Abstract
Epstein-Barr virus (EBV) is a well-known human herpesvirus associated with virtually all nasopharyngeal carcinoma (NPC) and ∼10% of gastric cancer (GC) worldwide. Increasing evidence shows that acquired genetic and epigenetic alterations lead to the initiation and progression of NPC and GC. However, even deep whole exome sequencing studies showed a relatively low frequency of gene mutations in NPC and EBV-associated GC (EBVaGC), suggesting a predominant role of epigenetic abnormities, especially promoter CpG methylation, in the pathogenesis of NPC and EBVaGC. High frequencies of promoter methylation of tumor suppressor genes (TSGs) have been frequently reported in NPC and EBVaGC, with several EBV-induced methylated TSGs identified. Further characterization of the epigenomes (genome-wide CpG methylation profile—methylome) of NPC and EBVaGC shows that these EBV-associated tumors display a unique high CpG methylation epigenotype with more extensive gene methylation accumulation, indicating that EBV acts as a direct epigenetic driver for these cancers. Mechanistically, oncogenic modulation of cellular CpG methylation machinery, such as DNA methyltransferases (DNMTs), by EBV-encoded viral proteins accounts for the EBV-induced high CpG methylation epigenotype in NPC and EBVaGC. Thus, uncovering the EBV-associated unique epigenotype of NPC and EBVaGC would provide new insight into the molecular pathogenesis of these unique EBV-associated tumors and further help to develop pharmacologic strategies targeting cellular methylation machinery in these malignancies.
Keywords: Epstein-Barr virus, CpG methylation, DNA methyltransferase, tumor suppressor gene, naso-pharyngeal carcinoma, gastric cancer
Epstein-Barr virus (EBV) is a human herpesvirus with life-long asymptomatic infection in >95% of the world's population. Importantly, it is involved in the pathogenesis of multiple malignancies of either epithelial or lymphoid origin, including nasopharyngeal carcinoma (NPC), gastric cancer (GC), Burkitt's lymphoma, Hodgkin lymphoma, natural killer (NK)-cell/T-cell lymphoma, and certain B-cell and T-cell lymphomas[1]–[4]. NPC is nearly 100% associated with EBV infection, and it is a major and prevalent tumor in South China (including Hong Kong, Guangdong, Guangxi, and Hunan) and Southeast Asia, with a reported annual incidence of ∼30 cases/100,000[5],[6]. Although GC is also a major tumor worldwide, EBV-associated GC (EBVaGC) comprises only ∼10% of all GC cases with random distribution worldwide, with ∼90,000 new cases per year[7],[8]. EBV infects B cells and epithelial cells mainly in a latent form with spontaneous lytic activation, whereas the viral infection in EBV-associated tumors including NPC and EBVaGC is exclusively latent with the expression of only a few latent genes[2].
Cancer is a disease driven by accumulated genetic and epigenetic alterations[9],[10]. In recent years, increasing evidence has shown that multiple epigenetic alterations, as a hallmark of cancers, also contribute critically to cancer initiation and progression with the same influence as genetic abnormalities[11]. Epigenetic regulation, including promoter CpG methylation, histone modifications, and chromatin remodeling, is the fundamental regulatory mechanism for gene expression modulation. Abnormal epigenetic regulation inactivates tumor suppressor genes (TSGs) and reactivates oncogenes, leading to aberrant gene expression and functions that initiate or promote tumorigenesis[10]. Importantly, epigenetic changes such as promoter CpG methylation can occur in the early stage of tumor pathogenesis[12], indicating an initiator-like function during tumorigenesis[13]. This has become an exciting new direction for cancer research.
EBV infection induces increased genome-wide gene methylation, resulting in the formation of a unique epigenotype with cellular high CpG methylation in tumor cells[14]. EBV-positive carcinomas have been shown to possess a unique epigenetic phenotype compared to EBV-negative ones, with significantly higher frequencies of TSG methylation[15]–[17]. EBV appears to act as a direct epigenetic driver for the pathogenesis of its associated tumors. This review aims to provide an overview of the high methylation phenotype induced by EBV in malignant epithelial cells and the aberrant TSG methylation caused by EBV infection through modulating the cellular CpG methylation machinery.
