Abstract
Motivation: Abstract shape analysis, first proposed in 2004, allows one to extract several relevant structures from the folding space of an RNA sequence, preferable to focusing in a single structure of minimal free energy. We report recent extensions to this approach.
Results: We have rebuilt the original RNAshapes as a repository of components that allows us to integrate several established tools for RNA structure analysis: RNAshapes, RNAalishapes and pknotsRG, including its recent extension pKiss. As a spin-off, we obtain heretofore unavailable functionality: e. g. with pKiss, we can now perform abstract shape analysis for structures holding pseudoknots up to the complexity of kissing hairpin motifs. The new tool pAliKiss can predict kissing hairpin motifs from aligned sequences. Along with the integration, the functionality of the tools was also extended in manifold ways.
Availability and implementation: As before, the tool is available on the Bielefeld Bioinformatics server at http://bibiserv.cebitec.uni-bielefeld.de/rnashapesstudio.
Contact: bibi-help@cebitec.uni-bielefeld.de
1 THE RNA SHAPES STUDIO
1.1 Integration of tools for RNA abstract shape analysis
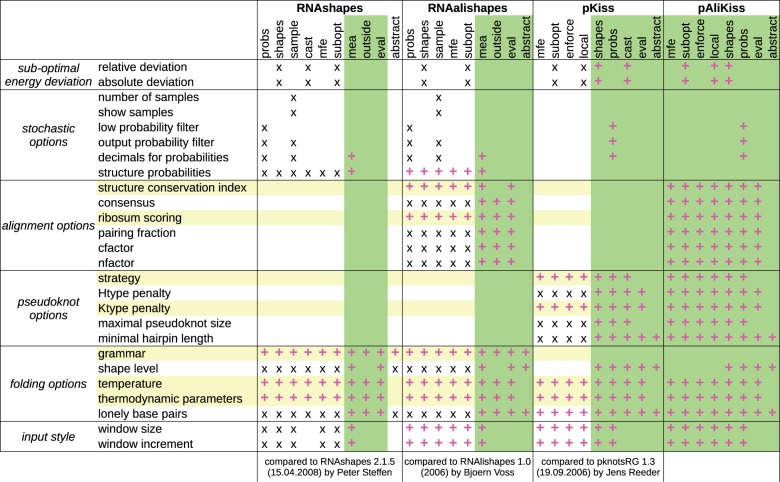
The framework of algebraic dynamic programming (ADP) allows us to express dynamic programming algorithms for sequence analysis on a high level of abstraction. They are composed from signatures, tree grammars and evaluation algebras (Giegerich et al., 2004a). Powerful product operations on algebras allow one to derive new types of analysis by the combination of available components, essentially with a single keystroke (Steffen and Giegerich, 2005). Relying on the recent Bellman’s GAP system (Sauthoff et al., 2013), which implements the ADP framework, we have built a repository of components that allows us to integrate several established tools for RNA structure analysis: RNAshapes, RNAalishapes and pknotsRG, including its recent extension pKiss. As a spin-off, we obtain heretofore unavailable functionality: e. g. with pKiss, we can now perform abstract shape analysis for structures holding pseudoknots up to the complexity of kissing hairpin motifs. The new tool pAliKiss can predict kissing hairpin motifs from aligned sequences. Along with the integration, the functionality of the tools was also extended in manifold ways. Figure 1 gives an overview.
Fig. 1.
Parameters for the RNA shapes studio. New features are indicated by +. New parameters are highlighted in yellow. New analysis modes are shaded in green
1.2 Integrated tools and their new functionality
1.2.1 Extensions to RNAshapes
It is generally agreed that predicting a single structure of minimal free energy does not adequately capture the subtlety and versatility of RNA structure formation. The RNAshapes tool introduced the notion of abstract shapes (Giegerich et al., 2004b; Voß et al., 2006)—a (mathematically precise) characterization of structures by their arrangement of helices. For example, ‘[[ ][ ][ ]]’ indicates a cloverleaf shape, and ‘[_[_[ ]_]]’ a single stem-loop with a 5′ bulge and an internal loop. Classical abstract shape analysis reports minimum free energy structures from different shape classes, or Boltzmann structure probabilities accumulated by shape. This gives synoptic information about the folding space of a given RNA sequence, without heuristics or sampling. Extending RNAshapes, we added different modes of treating dangling bases (consistent with RNAfold options −d0, −d1 and −d2) (Janssen et al., 2011; Lorenz et al., 2011), computation of base pair probabilities and maximum expected accuracy (MEA) folding (Lu et al., 2009).
