Abstract
The grain aphid Sitobion avenae (F.) is a major pest of wheat, acting as a virus vector as well as causing direct plant damage. Commonly grown wheat varieties in the UK have only limited resistance to this pest. The present study was carried out to investigate the potential of a diploid wheat line (ACC20 PGR1755), reported as exhibiting resistance to S. avenae, to serve as a source of resistance genes. The diploid wheat line was confirmed as partially resistant, substantially reducing the fecundity, longevity and growth rate of the aphid. Proteomic analysis showed that approximately 200 protein spots were reproducibly detected in leaf extracts from both the resistant line and a comparable susceptible line (ACC5 PGR1735) using two-dimensional gel electrophoresis and image comparison software. Twenty-four spots were significantly up-regulated (>2-fold) in the resistant line after 24 h of aphid feeding (13 and 11 involved in local and systemic responses, respectively). Approximately 50 % of all differentially expressed protein spots were identified by a combination of database searching with MS and MS/MS data, revealing that the majority of proteins up-regulated by aphid infestation were involved in metabolic processes (including photosynthesis) and transcriptional regulation. However, in the resistant line only, several stress response proteins (including NBS–LRR-like proteins) and oxidative stress response proteins were identified as up-regulated in response to aphid feeding, as well as proteins involved in DNA synthesis/replication/repair. This study indicates that the resistant diploid line ACC20 PGR1755 may provide a valuable resource in breeding wheat for resistance to aphids.
Electronic supplementary material
The online version of this article (doi:10.1007/s11032-015-0220-x) contains supplementary material, which is available to authorized users.
Keywords: Aphid resistance, Biotic stress, Diploid wheat, Oxidative stress, Stress response, Wheat proteomics
Introduction
Wheat, Triticum aestivum L., is currently second to rice as the main human food crop and is the leading source of vegetable protein in human nutrition (United Nations 2012). Aphids (Order Hemiptera) are major insect pests of world agriculture, damaging crops by removing photoassimilates and vectoring numerous plant viruses (Smith and Boyko 2007). The grain aphid, Sitobion avenae (Fabricius), is considered a serious pest of commercial wheat in the UK. Many aphid species have evolved resistance to insecticides (Devonshire and Field 1991), and restrictions on the availability of active ingredients for insecticide production in Europe (European Directives 91/414/EEC) have prioritised research on crop varieties with resistance to aphid pests (Smith 2005). Most commercial wheat varieties have very little resistance to aphid pests (Migui 2002; Migui and Lamb 2003), with at best partial antibiosis, antixenosis and tolerance in some winter varieties (Havlícková 1993). However, recently a synthetic wheat line (98-10-35) with moderate levels of resistance to the grain aphid has been developed, although the mechanism of this constitutive resistance to S. avenae is not known (Wang et al. 2013).
Aphids differ from many other pest insects in their mode of feeding in that they do not cause major tissue damage or induce plant wounding responses (Farmer and Ryan 1992; Gatehouse 2002) since they feed from plant phloem by inserting a stylet between the cells. As a result, plant responses to aphid feeding have been reported to be similar to those induced by pathogen attack and, in some cases, have been classified as gene-for-gene interactions characteristic of plant–pathogen interactions (Walling 2000; Moran and Thompson 2001; Moran et al. 2002; Smith and Boyko 2007). This response has been reported to be induced by aphid-derived elicitors with salicylic acid (SA) as the major signalling molecule (Walling 2000; Moran et al. 2002; Smith and Boyko 2007). However, aphids can also induce a non-specific defence response in the plant, resulting in a general stress response, which can be detected in both aphid-resistant and aphid-susceptible plants (Smith and Boyko 2007). Plants experience extensive transcriptome reprogramming when they are subjected to insect attack (Moran and Thompson 2001; Zhang et al. 2004; De Vos et al. 2005; Yuan et al. 2005; Wei et al. 2009). Ferry et al. (2011) demonstrated that the proteome of wheat, variety ‘Claire’ a commercial winter wheat cultivar commonly grown in the UK, changed following aphid infestation. These changes were more consistent with SA-induced responses than the jasmonic acid (JA)-induced wounding responses, although none of these were sufficient to confer resistance to the grain aphid. These findings confirm previous studies where rice brown planthopper (Nilaparvata lugens) caused changes in the expression of rice proteins involved in signalling pathways, oxidative stress/apoptosis, wound response, drought response and pathogen-related response (Zhang et al. 2004).
Whilst none of the commercial wheat varieties grown in Europe are resistant to S. avenae or other cereal aphids of European origin (Di Pietro et al. 1998; Migui 2002; Migui and Lamb 2003), some commercial wheat has partial resistance towards Russian wheat aphids, Diuraphis noxia (Mordvilko) (RWA). Ten RWA resistance genes have been identified in T. aestivum and named Dn genes (Ma et al. 1998). Microarray and real-time PCR analysis revealed that more than 180 genes up-regulated on attack by D. noxia are related to reactive oxygen species, signalling, pathogen defence and arthropod allelochemical and physical defence (Smith et al. 2010). In a further study, superoxide dismutase, glutathione reductase and ascorbate peroxidase were uniquely up-regulated in the RWA resistant wheat (T. aestivum) cultivar Tugela DN, compared to the RWA susceptible cultivar Tugela after D. noxia infestation. These findings suggest the involvement of antioxidative enzymes in the RWA–wheat resistance response to minimise toxic effects to plant cells (Moloi and van der Westhuizen 2006, 2008).
Modern wheat varieties are hexaploid and have low genetic diversity for insect resistance traits (Ogbonnaya et al. 2013; Donini et al. 2000). A screen of 87 ancient diploid wheat genotypes, including 67 Triticum monococcum L. genotypes, 13 Triticum boeoticum Bois genotypes, 7 Triticum urartu Tumanian ex Gandilyan genotypes, showed that many exhibited higher resistance to S. avenae than a cultivar of the modern wheat T. aestivum (variety ‘Arminda’; Di Pietro et al. 1998). Accessions of the diploid ancestral wheat—Einkorn wheat, T. monococcum with partial resistance to aphids—have been reported (Migui and Lamb 2004). Furthermore, accessions of wild wheat species, T. boeoticum (Bois), T. tauschii (Coss.) Schmal. and T. araraticum Jakubz., were also found to exhibit high levels of resistance to aphids (Migui and Lamb 2003). Thus, ancient diploid wheat may be a useful source of genes for improving resistance to S. avenae in modern wheat (Di Pietro et al. 1998). However, no previous studies have been carried out to investigate differential gene expression in these lines in response to aphid infestation. The aim of this study was therefore to identify putative defence genes in diploid wheat lines (T. monococcum) in response to grain aphid (S. avenae) feeding using differential proteomics to better understand the basis of this observed resistance/tolerance.
Materials and methods
Plant materials and treatment
Two diploid accessions of wild Einkorn wheat, ACC5 PGR1735 and ACC20 PGR1755, were selected for study, representing lines exhibiting aphid susceptibility and tolerance, respectively. These seeds were kindly donated by R. J. Lamb from seed collections of Agriculture and Agri-Food Canada. They are described as ‘accessions’ in Migui and Lamb (2004). Wheat seedlings were grown to the four-leaf stage in soil (John Innes, No. 2) under controlled environmental conditions in custom-built growth rooms (HA Davie Ltd, U.K.) under the following conditions (light intensity; photosynthetically active photon flux: 600 mol/m2/s, 16:8 LL:DD light regime) with a temperature regime of 18 °C (day):16 °C (night) and 70 % relative humidity.
Aphid bioassays
Grain aphids, S. avenae, were obtained from a laboratory culture and maintained at 20 °C, 55 % relative humidity (R.H.) under a 16:8 LL:DD light regime. The aphids were reared on oats (Avena sativa L., cv. Coast Black), and infested plants were kept in 45 × 45 × 50 cm Perspex cages with new plant material supplied weekly.
Growth, survival and fecundity of S. avenae were evaluated on thirteen accessions of Triticum monoccoccum diploid wheat (Fig. 1). Experiments were also set up using a hexaploid wheat cultivar (Claire) and oats (Coast Black as comparisons). Single seedlings, at the two-leaf stage, of each wheat cultivar and oats were planted in 9-cm pots containing John Innes No. 3 compost and allowed to establish for 2–3 days. To assess fecundity and longevity, two-leaf stage seedlings of the different wheat varieties were each infested with one neonate nymph and randomly arranged within a growth room 20 °C, 55 % R.H. under a 16:8 LL:DD light regime. On each day following infestation, the survival of aphids was monitored, and following ecdysis to the adult stage, the production of nymphs was recorded. Ten replicates were set up for each plant type. To determine mean relative growth rates (MRGRs), neonate nymphs were removed from culture using a fine camel hair paintbrush and weighed on a Mettler AT20 balance. The aphids were then transferred singly to a seedling of one of the wheat varieties or oats. Following infestation, the plants were enclosed in 25 × 9 cm ventilated plastic cylinders to prevent aphid escape and arranged randomly within a growth room maintained at the conditions detailed above. The nymphs were reweighed after 4 days and again when they had moulted to the adult. The mean relative growth rates of aphids developing on the different plants, and over different time scales, were calculated using the methods described by Leather and Dixon (1984).
Fig. 1.
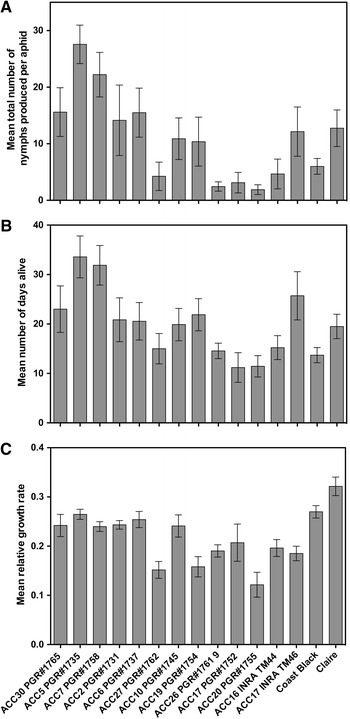
Thirteen accessions of diploid Einkorn wheat lines together with an oat line (cv. Coast Black) and a hexaploid wheat reference line (cv. Claire) were screened for aphid (Sitobion avenae) resistance using three parameters: mean fecundity (a), longevity (b) and relative growth rates (c). Based on this bioassay data, a resistant (R, ACC20 PGR1755) and a susceptible line (S, ACC5 PGR1735) were selected for further study to investigate potential genes involved in aphid tolerance/resistance using a reverse genetic approach
Data were analysed using one-way analysis of variance (ANOVA). Means were subsequently separated by Tukey–Kramer I tests. Analysis was conducted using GraphPad Prism 5.0.