The Unique Epigenotype of EBV-infected Carcinomas
The common feature of EBV-associated NPC and EBVaGC is that both tumors originate from the monoclonal proliferation of EBV-infected epithelial cells, indicating that EBV is most likely an oncogenic driver for the pathogenesis of both tumors[5],[18]. While only rare insertion events of EBV DNA into the host genome have been reported, recent deep whole-genome studies incorporating a whole exome sequencing approach also revealed infrequent genetic mutations in NPC and EBVaGC compared with other common cancers and their EBV-negative counterparts[19]–[21], suggesting a predominant role of EBV infection in driving the epigenetic deregulation of NPC and EBVaGC tumor cells.
A methylome study (CpG methylation profiling) of multiple GC samples using Illumina HumanMethylation27 BeadChip (analyzing 27,000 CpG sites per genome) uncovered the methylome of this tumor, albeit at low resolution, and revealed a high methylation subtype in EBV-positive GCs[15]. Another genome and methylome study of GCs was conducted on 295 gastric adenocarcinoma and 27 adjacent non-malignant tissue samples, using a combination of HumanMethylation27 (HM27) and HumanMethylation450 (analyzing 485,000 CpG sites per genome) BeadChips[20]. Similarly, a high methylation/EBV-positive epigenotype was identified in all EBVaGC tissues examined. A third study that also used the HumanMethylation450 platform analyzed 98 GCs and 31 paired normal samples[19]. In this study, high frequencies of genome-wide methylation were also observed in EBVaGC, consistent with the previous two reports. Through these genome-wide and other candidate gene-based studies, a series of frequently methylated genes have been identified in EBVaGC as well as EBV-negative GC but with much less frequent methylation of genes, such as runt-related transcription factor 3 (RUNX3), p73, p16, death-associated protein kinase 1 (DAPK1), phosphatase and tensin homolog (PTEN), Ras association domain family member 1 (RASSF1A), and glutathione S-transferase pi 1 (GSTP1)[22]–[25].
Unfortunately, the epigenome study of NPC lags significantly behind that of GC, although nearly all NPC cases are associated with latent EBV infection; hence, NPC is hypothesized to display a unique high methylation/EBV-positive epigenotype. We have analyzed the methylomes of NPC cell lines and tumor samples using methylated DNA immunoprecipitation coupled with promoter microarray hybridization (MeDIP-chip). We observed extensive, genome-wide methylation of cellular genes in NPC (Li et al., Epigenomics 2014, in press). Through epigenome and candidate gene-based studies, a series of functional TSGs frequently silenced by promoter methylation have been identified in NPC[16], including RAS protein activator like 1 (RASAL1)[26], RASSF1[27], zinc finger, MYND-type containing 10 (ZMYND10/BLU)[28], protocadherin 10 (PCDH10) [29], and zinc finger protein 382 (ZNF382)[30].
We also profiled the methylomes of EBV-infected immortalized normal nasopharyngeal epithelial cells and discovered a series of methylated genes that are candidate target genes of EBV-induced epigenetic alterations involved in early NPC pathogenesis. More functional and mechanistic studies of these critical genes are currently ongoing (Li et al., unpublished). A very recent study using reduced representation bisulfite sequencing (RRBS) profiled the methylome changes of EBV-infected telomerase-immortalized oral keratinocytes, and the results identified a high methylation/EBV-positive epigenotype in the infected clones[31]. Thus, EBV does act as a direct epigenetic driver in normal and malignant epithelial cells.
Mechanisms of EBV-induced High CpG Methylation Epigenotype
EBV-infected cells are characterized by different patterns of latent gene expression. There are three different forms of EBV latency (I, II, and III), each with a distinct pattern of gene expression[32]. However, these different types of latency are controlled through the epigenetic regulation of various EBV promoters[33]. NPC with type II latency only expresses EBV-encoded small RNA (EBER), EBV-associated nuclear antigen 1 (EBNA1), BamHI-A rightward transcripts (BARTs), latent membrane protein 1 and 2 (LMP1, LMP2A, and LMP2B), and BamHI- A rightward open-reading frame-1 (BARF1)[34]. EBVaGC shares the same type I latency as Burkitt's lymphoma, with the expression of EBER, EBNA1, BARTs, LMP2A, and BARF1[35]–[37]. However, LMP1 is often not expressed or only expressed at a very low level in EBVaGC[38]. Functional studies show that these viral genes are involved in the oncogenic modulation of host gene expression including components of the cellular CpG methylation machinery[17]. EBV also expresses a large number of microRNAs (miRNAs)[39],[40]; however, the exact biological functions of these complex miRNAs in the EBV life cycle or the pathogenesis of EBV-associated tumors is still essentially unknown.