1.2.2 Extensions to RNAalishapes
The work of Voß (2006) combines the ideas of RNAalifold and RNAshapes and performs shape analysis based on pre-aligned RNA sequences. We added the computation of a structure conservation index, different dangling base models, MEA folding and a window mode. RIBOSUM scoring (Bernhart et al., 2008) was added for evaluating sequence similarity.
1.2.3 Extensions to pKiss
In Theis et al. (2010) the ideas of pknotsRG (Reeder and Giegerich, 2004) are extended to predict (aside from unknotted structures and H-type pseudoknots) RNA structures that exhibit kissing hairpin motifs in an arbitrarily nested fashion, requiring time and space. We added shape analysis, probabilities, different folding strategies and different dangling base models. The -cast option provides comparative prediction of pseudoknotted structures as in the RNAcast approach (Reeder and Giegerich, 2005). A window mode was also included.
1.2.4 New tool pAliKiss
The program pAliKiss allows to predict pseudoknots, including kissing hairpins from aligned sequences. Being composed from the grammars and algebras of the other tools, it inherits all the features and options that make sense for it.
1.2.5 Utilities
All tools were augmented with utilities to compute folding energy or abstract shape for sequences that are provided with a structure from an outside source, in a way consistent with the tools’ energy model. The graphical motif description tool Locomotif (Reeder et al., 2007) now uses modules from the RNA shapes studio. The KnotInFrame (Theis et al., 2008) tool that predicts −1 ribosomal frameshifts has been updated as well.
2 APPLICATION CASE: A FRAMESHIFT STIMULATION ELEMENT IN MERS
The Corona virus family contains a frameshift stimulation element (Baranov et al., 2005), where the frameshift is facilitated by a slippery site together with either an H-type or a K-type pseudoknot. Rfam (Burge et al., 2013) holds the corresponding family model RF00507, although the tools of Rfam cannot explicitly model pseudoknots. pKiss (Theis et al., 2010) in -enforce mode reveals that for 11 family members, minimal free energy structures are H-types, another 11 are K-type pseudoknots and for only one member a purely nested structure has the best energy.
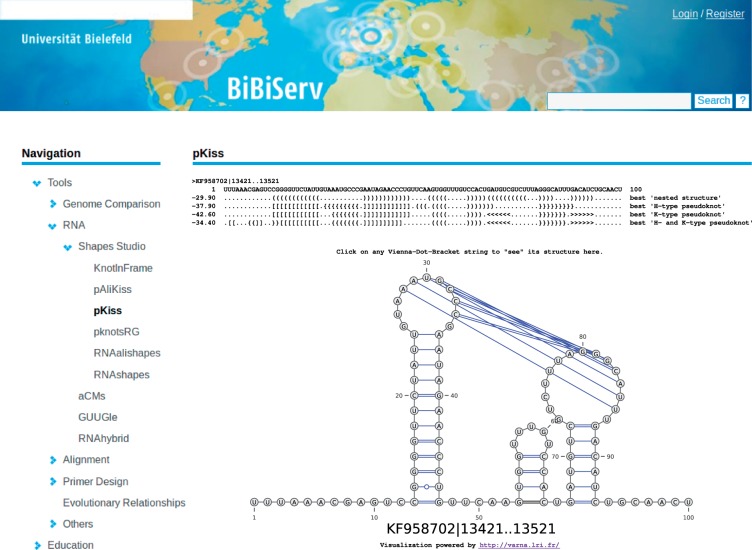
The recently sequenced MERS genome (KF958702.1) is annotated with a homologous frameshift site, whereas the structure of the triggering element remains unclear. Structure prediction with pKiss for a 100 bp stretch downstream the slippery site attests a most stable K-type pseudoknot (see Fig. 2). A second run of pKiss, this time in probability mode, shows that the shape class of this particular K-type pseudoknot has an overwhelming Boltzmann probability of ; leaving not much probability mass for any other shape classes.