Aphid infestation for proteomic experiments
The set-up and protocol for aphid infestation on both resistant and susceptible wheat plants (local and systemic treatments) were the same as the ones used by Ferry et al. (2011). In brief, wheat seedlings (four-leaf stage) were infested with S. avenae adults (20 aphids per leaf) confined to two leaves with clip cages (12 plants per treatment). There were 8 treatments in total in this study, as illustrated in the table:
| Time of infestation | Time of infestation | |
|---|---|---|
| Wheat line | 24 h | 8 days |
| Resistant accession | Local | Local |
| Systemic | Systemic | |
| Susceptible accession | Local | Local |
| Systemic | Systemic |
Aphids were left on the seedlings for either 24 h or 8 days after which time they were removed and leaf tissues immediately frozen in liquid nitrogen. Tissues exposed to aphids were designated ‘local treatment’ and the leaves not infested on the same plant designated ‘systemic treatment’. Corresponding tissues were taken from aphid-free control plants for all time points and treatments.
Protein extraction
Leaf samples from 12 experimental plants (4-leaf stage, divided into local and systemic infested tissues) and six control plants were ground to a fine powder using a mortar and a pestle with liquid nitrogen. Samples were pooled into three biological replicates per treatment; six technical replicates of the pooled samples were used in this work. Protein extraction, resuspension, quantification, IEF, SDS-PAGE and staining were carried out following the protocol used in Ferry et al. (2011). In brief, samples were incubated in 10 % (w/v) trichloroacetic acid/acetone with 0.07 % v/v 2-mercaptoethanol at -20 °C for 5 h and then centrifuged at 35,000×g for 20 min. The pellets were washed with ice-cold acetone (0.07 % 2-mercaptoethanol) incubated at −20 °C for 1 h and centrifuged at 12,000×g for 15 min. This wash was repeated six times. Pellets were vacuum dried, and the resultant powder suspended in 7 M urea, 2 M thiourea, 4 % CHAPS, 60 mM DTT, 0.5 % v/v pH 3–10 carrier ampholytes and 1 % v/v protease inhibitor mix (GE Healthcare, Bucks, UK) by sonication. Centrifugation at 20,000×g for 20 min at 4 °C removed insoluble debris, and the total protein content of the supernatant was determined using the 2-D Quant kit (GE Healthcare) method with BSA as standard. Isoelectric focussing (IEF) and SDS-PAGE were carried out essentially according to the manufacturers’ instructions on an Ettan IPGphor II and Ettan DALTsix system (GE Healthcare). For IEF, 18-cm IPG strips with a nonlinear gradient (pH 3–10) were actively rehydrated using 340 μl of DeStreak Rehydration solution (GE Healthcare) at 30 V for 10 h. Five hundred micrograms of protein from each pooled sample was loaded, and IEF conducted at 20 °C under the following conditions: 500 V for 500 Vh, followed by two gradients of 1,000 V for 800 Vh, 8,000 V for 13,500 Vh and finally 8,000 V for 20,000 Vh. The focussed strips were equilibrated for 15 min in 10 mL equilibration solution (75 mM Tris–HCl, 6 M urea, 30 % w/v glycerol, 2 % w/v SDS, 1 % w/v DTT) followed by equilibration in buffer containing 3 % w/v iodoacetic acid for another 15 min. Strips were transferred to a vertical SDS-PAGE gel, and the second dimension performed on a 12.5 % gel using the Ettan DALTsix system (GE Healthcare) at 4 °C. Protein spots were stained with colloidal Coomassie blue G-250 (Sigma).
Image and data analysis
Wet stained gels were scanned using a LabScan 5.0 (GE Healthcare) at a resolution of 600 dpi, bit depth 12. Image analysis, spot detection and quantification were carried out using the Progenesis SameSpots software package version 4.0 and 4.1 (Nonlinear Dynamics, Newcastle, UK). Image and data analysis were as previously described (Ferry et al. 2011). Total spot volume (from matched gel replicates) for each treatment was compared to control images, and a threshold level of ±2-fold change set and subjected to statistical analysis using ANOVA. Spots were determined to be significantly up- or down-regulated when p value was <0.05 (Tukey–Kramer post hoc test). Within group comparisons allowed quality control of images. The molecular masses of protein spots were determined by co-electrophoresis of Mark 12 standard protein markers (Invitrogen), and pI values estimated from the Immobiline DryStrip as per manufacturers recommendations and further calibrated by comparison to published wheat leaf proteomes (Ferry et al. 2011).
MALDI-TOF MS, MS/MS and database search
Selected protein spots from 2D gels with two changes in spot volume after aphid infestation were excised from gel, digested with sequencing grade trypsin (Promega, USA) and subjected to MALDI-TOF MS and MS/MS combined with database searching to assign putative identities to the proteins, as previously described (Ferry et al. 2011). Tryptic peptides were deposited onto 384-well stainless steel target plates manually with a calibration mix (Peptide calibration standard; Bruker Daltonics, Germany) spotted between every 4 sample wells and overlaid with 0.5 μl matrix (CHCA). Peptide mass spectra were recorded using a Bruker UltraFlex II MALDI mass spectrometer in positive reflectron mode over a m/z range of 700–4,500 and analysed using the instruments Flex Analysis software (v3.0). Monoisotopic peak selection was restricted to 50 peptides, and known trypsin autolysis peaks deleted, the peak list was searched against the nrNCBI and Swiss-Prot databases restricted to ‘Viridiplantae’ using the MASCOT (www.matrixscience.com) search engine or a local custom-built restricted database constructed from freely available EST libraries (8,530 wheat and 7,341 barley, http://trifldb.psc.riken.jp/index.pl) with the MASCOT 2.2 software allowing an m/z error of 50 ppm, maximum two missed cleavages, and possible modification of cysteines by carbamidomethylation as well as oxidation of methionine. Protein identification was accepted based on a significant MOWSE score and at least four matched peptide masses. Matching ESTs were queried to the nrNCBI database with a significance cut-off value of 1e-5 using BLASTX searches. Gene ontology (GO) phrases were collected using the PRO protein ontology search engine (http://pir.georgetown.edu/pro/) using the generic GO Slim. Putative protein ID and gene annotation are listed in Tables 1 and 2.
Table 1.
Identification by PMF of differentially regulated proteins showing a minimum twofold increase in abundance in response to S. avenae infestation of resistant or susceptible diploid lines at 24-h time point in local and systemic tissues
| Spot Name (a) | Ac. No. (b) | Predicted ID (c) | Score (d) | e-Value (e) | Th. pI (f) | Function and biological process/category (g) |
|---|---|---|---|---|---|---|
| RL versus RC 1 | Q56YM6 | Hypothetical protein—Arabidopsis thaliana | 69 | 0.0075 | 8.88 | 2-Polyprenyl-6-methoxyphenol hydroxylase and related FAD, involves in oxidation–reduction process—response to oxidative stress |
| RL versus RC 2 | Q6K6S5 | Heat stress transcription factor A-5—Oryza sativa | 52 | 0.015 | 5.3 | Regulation of transcription/response to stress—stress response (and transcriptional regulation) |
| RL versus RC 3 | Q84R82 | Hypothetical protein OSJNBb0041J20.14.—Oryza sativa | 53 | 0.39 | 11.19 | Unknown—Unknown |
| RL versus RC 4 | Q53P41 | Lipase class 3 family protein, putative, expressed—Oryza sativa | 63 | 0.031 | 8.62 | Triacylglycerol lipase activity—other metabolism (lipase) |
| RL versus RC 5 | Q7YMR6 | Ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit—Hordeum vulgare | 93 | 0.00012 | 6.22 | Ribulose bisphosphate carboxylase complex; ribulose bisphosphate carboxylase activity—photosynthesis |
| RL versus RC 6 | B5B1F8 | Chloroplast aspartate aminotransferase—Triticum aestivum | 58 | 0.97 | 6.69 | L-aspartate: 2-oxoglutarate aminotransferase activity; pyridoxal phosphate binding—photosynthesis |
| RL versus RC 7 | Q09QZ5 | ATPase alpha subunit—Tetraphis pellucida | 57 | 1.2 | 9.21 | Hydrogen ion transporting ATP synthase activity, rotational mechanism/proton-transporting ATPase activity—ATP synthesis |
| Dephosphorylation reaction, releases energy—ATP synthesis | ||||||
| RL versus RC 8 | Q8L5E7 | Dof zinc finger protein (Fragment)—Hordeum vulgare | 52 | 1.6 | 9.84 | Transcriptional factor (activator/repressor), may permit leaf-specific gene expression—transcriptional regulation |
| RL versus RC 9 | B23703 | Ribulose bisphosphate oxygenase carboxylase activase—Hordeum vulgare | 97 | 4.9E_05 | 5.64 | ATP binding, nucleotide binding, chloroplast component, ligase activity, have nucleotide-binding motif A (P-loop)—photosynthesis |
| C23703 | Ribulose bisphosphate oxygenase carboxylase activase—Hordeum vulgare | 95 | 7.7E_05 | 8.39 | ATP binding, nucleotide binding, chloroplast component, ligase activity, have nucleotide-binding motif A (P-loop)—photosynthesis | |
| Q40073 | Ribulose bisphosphate carboxylase/oxygenase activase A—Hordeum vulgare | 84 | 0.9E_03 | 8.62 | Chloroplast stroma component, ATP binding. Activation of RuBisC—photosynthesis | |
| RL versus RC 10 | Q10EH0 | Lipoxygenase—Oryza sativa | 59 | 0.33 | 6.82 | Lipoxygenase activity—other metabolism (Lipoxygenase) |