DNMTs are the key components of cellular CpG methylation machinery, including mainly DNMT1, DNMT3A, and DNMT3B, which are responsible for methylation maintenance and alteration in human cells. DNMT1 is a maintenance methyltransferase, whereas DNMT3A and DNMT3B are essential for de novo DNA methylation. In addition, a series of histone modifiers and chromatin remodelers can also modulate the activity of cellular CpG methylation machinery[41]. Polycomb group (PcG) proteins, as epigenetic regulators of tran-scription through the formation of polycomb repressive complexes containing BMI1 polycomb ring finger proto-oncogene (BMI1) or enhancer of zeste 2 polycomb repressive complex 2 subunit (EZH2), also modulate histone modification, chromatin structure, and CpG methylation levels[42],[43].
LMP1 and LMP2A are well-documented oncogenic EBV proteins that play critical roles in the tumor transformation of epithelial and lymphoid cells. LMP1 can activate multiple cellular signaling pathways, including nuclear factor of kappa light polypeptide gene enhancer in B cells (NF-κB), Janus kinase/signal transducers and activators of transcription 3 (JAK/STAT3), c-Jun N-terminal kinase and activator protein 1 (JNK/AP-1), and phosphatidylinositol 3-kinase (PI3K)/AKT signaling. LMP1 protein, via its carboxy terminal activating region-2—the last three amino acids (CTAR2-YYD) domain, can up-regulate the transcripts of DNMT1, DNMT3a, and DNMT3b through the activation of JNK signaling[44],[45]. LMP1 also promotes DNMTs to form transcriptional complexes with methyl CpG-binding protein 2 (MeCP2) and histone deacetylase 1 (HDAC1) on the E-cadherin promoter, whereas a JNK inhibitor prevents this complex formation[44],[45]. Activated DNMT1 then methylates and represses cellular promoters such as E-cadherin and docking protein 1 (DOK1) in LMP1-expressing cells[44]–[46]. DNMT enzyme activity is also elevated by 2-3 folds in LMP1-expressing epithelial cells[44].
LMP2A also activates multiple cellular signaling pathways, including JAK/STAT3 and PI3K/AKT signaling, which further regulates DNMTs and other epigenetic modifiers during NPC and EBVaGC pathogenesis. LMP2A could up-regulate DNMT1, DNMT3b, and BMI1 expression at the transcriptional and protein levels[47],[48]. LMP2A up-regulates DNMT1 expression by inducing STAT3 phosphorylation independent of interleukin-6 (IL-6) stimulation, which further causes PTEN methylation and silencing in EBVaGC[47]. A significant correlation between DNMT1 and STAT3 phosphorylation was revealed by immunochemistry in EBVaGC.
EBNA1 as a viral nuclear protein is consistently expressed in all EBV-associated tumors. EBNA1 binds to the latent origin of EBV replication (OriP), which is essential for EBV genome replication and maintenance during its latency[49],[50]. EBNA1 is a DNA-binding protein localized at cellular chromatin via its chromosome-binding domains[51]. Chromatin immunoprecipitation sequencing (ChIP-Seq) studies have uncovered the genome-wide binding profile of EBNA1 to its target genes including modulators of cellular methylation machinery such as histone deacetylase 3 (HDAC3), indicating that EBNA1 can directly interfere with the CpG methylation machinery[52],[53].
