Abstract
Since its discovery 50 years ago, Epstein-Barr virus (EBV) has been linked to the development of cancers originating from both lymphoid and epithelial cells. Approximately 95% of the world's population sustains an asymptomatic, life-long infection with EBV. The virus persists in the memory B-cell pool of normal healthy individuals, and any disruption of this interaction results in virus-associated B-cell tumors. The association of EBV with epithelial cell tumors, specifically nasopharyngeal carcinoma (NPC) and EBV-positive gastric carcinoma (EBV-GC), is less clear and is currently thought to be caused by the aberrant establishment of virus latency in epithelial cells that display premalignant genetic changes. Although the precise role of EBV in the carcinogenic process is currently poorly understood, the presence of the virus in all tumor cells provides opportunities for developing novel therapeutic and diagnostic approaches. The study of EBV and its role in carcinomas continues to provide insight into the carcinogenic process that is relevant to a broader understanding of tumor pathogenesis and to the development of targeted cancer therapies.
Keywords: Nasopharyngeal carcinoma, EBV, latency, gastric carcinoma
Infection is involved in the development of an estimated 20% of human cancers—approximately 2 million cases per year[1]. Studies on the role of infection in cancer continue to reveal the underlying mechanisms that drive the oncogenic process and to highlight opportunities for therapeutic and prophylactic intervention. Epstein-Barr virus (EBV) was the first human tumor virus to be discovered and has yielded significant insight into both the pathogenesis of cancer and the natural history of persistent herpesvirus infection.
Infection with EBV is very common, affecting all human popu-lations[2],[3], and is largely asymptomatic and life-long. Early in the course of primary infection, EBV infects B lymphocytes, though it is unknown where B lymphocytes are infected and whether this infection also involves epithelial cells of the upper respiratory tract. To achieve long-term persistence in vivo, EBV colonizes the memory B-cell pool, where it establishes latent infection characterized by the expression of a limited subset of viral genes known as latent genes[4]. There are several well-described forms of EBV latency, each of which the virus uses at different stages of the viral life cycle and which are also reflected in the patterns of latency observed in various EBV-associated malignancies[2],[5]. Furthermore, during its life cycle, EBV must periodically replicate to generate infectious virus for transmission to other susceptible hosts, with epithelial sites in the oropharynx and salivary glands appearing to be the major sites for viral replication. Thus, the natural history of persistent EBV infection appears to resemble that of other herpesviruses in requiring distinct cell lineages to manifest the latent and replicative forms of the viral life cycle.
The dual tropism of EBV infection is also reflected in the types of tumor associated with the virus. EBV can colonize the memory B-cell pool in vivo and efficiently transform resting B cells into permanent, latently infected lymphoblastoid cell lines (LCLs) in vitro—characteristics mirrored in the various malignancies of B-cell origin that are closely associated with EBV infection[2],[3],[5]. However, it is the undifferentiated form of nasopharyngeal carcinoma (NPC) that shows the most consistent worldwide association with EBV. Furthermore, a subset of gastric adenocarcinomas and certain salivary gland carcinomas are also infected with EBV. Hence, epithelial infection with EBV can result in malignant transformation, suggesting that targeted approaches should be considered for preventive and therapeutic intervention.
Historical Background
In 1964, Tony Epstein and Yvonne Barr identified herpesvirus-type particles by electron microscopy in a subpopulation of Burkitt lymphoma (BL)-derived tumor cells in vitro. Only two years later, Old et al.[6] described antibodies in the serum of African patients with BL that recognized antigens prepared from BL cell lines. Surprisingly, similar BL antigen-specific antibodies were also present in serum from patients with carcinomas of the post-nasal space, and these antibodies were found in a high proportion of patients from both Africa and the US. This led the authors to suggest, “the high frequency of positive sera among patients with carcinoma of the post-nasal space indicates the desirability of searching for similar particles in cultures of this class of tumor also.”[6]. A more specific immunofluorescence assay for detecting antibodies against EBV-encoded replicative antigens was developed by de Schryver et al.[7], and this assay confirmed the association of elevated antibody titers against EBV-encoded viral capsid antigen (VCA) and membrane antigen (MA) with NPC. In 1970, zur Hausen et al.[8] showed that EBV DNA was present in extracts from NPC tumors using DNA hybridization technology. Further serological analysis showed an association between EBV antibody titers and NPC tumor stage[9] and revealed VCA-specific IgA levels as a potential prognostic indicator[10]. In 1973, EBV DNA was detected by in situ hybridization in NPC tumor cells but not in infiltrating lymphoid cells[11]. Based on the apparent specificity of the association between VCA IgA levels and NPC, Zeng et al.[12] embarked on a mass serological screening program in 1980 in Wuzhou City in China, where these EBV-specific antibodies were found to be useful for early detection of NPC and where subsequent prospective analysis revealed the presence of IgA VCA up to 41 months prior to the clinical diagnosis of NPC. Analysis of terminal repeats (TRs) in the EBV genome in NPCs by Southern blot hybridization demonstrated that resident viral genomes were monoclonal, providing important evidence that EBV infection occurred before the clonal expansion of the malignant cell population[13].
EBV Infection and NPC
The World Health Organization (WHO) has classified NPC into two main histological types—keratinizing squamous cell carcinoma (type I) and non-keratinizing squamous cell carcinoma (types II and III)—based on the tumor cells' appearance under the light microscope. The non-keratinizing type is further subdivided into differentiated non-keratinizing carcinoma (type II) and undifferentiated carcinoma (type III), and these type II and type III tumors are predominantly EBV-positive[14],[15]. Well-differentiated keratinizing NPC (type I) accounts for less than 20% of all NPC cases worldwide, and this tumor type is relatively rare in southern China[15],[16]. However, the association of EBV with the more differentiated, WHO type I form of NPC has been found particularly in the geographical regions with a high incidence of undifferentiated NPC[14]. In NPC, the virus exists in a latent state, exclusively in the tumor cells and absent from the surrounding lymphoid infiltrate. However, the interaction between the prominent lymphoid stroma found in undifferentiated NPC and adjacent carcinoma cells appears to be crucial for the continued growth of malignant NPC cells.
Carcinomas with similar features to undifferentiated NPC have been described at other sites including the thymus, tonsils, lungs, stomach, skin, or uterine cervix and are often referred to as undifferentiated carcinomas of nasopharyngeal type (UCNT) or lymphoepitheliomas. The morphological similarities of UCNTs to undifferentiated NPC prompted several groups to examine UCNTs for the presence EBV. UCNTs of the stomach are consistently EBV-positive[17], whereas the association of the other UCNTs with EBV is less strong. EBV was found in thymic epithelial tumors from Chinese but not Western patients[18]. Salivary gland UCNTs are associated with EBV in Greenland Eskimos and Chinese but not in Caucasian patients[19], and several case reports have demonstrated the absence of EBV from UCNTs arising in the uterine cervix and breast[20], [21].
