransformation by the human cytochrome P450 system in vitro: prominent roles for CYP3A and CYP2D6. J Pharmacol Exp Ther. 2004;310:1062–1075. doi: 10.1124/jpet.104.065607. [DOI] [PubMed] [Google Scholar]
- Mürdter TE, Schroth W, Bacchus-Gerybadze L, Winter S, Heinkele G, Simon W, et al. Activity levels of tamoxifen metabolites at the estrogen receptor and the impact of genetic polymorphisms of phase I and II enzymes on their concentration levels in plasma. Clin Pharmacol Ther. 2011;89:708–717. doi: 10.1038/clpt.2011.27. [DOI] [PubMed] [Google Scholar]
- Madlensky L, Natarajan L, Tchu S, Pu M, Mortimer J, Flatt SW, et al. Tamoxifen metabolite concentrations, CYP2D6 genotype, and breast cancer outcomes. Clin Pharmacol Ther. 2011;89:718–725. doi: 10.1038/clpt.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvin WJ, Jr, Walko CM, Weck KE, Ibrahim JG, Chiu WK, Dees EC, et al. Genotype-guided tamoxifen dosing increases active metabolite exposure in women with reduced CYP2D6 metabolism: a multicenter study. J Clin Oncol. 2011;29:3232–3239. doi: 10.1200/JCO.2010.31.4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanger UM, Schwab M. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther. 2013;138:103–141. doi: 10.1016/j.pharmthera.2012.12.007. [DOI] [PubMed] [Google Scholar]
- Sistonen J, Sajantila A, Lao O, Corander J, Barbujani G, Fuselli S. CYP2D6 worldwide genetic variation shows high frequency of altered activity variants and no continental structure. Pharmacogenet Genomics. 2007;17:93–101. doi: 10.1097/01.fpc.0000239974.69464.f2. [DOI] [PubMed] [Google Scholar]
- Schroth W, Goetz MP, Hamann U, Fasching PA, Schmidt M, Winter S, et al. Association Between CYP2D6 polymorphisms and outcomes among women with early stage breast cancer treated with tamoxifen. JAMA. 2009;302:1429–1436. doi: 10.1001/jama.2009.1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz MP, Suman VJ, Hoskin TL, Gnant M, Filipits M, Safgren SL, et al. CYP2D6 metabolism and patient outcome in the Austrian Breast and Colorectal Cancer Study Group trial (ABCSG) 8. Clin Cancer Res. 2013;19:500–507. doi: 10.1158/1078-0432.CCR-12-2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Province MA, Goetz MP, Brauch H, Flockhart DA, Hebert JM, Whaley R, et al. CYP2D6 genotype and adjuvant tamoxifen: meta-analysis of heterogeneous study populations. Clin Pharmacol Ther. 2013;95:216–227. doi: 10.1038/clpt.2013.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratain MJ, Nakamura Y, Cox NJ. CYP2D6 genotype and tamoxifen activity: understanding interstudy variability in methodological quality. Clin Pharmacol Ther. 2013;94:185–187. doi: 10.1038/clpt.2013.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauch H, Schroth W, Goetz MP, Mürdter TE, Winter S, Ingle JN, et al. Tamoxifen use in postmenopausal breast cancer: CYP2D6 matters. J Clin Oncol. 2013;31:176–180. doi: 10.1200/JCO.2012.44.6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regan MM, Leyland-Jones B, Bouzyk M, Pagani O, Tang W, Kammler R, et al. CYP2D6 genotype and tamoxifen response in postmenopausal women with endocrine-responsive breast cancer: the breast international group 1-98 trial. J Natl Cancer Inst. 2012;104:441–451. doi: 10.1093/jnci/djs125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae JM, Drury S, Hayes DF, Stearns V, Thibert JN, Haynes BP, et al. CYP2D6 and UGT2B7 genotype and risk of recurrence in tamoxifen-treated breast cancer patients. J Natl Cancer Inst. 2012;104:452–460. doi: 10.1093/jnci/djs126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauch H, Schwab M. Prediction of tamoxifen outcome by genetic variation of CYP2D6 in postmenopausal women with early breast cancer. Br J Clin Pharmacol. 