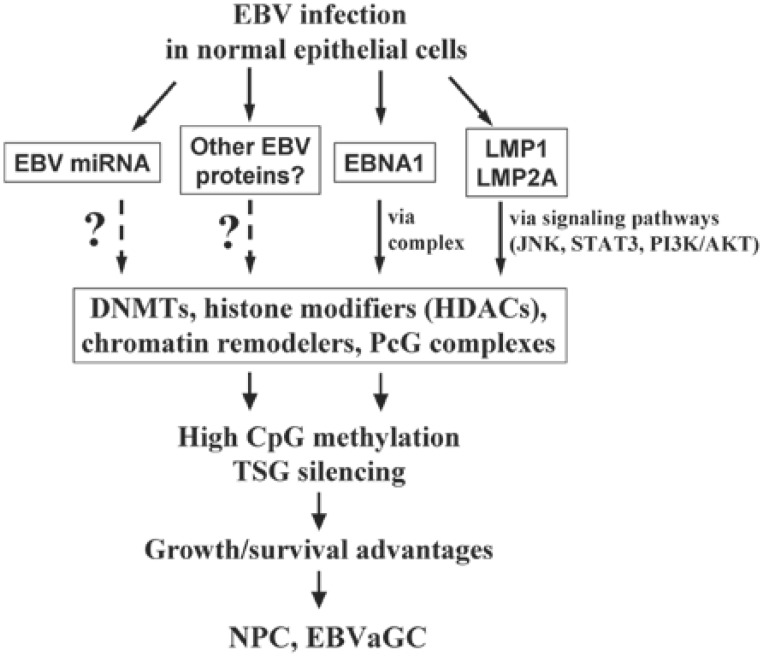
Thus, EBV-encoded proteins can regulate multiple components of the cellular CpG methylation machinery, including DNMTs, histone modifiers, chromatin remodelers, and PcG complexes (Figure 1), and they can further regulate aberrant TSG methylation and silencing in EBV-infected tumor cells. It is possible that other EBV proteins and miRNAs have similar functions, which requires further in-depth investigation. NPC and EBVaGC are thus typical tumor models of an EBV-induced aberrant high-methylation epigenotype. This unique biological feature has specific therapeutic implications for the clinical treatment of these tumors, as multiple epigenetic agents, such as DNMT inhibitors and HDAC inhibitors, are currently being developed, with some already being approved by the U.S. Food & Drug Administration (FDA) in clinical trials[54],[55]. It is thus predicted that further epigenetic therapy would greatly improve the modern treatment of these patients with EBV-positive tumor[56],[57].
Figure 1. Model of oncogenic modulation of cellular CpG methylation machinery by Epstein-Barr virus (EBV) and EBV-induced high methylation epigenotype in malignant epithelial cells.
EBV-encoded proteins regulate multiple components of the cellular methylation machinery through either cellular signaling pathways or transcription complexes, which finally leads to tumor suppressor gene (TSG) methylation and silencing and contributes to nasopharyngeal carcinoma (NPC) and EBV-associated gastric cancer (EBVaGC) pathogenesis. EBNA1, EBV-associated nuclear antigen 1; LMP, latent membrane protein; JNK, c-Jun N-terminal kinase; STAT3, signal transducers and activators of transcription 3; PI3K/AKT, phosphatidylinositol 3-kinase (PI3K)/AKT; DNMT, DNA methyltransferase; HDAC, histone deacetylase; PcG, poly-comb group; ?, unknown mechanism.
Acknowledgments
Supported by grants from Health and Medical Research Fund (HMRF) (No. 13120082), Hong Kong Research Grants Council (RGC) (No. 474710 and T12-401/13R), National Natural Science Foundation of China (NSFC) (No. 81372898 and 81172582), and The Chinese University of Hong Kong.