| RL versus RC 11 | B23703 | Ribulose bisphosphate oxygenase carboxylase activase—Hordeum vulgare | 104 | 9.9E_06 | 5.64 | ATP binding, nucleotide binding, chloroplast component, ligase activity, have nucleotide-binding motif A (P-loop)—photosynthesis |
| C23703 | Ribulose bisphosphate oxygenase carboxylase activase—Hordeum vulgare | 100 | 2.5E_05 | 8.39 | ATP binding, nucleotide binding, chloroplast component, ligase activity, have nucleotide-binding motif A (P-loop)—photosynthesis | |
| Q40073 | Ribulose bisphosphate carboxylase/oxygenase activase A-Hordeum vulgare | 86 | 0.62E_03 | 8.62 | Chloroplast stroma component, ATP binding. RuBisCo activase—photosynthesis | |
| RL versus RC 12 | Q76MG1 | Dehydrin—Nicotiana tabacum | 64 | 0.25 | 5.62 | Response to stress, response to water—stress response |
| RL versus RC 13 | A2WZ04 | Putative uncharacterised protein—Oryza sativa | 59 | 0.77 | 7.22 | Unknown—unknown |
| RS versus RC 2 | A9T3C3 | Predicted protein—Tetraphis pellucida | 63 | 0.35 | 7.05 | Unknown—unknown |
| RS versus RC 3 | Q69X41 | Putative uncharacterised protein P0429G06.11—Oryza sativa | 55 | 4.3 | 12.05 | Unknown—unknown |
| RS versus RC 5 | P12782 | Phosphoglycerate kinase, chloroplastic—Wheat | 59 | 0.32 | 6.58 | Kinase and transferase, involves in carbohydrate biosynthesis/calvin cycle—photosynthesis |
| O48991 | NBS–LRR type resistance protein—Oryza sativa | 53 | 2.7 | 5.68 | ATP binding, involves in apoptosis and defence response-Apoptosis and defence response | |
| RS versus RC 6 | Q7EZQ0 | OJ1005_b10.1 protein—Oryza sativa | 54 | 5.2 | 11.17 | Unknown—unknown |
| RS versus RC 7 | A9T2E0 | Predicted protein—Tetraphis pellucida | 58 | 0.98 | 10.02 | Unknown—unknown |
| RS versus RC 8 | B9IMQ9 | Predicted protein—Black cottonwood | 57 | 1.3 | 8.81 | Unknown—unknown |
| RS versus RC 9 | Q7F7Z1 | Hypothetical protein—Oryza sativa | 59 | 0.79 | 11.56 | Unknown—unknown |
| RS versus RC 10 | B6TSE5 | Ras-related protein Rab-18—Zea Mays | 52 | 3.7 | 6.34 | Small GTPase superfamily, Rab family. GTP binding and nucleotide binding—other metabolism (GTPase) |
| RS versus RC 12 | XP_002266213 | Hypothetical protein—Vitis vinifera | 67 | 0.13 | 9.22 | Unknown—unknown |
| RS versus RC 13 | Q6YTS9 | Os08g0484700 protein (proline-rich family protein-like)—Oryza sativa | 54 | 0.57 | 7.09 | Unknown—unknown |
| RS versus RC 15 | Q84YU2 | Putative uncharacterised protein P0045A07.118—Oryza sativa | 57 | 2.4 | 11.2 | Unknown—unknown |
| SL versus SC 5 | Q6L5A9 | Os05g0545500 protein—Oryza sativa | 75 | 8.60E−03 | 10.59 | Unknown—unknown |
| SL versus SC 7 | Q655S1 | Cell division protease ftsH homolog 2, chloroplastic—Oryza sativa | 86 | 5.50E−04 | 5.54 | Probable ATP-dependent zinc metallopeptidase, have hydrolase, metalloprotease and protease activities—cell division (peptidase/protease) |
| SL versus SC 8 | Q8S6F3 | ATP synthase subunit beta—Oryza sativa | 95 | 8.40E−05 | 5.38 | Hydrogen ion transporting ATP synthase activity, rotational mechanism—ATP synthesis |
| SL versus SC 9 | P00828 | ATP synthase subunit beta, chloroplastic—Hordeum vulgare | 102 | 1.60E−05 | 5.11 | Produces ATP from ADP in the presence of a proton gradient across the membrane—ATP synthesis |
| SL versus SC 13 | Q6Z9W5 | Epstein-Barr virus EBNA-1-like protein—Oryza sativa | 68 | 0.04 | 11.42 | Unknown—unknown |
| SL versus SC 14 | Q3T5J7 | Ribulose bisphosphate carboxylase large chain—Sapium glandulosum | 72 | 0.014 | 6.05 | Calvin cycle, carbon dioxide fixation and photosynthesis, have lyase, monooxygenase and oxidoreductase activities—photosynthesis |
| SL versus SC 18 | A9TYL4 | Predicted protein—Physcomitrella patens | 82 | 4.70E−03 | 6.52 | Unknown—unknown |
| SL versus SC 22 | Q75K85 | Putative uncharacterised protein—Oryza sativa | 73 | 0.012 | 6.28 | Chromosome organisation, chromosome, ATP-binding and protein binding activities—chromosome organisation |
| SL versus SC 25 | Q9SEU4 | At1g55310—Arabidopsis thaliana | 82 | 1.60E−03 | 11.73 | Nuclear mRNA splicing, via spliceosome, identical protein binding, nucleic acid-binding and nucleotide-binding activities—mRNA processing |
| SL versus SC 27 | Q9C7P1 | Putative zinc finger CCCH domain-containing protein 10—Arabidopsis thaliana | 64 | 0.011 | 8.74 | DNA binding and zinc ion binding—unknown |
| SL versus SC 15 | P12782 | Phosphoglycerate kinase, chloroplastic—Triticum aestivum | 74 | 0.026 | 6.58 | Involves in glycolysis; ATP-binding and phosphoglycerate kinase activities—other metabolism (glycolysis) |
| SL versus SC 26 | Q75G97 | Putative uncharacterised protein OSJNBa0042F15.18—Oryza sativa | 81 | 0.006 | 7.61 | DNA integration and RNA-dependent DNA replication, DNA-binding, RNA-binding and RNA-directed DNA polymerase activities—DNA processing |
| SS versus SC 9 | P00828 | ATP synthase subunit beta, chloroplastic—Arabidopsis thaliana | 74 | 9.40E−03 | 5.11 | ATP synthesis, hydrogen ion transport, hydrolase activity (catalyse hydrolysis)—ATP synthesis |
| SS versus SC 13 | Q9FQ02 | 5′-3′ exoribonuclease 2—Arabidopsis thaliana | 67 | 6.30E−03 | 9.04 | mRNA processing and miRNA catabolic process, 5′-3′ exoribonuclease activity, nucleic acid binding and zinc ion binding—mRNA and miRNA processing |
| SS versus SC 4 | Q69LV5 | Putative uncharacterised protein OSJNBa0057D11.41—Oryza sativa | 75 | 7.80E−03 | 11.72 | Unknown—unknown |
| SS versus SC 8 | Q67H50 | ATP synthase beta subunit (Fragment)—Sisyrinchium montanum | 72 | 0.017 | 5.28 | Produces ATP from ADP in the presence of a proton gradient across the membrane—ATP synthesis |
Keys: (above proteins are arranged by treatments)
Italic: proteins up-regulated in Local tissues of Resistant line to 24-h aphid infestation
Normal: proteins up-regulated in Systemic tissues of Resistant line to 24-h aphid infestation
Bold: proteins up-regulated in Local tissues of Susceptible line to 24-h aphid infestation
Bold italic: proteins up-regulated in Systemic tissues of Susceptible line to 24-h aphid infestation
Some proteins have more than one eligible ID, and it is not possible to distinguish which ID is true due to limitations of the approach
Table 2.
Identification by PMF of differentially regulated proteins showing a minimum twofold increase in abundance in response to S. avenae infestation of resistant or susceptible diploid lines at 8-day time point in local and systemic tissues
| RL versus RC 1Spot Name (a) | Ac. No. (b) | Predicted ID (c) | Score (d) | e-Value (e) | Th. pI (f) | Function and biological process/category (g) |
|---|---|---|---|---|---|---|
| RL versus RC 1 | Q5NB00 | Putative uncharacterised protein P0483F08.25O—Oryza sativa | 57 | 4.8 | 11.92 | Unknown—unknown |
| RL versus RC 2 | Q94HR4 | Hypothetical protein OSJNBa0065C16.5—Oryza sativa | 66 | 2 | 11.95 | Unknown—unknown |
| RL versus RC 3 | Q7XYU5 | Integrase (fragment)—Gossypium herbaceum | 62 | 0.16 | 9.24 | DNA binding—unknown |
| RL versus RC 4 | Q6RH10 | 60S ribosomal protein L13a—Capsicum annuum | 68 | 0.041 | 10.83 | Translation, ribonucleoprotein, structural constituent of ribosome—protein synthesis |
| RL versus RC 5 | Q1SZU6 | RNA-directed DNA polymerase—Drosophila melanogaster | 63 | 0.12 | 10.06 | Unknown—unknown |
| RL versus RC 6 | Q6XRD3 | Ribosomal protein S4 (Fragment)—Passiflora morifolia | 77 | 5.10E−03 | 10.28 | Translation, ribonucleoprotein, structural constituent of ribosome, chloroplast—protein synthesis |
| RL versus RC 7 | Q9MBH3 | Arabidopsis thaliana genomic DNA—Arabidopsis thaliana | 59 | 0.033 | 9.74 | Unknown—unknown |
| RL versus RC 8 | Q7XXI2 | OSJNBa0059H15.7 protein P- Oryza sativa | 135 | 7.8E−09 | 8.94 | DNA integration, DNA binding—unknown |
| Q7Y008 | Putative RNA-binding protein—Oryza sativa | 135 | 7.8E−09 | 9.05 | nucleic acid binding, Zn ion binding—unknown | |
| Q1S1F3 | Polyprotein—Medicago truncatula | 68 | 0.041 | 9.84 | Unknown—unknown | |
| RL versus RC 9 | Q9FNL5 | Similarity to guanylate binding protein—Arabidopsis thaliana | 86 | 6.40E−04 | 6.25 | Immune response/beta-lactamase activity, GTP binding—immune response |
| RL versus RC 11 | Q3HRW1 | 60S ribosomal protein L13a-like protein—Solanum tuberosum | 66 | 0.064 | 10.54 | Translation, ribonucleoprotein, structural constituent of ribosome—protein synthesis |
| RL versus RC 12 | Q9SL02 | DNA repair protein RAD50—Arabidopsis thanliana | 57 | 0.53 | 5.98 | DNA repair, Mre11 complex, ATP binding, nuclease activity—DNA processing |
| RL versus RC 13 | Q8H636 | Os06g0128100 protein—Oryza sativa | 129 | 3.10E−08 | 7.85 | Putative methyltransferases—other metabolism (Methyltransferase) |
| Q10JM0 | Retrotransposon protein, putative, Ty3-gypsy subclass—Oryza sativa | 129 | 3.10E−08 | 8.92 | RNA-dependent DNA replication—DNA processing | |
| RL versus RC 14 | Q6H660 | Putative stress-induced protein sti1—Oryza sativa | 58 | 0.41 | 6.03 | Heat-shock chaperonin-binding—stress response |
| RL versus RC 15 | Q8GWT6 | Putative uncharacterised protein At1g53885/T18A20.20 (At1g53885)—Arabidopsis thaliana | 74 | 0.009 | 9.57 | Unknown—unknown |
| RL versus RC 16 | Q2QLW7 | Retrotransposon protein, putative—Oryza sativa | 62 | 0.15 | 8.88 | RNA-dependent DNA replication, RNA-directed DNA polymerase activity—DNA processing |
| RL versus RC 17 | S43767 | Ribosomal protein S3—evening primrose mitochondrion—Oenothera villaricae | 69 | 0.03 | 10.24 | Unknown—unknown |