EBV Latent Gene Expression in Virus-associated Tumors
Much of our understanding of the biology of EBV relates to its interaction with B lymphocytes. EBV can readily infect and transform normal resting B lymphocytes in vitro, a model system that has provided important insight into the virus' biology and behavior. The LCLs generated by in vitro infection of B lymphocytes carry multiple copies of circular, extrachromosomal viral DNA (episomes) in every cell and express a limited set of latent proteins, including 6 nuclear antigens (EBNAs 1, 2, 3A, 3B, 3C, and -LP) and 3 latent membrane proteins (LMPs 1, 2A, and 2B)[2],[3],[5]. In addition to the latent proteins, LCLs also show abundant expression of the small non-polyadenylated EBV-encoded small RNAs (EBER) 1 and EBER2; these are expressed in all forms of latent EBV infection and have served as excellent targets to detect EBV in tumors. Transcripts from the BamHI-A region (Bam A) of the viral genome, originally referred to as BamHI-A rightward transcripts (BARTs), are also detected in LCLs, and these, along with transcripts from the BamHI H rightward reading frame 1 (BHRF1) region, encode microRNAs (miRNAs). This pattern of latent EBV gene expression is referred to as the Latency III form of EBV infection and is characteristic in the majority of EBV-associated lymphomas arising in immunosuppressed patients.
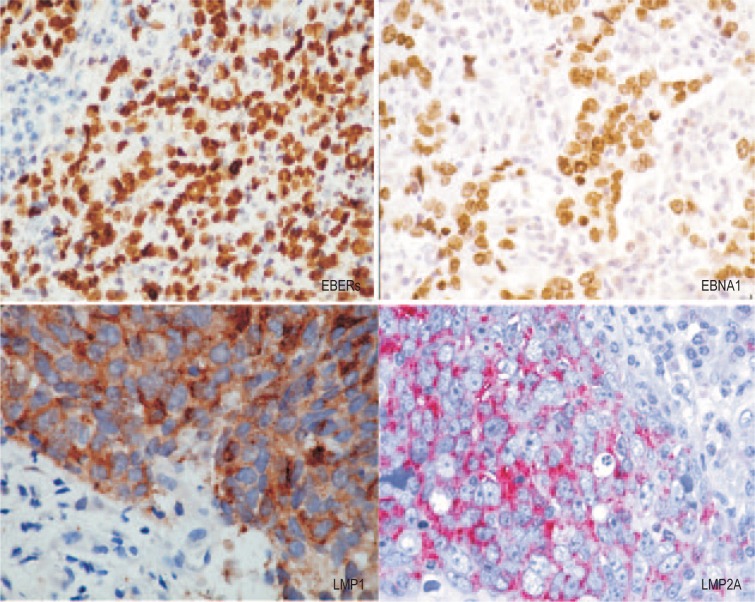
Studies examining EBV latent gene expression in virus-associated tumors and cell lines derived from BL biopsies revealed at least 2 additional forms of EBV latency. EBNA1 is the only EBV protein consistently observed in EBV-positive BL tumors, along with the EBER and BamHI-A transcripts; this form of latency is referred to Latency I[2],[3],[5]. A variant of Latency I, with expression of BHRF1 and EBNAs 3A, 3B, and 3C, is observed in approximately 5%–10% of EBV-positive BL tumors[22]. It appears that in these tumors, the selective pressure to down-regulate EBNA2 expression has occurred via deletion of the EBNA2 gene rather than through the switch in viral promoter usage observed in the conventional BL scenario. Another form of EBV latency, Latency II, was originally identified in biopsies of NPC and subsequently found in cases of EBV-associated Hodgkin lymphoma (HL) (Figure 1)[23]–[26]. Here, expression of the EBERs, EBNA1, and BamHI-A transcripts is accompanied by expression of LMP1 and LMP2A/B. While this Latency II pattern of EBV latent gene expression is a consistent feature of virus-associated HL, LMP1 expression in NPC is variable, with only approximately 20% of biopsies being unequivocally positive for LMP1 at the protein level[3]. The mechanisms underlying differential LMP1 expression in NPC and the consequent effects on the NPC phenotype remain unknown.
Figure 1. Epstein-Barr virus (EBV) latent gene expression in nasopharyngeal carcinoma (NPC).
In situ hydridization to the abundant EBV-encoded small RNAs (EBER) transcripts (left, upper panel) is the standard approach to detect EBV infection in cells and tissues. Immunohistochemical staining of NPC confirms EBNA1 expression in every tumor cell (right, upper panel). The expression of latent membrane protein 1 (LMP1) and LMP2A in NPC biopsies (lower panels) is more variable. The prominent lymphoid infiltrate in NPC is believed to contribute to the growth and survival of the tumor cells. (magnification, × 200)
These different forms of EBV latent infection are a reflection of the cellular environment and the complex interplay between host regulatory factors and viral promoters that drive EBV latent gene expression. In Latency III, the different EBNAs are encoded by individual mRNAs generated by differential splicing of the same long (over 100 kb), rightward primary transcript expressed from one of two promoters (Cp or Wp) located close together in the BamHI-C and BamHI-W regions of the genome[27]. A switch from Wp to Cp occurs early in B-cell infection as a consequence of the transactivating effects of both EBNA1 and EBNA2 on Cp. LMP transcripts are expressed from separate promoters in the BamHI-N region of the EBV genome, with the leftward LMP1 and rightward LMP2B mRNAs apparently controlled by the same bidirectional promoter sequence (ED-L1) that also responds to transactivation by EBNA2[2],[27]. The LMP2A promoter is also regulated by EBNA2. Both LMP2A and LMP2B transcripts cross the TRs into the U1 region, thus requiring the circularization of the genome for transcription. Circularization occurs by homologous recombination of the TRs, resulting in fused termini of unique length—a feature that has been used as a measure of EBV clonality, on the assumption that fused TRs with an identical number of repeats denote expansions of a single infected progenitor cell[13]. However, EBV clonality postinfection may be a consequence of the selective growth advantage achieved by optimal LMP2A expression over a minimal number of TRs[28]. In the more restricted forms of EBV latency observed in NPC and HL, EBNA1 transcription is driven from the TATA-less Qp promoter[29], and an alternative promoter, L1-TR, located in the terminal repeats is responsible for LMP1 expression[30]. These forms of EBV latency reflect the different transcription programs that EBV employs as the virus transits through the germinal center B-cell reaction to finally colonize the resting memory B-cell compartment[4]. The adoption of these forms of latency in EBV-associated tumors is therefore assumed to represent the manifestation of inappropriate viral transcription programs as a consequence of both the host cell environment and the selection pressures associated with oncogenic progression.
The Function of EBV Latent Genes Expressed in NPC
Understanding the function of the EBV latent genes expressed in NPC is clearly essential in determining the role of viral infection in the carcinogenic process. Furthermore, a detailed appreciation of the function of these EBV latent genes could provide opportunities to develop novel diagnostic and therapeutic approaches.
EBNA1 and the LMPs
While the EBNA1 protein has an essential role in replication and segregation of the EBV episome, it has recently been found to protect cells from apoptosis, enhance cell survival, and directly contribute to the tumorigenic phenotype[31]. These effects manifest through a variety of mechanisms including destabilization of p53, disruption of promyelocytic leukaemia (PML) nuclear bodies, and modulation of various signaling pathways.