Reference
- 1.Young LS, Rickinson AB. Epstein-Barr virus: 40 years on. Nat Rev Cancer. 2004;4:757–768. doi: 10.1038/nrc1452. [DOI] [PubMed] [Google Scholar]
- 2.Kieff E. Current perspectives on the molecular pathogenesis of virus-induced cancers in human immunodeficiency virus infection and acquired immunodeficiency syndrome. J Natl Cancer Inst Monogr. 1998;23:7–14. doi: 10.1093/oxfordjournals.jncimonographs.a024177. [DOI] [PubMed] [Google Scholar]
- 3.Raab-Traub N. Epstein-Barr virus in the pathogenesis of NPC. Semin Cancer Biol. 2002;12:431–441. doi: 10.1016/s1044579x0200086x. [DOI] [PubMed] [Google Scholar]
- 4.Tao Q, Young LS, Woodman CB, et al. Epstein-Barr virus (EBV) and its associated human cancers—genetics, epigenetics, pathobiology and novel therapeutics. Front Biosci. 2006;11:2672–2713. doi: 10.2741/2000. [DOI] [PubMed] [Google Scholar]
- 5.Tao Q, Chan AT. Nasopharyngeal carcinoma: molecular pathogenesis and therapeutic developments. Expert Rev Mol Med. 2007;9:1–24. doi: 10.1017/S1462399407000312. [DOI] [PubMed] [Google Scholar]
- 6.Lo KW, To KF, Huang DP. Focus on nasopharyngeal carcinoma. Cancer Cell. 2004;5:423–428. doi: 10.1016/s1535-6108(04)00119-9. [DOI] [PubMed] [Google Scholar]
- 7.Parkin DM, Bray F, Ferlay J, et al. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 8.Osato T, Imai S. Epstein-Barr virus and gastric carcinoma. Semin Cancer Biol. 1996;7:175–182. doi: 10.1006/scbi.1996.0024. [DOI] [PubMed] [Google Scholar]
- 9.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 11.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 12.Baylin SB, Ohm JE. Epigenetic gene silencing in cancer—a mechanism for early oncogenic pathway addiction? Nat Rev Cancer. 2006;6:107–116. doi: 10.1038/nrc1799. [DOI] [PubMed] [Google Scholar]
- 13.Yu DH, Waterland RA, Zhang P, et al. Targeted p16ink4a epimutation causes tumorigenesis and reduces survival in mice. J Clin Invest. 2014;124:3708–3712. doi: 10.1172/JCI76507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaneda A, Matsusaka K, Aburatani H, et al. Epstein-Barr virus infection as an epigenetic driver of tumorigenesis. Cancer Res. 2012;72:3445–3450. doi: 10.1158/0008-5472.CAN-11-3919. [DOI] [PubMed] [Google Scholar]
- 15.Matsusaka K, Kaneda A, Nagae G, et al. Classification of Epstein-Barr virus-positive gastric cancers by definition of DNA methylation epigenotypes. Cancer Res. 2011;71:7187–7197. doi: 10.1158/0008-5472.CAN-11-1349. [DOI] [PubMed] [Google Scholar]
- 16.Li LL, Shu XS, Wang ZH, et al. Epigenetic disruption of cell signaling in nasopharyngeal carcinoma. Chin J Cancer. 2011;30:231–239. doi: 10.5732/cjc.011.10080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li HP, Leu YW, Chang YS. Epigenetic changes in virus-associated human cancers. Cell Res. 2005;15:262–271. doi: 10.1038/sj.cr.7290295. [DOI] [PubMed] [Google Scholar]
- 18.Raab-Traub N, Flynn K. The structure of the termini of the Epstein-Barr virus as a marker of clonal cellular proliferation. Cell. 1986;47:883–889. doi: 10.1016/0092-8674(86)90803-2. [DOI] [PubMed] [Google Scholar]
- 19.Wang K, Yuen ST, Xu J, et al. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat Genet. 2014;46:573–582. doi: 10.1038/ng.2983. [DOI] [PubMed] [Google Scholar]
- 20.The Cancer Genome Atlas Research Network Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin DC, Meng X, Hazawa M, et al. The genomic landscape of nasopharyngeal carcinoma. Nat Genet. 2014;46:866–871. doi: 10.1038/ng.3006. [DOI] [PubMed] [Google Scholar]
- 22.Kang GH, Lee S, Kim WH, et al. Epstein-Barr virus-positive gastric carcinoma demonstrates frequent aberrant methylation of multiple genes and constitutes CpG island methylator phenotype-positive gastric carcinoma. Am J Pathol. 2002;160:787–794. doi: 10.1016/S0002-9440(10)64901-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fukayama M, Hino R, Uozaki H. Epstein-Barr virus and gastric carcinoma: virus-host interactions leading to carcinoma. Cancer Sci. 2008;99:1726–1733. doi: 10.1111/j.1349-7006.2008.00888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang MS, Uozaki H, Chong JM, et al. CpG island methylation status in gastric carcinoma with and without infection of Epstein-Barr virus. Clin Cancer Res. 2006;12:2995–3002. doi: 10.1158/1078-0432.CCR-05-1601. [DOI] [PubMed] [Google Scholar]