| RL versus RC 18 | P36688 | 50S ribosomal protein L12, chloroplastic—Nicotiana sylvestris | 54 | 2.1 | 5.99 | Translation, ribonucleoprotein, structural constituent of ribosome—protein synthesis |
| RL versus RC 19 | Q84M91 | Putative uncharacterised protein At3g05330—Arabidopsis thaliana | 63 | 0.12 | 11.7 | Cytokinesis, initiation of separation, microtubule associated complex—cytokinesis |
| RL versus RC 20 | Q7XPG9 | OSJNBb0003B01.14 protein—Oryza sativa | 68 | 0.035 | 6.02 | Cysteine-type peptidase activity—peptidase |
| RL versus RC 21 | Q5Z660 | Putative viral resistance protein—Oryza sativa | 53 | 1.4 | 6.17 | Apoptosis, ATP binding—apoptosis and defence response to virus |
| RL versus RC 22 | Q8H3G8 | Myosin heavy chain-like protein—Oryza sativa | 66 | 0.7 | 5.55 | Unknown—unknown |
| RL versus RC 23 | Q9FVL0 | Non-symbiotic haemoglobin 1, MEDsa GLB1—Medicago sativa | 59 | 0.34 | 9.08 | Stress response/NsHb/non-symbiotic haemoglobin, heme-binding, oxygen-binding—stress response |
| RL versus RC 24 | Q9AUR0 | Putative uncharacterised protein OSJNBb0033N16.14—Oryza sativa | 71 | 0.021 | 9.08 | Cell division cycle-associated protein—cell division |
| RL versus RC 25 | Q6F367 | Putative uncharacterised protein OJ1268_B08.14—Oryza sativa | 60 | 0.27 | 11.94 | Unknown—unknown |
| RL versus RC 26 | P46801 | 60S ribosomal protein L16, mitochondrial—Oryza sativa | 64 | 0.08 | 10.67 | Translation, ribonucleoprotein, structural constituent of ribosome—protein synthesis |
| RL versus RC 27 | Q93WC5 | Pentatricopeptide repeat-containing protein At4g01990/T7B11_26, mitochondrial—Arabidopsis thaliana | 60 | 0.22 | 6.59 | Mitochondrion component, binding ability—mitochondrion component |
| RL versus RC 28 | Q6UCC4 | Maturase K (Fragment)—Planocarpa nitida | 61 | 0.18 | 9.74 | mRNA processing, chloroplast—mRNA processing |
| RL versus RC 29 | Q8RVC8 | Putative polyprotein (transposable element protein, putative)—Oryza sativa | 61 | 0.22 | 8.69 | Aspartic-type endopeptidase activity—peptidase |
| RL versus RC 30 | Q3HVK7 | Glycoprotein-like protein—Solanum tuberosum | 82 | 0.017 | 10.44 | Translation, ribosome, structural constituent of ribosome—protein synthesis |
| RL versus RC 31 | Q2MGR2 | SGS; HSP20-like chaperone—Medicago truncatula | 56 | 0.55 | 6.98 | Unknown—unknown |
| RL versus RC 32 | Q9STH1 | Stress-induced protein sti1-like protein—Arabidopsis thaliana | 64 | 0.092 | 6 | Response to hydrogen peroxide, heat and high light—response to oxidative stress, |
| RL versus RC 33 | Q1SMU7 | Hypothetical protein—Medicago truncatula | 58 | 0.38 | 4.84 | Unknown—unknown |
| RL versus RC 34 | Q93WC5 | Pentatricopeptide repeat-containing protein At4g01990/T7B11.26, mitochondrial—Arabidopsis thaliana | 56 | 0.58 | 6.59 | Mitochondrion component, binding ability—mitochondrion component |
| RL versus RC 10a | Q1SCC2 | Cullin—Medicago truncatula | 57 | 0.51 | 6.83 | Unknown—unknown |
| RL versus RC 10b | Q3SC85 | 60S ribosomal protein L10—Lycopersicon esculentum | 77 | 4.50E − 03 | 10.45 | Translation, ribonucleoprotein, structural constituent of ribosome—protein synthesis |
| RL versus RC 10c | Q9MTH5 | Putative membrane protein ycf1—Oenothera elata | 55 | 0.78 | 9.92 | Chloroplast membrane protein—photosynthesis |
| RL versus RC 18b | Q9MTH5 | Putative membrane protein ycf1—Oenothera elata | 68 | 0.041 | 9.92 | Chloroplast membrane protein—photosynthesis |
| RS versus RC 168 | B0BEM8 | Cytochrome c biogenesis protein—Muraltia aspalatha | 55 | 5.7 | 9.25 | Chloroplast component, involves in respiratory chain complex IV assembly—respiratory |
| RS versus RC 214 | Q6EPI0 | Putative uncharacterised protein OSJNBb0057I13.23—Oryza sativa | 57 | 2.2 | 4.47 | Unknown—unknown |
| RS versus RC 242 | A2T315 | Photosystem I protein M—Angiopteris evecta | 59 | 9.4E+02 | 5.8 | Chloroplast component, plasma membrane-derived photosystem I component, involves in photosynthesis—photosynthesis |
| RS versus RC 296 | C7A7K7 | NBS-containing resistance-like protein—Corylus avellana | 64 | 0.29 | 5.5 | Have ATP-binding ability, involves in apoptosis and defence response (pathogen)—apoptosis and defence response to pathogen |
| C5WNH9 | Putative uncharacterised protein Sb01g038360—Sorghum bicolour | 66 | 0.054 | 5.95 | Have carboxy-lyase, magnesium ion binding and thiamin pyrophosphate binding activities—other metabolism (carboxy-lyase) | |
| RS versus RC 298 | Q49KX3 | ATP-dependent Clp protease proteolytic subunit—Eucalyptus globulus | 58 | 0.16 | 4.72 | Cleaves peptides in various protein, play a role in degradation of misfolded proteins. Has a chymotrypsin-like activity—protease |
| SL versus SC 43 | B6SSP2 | EF hand family protein—Zea mays | 52 | 4.9 | 5.63 | Calcium ion binding—unknown |
| SL versus SC 475 | B9T620 | Putative uncharacterised protein—Ricinus communis | 66 | 0.19 | 10.13 | Unknown—unknown |
| SL versus SC 116 | D7M917 | Hypothetical protein ARALYDRAFT_914223—Arabidopsis lyrata | 53 | 4.7 | 6.15 | Protein phosphorylation—post-translational modification |
| SL versus SC 707a | C5X6P9 | Hypothetical protein SORBIDRAFT_02g012940—Sorghum bicolour | 70 | 0.087 | 6.54 | GTPase activity—other metabolism (GTPase) |
| SL versus SC 707b | 1GHSA(MSDB) | Glucan endo-1,3-beta-D-glucosidase (EC 3.2.1.39) II, chain A—Hordeum vulgare | 53 | 12 | 8.83 | Endoglucosidase—other metabolism (endoglucosidase) |
| SL versus SC 75 | Q6Z0C7 | Hypothetical protein OSJNBa0086M15.4 (Hypothetical protein OJ1465_C11.25)—Oryza sativa | 56 | 0.58 | 11.2 | Unknown—unknown |
| SL versus SC 37 | C1FGT1 | Predicted protein—Micromonas sp. (strain RCC299/NOUM17) | 61 | 0.74 | 5.86 | Nuclease activity, involves in nucleotide excision repair—DNA processing (nuclease/nucleotide excision repair) |
| SL versus SC 69 | B9SAI3 | Conserved hypothetical protein—Ricinus communis | 59 | 10 | 5.8 | Unknown—unknown |
| SS versus SC 43 | Q8GUW5 | Trehalose-6-phosphate synthase/phosphatase (Fragment)—Cypripedium parviflorum | 65 | 0.078 | 7.84 | Catalyse UDP-glucose and d-glucose 6-phosphate into UDP and alpha, alpha-trehalose 6-phosphate—regulates cell shape |
| SS versus SC 179 | B8B946 | Hypothetical protein OsI_30043—Oryza sativa | 61 | 0.62 | 7.73 | Unknown—unknown |
| SS versus SC 692 | A9T965 | Predicted protein—Physcomitrella patens | 63 | 0.41 | 8.43 | Have nucleic acid and zinc ion binding activities, bioprocess involved Unknown—unknown |
| SS versus SC 159 | A2Q630 | Phosphatidylinositol 3- and 4-kinase, catalytic—Medicago truncatula | 56 | 1.9 | 5.76 | Have kinase activity and phosphotransferase activity, alcohol group as acceptor—other metabolism (phosphotransferase) |
Keys: (above proteins are arranged by treatments)
Italic: proteins up-regulated in Local tissues of Resistant line after 8-day aphid infestation
Normal: proteins up-regulated in Systemic tissues of Resistant line after 8-day aphid infestation
Bold: proteins up-regulated in Local tissues of Susceptible line after 8-day aphid infestation
Bold italic: proteins up-regulated in Systemic tissues of Susceptible line after 8-day aphid infestation
Please note: some proteins have more than one eligible ID, and it is not possible to distinguish which ID is true due to limitations of the approach
Results
Resistance of diploid wheat lines to grain aphid
A significant impact on the reproductive capacity of S. avenae was recorded when developing on different lines of T. monoccoccum, oats or hexaploid wheat (F 14,111 = 4.67, P < 0.0001) (Fig. 1a). Whilst two diploid lines allowed aphids to produce in excess of 20 offspring per female, five lines only allowed for mean fecundities of less than 5 nymphs per female (Fig. 1a). By comparison, the hexaploid cultivar Claire was intermediate, with aphids producing an average of ca. 13 nymphs per female. Longevity closely reflected fecundity and was also significantly affected by variety (F 14,112 = 3.78, P < 0.0001), such that aphids that produced the largest numbers of offspring typically exhibited the longest lifespans (Fig. 1b). Mean relative growth rate was similarly significantly affected by variety (Fig. 1c), although less markedly than fecundity or longevity (F 14,113 = 8.32, P < 0.0001). Notably, the growth rate was highest on the commercial hexaploid wheat variety (Claire) and the habitual laboratory host, oats (Coast Black). These bioassays thus identified the following as exhibiting resistance to S. avenae: ACC16 INRA TM44, ACC17 PGR1752, ACC20 PGR1755, ACC26 PGR1761 and ACC27PGR1762, whilst ACC5 PGR1735 and ACC PGR1758 showed susceptibility to S. avenae. Based on these bioassays, the resistant diploid line (ACC20 PGR1755; #11) and the susceptible line ACC5 PGR1735 were selected in the present study to investigate genes and proteins potentially involved in the observed aphid resistance/tolerance.
Proteome responses of wheat seedlings to aphid infestation
Local and systemic changes in the wheat leaf proteome in both a resistant diploid line (ACC20 PGR1755) and a susceptible diploid line (ACC5 PGR1735) in response to aphid infestation were investigated. Two time points were selected, 24 h (early response) and 8 days (late response). The proteome maps for 24 h and 8 days post-treatment control gels were consistent with previous 2-D gel separations reported for wheat and barley leaves (Geddes et al. 2008; Jiang et al. 2008; Ferry et al. 2011).