Fig. 2.
RNA shapes studio result page for folding the MERS example with pKiss. Illustration by VARNA (Darty et al., 2009)
3 AVAILABILITY
The RNA shapes studio is available at http://bibiserv.cebitec.uni-bielefeld.de/rnashapesstudio. Users can access the Bellman’s GAP source code of all components in the repository, and combine or extend them according to their own goals. This has been done, for example, in Reinkensmeier et al. (2011) for defining the CCUCCUCC-motif family in the Rhizobiales.
Conflict of interest: none declared.
REFERENCES
- Baranov PV, et al. Programmed ribosomal frameshifting in decoding the sars-cov genome. Virology. 2005;332:498–510. doi: 10.1016/j.virol.2004.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhart S, et al. RNAalifold: improved consensus structure prediction for RNA alignments. BMC Bioinformatics. 2008;9:474. doi: 10.1186/1471-2105-9-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge SW, et al. Rfam 11.0: 10 years of RNA families. Nucleic Acids Res. 2013;41:D226–D232. doi: 10.1093/nar/gks1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darty K, et al. VARNA: Interactive drawing and editing of the RNA secondary structure. Bioinformatics. 2009;25:1974–1975. doi: 10.1093/bioinformatics/btp250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giegerich R, et al. A discipline of dynamic programming over sequence data. Sci. Comput. Program. 2004a;51:215–263. [Google Scholar]
- Giegerich R, et al. Abstract shapes of RNA. Nucleic Acids Res. 2004b;32:4843–4851. doi: 10.1093/nar/gkh779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen S, et al. Lost in folding space? Comparing four variants of the thermodynamic model for RNA secondary structure prediction. BMC Bioinformatics. 2011;12:429. doi: 10.1186/1471-2105-12-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz R, et al. ViennaRNA package 2.0. Algorithms Mol. Biol. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu ZJ, et al. Improved RNA secondary structure prediction by maximizing expected pair accuracy. RNA. 2009;15:1805–1813. doi: 10.1261/rna.1643609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder J, Giegerich R. Design, implementation and evaluation of a practical pseudoknot folding algorithm based on thermodynamics. BMC Bioinformatics. 2004;5:104. doi: 10.1186/1471-2105-5-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder J, Giegerich R. Consensus shapes: an alternative to the Sankoff algorithm for RNA consensus structure prediction. Bioinformatics. 2005;21:3516–3523. doi: 10.1093/bioinformatics/bti577. [DOI] [PubMed] [Google Scholar]
- Reeder J, et al. Locomotif: from graphical motif description to RNA motif search. Bioinformatics. 2007;23:i392. doi: 10.1093/bioinformatics/btm179. [DOI] [PubMed] [Google Scholar]
- Reinkensmeier J, et al. Conservation and occurrence of trans-encoded sRNAs in the rhizobiales. Genes. 2011;2:925–956. doi: 10.3390/genes2040925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauthoff G, et al. Bellman’s GAP - a language and compiler for dynamic programming in sequence analysis. Bioinformatics. 2013;29:551–556. doi: 10.1093/bioinformatics/btt022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen P, Giegerich R. Versatile and declarative dynamic programming using pair algebras. BMC Bioinformatics. 2005;6:224. doi: 10.1186/1471-2105-6-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theis C, et al. Knotinframe: prediction of -1 ribosomal frameshift events. Nucleic Acids Res. 2008;36:6013–6020. doi: 10.1093/nar/gkn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theis C, et al. Prediction of RNA secondary structure including kissing hairpin motifs. In: Moulton V, Singh M, editors. Algorithms in Bioinformatics, Vol. 6293 of Lecture Notes in Computer Science. Springer: Berlin Heidelberg; 2010. pp. 52–64. [Google Scholar]
- Voß B. Structural analysis of aligned RNAs. Nucleic Acids Res. 2006;34:5471–5481. doi: 10.1093/nar/gkl692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voß B, et al. Complete probabilistic analysis of RNA shapes. BMC Biol. 2006;4:5. doi: 10.1186/1741-7007-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]