- 25.Zhao J, Liang Q, Cheung KF, et al. Genome-wide identification of Epstein-Barr virus-driven promoter methylation profiles of human genes in gastric cancer cells. Cancer. 2013;119:304–312. doi: 10.1002/cncr.27724. [DOI] [PubMed] [Google Scholar]
- 26.Jin H, Wang X, Ying J, et al. Epigenetic silencing of a Ca(2+)-regulated Ras GTPase-activating protein RASAL defines a new mechanism of Ras activation in human cancers. Proc Natl Acad Sci U S A. 2007;104:12353–12358. doi: 10.1073/pnas.0700153104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lo KW, Kwong J, Hui AB, et al. High frequency of promoter hypermethylation of RASSF1A in nasopharyngeal carcinoma. Cancer Res. 2001;61:3877–3881. [PubMed] [Google Scholar]
- 28.Qiu GH, Tan LK, Loh KS, et al. The candidate tumor suppressor gene BLU, located at the commonly deleted region 3p21.3, is an E2F-regulated, stress-responsive gene and inactivated by both epigenetic and genetic mechanisms in nasopharyngeal carcinoma. Oncogene. 2004;23:4793–4806. doi: 10.1038/sj.onc.1207632. [DOI] [PubMed] [Google Scholar]
- 29.Ying J, Li H, Seng TJ, et al. Functional epigenetics identifies a protocadherin PCDH10 as a candidate tumor suppressor for nasopharyngeal, esophageal and multiple other carcinomas with frequent methylation. Oncogene. 2006;25:1070–1080. doi: 10.1038/sj.onc.1209154. [DOI] [PubMed] [Google Scholar]
- 30.Cheng Y, Geng H, Cheng SH, et al. KRAB zinc finger protein ZNF382 is a proapoptotic tumor suppressor that represses multiple oncogenes and is commonly silenced in multiple carcinomas. Cancer Res. 2010;70:6516–6526. doi: 10.1158/0008-5472.CAN-09-4566. [DOI] [PubMed] [Google Scholar]
- 31.Birdwell CE, Queen KJ, Kilgore P, et al. Genome-wide DNA methylation as an epigenetic consequence of Epstein-Barr virus infection of immortalized keratinocytes. J Virol. 2014;88:11442–11458. doi: 10.1128/JVI.00972-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Young LS, Murray PG. Epstein-Barr virus and oncogenesis: from latent genes to tumours. Oncogene. 2003;22:5108–5121. doi: 10.1038/sj.onc.1206556. [DOI] [PubMed] [Google Scholar]
- 33.Ambinder RF, Robertson KD, Tao Q. DNA methylation and the Epstein-Barr virus. Semin Cancer Biol. 1999;9:369–375. doi: 10.1006/scbi.1999.0137. [DOI] [PubMed] [Google Scholar]
- 34.Seto E, Ooka T, Middeldorp J, et al. Reconstitution of naso-pharyngeal carcinoma-type EBV infection induces tumorigenicity. Cancer Res. 2008;68:1030–1036. doi: 10.1158/0008-5472.CAN-07-5252. [DOI] [PubMed] [Google Scholar]
- 35.Fukayama M, Hayashi Y, Iwasaki Y, et al. Epstein-Barr virus-associated gastric carcinoma and Epstein-Barr virus infection of the stomach. Lab Invest. 1994;71:73–81. [PubMed] [Google Scholar]
- 36.Imai S, Koizumi S, Sugiura M, et al. Gastric carcinoma: monoclonal epithelial malignant cells expressing Epstein-Barr virus latent infection protein. Proc Natl Acad Sci U S A. 1994;91:9131–9135. doi: 10.1073/pnas.91.19.9131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uozaki H, Fukayama M. Epstein-Barr virus and gastric carcinoma—viral carcinogenesis through epigenetic mechanisms. Int J Clin Exp Pathol. 2008;1:198–216. [PMC free article] [PubMed] [Google Scholar]
- 38.Strong MJ, Xu G, Coco J, et al. Differences in gastric carcinoma microenvironment stratify according to EBV infection intensity: implications for possible immune adjuvant therapy. PLoS Pathog. 2013;9:e1003341. doi: 10.1371/journal.ppat.1003341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pfeffer S, Zavolan M, Grasser FA, et al. Identification of virus-encoded microRNAs. Science. 2004;304:734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- 40.Cai X, Schafer A, Lu S, et al. Epstein-Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog. 2006;2:e23. doi: 10.1371/journal.ppat.0020023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.You JS, Jones PA. Cancer genetics and epigenetics: two sides of the same coin? Cancer Cell. 2012;22:9–20. doi: 10.1016/j.ccr.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schuettengruber B, Cavalli G. Recruitment of polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development. 2009;136:3531–3542. doi: 10.1242/dev.033902. [DOI] [PubMed] [Google Scholar]