Proteome maps of wheat leaf tissues from each treatment (a resistant and a susceptible line) were compared with their respective controls using Progenesis SameSpots software (S Fig 1) and differentially regulated proteins identified by MS and MS/MS searches of nrNCBI/Swiss-Prot and a locally restricted wheat EST database. Differentially expressed proteins identified after 24 h and 8 days aphid infestation are presented in Tables 1 and 2, respectively. For both time points, both local and systemic responses were investigated. Approximately 200 protein spots were detected on each gel, with the differences in protein expression levels between a given line/time point/feeding site with its respective control varying from 4 to 34 protein spots. In the susceptible line, 16 protein spots were significantly up-regulated after 24-h aphid feeding, with 12 being involved in the local response and 4 involved in the systemic response, but after 8 days the up-regulated spots had decreased to 12 (8 locally and 4 systemically). In contrast, in the resistant line, 28 protein spots were significantly up-regulated after 24-h aphid feeding (17 locally and 11 systemically), and the number of up-regulated spots increased to 41 (37 locally and 4 systemically) after 8 days. Thus, the data show that more proteins were observed to be up-regulated in the resistant line at day 8 compared to the other treatments.
Differentially expressed proteins in response to aphid infestation over time
Identified up-regulated protein spots listed in Tables 1 and 2 are grouped by functional categories in Fig. 2a, b. The results clearly show that the majority of identified proteins that were differentially regulated in both the resistant (ACC20 PGR1755) and susceptible (ACC5 PGR1735) diploid line following aphid infestation were involved in photosynthesis or other metabolic processes or were classified as being uncharacterised/unknown. Other proteins identified in both lines were shown to be involved in: ATP synthesis, proteolysis, post-translational modification, immune response, nuclear mRNA splicing, mRNA and miRNA processing, chromosome organisation, cell division and cytokinesis (Tables 1, 2). However, there were notable differences in the response of these two lines, particularly in respect of the up-regulation of general and specific stress response proteins. Importantly, these results show that proteins known to be involved in the stress response and the defence response were clearly induced in the resistant line, but not the susceptible line, in response to aphid infestation (Fig. 3a, b) and that different proteins were up-regulated at different time points. Thus, both spatial and temporal effects were observed. For example, at 24 h (representing an early response), a heat stress TF and dehydrin were up-regulated as part of the local response, whereas NBS–LRR type resistant protein was up-regulated as part of the systemic response. At 8 days (late response), a different set of proteins (two putative stress-induced protein sti1, putative viral resistance protein, MEDsa GLB1 and HSP20-like chaperone) was up-regulated as part of the local response, whilst a NBS-containing resistance-like protein was up-regulated as part of the systemic response. Additionally, at 24 h post-infestation, transcription factors and proteins with roles in signalling were shown to be clearly induced in the resistant line; by day 8, proteins involved in protein synthesis were shown to be up-regulated in this line. Interestingly, DNA repair proteins were also up-regulated in the resistant line. These results also show that the majority of the up-regulated proteins (in terms of number) were induced locally rather than systemically (Fig. 2a, b).
Fig. 2.
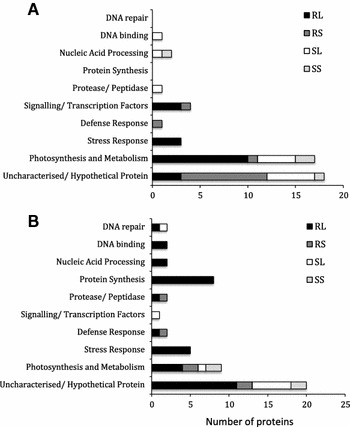
Changes in diploid wheat leaf proteins following aphid infestation for 24 h (a) or for 8 days (b). Proteins identified were assigned to categories based on biological process GO terms, shown as number of total response in each category for each treatment. Proteins that could not be identified were not included. First character: R resistant line, S susceptible line. Second character: L local tissues, S systemic tissues
Fig. 3.
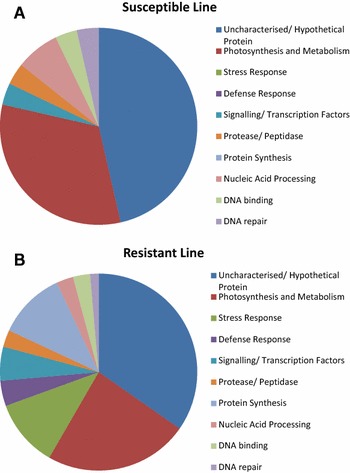
Global changes in wheat leaf proteins following aphid infestation of (a) the susceptible diploid line (ACC5 PGR#1735) and b the resistant diploid line (ACC20 PGR1755). Proteins identified were assigned to categories based on biological process GO terms, shown as number of total response in each category for each line at all time points tested. The results show that proteins involved in the stress response and the defence response were up-regulated in the resistant line, but only in response to aphid feeding. No such proteins were identified in the susceptible line either prior to, or following aphid infestation. (Color figure online)
General stress response proteins
The fold change (in normalised spot volume) for selected stress response proteins up-regulated in the resistant line in response to aphid infestation is presented in Table 3. The heat stress transcription factor A-5 (Oryza sativa) and dehydrin (Nicotiana tabacum) were up-regulated 2.6- and 2.9-fold, respectively, in the resistant line after 24-h aphid infestation (local response), as was a lipoxygenase (Oryza sativa), a compound with multiple roles including plant response to stress, where the normalised spot volume showed a 3.2-fold increase. A putative stress-induced protein Sti1 (Oryza sativa and Arabidopsis thaliana), a non-symbiotic haemoglobin 1 MEDsa GLB1 (Medicago sativa) and a protein with similarity to guanylate binding proteins from Arabidopsis were also up-regulated in this line (3.5-, 3.7-, 2.8- and 2.2-fold increases, respectively), but only after 8 days aphid infestation (local response). In addition to these general stress response proteins, proteins with known roles in plant defence and apoptosis, such as NBS–LRR type resistance protein (Oryza sativa), were also shown to be systemically up-regulated in the resistant line after 24-h aphid infestation (2.4-fold increase). After 8 days of aphid feeding, a putative viral resistance protein (Oryza sativa) was identified as part of the local response in this resistant line (4.3-fold increase), whilst an NBS-containing resistance-like protein was identified as part of the systemic response (2.7-fold increase). Interestingly, proteins involved in protein synthesis and transcriptional regulation (ras-related protein Rab-18, Zea mays, 2.1-fold increase) were also up-regulated in the resistant line, but none were identified as being up-regulated in the susceptible line in response to aphid infestation (Fig. 2a, b; Table 3). Other proteins such as chaperones (SGS; HSP20-like chaperone, Medicago truncatula, 4.1-fold increase) and heat-shock proteins (sti 1) were also shown to be induced in the resistant line after 8 days post-aphid infestation. No stress/defence proteins were identified in the susceptible line in response to aphid infestation.
Table 3.
Selected proteins (putative identity) known to be stress responsive
| Predicted ID | Fold change |
|---|---|
| Treatment | |
| Resistant line (Local) 24 h | |
| Heat stress transcription factor A-5—Oryza sativa | 2.6 |
| Lipoxygenase—Oryza sativa | 3.2 |
| Dehydrin—Nicotiana tabacum | 2.9 |
| Treatment | |
| Resistant line (Systemic) 24 h | |
| NBS–LRR type resistance protein—Oryza sativa | 2.4 |
| Ras-related protein Rab-18—Zea Mays | 2.1 |
| Treatment | |
| Resistant line (Local) 8 days | |
| Similarity to guanylate binding protein—Arabidopsis thaliana | 2.2 |
| Putative stress-induced protein sti1—Oryza sativa | 3.5 |
| Putative viral resistance protein—Oryza sativa | 4.3 |
| Non-symbiotic haemoglobin 1, MEDsa GLB1—Medicago sativa | 2.8 |
| SGS; HSP20-like chaperone—Medicago truncatula | 4.1 |
| Stress-induced protein sti1-like protein—Arabidopsis thaliana | 3.7 |
| Treatment | |
| Resistant line (Systemic) 8 days | |
| NBS-containing resistance-like protein—Corylus avellana | 2.7 |
Fold change relative to the appropriate control is shown
Oxidative stress response proteins
Of particular interest, proteins associated with oxidative stress caused by insect herbivores were detected. After 24-h aphid infestation (local response), one stress response protein, called dehydrin from cultivated tobacco, and one redox protein from Arabidopsis (Table 1, spot RL v RC 1), were identified putatively in resistant plants, whilst after 8 days, a further three stress response proteins, putative stress-induced protein sti1 (from rice), non-symbiotic haemoglobin 1 (from alfalfa) and a stress-induced protein sti1-like protein (from Arabidopsis), which is involved in response to hydrogen peroxide and oxidative stress, were identified in these resistant plants. Surprisingly, no such protein was identified in the resistant plants as a systemic response, and neither were any such proteins identified in the susceptible plant line, irrespective of the time frame (24 h or 8 days).
Proteins involved in photosynthesis and metabolism
Proteins involved in photosynthesis and metabolism formed the majority of proteins differentially expressed in either the susceptible or the resistant line in response to aphid infestation; interestingly, more proteins in these categories were detected after 24-h than 8-day aphid infestation (Tables 1, 2). Putative proteins involved in photosynthesis that were shown to be up-regulated locally in the resistant line at 24 h post-infestation included the large subunit of ribulose-1, 5-bisphosphate carboxylase/oxygenase and two ribulose bisphosphate oxygenase carboxylase activase from barley. Putative proteins involved in metabolism that were up-regulated locally at this same time point included a lipase class 3 family protein from rice (lipid metabolism), whilst a methyltransferase (Os06g0128100 protein from rice) was identified locally in these resistant plants, but after 8 days. Proteins with roles in protein degradation were also noted to be differentially regulated. For example, a protein with cysteine-type peptidase activity (OSJNBb0003B01.14 protein from rice) was identified putatively in the resistant line after 8 days aphid infestation as part of the local response, whilst a cell division protease (ftsH homolog 2 from rice), which is a probable ATP-dependent zinc metallopeptidase with hydrolase, metalloprotease and protease activities, was identified putatively as part of the local response in susceptible plants after 24-h aphid infestation.
Proteins involved in transcriptional regulation
Proteins with roles in transcriptional regulation were also changed in response to aphid infestation, but predominantly in the susceptible wheat genotype. For example, a protein called At1g55310 from Arabidopsis, which is involved in nuclear mRNA splicing, was identified putatively in the susceptible line at 24 h post-local aphid infestation, whilst 5′-3′ exoribonuclease 2 (from Arabidopsis), which is involved in mRNA processing and miRNA catabolic processes, was also identified putatively in these plants after 24 h, but only as part of the systemic response. Furthermore, a putative uncharacterised protein, OSJNBa0042F15.18 from rice, which is involved in DNA integration and RNA-dependent DNA replication, was identified putatively in the susceptible line after 24-h aphid local infestation. After 8d aphid infestation (local), two retrotransposon proteins, known to be involved in RNA-dependent DNA replication, were identified putatively in the resistant genotype after 24-h aphid infestation as part of the local response.