LMP1 is the major transforming protein of EBV, behaving as a classical oncogene in rodent fibroblast transformation assays and being essential for EBV-induced B-cell transformation in vitro[32]. LMP1 has pleiotropic effects when expressed in cells, resulting in induction of cell surface adhesion molecules and activation antigens, up-regulation of antiapoptotic proteins (Bcl-2, A20), and stimulation of cytokine production [interleukin (IL)-6, IL-8][33]. LMP1 functions as a constitutively activated member of the tumor necrosis factor receptor (TNFR) superfamily, activating a number of signaling pathways (NF-κB, ERK-MAPK, JNK/AP1, PI3K) in a ligand-independent manner[34]. The expression of LMP1 in epithelial cells is associated with a range of phenotypic effects including hyperproliferation, induction of proinflammatory cytokines, resistance to apoptosis, and enhanced motility[33]. The role of LMP1 in the pathogenesis of NPC remains speculative. There is considerable variation in the reported expression of LMP1 in NPC biopsies and a general consensus that approximately 20%–40% of tumors express LMP1 at the protein level[3]. In one study, all 6 early, preinvasive NPC (NPC in situ) lesions analyzed expressed the LMP1 protein, arguing for a crticial role of LMP1 in the early pathogenesis of NPC[35]. The possibility of geographical variation in LMP1 expression in NPC has been suggested and is supported by studies in North Africa, where LMP1 is more prevalent in the juvenile form of NPC[36].
The two proteins encoded by the LMP2 gene, LMP2A and LMP2B, share 12 hydrophobic membrane-spanning domains and a short cytoplasmic C-terminus, but it is the unique immunoreceptor tyrosine-based activation motif (ITAM) within the cytoplasmic N-terminal domain of LMP2A that seems responsible for this protein's functional effects[37]. LMP2A appears to drive B-cell proliferation and survival in the absence of signaling through the B-cell receptor (BCR), an effect that may be relevant to the ability of EBV to colonize the memory B-cell pool[38],[39]. LMP2A is required for the successful outgrowth of EBV-infected epithelial cells in vitro[28] and can induce anchorage-independent growth, enhance cell adhesion and cell motility, and inhibit epithelial cell differentiation[40]–[44]. Many of these effects are a consequence of LMP2A's ability to activate the PI3K/Akt and β-catenin pathways. LMP2A can also induce the epithelial-to-mesenchymal transition (EMT), which is associated with the acquisition of stem cell–like properties[45]. LMP2A and LMP2B play a role in resistance to the antiviral effects of interferons α and β in epithelial cells[46], and LMP2A modulates NF-κB and STAT signaling in EBV-infected epithelial cells through effects on IL-6 transcription and secretion[47].
Unlike LMP1, LMP2A expression is more consistent in NPC. Studies using reverse transcription-polymerase chain reaction (RT-PCR) confirmed expression of LMP2A mRNA in more than 98% of NPC cases, whereas expression of LMP2B was lower and mirrored LMP1[23],[48]. Immunohistochemical staining has confirmed expression of LMP2A protein in more than 50% of NPC cases[45],[49]. Interestingly, this consistent expression of LMP2 in NPC is reflected in serological responses such that the majority of NPC patients have detectable IgG responses to LMP2A/2B[50].
EBERs and the BamHI-A region
Two small non-polyadenylated (non-coding) RNAs, EBERs 1 and 2, are highly expressed in all forms of EBV latency and serve as sensitive targets for detecting EBV infection in cells and tissues[2],[3],[5]. The EBERs assemble into stable ribonucleoprotein particles with the auto-antigen La and ribosomal protein L22 and bind the interferon-inducible, double-stranded RNA-activated protein kinase protein kinase RNA-activated (PKR)[51]. The interaction of EBERs with retinoic acid–inducible gene 1 (RIG-1) induces type I interferon production, an effect that may be counteracted by other viral genes such as LMP1 and LMP2A/B[52]. EBERs are also reported to induce insulin-like growth factor 1 (IGF-1) in NPC cell lines[53]. This effect is associated with enhanced growth of NPC cell lines and its relevance to NPC is supported by the frequent expression of IGF-1 in tumor biopsies.
A group of abundantly expressed RNAs encoded by the BamHI-A region of the EBV genome were originally identified in NPC but were subsequently found to be expressed in other EBV-associated malignancies and in the peripheral blood of healthy individuals[2],[3],[5],[54],[55]. These highly spliced transcripts, commonly referred to as the BARTs (BamHI-A rightward transcripts), are a cluster of 22 miRNAs that generate 44 mature miRNAs[56]. These miRNAs target and thereby regulate a variety of viral and cellular transcripts[55], [57]. Thus, specific BART miRNAs have been found to protect against apoptosis and contribute to immune evasion. The BART miRNAs also appear crucial in the regulation of EBV gene expression, targeting both latent (LMP1, LMP2A) and lytic genes (BALF5). Because of their stability in serum, miRNAs have been recognized as potential cancer biomarkers, and recent studies support the possible use of serum BART miRNAs in the detection and prognosis of NPC[58], [59]. It appears that the BART miRNAs in serum reside in cell-derived exosomes that are able to deliver miRNAs to other non-EBV infected cells[60], [61]. This raises the intriguing possibility that BART miRNAs may contribute to the development of NPC by influencing the behavior of stromal and immune cells.
Another transcript generated from the BamHI-A region is BARF1, which encodes a 31-kDa protein that was originally identified as an early antigen expressed upon induction of the EBV lytic cycle. However, BARF1 is a secreted protein expressed during latency in EBV-associated NPC and gastric carcinoma (GC)[62],[63]. BARF1 shares limited homology with the human colony stimulating factor 1 (CSF-1) receptor (c-fms oncogene) and competes for its natural ligand, macrophage colony-stimulating factor (M-CSF or CSF-1), thereby modulating immune cell growth and function[64]. BARF1 displays oncogenic activity when expressed in rodent fibroblasts and simian primary epithelial cells[65]. Engineered expression of BARF1 in the context of a recombinant EBV enhanced the growth and tumorigenicity of virus-negative NPC cell lines, implicating BARF1 in the pathogenesis of NPC[66]. In addition, serum BARF1 may be a useful diagnostic marker for NPC[67].
EBV Strain Variation
EBV isolates from different regions of the world or from patients with different virus-associated diseases are remarkably similar when their genomes are compared with restriction fragment length polymorphism analysis[3],[5],[68]. However, variations in repeat regions of the EBV genome are observed among different EBV isolates. Analysis of the EBV genome in a number of BL cell lines revealed gross deletions, some of which account for biological differences. More specifically, P3HR-1 virus, which is non-transforming, has a deletion of the EBNA2-encoding gene[3], [5], [68]. Strain variation over the EBNA2-encoding (BamHI-WYH) region of the EBV genome permits all virus isolates to be classified as either type 1 (EBV-1, B95.8-like) or type 2 (EBV-2, Jijoye-like)[68]. This genomic variation results in the production of two antigenically distinct forms of the EBNA2 protein, which share only 50% amino acid homology. Similar allelic polymorphisms (with 50%–80% sequence homology, depending on the locus) related to the EBV type occur in a subset of latent genes, namely those encoding EBNA-LP, EBNA3A, EBNA3B, and EBNA3C[69]. These differences have functional consequences as EBV-2 isolates are less efficient in B-lymphocyte transformation assays in vitro compared with EBV-1 isolates[70]. A combination of virus isolation and sero-epidemiological studies suggests that type 1 virus isolates are predominant (but not exclusively so) in many Western countries, whereas both types are widespread in equatorial Africa, New Guinea, and perhaps certain other regions[68], [71].