- 43.Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Develop. 2011;25:1010–1022. doi: 10.1101/gad.2037511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsai CN, Tsai CL, Tse KP, et al. The Epstein-Barr virus oncogene product, latent membrane protein 1, induces the downregulation of E-cadherin gene expression via activation of DNA methyl-transferases. Proc Natl Acad Sci U S A. 2002;99:10084–10089. doi: 10.1073/pnas.152059399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsai CL, Li HP, Lu YJ, et al. Activation of DNA methyltransferase 1 by EBV LMP1 involves c-Jun NH(2)-terminal kinase signaling. Cancer Res. 2006;66:11668–11676. doi: 10.1158/0008-5472.CAN-06-2194. [DOI] [PubMed] [Google Scholar]
- 46.Siouda M, Frecha C, Accardi R, et al. Epstein-Barr virus down-regulates tumor suppressor DOK1 expression. PLoS Pathog. 2014;10:e1004125. doi: 10.1371/journal.ppat.1004125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hino R, Uozaki H, Murakami N, et al. Activation of DNA methyltransferase 1 by EBV latent membrane protein 2A leads to promoter hypermethylation of PTEN gene in gastric carcinoma. Cancer Res. 2009;69:2766–2774. doi: 10.1158/0008-5472.CAN-08-3070. [DOI] [PubMed] [Google Scholar]
- 48.Kong QL, Hu LJ, Cao JY, et al. Epstein-Barr virus-encoded LMP2A induces an epithelial-mesenchymal transition and increases the number of side population stem-like cancer cells in nasopharyngeal carcinoma. PLoS Pathog. 2010;6:e1000940. doi: 10.1371/journal.ppat.1000940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leight ER, Sugden B. EBNA-1: a protein pivotal to latent infection by Epstein-Barr virus. Rev Med Virol. 2000;10:83–100. doi: 10.1002/(sici)1099-1654(200003/04)10:2<83::aid-rmv262>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 50.Sugden B, Warren N. A promoter of Epstein-Barr virus that can function during latent infection can be transactivated by EBNA-1, a viral protein required for viral DNA replication during latent infection. J Virol. 1989;63:2644–2649. doi: 10.1128/jvi.63.6.2644-2649.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kanda T, Horikoshi N, Murata T, et al. Interaction between basic residues of Epstein-Barr virus EBNA1 protein and cellular chromatin mediates viral plasmid maintenance. J Biol Chem. 2013;288:24189–24199. doi: 10.1074/jbc.M113.491167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lu F, Wikramasinghe P, Norseen J, et al. Genome-wide analysis of host-chromosome binding sites for Epstein-Barr virus nuclear antigen 1 (EBNA1) Virol J. 2010;7:262. doi: 10.1186/1743-422X-7-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Canaan A, Haviv I, Urban AE, et al. EBNA1 regulates cellular gene expression by binding cellular promoters. Proc Natl Acad Sci U S A. 2009;106:22421–22426. doi: 10.1073/pnas.0911676106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 55.Baylin SB, Jones PA. A decade of exploring the cancer epigenome—biological and translational implications. Nat Rev Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chan AT, Tao Q, Robertson KD, et al. Azacitidine induces demethylation of the Epstein-Barr virus genome in tumors. J Clin Oncol. 2004;22:1373–1381. doi: 10.1200/JCO.2004.04.185. [DOI] [PubMed] [Google Scholar]
- 57.Juergens RA, Wrangle J, Vendetti FP, et al. Combination epigenetic therapy has efficacy in patients with refractory advanced non-small cell lung cancer. Cancer Discov. 2011;1:598–607. doi: 10.1158/2159-8290.CD-11-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]