Discussion
The preliminary bioassays carried out with diploid wheat lines and a standard commercial cultivar showed clear differences in susceptibility towards cereal aphids, although none of the lines tested showed strong resistance in the sense of causing significant levels of mortality of the pest. However, the partial resistance observed is sufficient to be useful in minimising cereal aphid outbreaks in the field by slowing the rate of population increase and thus provide a useful basis for further investigation. These results allowed two diploid wheat lines to be selected for a ‘resistant’ versus ‘susceptible’ comparison by proteomic analyses. The proteomics results showed that there were notable differences in terms of proteins up-regulated between the resistant and susceptible diploid wheat lines in response to aphid feeding. In this study, only up-regulated proteins were identified, as they are more likely to be directly involved in plant resistance to aphid infestation. Down-regulated proteins are likely to be the result of aphid manipulation or sabotage of plant defence (Smith and Boyko 2007).
Previous studies demonstrated that two-dimensional gel protein electrophoresis followed MALDI-TOF mass spectrometry (MS and MS/MS) was an effective initial tool for identifying differentially expressed proteins in wheat in response to aphid infestation (Ferry et al. 2011). Although the wheat genome has not been fully annotated, over 50 % of the protein spots excised from gels could be putatively identified as showing significant similarity to known wheat or other cereal/plant genes using NCBI, Swiss-Prot and wheat EST databases. Results from the previous study using the commercial line Claire (T. aestivum, hexaploid, AuAuBBDD) showed little resistance to aphid feeding, and thus, the diploid accessions were investigated as potential sources of aphid resistance genes/gene products (Diploid AmAm; T. monococcum) for exploitation in subsequent breeding programmes.
The present study could not definitively identify any specific gene-for-gene interactions between the diploid wheat T. monococcum and the grain aphid S. avenae, although proteins with similarity to NBS–LRR (nucleotide-binding site leucine-rich repeat) resistance proteins were identified as being systemically induced in the resistant line (ACC20 PGR1755) (Table 3). Most of the disease resistance genes (R genes) in plants cloned to date encode nucleotide-binding site leucine-rich repeat (NBS–LRR) proteins characterised by nucleotide-binding site (NBS) and leucine-rich repeat (LRR) domains. These abundant proteins are involved in the detection of pathogens (bacteria, viruses, fungi), nematodes and insects (McHale et al. 2006). Plant NBS–LRR proteins are numerous and are encoded by one of the largest gene families known in plants (Monosi et al. 2004). Disease resistance is the only function so far demonstrated for NBS–LRR proteins; however, a role in resistance has yet to be confirmed for most (Deslandes et al. 2002). Little is known about the regulation of the plant genes that encode NBS–LRRs. Consistent with the need for a rapid response to pathogen attack, many NBS–LRR-encoding genes are constitutively expressed at low levels in healthy, unchallenged tissue, although some show tissue-specific expression (McHale et al. 2006). They are upregulated, however, in response to bacterial flagellin, which induces basal resistance, suggesting that plants can establish a state of heightened sensitivity to pathogen attack (Zipfel et al. 2004). The tomato Mi-1 gene encoding an NBS–LRR-like protein confers resistance to both root-knot nematodes and potato aphids (Vos et al. 1998; Li et al. 2006), and aphid resistance in Medicago truncatula involves antixenosis and phloem-specific, inducible antibiosis, which maps to a single locus flanked by NBS–LRR resistance gene analogues (Klingler et al. 2005). Thus, findings from the present study provide the basis for a future molecular analysis of aphid resistance.
The diploid resistant line also responded to aphid infestation by significant up-regulation of stress response proteins, oxidative stress response proteins, defensive proteins and transcriptional regulators; interestingly, these same proteins were not detected in the susceptible line ACC5 PGR1735 in response to aphid infestation (Fig. 2a, b). This result suggests that the above proteins are playing a role in the observed resistance/tolerance of line ACC20 PGR1755 to aphid infestation. Furthermore, the diploid resistant wheat exhibited greater up-regulation of DNA synthesis/replication/repair proteins, which could have potential impact on plant resistance/tolerance to aphids and plant survival under aphid attack and oxidative stress.
Overall, the aphid responsive changes in the diploid wheat lines investigated were both spatially and temporally regulated, with differences between the two time points (24 h and 8 days), as well as differences in local and systemic responses (Tables 1, 2, 3; Fig. 2a, b). These proteins are grouped by functional categories (Fig. 3) to allow comparison between the resistant and susceptible lines and their potential roles discussed below.
Metabolism related proteins
Plants experience a metabolic re-programming when attacked by herbivores; genes that are involved in photosynthesis may be up-regulated to compensate for nutritional loss caused by aphid attack (Smith and Boyko 2007). Photosynthetic adjustments in wheat can significantly contribute to its tolerance to damage/metabolic drain caused by Russian wheat aphid (RWA; D. noxia; Haile et al. 1999). In the present study, several enzymes involved in photosynthesis and ATP synthesis were up-regulated in response to aphid infestation (ribulose-1, 5-bisphosphate carboxylase/oxygenase large subunit, ribulose bisphosphate oxygenase carboxylase activase and ATPase alpha subunit) after 24-h infestation. Similar changes in metabolic processes were detected in both rice and barley following insect or pathogen attack, respectively (Wei et al. 2009; Geddes et al. 2008). Differential expression of photosynthetic or photorespiration genes have also been observed for Myzus persicae feeding on leaves of celery, D. noxia feeding on wheat foliage, M. nicotianae feeding on Nicotiana attenuata foliage, Schizaphis graminum feeding on N. attenuata foliage (Smith and Boyko 2007) and Bemisia tabaci feeding on Arabidopsis (Yin et al. 2012). These changes indicate the potential switch of plant metabolic resources from normal growth to defensive functions, when it is subjected to attack by phloem feeders. Thus, we also observe changes in protein turnover as plants adapt to different environmental conditions, including the challenge raised by insect pests (Pickart and Eddins 2004; Dreher and Callis 2007).
In the present study, a protein, OSJNBb0003B01.14 (Table 2, spot 20), with proteolysis/cysteine-type peptidase activity was found to be up-regulated locally after 8 days aphid infestation in the resistant line. Feng et al. (2003) showed similar results of regulated proteolysis of endogenous proteins can be a contributing factor to plant defence against insect pests. Similarly, changes in transcriptional regulators are essential to plants under different abiotic and biotic stresses. In this study, heat stress transcription factor A-5 (Table 1, spot RLvsRC2) and a Dof zinc finger protein (Fragment) (Table 1, spot RLvsRC8) were up-regulated locally in the resistant line 24-h post-aphid infestation. These findings are in agreement with those of other studies that suggest that transcription factors play an important role in plant response to environmental changes through regulation of plant signalling pathways (Lucyshyn and Wigge 2009; Koini et al. 2009; Casson et al. 2009). Metabolic reprogramming is further illustrated by results showing that aphid feeding up-regulates proteins involved in mRNA and miRNA processing (Fig. 2a, b), which in turn leads to changes in protein synthesis. Similar results have been shown for RWA feeding on wheat leading to the up-regulation of transcripts that are involved in protein synthesis (Boyko et al. 2006; Smith et al. 2010).
Stress response and oxidative stress response proteins
Components of aphid salivary secretions are known to generate local and systemic production of reactive oxygen species (ROS; Tjallingii 2006), which is a commonly known early plant response to biotic stress (Apel and Hirt 2004; Bolwell and Wojtaszek 1997). ROS are important for plant signalling under insect attack, but can cause oxidative damage of membrane integrity due to lipid peroxidation, and can also generate highly cytotoxic compounds in the process. Therefore, plants have to enhance their resistance mechanisms, such as ROS scavenging and cell defence, to maintain homeostasis under stress conditions.
In this study, stress response proteins were identified as up-regulated in the resistant line after both 24-h and 8-day aphid infestation. A heat stress transcription factor A-5 (Oryza sativa, 24 h RL) and dehydrin (Nicotiana tabacum, 24 h RL) were induced early in the response, whilst after 8 days other known stress response proteins were induced including non-symbiotic haemoglobin 1, MEDsa GLB1 (Medicago sativa, 8 days RL) and two putative stress-induced protein sti1 (O. sativa and Arabidopsis, 8 days RL). The stress-induced protein sti1-like proteins (Table 3, induced in the resistant line as part of the local response, 8 days after infestation) are also thought to be induced in response to hydrogen peroxide and oxidative stress (Sano et al. 2013). Additionally, a hypothetical protein (Arabidopsis, 24 h RL, spot 1) is also thought to have a role in oxidative stress. These results indicate that diploid wheat plants recognised aphid infestation as a threat and a general stress response was triggered at or near the site of feeding, which was shown by the up-regulation of multiple stress-related proteins in leaves of the resistant line (24 h RL or 8 days RL). Interestingly, this stress response was not detected in the susceptible wheat line under the same treatment or the systemically infested resistant wheat leaves. This conclusion is further supported by the identification of induction of heat-shock proteins (HSPs) in the resistant line in response to aphid infestation. Stresses usually cause protein dysfunction, and maintaining functional proteins is particularly important for cell survival under stress. Many HSPs act as chaperones during protein folding, assembly, translocation and degradation, and can assist in protein refolding under stress conditions: for example, the HSP-70 chaperone system is crucial during both the stress response and development because protein misfolding and aggregation disrupt cellular homeostasis (Koizumi et al. 2014). They thus protect plants against stress by re-establishing normal protein conformation and hence cellular homeostasis (Wang et al. 2004). Similar results have been reported in other systems, for example, several proteins with putative functions in stress responses were identified in barley in response to Rhopalosiphum padi infestation (Gaupels et al. 2008). However, some chaperones are also multifunctional and may show enzymatic functions besides roles in protein folding (Scranton et al. 2012).
The induction of oxidative stress following aphid feeding is suggested by the identification of two stress-induced protein sti1-like proteins (Table 3) in this study and indeed in other aphid studies. Previous studies have shown that D. noxia infestation significantly induced an early accumulation of hydrogen peroxide and not only increased NADPH oxidase activity in a resistant (cv. Tugela DN) wheat line (Moloi and van der Westhuizen 2006), but also antioxidative enzyme activity in this same resistant wheat line compared to a susceptible wheat line (Moloi and van der Westhuizen 2008). Thus, the ability to control ROS levels in the wheat plant is closely linked to its resistance to aphid infestation. Oxidative stress is one of the first general reactions to the damage caused by phloem-feeding insects when they penetrate the plant (Wei et al. 2009). The expression of oxidative stress-related proteins, including a catalase, an ascorbate peroxidase and five extracellular class III peroxidases, was significantly increased in rice (O. sativa) in response to the brown plant hopper (Nilaparvata lugens; Wei et al. 2009). Proteomic studies carried out by Collins et al. (2010) revealed that proteins known to be involved in limiting ROS were more abundant in the resistant rather than susceptible varieties of Arabidopsis when under insect stress. Oxidative stress may also explain why proteins involved in DNA repair were identified in the present study. Interestingly, there are more DNA processing proteins up-regulated in the resistant line than the susceptible line following aphid feeding, suggesting some involvement of these proteins in the observed resistance/tolerance of line ACC20 PGR1755 to aphid infestation. Micro-array studies have similarly identified transcripts involved in DNA repair in Arabidopsis following aphid infestation, for example the increased expression of a nucleoside diphosphate-linked moiety transcript (Couldridge et al. 2007). Filkowski et al. (2004) found that mutant Arabidopsis lines impaired in certain aspects of protection against elevated levels of free radicals induce the production of scavenging enzymes earlier than wild-type plants. The higher levels of radical species resulted in the increased incidence of spontaneous double-strand breaks resulting in a higher expression of DNA repair genes. Gene sequences putatively involved in DNA synthesis were identified in wheat plants containing Dn genes in response to RWA biotype I attack (Boyko et al. 2006).