2013;77:695–703. doi: 10.1111/bcp.12229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae JM. CYP2D6 genotype should not be used to determine endocrine therapy in postmenopausal breast cancer patients. Clin Pharmacol Ther. 2013;94:183–185. doi: 10.1038/clpt.2013.102. [DOI] [PubMed] [Google Scholar]
- Abraham JE, Maranian MJ, Driver KE, Platte R, Kalmyrzaev B, Baynes C, et al. CYP2D6 gene variants: association with breast cancer specific survival in a cohort of breast cancer patients from the United Kingdom treated with adjuvant tamoxifen. Breast Cancer Res. 2010;12:R64. doi: 10.1186/bcr2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dezentjé VO, van Schaik RHN, Vletter-Bogaartz JM, van der Straaten T, Wessels JAM, Kranenbarg EM-K, et al. CYP2D6 genotype in relation to tamoxifen efficacy in a Dutch cohort of the tamoxifen exemestane adjuvant multinational (TEAM) trial. Breast Cancer Res Treat. 2013;140:363–373. doi: 10.1007/s10549-013-2619-6. [DOI] [PubMed] [Google Scholar]
- Maximov PY, McDaniel RE, Jordan VC. Tamoxifen Pioneering Medicine In Breast Cancer. Springer: Basel: New York; 2013. [Google Scholar]
- Maximov PY, McDaniel RE, Fernandes DJ, Korostyshevskiy VR, Bhatta P, et al. Simulation with cells in vitro of tamoxifen treatment in premenopausal breast cancer patients with different CYP2D6 genotypes Br J Pharmacol(in press). [DOI] [PMC free article] [PubMed]
- Lim JSL, Chen XA, Singh O, Yap YS, Ng RCH, Wong NS, et al. Impact of CYP2D6, CYP3A5, CYP2C9 and CYP2C19 polymorphisms on tamoxifen pharmacokinetics in Asian breast cancer patients. Br J Clin Pharmacol. 2011;71:737–750. doi: 10.1111/j.1365-2125.2011.03905.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awada Z, Haider S, Tfayli A, Bazarbachi A, Saghir NE, Salem Z, et al. Pharmacogenomics variation in drug metabolizing enzymes and transporters in relation to docetaxel toxicity in Lebanese breast cancer patients: paving the way for OMICs in low and middle income countries. OMICS. 2013;17:353–367. doi: 10.1089/omi.2013.0019. [DOI] [PubMed] [Google Scholar]
- Eccles D, Gerty S, Simmonds P, Hammond V, Ennis S, Altman D, et al. Prospective study of Outcomes in Sporadic versus Hereditary breast cancer (POSH): study protocol. BMC Cancer. 2007;7:160. doi: 10.1186/1471-2407-7-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copson E, Eccles B, Maishman T, Gerty S, Stanton L, Cutress RI, et al. Prospective observational study of breast cancer treatment outcomes for UK women aged 18-40 years at diagnosis: the POSH study. J Natl Cancer Inst. 2013;105:978–988. doi: 10.1093/jnci/djt134. [DOI] [PubMed] [Google Scholar]
- McCague R, Leclercq G, Jordan VC. Nonisomerizable analogues of (Z)- and (E)-4-hydroxytamoxifen. Synthesis and endocrinological properties of substituted diphenylbenzocycloheptenes. J Med Chem. 1988;31:1285–1290. doi: 10.1021/jm00402a005. [DOI] [PubMed] [Google Scholar]
- Zineh I, Beitelshees AL, Gaedigk A, Walker JR, Pauly DF, Eberst K, et al. Pharmacokinetics and CYP2D6 genotypes do not predict metoprolol adverse events or efficacy in hypertension. Clin Pharmacol Ther. 2004;76:536–544. doi: 10.1016/j.clpt.2004.08.020. [DOI] [PubMed] [Google Scholar]