In addition to this broad distinction between EBV types 1 and 2, there is also minor heterogeneity within each virus type. Individual strains have been identified on the basis of changes compared with B95.8, ranging from single base mutations to extensive deletions[68]. While infection with multiple strains of EBV was originally thought to be confined to immunologically compromised patients, more recent studies demonstrate that normal, healthy patients who are seropositive can be infected with multiple EBV isolates that vary in abundance and presence over time[72]. Co-infection of the host with multiple virus strains could have evolutionary benefit to EBV, creating diversity through genetic recombination. Such intertypic recombination has been reported in HIV-infected patients and in the Chinese population, and appears to arise via recombination of multiple EBV strains during the intense EBV replication that results from immunosuppression[73].
The possible contribution of EBV strain variation to virus-associated tumors remains contentious. Many studies have failed to establish an epidemiological association between EBV strains and disease and suggest that the specific EBV gene polymorphisms detected in virus-associated tumors occur with similar frequencies in EBV isolates from healthy virus carriers from the same geographic region[3],[71]. However, these studies focused on specific regions of the EBV genome rather than on comparing the entire viral DNA sequence. More recent work has used next-generation sequencing (NGS) to analyze EBV isolates from NPC biopsies. These studies have confirmed that, while there is a high level of overall similarity among the NPC-derived virus strains with the prototypical EBV genome, variation exists in viral genes that might result in functional differences[74],[75]. In this regard, an LMP1 variant containing a 10 amino acid deletion (residues 343 to 352) was originally identified in Chinese NPC biopsies and has oncogenic and other functional properties distinct from those of the B95.8 LMP1 gene[76]–[78]. It is therefore likely that variation in LMP1 and other EBV genes can contribute to the risk of developing virus-associated tumors. However, more biological studies using well-defined EBV variants are required, along with more detailed NGS comparisons of tumor-derived EBV strains versus those derived from healthy donors.
NPC Pathogenesis
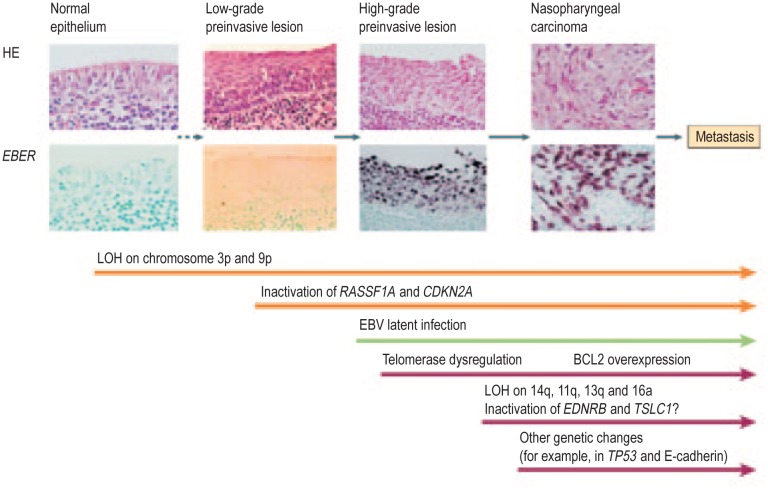
The presence of monoclonal EBV episomes in NPC indicates that viral infection precedes the clonal expansion of malignant cells[13]. However, epithelial infection may not be the initiating event in virus-associated carcinogenesis, as tonsils from patients with infectious mononucleosis (IM) and normal nasopharyngeal biopsies from individuals at high risk of developing NPC lack evidence of epithelial EBV infection[2]. EBV infection as detected by in situ hybridization to the abundantly expressed non-polyadenylated EBER RNAs has been demonstrated in high-grade (severe dysplastic and carcinoma in situ), preinvasive lesions in the nasopharynx, but not in low-grade disease or histologically normal nasopharyngeal epithelium[35],[79]. Both high-grade and carcinoma in situ lesions carry monoclonal EBV genomes[35]. Multiple genetic changes have been found in NPC, with frequent deletion of regions on chromosomes 3p, 9p, 11q, 13q, and 14q and promoter hypermethylation of specific genes on chromosomes 3p (RASSF1A, RARβ2) and 9p (p16, p15, p14, DAP-kinase)[80],[81]. Both 3p and 9p deletions have been identified in the absence of EBV infection in low-grade dysplastic lesions and in normal nasopharyngeal epithelium from individuals at high risk of developing NPC, suggesting that these genetic events occur early in NPC pathogenesis and may predispose to subsequent EBV infection[79], [82]. This possibility is supported by in vitro data demonstrating that stable EBV infection of epithelial cells requires an altered, undifferentiated cellular environment[83] and that cyclin D1 overexpression (a consequence of p16 deletion on chromosome 9p and amplification of the cyclin D1 locus on chromosome 11q) facilitates persistent EBV infection of immortalized nasopharyngeal epithelial cells[84]. Thus, a scheme has been proposed whereby loss of heterozygosity occurs early in NPC pathogenesis because of exposure to environmental cofactors such as dietary components (i.e., salted fish), creating low-grade, preinvasive lesions that become susceptible to EBV infection after additional genetic and epigenetic events (Figure 2). Once infected, EBV latent genes provide growth and survival benefits, resulting in the development of NPC. Additional genetic and epigenetic changes occur after EBV infection.
Figure 2. Schematic representation of NPC pathogenesis.
This model proposes that loss of heterozygosity (LOH) occurs early in NPC pathogenesis, possibly as a result of exposure to environmental cofactors such as dietary components (e.g., salted fish). This results in low-grade, preinvasive lesions that, after additional genetic and epigenetic events, become susceptible to stable EBV infection. Once cells have become infected, EBV latent genes provide growth and survival benefits, resulting in the development of NPC. Additional genetic and epigenetic changes occur after EBV infection.
EBV and GC
GC remains the second leading cause of death from cancer. Although incidence and mortality have been falling for over 50 years, the number of deaths from this tumor will inevitably continue to increase globally because of population growth and aging in high-risk countries. As approximately 10% of GC comprise cells latently infected with EBV, EBV-associated GC (EBV-GC) may represent the most common form of EBV-associated malignancy[85], [86]. These EBV-GC tumors display a restricted pattern of EBV latent gene expression (EBERs, EBNA1, LMP2A, the BART miRNAs, BARF1), similar to NPC[63],[87]. The association of EBV and GC varies significantly with geography, which may be attributed to ethnic and genetic differences[85],[86]. EBV-GCs have distinct phenotypic and clinical characteristics compared to EBV-negative GC tumors, including loss of p16 expression, promoter methylation of p73, expression of wild-type p53, distinct patterns of allelic loss, and improved overall survival time[88]–[91]. Comprehensive genomic and molecular characterization of GC has confirmed that EBV-GC is a distinct entity with a high CpG island methylator (CIMP) phenotype and apparent driver mutations in the PIK3CA and ARID1A genes[92], [93].
As in NPC, the precise role of EBV in the pathogenesis of GC remains to be determined, but the absence of EBV infection in premalignant gastric lesions supports the contention that viral infection is a relatively late event in gastric carcinogenesis[94]. Epidemiological studies suggest that EBV-GC is related to birth order, high salt intake, and exposure to metal dust, but these factors may vary geographically (e.g., between Japan and Colombia), supporting the need for more detailed investigation[85], [86]. Distinct EBV strains are suggested to play a role in EBV-GC development, though this could reflect geographical variation in the prevalence of EBV strains in the population[95].