In addition to investigating both local and systemic responses in diploid wheat lines to aphid infestation, the present study also investigated this response over time to represent both an early response (24 h) and late response (8 days). Whilst stress and defence proteins were up-regulated at both time points, these represented different stress proteins, with more occurring during the late response phase. Interestingly, transcription factors and proteins with roles in signalling were shown to be induced during the early phase, as opposed to the late phase. Few studies have attempted to investigate these responses either over time or at the proteome level. Previous studies have shown that changes in transcript expression in wheat in response to RWA, where tolerance has previously been well characterised, comprise two phases: an immediate response (i.e. hypersensitive response) 24 h after infestation with RWA and a second prolonged response that prevails in the tissue for an extended period of time (i.e. systemic acquired resistance) (Botha et al. 2006), although differences in expression of transcripts were not reported. However, recently this same group has reported the differential regulation of transcripts involved in stress, signal transduction, photosynthesis, metabolism and gene regulation in susceptible wheat lines in response to RWA to better understand the different modes of resistance in wheat to this aphid species and the role of Dn genes (Botha et al. 2014). The ability of these genes to confer tolerance to S. avenae is currently being investigated.
Ultimately, the aim of the present study was to try to identify potential aphid resistance proteins. An NBS–LRR type resistance protein, types of which are known to be involved in apoptosis and plant defence (Takken and Joosten 2000; De Young and Innes 2006), was found to be up-regulated systemically in the resistant line following 24-h aphid infestation (Table 3). Furthermore, a putative viral resistance protein was shown to be up-regulated locally in the resistant line post 8 days aphid infestation as was the NBS-containing resistance-like protein (Table 3); however, in this case, the protein was systemically up-regulated. Both proteins have previously been shown to be involved in apoptosis and plant defence responses (Takken and Joosten 2000; De Young and Innes 2006; Rossignol et al. 2006). Possible involvement of these proteins in aphid resistance may occur, although further study is required to confirm and better understand their contributions to the observed resistance.
Currently, there is a major focus to identify potential aphid resistance genes through the study of aphid-induced gene expression (Thompson and Goggin 2006; De Vos et al. 2007; Smith et al. 2010). However, previous studies with commercial wheat lines have shown that this crop has a relatively poor inducible defence system (Ferry et al. 2011). Whilst there was very little evidence of any local induction of defensive proteins, e.g. proteinase inhibitors or other wound-responsive genes in response to grain aphid infestation (Thompson and Goggin 2006; Ferry et al. 2011) in this study, it does give new insights into the aphid-stress response in diploid wheat. The results suggest that stress response, oxidative stress response, defence response, apoptosis and DNA synthesis/replication/repair proteins play an important role in conferring resistance in the diploid line studied to S. avenae. This study indicates that the resistant diploid line ACC20 PGR1755 may provide a valuable resource in breeding wheat for resistance to aphids.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary material 1 Leaf proteome of resistant wheat (R, ACC20 PGR1755) following 24-h aphid infestation showing the local response versus non-infested leaf proteome of same aged resistant wheat plants. Up-regulated protein spots of twofold change or greater are indicated by O; down-regulated protein spots of twofold change or greater are indicated by O (PPT 3092 kb)
Acknowledgments
Funding to support the work described in this paper was provided by the BBSRC (Crop Science Initiative; Grant BB/E006280/1) and is gratefully acknowledged. The authors would like to thank R. J. Lamb for the diploid wheat seeds that were kindly donated from the seed collections of Agriculture and Agri-Food Canada and for critically commenting on the MS. The authors also thank the British Wheat Breeders for advice and support throughout the programme.
Footnotes
Wenzhu Guan and Natalie Ferry have equally contributed to this work.
References
- Apel K, Hirt H. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol. 2004;55:373–399. doi: 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- Bolwell GP, Wojtaszek P. Mechanisms for the generation of reactive oxygen species in plant defence—a broad perspective. Phys Mol Plant Path. 1997;51:347–366. doi: 10.1006/pmpp.1997.0129. [DOI] [Google Scholar]
- Botha A-M, Lacock L, van Niekerk C, Matsioloko MT, et al. Is photosynthetic transcriptional regulation in Triticum aestivum L. cv. ‘TugelaDN’ a contributing factor for tolerance to Diuraphis noxia (Homopera: Aphididae)? Plant Cell Rep. 2006;25:41–54. doi: 10.1007/s00299-005-0001-9. [DOI] [PubMed] [Google Scholar]
- Botha A-M, van Eck L, Burger NFV, Swanevelder ZH. Near-isogenic lines of Triticum aestivum with distinct modes of resistance exhibit dissimilar transcriptional regulation during Diuraphis noxia feeding. Biol Open. 2014 doi: 10.1242/bio.201410280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyko EV, Smith CM, Thara VK, Bruno JM, Deng YP, Starkey SR, Klaahsen DL. Molecular basis of plant gene expression during aphid invasion: wheat Pto- and Pti-like sequences are involved in interactions between wheat and Russian wheat aphid (Homoptera: Aphididae) J Econ Entomol. 2006;99:1430–1445. doi: 10.1093/jee/99.4.1430. [DOI] [PubMed] [Google Scholar]
- Casson SA, Franklin KA, Gray JE, Grierson CS, et al. Phytochrome B and PIF4 regulate stomatal development in response to light quantity. Curr Biol. 2009;19:229–234. doi: 10.1016/j.cub.2008.12.046. [DOI] [PubMed] [Google Scholar]
- Collins RM, Afzal M, Ward DA, Prescott MC et al (2010) Differential proteomic analysis of arabidopsis thaliana genotypes exhibiting resistance or susceptibility to the insect herbivore, Plutella xylostella. PLos One (open access publishing) [DOI] [PMC free article] [PubMed]
- Couldridge CE, Newbury HJ, Ford-Lloyd BV, Bale JS, Pritchard J. Exploring plant responses to aphid feeding using a full Arabidopsis microarray reveals a small number of genes with significantly altered expression. Bull Entomol Res. 2007;97:523–532. doi: 10.1017/S0007485307005160. [DOI] [PubMed] [Google Scholar]
- De Vos M, Van Oosten VR, van Poecke RMP, Van Pelt JA, et al. Signal signature and transcriptome changes of Arabidopsis during pathogen and insect attack. Mol Plant Microbe. 2005;18:923–937. doi: 10.1094/MPMI-18-0923. [DOI] [PubMed] [Google Scholar]
- De Vos M, Kim J-H, Jander G. Biochemistry and molecular biology of Arabidopsis-aphid interactions. BioEssays. 2007;29:871–883. doi: 10.1002/bies.20624. [DOI] [PubMed] [Google Scholar]
- De Young BJ, Innes RW. Plant NBS–LRR proteins in pathogen sensing and host defense. Nat Immunol. 2006;7:1243–1249. doi: 10.1038/ni1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslandes L, Olivier J, Theulieres F, Hirsch J, Feng DX, Bittner-Eddy P, Beynon J, Marco Y. Resistance to Ralstonia solanacearum in Arabidopsis thaliana is conferred by the recessive RRS1-R gene, a member of a novel family of resistance genes. Proc Natl Acad Sci USA. 2002;99:2404–2409. doi: 10.1073/pnas.032485099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devonshire AL, Field LM. Gene amplification and insecticide resistance. Ann Rev Entomol. 1991;36:1–23. doi: 10.1146/annurev.en.36.010191.000245. [DOI] [PubMed] [Google Scholar]
- Di Pietro JP, Caillaud CM, Chaubet B, Pierre JS, Trottet M. Variation in resistance to the grain aphid, Sitovion avenae (Sternorhynca: Aphididae), among diploid wheat genotypes: multivariate analysis of agronomic data. Plant Breed. 1998;117:407–412. doi: 10.1111/j.1439-0523.1998.tb01964.x. [DOI] [Google Scholar]
- Donini P, Law JR, Koebner RMD, Reeves J, Cooke RJ. Temporal trends in the diversity of UK wheat. Theor Appl Genet. 2000;100:912–917. doi: 10.1007/s001220051370. [DOI] [Google Scholar]
- Dreher KA, Callis J. Ubiquitin, hormones and biotic stress in plants. Ann Bot. 2007;9:787–822. doi: 10.1093/aob/mcl255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer EE, Ryan CA. Octadecanoid precursors of jasmonic acid activate the synthesis of wound-inducible proteinase inhibitors. Plant Cell. 1992;4:129–134. doi: 10.1105/tpc.4.2.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, Shi Y, Li M, Zhang M. Tandem PDZ repeats in glutamate receptor-interacting proteins have a novel mode of PDZ domain-mediated target binding. Nat Struct Biol. 2003;10:972–978. doi: 10.1038/nsb992. [DOI] [PubMed] [Google Scholar]
- Ferry N, Stavroulakis S, Guan W, Davison GM, Bell HA, Weaver RJ, Down RE, Gatehouse JA, Gatehouse AMR. Molecular interactions between wheat and cereal aphid (Sitobion avenae); analysis of changes to the wheat proteome. Proteomics. 2011;11:1985–2002. doi: 10.1002/pmic.200900801. [DOI] [PubMed] [Google Scholar]
- Filkowski J, Kovalchuk O, Kovalchuk I. Genome stability of vtc1, tt4, and tt5Arabidopsis thaliana mutants impaired in protection against oxidative stress. Plant J. 2004;38:60–69. doi: 10.1111/j.1365-313X.2004.02020.x. [DOI] [PubMed] [Google Scholar]
- Gatehouse JA. Plant resistance towards insect herbivores: a dynamic interaction. New Phytol. 2002;156:145–169. doi: 10.1046/j.1469-8137.2002.00519.x. [DOI] [PubMed] [Google Scholar]