- Jin Y, Desta Z, Stearns V, Ward B, Ho H, Lee KH, et al. CYP2D6 genotype, antidepressant use, and tamoxifen metabolism during adjuvant breast cancer treatment. J Natl Cancer Inst. 2005;97:30–39. doi: 10.1093/jnci/dji005. [DOI] [PubMed] [Google Scholar]
- Schroth W, Hamann U, Fasching PA, Dauser S, Winter S, Eichelbaum M, et al. CYP2D6 polymorphisms as predictors of outcome in breast cancer patients treated with tamoxifen: expanded polymorphism coverage improves risk stratification. Clin Cancer Res. 2010;16:4468–4477. doi: 10.1158/1078-0432.CCR-10-0478. [DOI] [PubMed] [Google Scholar]
- Love RR, Desta Z, Flockhart D, Skaar T, Ogburn ET, Ramamoorthy A, et al. CYP2D6 genotypes, endoxifen levels, and disease recurrence in 224 Filipino and Vietnamese women receiving adjuvant tamoxifen for operable breast cancer. Springerplus. 2013;2:52. doi: 10.1186/2193-1801-2-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chlebowski RT, Kim JS, Haque R. Adherence to endocrine therapy in breast cancer adjuvant and prevention settings. Cancer Prev Res (Phila) 2014;7:378–387. doi: 10.1158/1940-6207.CAPR-13-0389. [DOI] [PubMed] [Google Scholar]
- McCowan C, Wang S, Thompson AM, Makubate B, Petrie DJ. The value of high adherence to tamoxifen in women with breast cancer: a community-based cohort study. Br J Cancer. 2013;109:1172–1180. doi: 10.1038/bjc.2013.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makubate B, Donnan PT, Dewar JA, Thompson AM, McCowan C. Cohort study of adherence to adjuvant endocrine therapy, breast cancer recurrence and mortality. Br J Cancer. 2013;108:1515–1524. doi: 10.1038/bjc.2013.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershman DL, Shao T, Kushi LH, Buono D, Tsai WY, Fehrenbacher L, et al. Early discontinuation and non-adherence to adjuvant hormonal therapy are associated with increased mortality in women with breast cancer. Breast Cancer Res Treat. 2011;126:529–537. doi: 10.1007/s10549-010-1132-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershman DL, Kushi LH, Shao T, Buono D, Kershenbaum A, Tsai W-Y, et al. Early discontinuation and nonadherence to adjuvant hormonal therapy in a cohort of 8,769 early-stage breast cancer patients. J Clin Oncol. 2010;28:4120–4128. doi: 10.1200/JCO.2009.25.9655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanger UM, Raimundo S, Eichelbaum M. Cytochrome P450 2D6: overview and update on pharmacology, genetics, biochemistry. Naunyn-Schmiedebergs Arch Pharmacol. 2004;369:23–37. doi: 10.1007/s00210-003-0832-2. [DOI] [PubMed] [Google Scholar]
- Coller JK, Krebsfaenger N, Klein K, Endrizzi K, Wolbold R, Lang T, et al. The influence of CYP2B6, CYP2C9 and CYP2D6 genotypes on the formation of the potent antioestrogen Z-4-hydroxy-tamoxifen in human liver. Br J Clin Pharmacol. 2002;54:157–167. doi: 10.1046/j.1365-2125.2002.01614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu WJ, Xu C, Pei Z, Mayhoub AS, Cushman M, Flockhart DA. The tamoxifen metabolite norendoxifen is a potent and selective inhibitor of aromatase (CYP19) and a potential lead compound for novel therapeutic agents. Breast Cancer Res Treat. 2012;133:99–109. doi: 10.1007/s10549-011-1699-4. [DOI] [PubMed] [Google Scholar]
- Lash TL, Lien EA, Sørensen HT, Hamilton-Dutoit S. Genotype-guided tamoxifen therapy: time to pause for reflection. Lancet Oncol. 2009;10:825–833. doi: 10.1016/S1470-2045(09)70030-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowsett M, Haynes BP. Hormonal effects of aromatase inhibitors: focus on premenopausal effects and interaction with tamoxifen. J Steroid Biochem Mol Biol. 2003;86:255–263. doi: 10.1016/s0960-0760(03)00365-0. [DOI] [PubMed] [Google Scholar]