Conclusions
EBV was discovered over 50 years ago, and its DNA was fully sequenced in 1984.
The widespread nature of EBV infection testifies to the intimate interaction between the virus and the immune system that is characterised by an asymptomatic, life-long infection resident within the memory B-cell compartment. Perturbing this interaction, as occurs in various forms of immunosuppression, results in EBV-associated B-cell tumors. As for the association of EBV with NPC and EBV-GC, the contribution of the virus is less clear but appears to be a consequence of the aberrant establishment of virus latency in epithelial cells that have already undergone premalignant genetic changes. Whatever the nature of these interactions and the precise role of EBV in the carcinogenic process, there is clearly the opportunity to exploit this association for the clinical benefit of patients. Novel therapeutic approaches using virus reactivation[96], gene therapy[97], or therapeutic vaccination[98] augur well for our ability to effectively target EBV-associated carcinomas, and mass screening programs to identify patients with early-stage NPC are already underway in Hong Kong[99]. These studies are paradigms for the development of targeted cancer therapies and diagnostics and further confirm the far-reaching value of tumor virology to the whole cancer field.
Acknowledgments
The authors' studies are supported by Cancer Research UK and the European Commission's FP6 Life-Sciences-Health Programme (INCA Project: LSHC-CT-2005-018704). We thank Mu-Sheng Zeng for helpful comments and for providing the EBER in situ hybridisation panel in Figure 1.
References
- 1.de Martel C, Ferlay J, Franceschi S, et al. Global burden of cancers attributable to infections in 2008: a review and synthetic analysis. Lancet Oncol. 2012;13:607–615. doi: 10.1016/S1470-2045(12)70137-7. [DOI] [PubMed] [Google Scholar]
- 2.Young LS, Rickinson AB. Epstein-Barr virus: 40 years on. Nat Rev Cancer. 2004;4:757–768. doi: 10.1038/nrc1452. [DOI] [PubMed] [Google Scholar]
- 3.Tao Q, Young LS, Woodman CB, et al. Epstein-Barr virus (EBV) and its associated human cancers—genetics, epigenetics, pathobiology and novel therapeutics. Front Biosci. 2006;11:2672–2713. doi: 10.2741/2000. [DOI] [PubMed] [Google Scholar]
- 4.Thorley-Lawson DA. Epstein-Barr virus: exploiting the immune system. Nat Rev Immunol. 2001;1:75–82. doi: 10.1038/35095584. [DOI] [PubMed] [Google Scholar]
- 5.Rickinson AB, Kieff E. Epstein-Barr Virus. In: Knipe DM, Howley PM, editors. Fields Virology. Philadelphia: Lippincott Williams and Wilkins; 2001. pp. 2575–2627. [Google Scholar]
- 6.Old LJ, Boyse EA, Oettgen E, et al. Precipitating antibody in human serum to an antigen prersent in cultured Burkitt's lymphoma cells. Proc Natl Acad Sci U S A. 1966;56:1699–1704. doi: 10.1073/pnas.56.6.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Schryver A, Friberg S, Klein G, et al. Epstein-Barr virus-associated antibody patterns in carcinoma of the post-nasal space. Clin Exp Immunol. 1969;5:443–459. [PMC free article] [PubMed] [Google Scholar]
- 8.zur Hausen H, Schulte-Holthausen H, Klein G, et al. EBV DNA in biopses of Burkitt tumours and anaplastic carcinomas of the nasopharynx. Nature. 1970;228:1056–1058. doi: 10.1038/2281056a0. [DOI] [PubMed] [Google Scholar]
- 9.Henle W, Henle G, Ho HC, et al. Antibodies to Epstein-Barr virus in nasopharyngeal carcinoma, other head and neck neoplasms, and control groups. J Natl Cancer Inst. 1970;44:225–231. [PubMed] [Google Scholar]
- 10.Henle W, Ho JH, Henle G, et al. Nasopharyngeal carcinoma: significance of changes in Epstein-Barr virus-related antibody patterns following therapy. Int J Cancer. 1977;20:663–672. doi: 10.1002/ijc.2910200504. [DOI] [PubMed] [Google Scholar]
- 11.Wolf H, zur Hausen H, Becker V. EB-viral genomes in epitelial nasopharyngeal carcinoma cells. Nature New Biol. 1973;244:245–247. doi: 10.1038/newbio244245a0. [DOI] [PubMed] [Google Scholar]
- 12.Zeng Y, Zhang LG, Wu YC, et al. Prospective studies on nasopharyngeal carcinoma in Epstein-Brr vírus IgA/VCA antibody-positive persons in Wuzhou City, China. Int J Cancer. 1985;36:545–547. doi: 10.1002/ijc.2910360505. [DOI] [PubMed] [Google Scholar]
- 13.Raab-Traub N, Flynn K. The structure of the termini of the Epstein-Barr virus as a marker of clonal cellular proliferation. Cell. 1986;47:883–889. doi: 10.1016/0092-8674(86)90803-2. [DOI] [PubMed] [Google Scholar]
- 14.Pathmanathan R, Prasad U, Chandrika G, et al. Undifferentiated, nonkeratinizing, and squamous cell carcinoma of the nasopharynx. Variants of Epstein-Barr virus neoplasia. Am J Pathol. 1995;146:1355–1367. [PMC free article] [PubMed] [Google Scholar]
- 15.Wei K, Xu Y, Liu J, et al. Histopathological classification of nasopharyngeal carcinoma. Asian Pacific J Cancer Prev. 2011;12:1141–1147. [PubMed] [Google Scholar]
- 16.Ji X, Zhang W, Xie C, et al. Nasopharyngeal carcinoma risk by histologic type in central China: impact of smoking, alcohol and family history. Int J Cancer. 2011;129:724–732. doi: 10.1002/ijc.25696. [DOI] [PubMed] [Google Scholar]
- 17.Shibata D, Tokunaga M, Uemura Y, et al. Association of Epstein-Barr virus with undifferentiated gastric carcinomas with intense lymphoid infiltration. Lymphoepithelioma-like carcinoma. Am J Pathol. 1991;139:469–474. [PMC free article] [PubMed] [Google Scholar]
- 18.Fujii T, Kawai T, Saito K, et al. EBER-1 expression in thymic carcinoma. Acta Pathol Jpn. 1993;43:107–110. doi: 10.1111/j.1440-1827.1993.tb01118.x. [DOI] [PubMed] [Google Scholar]
- 19.Raab-Traub N, Rajadurai P, Flynn K, et al. Epstein-Barr virus infection in carcinoma of the salivary gland. J Virol. 1991;65:7032–7036. doi: 10.1128/jvi.65.12.7032-7036.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weinberg E, Hoisington S, Eastman AY, et al. Uterine cervical lymphoepithelial-like carcinoma. Absence of Epstein-Barr virus genomes. Am J Clin Pathol. 1993;99:195–199. doi: 10.1093/ajcp/99.2.195. [DOI] [PubMed] [Google Scholar]
- 21.Dadmanesh F, Peterse JL, Sapino A, et al. Lymphoepithelioma-like carcinoma of the breast: lack of evidence of Epstein-Barr virus infection. Histopathology. 2001;38:54–61. doi: 10.1046/j.1365-2559.2001.01055.x. [DOI] [PubMed] [Google Scholar]