- Gaupels F, Buhtz A, Knauer T, Deshmukh S, et al. Adaptation of aphid stylectomy for analyses of proteins and mRNAs in barley phloem sap. J Exp Bot. 2008;59(12):3297–3306. doi: 10.1093/jxb/ern181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geddes J, Eudes F, Laroche A, Selinger LB. Differential expression of proteins in response to the interaction between the pathogen Fusarium graminearum and its host. Hordeum Vulgare Proteomics. 2008;8:545–554. doi: 10.1002/pmic.200700115. [DOI] [PubMed] [Google Scholar]
- Haile FJ, Higley LG, Ni XZ, Quisenberry SS. Physiological and growth tolerance in wheat to Russian wheat aphid (Hopoptera: Aphididae) injury. Environ Entomol. 1999;28(5):787–794. doi: 10.1093/ee/28.5.787. [DOI] [Google Scholar]
- Havlícková H. Level and nature of the resistance to the cereal aphid, Sitobion avenae (F.), in thirteen winter wheat cultivars. J Agron Crop Sci. 1993;171:33–137. doi: 10.1111/j.1439-037X.1993.tb00122.x. [DOI] [Google Scholar]
- Jiang Q, Chen H, Pan X, Shi Y, et al. Proteomic analysis of wheat (Triticum aestivum L.) hybrid necrosis. Plant Sci. 2008;175:394–401. doi: 10.1016/j.plantsci.2008.05.017. [DOI] [Google Scholar]
- Klingler J, Creasy R, Gao L, Nair RM, Calix AS, Jacob HS, Singh KB. Aphid resistance in Medicago truncatula involves antixenosis and phloem-specific, inducible antibiosis, and maps to a single locus flanked by NBS–LRR resistance gene analogs. Plant Physiol. 2005;137(4):1445–1455. doi: 10.1104/pp.104.051243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koini MA, Alvey L, Allen T, Tilley CA, et al. High temperature-mediated adaptations in plant architecture require the phytochrome-interacting bHLH factor PIF4. Curr Biol. 2009;19:408–413. doi: 10.1016/j.cub.2009.01.046. [DOI] [PubMed] [Google Scholar]
- Koizumi S, Ohama N, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K. Functional analysis of the Hikeshi-like protein and its interaction with HSP70 in Arabidopsis. Biochem Biophys Res Commun. 2014;450:396–400. doi: 10.1016/j.bbrc.2014.05.128. [DOI] [PubMed] [Google Scholar]
- Leather SR, Dixon AFG. Aphid growth and reproductive rates. Ent Exp Appl. 1984;35:137–140. doi: 10.1111/j.1570-7458.1984.tb03373.x. [DOI] [Google Scholar]
- Li Q, Xie QG, Smith-Becker J, Navarre DA, et al. Mi-1-mediated aphid resistance involves salicylic acid and mitogen-activated protein kinase signalling cascades. Mol Plant Microbe Interact. 2006;19:655–664. doi: 10.1094/MPMI-19-0655. [DOI] [PubMed] [Google Scholar]
- Lucyshyn D, Wigge PA. Plant development: PIF4 integrates diverse environmental signals. Curr Biol. 2009;19:265–266. doi: 10.1016/j.cub.2009.01.051. [DOI] [PubMed] [Google Scholar]
- Ma Z-Q, Saidi A, Quick JS, Lapitan NLV. Genetic mapping of Russian wheat aphid resistance genes Dn2 and Dn4 in wheat. Genome. 1998;41(2):303–306. doi: 10.1139/gen-41-2-303. [DOI] [Google Scholar]
- McHale L, Tan X, Koehl P, Michelmore RW. Plant NBS–LRR proteins: adaptable guards. Genome Biol. 2006;7:212. doi: 10.1186/gb-2006-7-4-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migui SM (2002) Host relationships of three aphid species on wheat in the genus Triticum: potential for crop resistance in spring wheat. PhD thesis, University of Manitoba, Winnipeg, Canada
- Migui SM, Lamb RJ. Patterns of resistance to three cereal aphids among wheats in the genus Triticum(Poaceae) Bull Entomol Res. 2003;93(4):323–333. doi: 10.1079/BER2003246. [DOI] [PubMed] [Google Scholar]
- Migui SM, Lamb RJ. Seedling and adult plant resistance to Sitobion avenae (Hemiptera: Aphididae) in Triticum monococcum (Poaceae), an ancestor of wheat. Bull Entomol Res. 2004;94:35–46. doi: 10.1079/BER2003278. [DOI] [PubMed] [Google Scholar]
- Moloi MJ, van der Westhuizen AJ. The reactive oxygen species are involved in resistance responses of wheat to the Russian wheat aphid. J Plant Physiol. 2006;163(11):1118–1125. doi: 10.1016/j.jplph.2005.07.014. [DOI] [PubMed] [Google Scholar]
- Moloi MJ, van der Westhuizen AJ. Antioxidative enzymes and the Russian wheat aphid (Diuraphis noxia) resistance response in wheat (Triticum aestivum) Plant Biol. 2008;10(3):403–407. doi: 10.1111/j.1438-8677.2008.00042.x. [DOI] [PubMed] [Google Scholar]
- Monosi B, Wisser RJ, Pennill L, Hulbert SH. Full-genome analysis of resistance gene homologues in rice. Theor Appl Genet. 2004;109:1434–1447. doi: 10.1007/s00122-004-1758-x. [DOI] [PubMed] [Google Scholar]
- Moran PJ, Thompson GA. Molecular responses to aphid feeding in Arabidopsis in relation to plant defence pathways. Plant Physiol. 2001;125:1074–1085. doi: 10.1104/pp.125.2.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran P, Cheng Y, Cassell JL, Thompson GA. Gene expression profiling of Arabidopsis thaliana in compatible plant-aphid interactions. Arch Insect Biochem Phys. 2002;51:182–203. doi: 10.1002/arch.10064. [DOI] [PubMed] [Google Scholar]
- Ogbonnaya FC, Abdalla O, Mujeeb-Kazi A, Kazi AG, Xu SS, Gosman N, Lagudah ES, Bonnett D, Sorrells ME, Tsujimoto H (2013) Synthetic Hexaploids: Harnessing Species of the Primary Gene Pool for Wheat Improvement. In: Janick J (ed) Plant Breeding Reviews, vol 37. Wiley, New York, pp 35–122. doi:10.1002/9781118497869.ch2
- Pickart CM, Eddins MJ. Ubiquitin: structures, functions, mechanisms. Biochim Biophys Acta. 2004;695:55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- Rossignol M, Peltier J-B, Mock H-P, Matros A, et al. Plant proteome analysis: a 2004–2006 update. Proteomics. 2006;6(20):5529–5548. doi: 10.1002/pmic.200600260. [DOI] [PubMed] [Google Scholar]
- Sano N, Masaki S, Tananari T, Yamada T, Hirasawa T, Kanekatsu M. Proteomic analysis of stress-related proteins in rice seeds during the desiccation phase of grain filling. Plant Biotechnol. 2013;30:147–156. doi: 10.5511/plantbiotechnology.13.0207a. [DOI] [Google Scholar]
- Scranton MA, Yee A, Park SY, Walling LL. Plant leucine aminopeptidases moonlight as molecular chaperones to alleviate stress-induced damage. J Biol Chem. 2012;22:18408–18417. doi: 10.1074/jbc.M111.309500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CM. Plant resistance to arthropods—molecular and conventional approaches. Berlin: Springer; 2005. [Google Scholar]
- Smith CM, Boyko EV. The molecular bases of plant resistance and defence responses to aphid feeding: current status. Entomol Exp Appl. 2007;122:1–16. doi: 10.1111/j.1570-7458.2006.00503.x. [DOI] [Google Scholar]
- Smith CM, Liu X, Wang LJ, Liu X, Chen M-S, Starkey S, Bai J. Aphid feeding activates expression of transcriptome of oxylipin-based signals in wheat involved in resistance to herbivory. J Chem Ecol. 2010;36:260–276. doi: 10.1007/s10886-010-9756-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takken FLW, Joosten MHAJ. Plant resistance genes: their structure, function and evolution. Eur J Plant Pathol. 2000;106:699–713. doi: 10.1023/A:1026571130477. [DOI] [Google Scholar]
- Thompson GA, Goggin FL. Transcriptomics and functional genomics of plant defence induction by phloem-feeding insects. J Exp Bot. 2006;57:755–766. doi: 10.1093/jxb/erj135. [DOI] [PubMed] [Google Scholar]
- Tjallingii WF. Salivary secretions by aphids interacting with proteins of phloem wound responses. J Exp Bot. 2006;57(4):739–745. doi: 10.1093/jxb/erj088. [DOI] [PubMed] [Google Scholar]
- United Nations (2012) Searchable online statistical database from Food and Agriculture Division of the United Nations. FAOSTAT
- Vos P, Simons G, Jesse T, Wijbrandi J, Heinen L, Hogers R, Zabeau M. The tomato Mi-1 gene confers resistance to both root-knot nematodes and potato aphids. Nat Biotechnol. 1998;16(13):1365–1369. doi: 10.1038/4350. [DOI] [PubMed] [Google Scholar]
- Walling LL. The myriad plant responses to herbivores. J Plant Growth Regul. 2000;19:195–216. doi: 10.1007/s003440000026. [DOI] [PubMed] [Google Scholar]
- Wang W, Vinocur B, Shoseyov O, Altman A. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 2004;9:244–252. doi: 10.1016/j.tplants.2004.03.006. [DOI] [PubMed] [Google Scholar]
- Wang C-P, Wang Z-H, Zhao H-Y, Zhu Q-D, Luo K, Wang L-M, Dong P-H. Expression of potential resistance genes to the english grain aphid, Sitobion avenae, in wheat, Triticum aestivum. J Insect Sci. 2013;13:90. doi: 10.1673/031.013.9001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z, Hu W, Lin QS, Cheng XY, et al. Understanding rice plant resistance to the Brown Planthopper (Nilaparvata lugens): a proteomic approach. Proteomics. 2009;9:2798–2808. doi: 10.1002/pmic.200800840. [DOI] [PubMed] [Google Scholar]
- Yin H, Yan F, Ji J, Li Y, Wang R, Xu C. Proteomic analysis of Arabidopsis thaliana leaves infested by tobacco whitefly Bemisia tabaci (Gennadius) B biotype. Plant Mol Biol Rep. 2012;30:379–390. doi: 10.1007/s11105-011-0351-0. [DOI] [Google Scholar]
- Yuan H, Chen X, Zhu L, He G. Identification of genes responsive to brown planthopper Nilaparvata lugens Stål (Homoptera: Delphacidae) feeding in rice. Planta. 2005;221:105–112. doi: 10.1007/s00425-004-1422-3. [DOI] [PubMed] [Google Scholar]
- Zhang F, Zhu L, He G. Differential gene expression in response to brown planthopper feeding in rice. J Plant Physiol. 2004;161:53–62. doi: 10.1078/0176-1617-01179. [DOI] [PubMed] [Google Scholar]
- Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JD, Felix G, Boller T. Bacterial disease resistance in Arabidopsis through flagellin perception. Nature. 2004;428:764–767. doi: 10.1038/nature02485. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1 Leaf proteome of resistant wheat (R, ACC20 PGR1755) following 24-h aphid infestation showing the local response versus non-infested leaf proteome of same aged resistant wheat plants. Up-regulated protein spots of twofold change or greater are indicated by O; down-regulated protein spots of twofold change or greater are indicated by O (PPT 3092 kb)