- Kiyotani K, Mushiroda T, Imamura CK, Hosono N, Tsunoda T, Kubo M, et al. Significant effect of polymorphisms in CYP2D6 and ABCC2 on clinical outcomes of adjuvant tamoxifen therapy for breast cancer patients. J Clin Oncol. 2010;28:1287–1293. doi: 10.1200/JCO.2009.25.7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyotani K, Mushiroda T, Hosono N, Tsunoda T, Kubo M, Aki F, et al. Lessons for pharmacogenomics studies: association study between CYP2D6 genotype and tamoxifen response. Pharmacogenet Genomics. 2010;20:565–568. doi: 10.1097/FPC.0b013e32833af231. [DOI] [PubMed] [Google Scholar]
- Ravdin PM, Fritz NF, Tormey DC, Jordan VC. Endocrine status of premenopausal node-positive breast cancer patients following adjuvant chemotherapy and long-term tamoxifen. Cancer Res. 1988;48:1026–1029. [PubMed] [Google Scholar]
- Jordan VC, Fritz NF, Langan-Fahey S, Thompson M, Tormey DC. Alteration of endocrine parameters in premenopausal women with breast cancer during long-term adjuvant therapy with tamoxifen as the single agent. J Natl Cancer Inst. 1991;83:1488–1491. doi: 10.1093/jnci/83.20.1488. [DOI] [PubMed] [Google Scholar]
- Gaedigk A, Bradford LD, Alander SW, Leeder JS. Cyp2d6*36 gene arrangements within the Cyp2d6 locus: association of Cyp2d6*36 with poor metabolizer status. Drug Metab Dispos. 2006;34:563–569. doi: 10.1124/dmd.105.008292. [DOI] [PubMed] [Google Scholar]
- Saladores PH, Schroth W, Eccles D, Tapper W, Gerty S, Brauch HB. Abstract 2671: CYP2D6 polymorphisms are not associated with tamoxifen outcome in premenopausal women with ER positive breast cancer of the POSH cohort. Cancer Res. 2012;72:2671–2671. [Google Scholar]
- Kiyotani K, Mushiroda T, Imamura CK, Tanigawara Y, Hosono N, Kubo M, et al. Dose-adjustment study of tamoxifen based on CYP2D6 genotypes in Japanese breast cancer patients. Breast Cancer Res Treat. 2012;131:137–145. doi: 10.1007/s10549-011-1777-7. [DOI] [PubMed] [Google Scholar]
- Barginear MF, Jaremko M, Peter I, Yu C, Kasai Y, Kemeny M, et al. Increasing tamoxifen dose in breast cancer patients based on CYP2D6 genotypes and endoxifen levels: effect on active metabolite isomers and the antiestrogenic activity score. Clin Pharmacol Ther. 2011;90:605–611. doi: 10.1038/clpt.2011.153. [DOI] [PubMed] [Google Scholar]
- Dickschen K, Eissing T, Mürdter T, Schwab M, Willmann S. Overcoming CYP2D6-mediated tamoxifen resistance: phenotype-specific tamoxifen-endoxifen combinations. Breast. 2013;22:S86. [Google Scholar]
- Ahmad A, Shahabuddin S, Sheikh S, Kale P, Krishnappa M, Rane RC, et al. Endoxifen, a new cornerstone of breast cancer therapy: demonstration of safety, tolerability, and systemic bioavailability in healthy human subjects. Clin Pharmacol Ther. 2010;88:814–817. doi: 10.1038/clpt.2010.196. [DOI] [PubMed] [Google Scholar]
- Regan MM, Pagani O, Fleming GF, Walley BA, Price KN, Rabaglio M, et al. Adjuvant treatment of premenopausal women with endocrine-responsive early breast cancer: design of the TEXT and SOFT trials. Breast. 2013;22:1094–1100. doi: 10.1016/j.breast.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagani O, Regan MM, Walley BA, Fleming GF, Colleoni M, Láng I, et al. Adjuvant exemestane with ovarian suppression in premenopausal breast cancer. N Engl J Med. 2014;371:107–118. doi: 10.1056/NEJMoa1404037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.