- 22.Kelly G, Bell A, Rickinson AB. Epstein-Barr virus-associated Burkitt lymphomagenesis selects for downregulation of the nuclear antigen EBNA1. Nat Med. 2002;8:1098–1104. doi: 10.1038/nm758. [DOI] [PubMed] [Google Scholar]
- 23.Brooks L, Yao QY, Rickinson AB, et al. Epstein-Barr virus latent gene transcription in nasopharyngeal carcinoma cells: coexpression of EBNA1, LMP1, and LMP2 transcripts. J Virol. 1992;66:2689–2697. doi: 10.1128/jvi.66.5.2689-2697.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deacon EM, Pallesen G, Niedobitek G, et al. Epstein-Barr virus and Hodgkin's disease: transcriptional analysis of virus latency in the malignant cells. J Exp Med. 1993;177:339–349. doi: 10.1084/jem.177.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pallesen G, Hamilton-Dutoit SJ, Rowe M, et al. Expression of Epstein-Barr virus latent gene products in tumour cells of Hodgkin's disease. Lancet. 1991;337:320–322. doi: 10.1016/0140-6736(91)90943-j. [DOI] [PubMed] [Google Scholar]
- 26.Young LS, Dawson CW, Clark D, et al. Epstein-Barr virus gene expression in nasopharyngeal carcinoma. J Gen Virol. 1988;69:1051–1065. doi: 10.1099/0022-1317-69-5-1051. [DOI] [PubMed] [Google Scholar]
- 27.Speck SH, Strominger JL. Transcription of Epstein-Barr virus in latently infected, growth-transformed lymphocytes. In: Klein G, editor. Advances in viral oncology. New York: Raven Press; 1989. pp. 133–150. [Google Scholar]
- 28.Moody CA, Scott RS, Su T, et al. Length of Epstein-Barr virus termini as a determinant of epithelial cell clonal emergence. J Virol. 2003;77:8555–8561. doi: 10.1128/JVI.77.15.8555-8561.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tempera I, Klichinsky M, Lieberman PM. EBV latency types adopt alternative chromatin conformations. PLoS Pathog. 2011;7:e1002180. doi: 10.1371/journal.ppat.1002180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen H, Lee JM, Zong Y, et al. Linkage between STAT regulation and Epstein-Barr virus gene expression. J Virol. 2001;75:2929–2937. doi: 10.1128/JVI.75.6.2929-2937.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frappier L. Role of EBNA1 in NPC tumourigenesis. Sem Cancer Biol. 2012;22:154–161. doi: 10.1016/j.semcancer.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 32.Wang D, Liebowitz D, Kieff E. An EBV membrane protein expressed in immortalised lymphocytes transforms established rodent cells. Cell. 1985;43:831–840. doi: 10.1016/0092-8674(85)90256-9. [DOI] [PubMed] [Google Scholar]
- 33.Dawson CW, Port RJ, Young LS. The role of EBV-encoded latent membrane proteins LMP1 and LMP2 in the pathogenesis of nasopharyngeal carcinoma. Sem Cancer Biol. 2012;22:144–153. doi: 10.1016/j.semcancer.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 34.Mosialos G, Birkenbach M, Yalamanchili R, et al. The Epstein-Barr virus transforming protein LMP1 engages signaling proteins for the tumor necrosis factor receptor family. Cell. 1995;80:389–399. doi: 10.1016/0092-8674(95)90489-1. [DOI] [PubMed] [Google Scholar]
- 35.Pathmanathan R, Prasad U, Sadler R, et al. Clonal proliferations of cells infected with Epstein-Barr virus in preinvasive lesions related to nasopharyngeal carcinoma. N Engl J Med. 1995;333:693–698. doi: 10.1056/NEJM199509143331103. [DOI] [PubMed] [Google Scholar]
- 36.Khabir A, Karray H, Rodriguez S, et al. EBV latent membrane protein 1 abundance correlates with patient age but not with metastatic behavior in North African nasopharyngeal carcinomas. Virol J. 2005;2:39. doi: 10.1186/1743-422X-2-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fruehling S, Longnecker R. The immunoreceptor tyrosine-based activation motif of Epstein-Barr virus LMPA2 is essential for blocking BCR-mediated signal transduction. J Virol. 1997;235:241–251. doi: 10.1006/viro.1997.8690. [DOI] [PubMed] [Google Scholar]
- 38.Miller CL, Burkhardt AL, Lee JH, et al. Integral membrane protein 2 of Epstein-Barr virus regulates reactivation from latency through dominant negative effects on protein-tyrosine kinases. Immunity. 1995;2:155–166. doi: 10.1016/s1074-7613(95)80040-9. [DOI] [PubMed] [Google Scholar]
- 39.Caldwell RG, Wilson JB, Anderson SJ, et al. Epstein-Barr virus LMP2A drives B-cell development and survival in the absence of normal B cell receptor signals. Immunity. 1998:405–411. doi: 10.1016/s1074-7613(00)80623-8. [DOI] [PubMed] [Google Scholar]
- 40.Scholle F, Bendt KM, Raab-Traub N. Epstein-Barr virus LMP2A transforms epithelial cells, inhibits cell differentiation, and activates Akt. J Virol. 2000;74:10681–10689. doi: 10.1128/jvi.74.22.10681-10689.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morrison JA, Raab-Traub N. Roles of the ITAM and PY motifs of Epstein-Barr virus latent membrane protein 2A in the inhibition of epithelial cell differentiation and activation of beta-catenin signaling. J Virol. 2005;79:2375–2382. doi: 10.1128/JVI.79.4.2375-2382.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fukuda M, Longnecker R. Epstein-Barr virus latent membrane protein 2A mediates transformation through constitutive activation of the Ras/PI3-K/Akt pathway. J Virol. 2007;81:9299–9306. doi: 10.1128/JVI.00537-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Allen MD, Young LS, Dawson CW. The Epstein-Barr virus-encoded LMP2A and LMP2B proteins promote epithelial cell spreading and motility. J Virol. 2005;79:1789–1802. doi: 10.1128/JVI.79.3.1789-1802.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu J, Lin WH, Chen SY, et al. Syk tyrosine kinase mediates Epstein-Barr virus latent membrane protein 2A-induced cell migration in epithelial cells. J Biol Chem. 2006;281:8806–8814. doi: 10.1074/jbc.M507305200. [DOI] [PubMed] [Google Scholar]
- 45.Kong QL, Hu LJ, Cao JY, et al. Epstein-Barr virus-encoded LMP2A induces an epithelial-mesenchymal transition and increases the number of side population stem-like cancer cells in nasopharyngeal carcinoma. PLoS Pathog. 2010;6:e1000940. doi: 10.1371/journal.ppat.1000940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shah KM, Stewart SE, Wei W, et al. The EBV-encoded latent membrane proteins, LMP2A and LMP2B, limit the actions of interferon by targeting interferon receptors for degradation. Oncogene. 2009;28:3903–3914. doi: 10.1038/onc.2009.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stewart S, Dawson CW, Takada K, et al. Epstein-Barr virus-encoded LMP2A regulates viral and cellular gene expression by modulation of the NF-kappaB transcription factor pathway. Proc Natl Acad Sci U S A. 2004;101:15730–15735. doi: 10.1073/pnas.0402135101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Busson P, McCoy R, Sadler R, et al. Consistent transcription of the Epstein-Barr virus LMP2 gene in nasopharyngeal carcinoma. J Virol. 1992;66:3257–3262. doi: 10.1128/jvi.66.5.3257-3262.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heussinger N, Büttner M, Ott G, et al. Expression of the Epstein-Barr virus (EBV)-encoded latent membrane protein 2A (LMP2A) in EBV-associated nasopharyngeal carcinoma. J Pathol. 2004;203:696–699. doi: 10.1002/path.1569. [DOI] [PubMed] [Google Scholar]
- 50.Paramita DK, Fatmawati C, Juwana H, et al. Humoral immune responses to Epstein-Barr virus encoded tumor associated proteins and their putative extracellular domains in nasopharyngeal carcinoma patients and regional controls. J Med Virol. 2011;4:665–678. doi: 10.1002/jmv.21960. [DOI] [PubMed] [Google Scholar]
- 51.Takada K. Role of EBER and BARF1 in nasopharyngeal carcinoma (NPC) tumorigenesis. Sem Cancer Biol. 2012;22:162–165. doi: 10.1016/j.semcancer.2011.12.007. [DOI] [PubMed] [Google Scholar]
- 52.Samanta M, Iwakiri D, Kanda T, et al. EB virus-encoded RNAs are recognised by RIG-1 and activate signalling to induce type I IFN. EMBO J. 2006;25:4207–4214. doi: 10.1038/sj.emboj.7601314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Iwakiri D, Sheen TS, Chen JY, et al. Epstein-Barr virus-encoded small RNA induces insulin-like growth factor 1 and supports growth of nasopharyngeal carcinoma-derived cell lines. Oncogene. 2005;24:1767–1773. doi: 10.1038/sj.onc.1208357. [DOI] [PubMed] [Google Scholar]
- 54.Hitt MM, Allday MJ, Hara T, et al. EBV gene expression in an NPC-related tumour. EMBO J. 1989;8:2639–2651. doi: 10.1002/j.1460-2075.1989.tb08404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marquitz AR, Raab-Traub N. The role of miRNAs and EBV BARTS in NPC. Sem Cancer Biol. 2012;22:166–172. doi: 10.1016/j.semcancer.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pfeffer S, Zavolan M, Grasser FA, et al. Identification of virus-encoded microRNAs. Science. 2004;304:734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- 57.Lo AKF, Dawson CW, Jin DY, et al. The pathological roles of BART miRNAs in nasopharyngeal carcinoma. J Pathol. 2012;227:392–403. doi: 10.1002/path.4025. [DOI] [PubMed] [Google Scholar]
- 58.Wong AM, Kong KL, Tsang JW, et al. Profiling of Epstein-Barr virus-encoded microRNAs in nasopharyngeal carcinoma reveals potential biomarkers and oncomirs. Cancer. 2012;118:698–710. doi: 10.1002/cncr.26309. [DOI] [PubMed] [Google Scholar]
- 59.Chan JY, Gao W, Ho WK, et al. Overexpression of Epstein-Barr virus-encoded microRNA-BART7 in undifferentiated naso-pharyngeal carcinoma. Anticancer Res. 2012;32:3201–3210. [PubMed] [Google Scholar]
- 60.Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, et al. Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci U S A. 2010;107:6328–6333. doi: 10.1073/pnas.0914843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meckes DG, Shair KHY, Marquitz AR, et al. Human tumor virus utilizes exosomes for intercellular communication. Proc Natl Acad Sci U S A. 2010;107:20370–20375. doi: 10.1073/pnas.1014194107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Decaussin G, Sbih-Lammali F, de Turenne-Tessier M, et al. Expression of BARF1 gene encoded by Epstein-Barr virus in naso-pharyngeal carcinoma biopsies. Cancer Res. 2000;60:5584–5588. [PubMed] [Google Scholar]
- 63.zur Hausen A, Brink AA, Craanen ME, et al. Unique transcription pattern of Epstein-Barr virus (EBV) in EBV-carrying gastric adeno-carcinomas: expression of the transforming BARF1 gene. Cancer Res. 2000;60:2745–2748. [PubMed] [Google Scholar]
- 64.Shim AHR, Chang RA, Chen X, et al. Multipronged attenuation of macrophage-colony stimulating factor signaling by Epstein-Barr virus BARF1. Proc Natl Acad Sci U S A. 2012;109:12962–12967. doi: 10.1073/pnas.1205309109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sheng W, Decaussin G, Sumner S, et al. N-terminal domain of BARF1 gene encoded by Epstein-Barr virus is essential for malignant transformation of rodent fibroblasts and activation of BCL-2. Oncogene. 2001;20:1176–1185. doi: 10.1038/sj.onc.1204217. [DOI] [PubMed] [Google Scholar]
- 66.Seto E, Ooka T, Middeldorp J, et al. Reconstruction of naso-pharyngeal carcinoma-type infection induced tumorigenicity. Cancer Res. 2008;68:1030–1036. doi: 10.1158/0008-5472.CAN-07-5252. [DOI] [PubMed] [Google Scholar]
- 67.Houali K, Wang X, Shimizu Y, et al. A new diagnostic marker for secreted Epstein-Barr virus-encoded LMP1 and BARF1 oncoproteins in the serum and saliva of patients with naso-pharyngeal carcinoma. Clin Cancer Res. 2007;13:4493–5000. doi: 10.1158/1078-0432.CCR-06-2945. [DOI] [PubMed] [Google Scholar]
- 68.Tzellos S, Farrell PJ. Epstein-Barr virus sequence variation—biology and disease. Pathogens. 2012;1:156–74. doi: 10.3390/pathogens1020156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sample J, Young L, Martin B, et al. Epstein-Barr virus types 1 and 2 differ in their EBNA-3A, EBNA-3B, and EBNA-3C genes. J Virol. 1990;64:4084–4092. doi: 10.1128/jvi.64.9.4084-4092.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rickinson AB, Young LS. Rowe M. Influence of the Epstein-Barr vírus nuclear antigen EBNA2 on the growth phenotype of virus-transformed B cells. J Virol. 1987;61:1310–1317. doi: 10.1128/jvi.61.5.1310-1317.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Khanim F, Yao QY, Niedobitek G, et al. Analysis of Epstein-Barr virus gene polymorphisms in normal donors and in virus-asssociated tumors from different geographic locations. Blood. 1996;88:3491–3501. [PubMed] [Google Scholar]
- 72.Tierney RJ, Edwards RH, Sitki-Green D, et al. Multiple Epstein-Barr virus strains in patients with infectious mononucleosis: comparison of ex vivo samples with in vitro isolates by use of heteroduplex tracking assays. J Infect Dis. 2006;193:287–297. doi: 10.1086/498913. [DOI] [PubMed] [Google Scholar]
- 73.Midgley RS, Blake NW, Yao QY, et al. Novel intertypic recombinants of Epstein-Barr virus in the Chinese population. J Virol. 2000;74:1544–1548. doi: 10.1128/jvi.74.3.1544-1548.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu P, Fang X, Feng Z, et al. Direct sequencing and characterization of a clinical isolate of Epstein-Barr virus from nasopharyngeal carcinoma tissue by using next-generation sequencing technology. J Virol. 2011;85:11291–11299. doi: 10.1128/JVI.00823-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kwok H, Wu CW, Palser AL, et al. Genomic diversity of Epstein-Barr virus genomes isolated from primary nasopharyngeal carcinoma biopsy samples. J Virol. 2014;88:10662–10672. doi: 10.1128/JVI.01665-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Edwards RH, Seillier-Moiseiwitsch F, Raab-Traub N. Signature amino acid changes in latent membrane protein 1 distinguish Epstein-Barr virus strains. Virology. 1999;261:79–95. doi: 10.1006/viro.1999.9855. [DOI] [PubMed] [Google Scholar]
- 77.Dawson CW, Eliopoulos AG, Blake SM, et al. Identification of functional differences between prototype Epstein-Barr virus-encoded LMP1 and a nasopharyngeal carcinoma-derived LMP1 in human epithelial cells. Virology. 2000;272:204–217. doi: 10.1006/viro.2000.0344. [DOI] [PubMed] [Google Scholar]
- 78.Fielding CA, Sandvej K, Mehl AM, et al. Epstein-Barr virus LMP-1 natural sequence variants differ in their potential to activate cellular signaling pathways. J Virol. 2001;75:9129–9141. doi: 10.1128/JVI.75.19.9129-9141.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chan ASC, To KF, Lo KW, et al. High frequency of chromosome 3p deletion in histologically normal nasopharyngeal epithelia from Southern Chinese. Cancer Res. 2000;60:5365–5370. [PubMed] [Google Scholar]
- 80.Lo KW, Chung GTY, To KF. Deciphering the molecular genetic basis of NPC through molecular, cytogenetic, and epigenetic approaches. Sem Cancer Biol. 2012;22:79–86. doi: 10.1016/j.semcancer.2011.12.011. [DOI] [PubMed] [Google Scholar]
- 81.Lung HL, Cheung AKL, Ko JMY, et al. Deciphering the molecular genetic basis of NPC through functional approaches. Sem Cancer Biol. 2012;22:87–95. doi: 10.1016/j.semcancer.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 82.Chan ASC, To KF, Lo KW, et al. Frequent chromosome 9p losses in histologically normal nasopharyngeal epithelia from Southern Chinese. Int J Cancer. 2002;102:300–303. doi: 10.1002/ijc.10689. [DOI] [PubMed] [Google Scholar]
- 83.Knox PG, Li QX, Rickinson AB, Young LS. In vitro production of stable Epstein-Barr virus-positive epithelial cell clones which resemble the virus: cell interaction observed in nasopharyngeal carcinoma. Virology. 1996;215:40–50. doi: 10.1006/viro.1996.0005. [DOI] [PubMed] [Google Scholar]
- 84.Tsang CM, Yip YL, Lo KW, et al. Cyclin D1 overexpression supports stable EBV infection in nasopharyngeal epithelial cells. Proc Natl Acad Sci U S A. 2012;109:E3473–E3482. doi: 10.1073/pnas.1202637109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lee JH, Kim SH, Han SH, et al. Clinicopathological and molecular characteristics of Epstein-Barr virus-associated gastric carcinoma: a meta-analysis. J Gastroent Hepatol. 2009;24:354–365. doi: 10.1111/j.1440-1746.2009.05775.x. [DOI] [PubMed] [Google Scholar]
- 86.Murphy G, Pfeiffer R, Camargo MC, et al. Meta-analysis show that prevalence of Epstein-Barr virus-positive gastric cancer differs based on sex and anatomic location. Gastroenterology. 2009;137:824–833. doi: 10.1053/j.gastro.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Imai S, Koizumi S, Sugiura M, et al. Gastric carcinoma: monoclonal epithelial malignant cells expressing Epstein-Barr virus latent infection protein. Proc Natl Acad Sci U S A. 1994;91:9131–9135. doi: 10.1073/pnas.91.19.9131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lee HS, Chang MS, Yang HK, et al. Epstein-Barr virus-positive gastric carcinoma has a distinct protein expression profile in comparison with Epstein-Barr virus-negative carcinoma. Clin Cancer Res. 2004;10:1698–1705. doi: 10.1158/1078-0432.ccr-1122-3. [DOI] [PubMed] [Google Scholar]
- 89.Schneider BG, Gulley ML, Eagan P, et al. Loss of p16/CDKN2A tumour suppressor protein in gastric adenocarcinoma is associated with Epstein-Barr virus and anatomic location in the body of the stomach. Hum Pathol. 2000;31:45–50. doi: 10.1016/s0046-8177(00)80197-5. [DOI] [PubMed] [Google Scholar]
- 90.Ushiku T, Chong JM, Uozaki H, et al. p73 gene promoter methylation in Epstein-Barr virus-associated gastric carcinoma. Int J Cancer. 2006;120:60–66. doi: 10.1002/ijc.22275. [DOI] [PubMed] [Google Scholar]
- 91.van Rees BP, Caspers E, zur Hausen A, et al. Different pattern of allelic loss in Epstein-Barr virus-positive gastric cancer with emphasis on the p53 tumour suppressor pathway. Am J Pathol. 2002;161:1207–1213. doi: 10.1016/S0002-9440(10)64397-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang K, Yuen ST, Lee SP, et al. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat Genet. 2014;46:573–582. doi: 10.1038/ng.2983. [DOI] [PubMed] [Google Scholar]
- 93.Cancer Genome Atlas Research Network Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.zur Hausen A, van Rees BP, van Beek J, et al. Epstein-Barr virus in gastric carcinomas and gastric stump carcinomas: a late event in gastric carcinogenesis. J Clin Pathol. 2004;57:487–491. doi: 10.1136/jcp.2003.014068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Corvalan A, Ding S, Koriyama C, et al. Association of a distinctive strain of Epstein-Barr virus with gastric cancer. Int J Cancer. 2006;118:1736–1742. doi: 10.1002/ijc.21530. [DOI] [PubMed] [Google Scholar]
- 96.Wildeman MA, Novalic Z, Verkuijlen SA, et al. Cytolytic virus activation therapy for Epstein-Barr virus-driven tumors. Clin Cancer Res. 2012;18:5061–5070. doi: 10.1158/1078-0432.CCR-12-0574. [DOI] [PubMed] [Google Scholar]
- 97.Li JH, Shi W, Chia M, et al. Efficacy of targeted FasL in nasopharyngeal carcinoma. Mol Ther. 2003;8:964–973. doi: 10.1016/j.ymthe.2003.08.018. [DOI] [PubMed] [Google Scholar]
- 98.Taylor GS, Jia H, Harrington K, et al. A recombinant modified vaccinia ankara vaccine encoding Epstein-Barr virus (EBV) target antigens: a phase I trial in UK patients with EBV-positive cancer. Clin Cancer Res. 2014;20:5009–5022. doi: 10.1158/1078-0432.CCR-14-1122-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chan KC, Hung EC, Woo JK, et al. Early detection of naso-pharyngeal carcinoma by plasma Epstein-Barr virus DNA analysis in a surveillance program. Cancer. 2013;119:1838–1844. doi: 10.1002/cncr.28001. [DOI] [PubMed] [Google Scholar]