Abstract
Summary: An important computational step following genome-wide association studies (GWAS) is to assess whether disease or trait-associated single-nucleotide polymorphisms (SNPs) enrich for particular biological annotations. SNP-based enrichment analysis needs to account for biases such as co-localization of GWAS signals to gene-dense and high linkage disequilibrium (LD) regions, and correlations of gene size, location and function. The SNPsnap Web server enables SNP-based enrichment analysis by providing matched sets of SNPs that can be used to calibrate background expectations. Specifically, SNPsnap efficiently identifies sets of randomly drawn SNPs that are matched to a set of query SNPs based on allele frequency, number of SNPs in LD, distance to nearest gene and gene density.
Availability and implementation: SNPsnap server is available at http://www.broadinstitute.org/mpg/snpsnap/.
Contact: joelh@broadinstitute.org
Supplementary information: Supplementary data are available at Bioinformatics online.
1 MOTIVATION
Genetic loci identified by genome-wide association studies (GWAS) point to biology that help us to understand the etiology of complex traits and diseases (Hirschhorn, 2009). A typical first step following GWAS is to assess whether associated loci as a group implicate biological pathways (Wang et al., 2010), or whether associated single-nucleotide polymorphisms (SNPs) are enriched for annotations such as non-coding functional elements (Ward and Kellis, 2012) or missense variants (Lango Allen et al., 2010). The simple approach, comparison of associated SNPs with random sets of SNPs, is susceptible to bias from non-random clustering of functionally related genes, and the greater likelihood of associated SNPs to be within (large) genes and regions of strong linkage disequilibrium (LD) and other potential confounders (Hindorff et al., 2009). If not properly accounted for by appropriate matching of random sets of SNPs, these biases may lead to spurious enrichments; for instance, brain pathways (typically containing large genes that are more likely to harbor associated SNPs) will appear to be overrepresented in most sets of GWAS loci (Raychaudhuri et al., 2010).
The SNPsnap Web server identifies randomly selected SNPs with similar genetic properties as a set of query (associated) SNPs. Random SNPs are matched based on minor allele frequency, number of SNPs in LD (LD buddies), distance to nearest gene and number of nearby genes (gene density). By using sets of random but matched SNPs, investigators can compute enrichment statistics on more appropriate negative controls to get an unbiased empirical estimate of the significance of enrichment results obtained with associated SNPs. We and others have previously used a similar approach (Gamazon et al., 2010; Lango Allen et al., 2010; Nicolae et al., 2010; Maurano et al., 2012; Schaub et al., 2012; Gamazon et al., 2013; Wood et al., 2014), however currently there is no software tool that formally implements this approach.
2 IMPLEMENTATION
We used biallelic, uniquely mapped SNPs from the 1000 Genomes Project (Abecasis et al., 2012) genotype data and computed the following properties (see also Supplementary Material):
Minor allele frequency: we partitioned SNPs into minor allele frequency bins (using 1–2, 2–3, … , 49–50% strata).
LD buddies: for each SNP, we counted the number of ‘buddy’ SNPs in LD at various thresholds (r2 > 0.1, 0.2, … , 0.9) [using PLINK v.1.07 (Purcell et al., 2007) to compute LD].
Distance to nearest gene: we computed the distance to the nearest 5′ start site using Ensembl gene coordinates (Flicek et al., 2014). If the SNP was within a gene, we used the distance to that gene’s start site.
Gene density: we counted the number of genes in loci around the SNP, using LD (r2 > 0.1, 0.2, … , 0.9) and physical distance (100, 200, … , 1000 kb) to define loci.
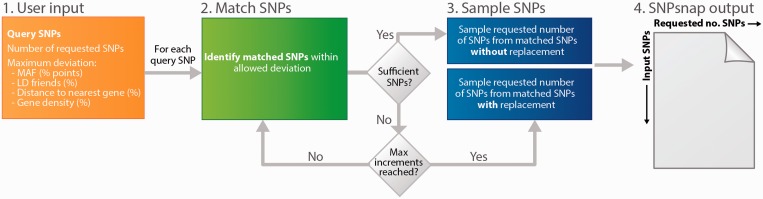
Next, we developed an algorithm to sample the best matching SNPs given the genetic properties of the query SNPs (Fig. 1). Because sampling of a sufficient number of SNPs exactly matched to a given query SNP is infeasible, we allowed deviations between the query and matched SNP for the properties used in matching. The SNPsnap algorithm identifies sets of matched SNPs for each query SNP as follows:
In five uniformly spaced increments, increase the allowable deviation for each of the properties, ending with the prespecified maximum allowable deviation. For each increment, identify matching SNPs, defined as SNPs with genetic properties within the allowable deviations.
If there are at least as many matching SNPs as requested, sample without replacement the requested number of SNPs from the matching SNPs and proceed to step 5.
If the number of matching SNPs is less than the number of requested SNPs, increment the allowable deviation and return to step 1; if the maximum allowable deviation has been reached, proceed to step 4.
Sample with replacement from the matched SNPs identified in step 1.
Proceed to the next query SNP.
Fig. 1.
Workflow of SNPsnap. SNPsnap takes as input a set of query SNPs, the number of requested matched SNPs and the allowed deviation for each of the four genetic properties. Using an incremental matching algorithm, SNPsnap identifies SNPs that match the properties and allowed deviations of the query SNPs. If less than the requested number of SNPs could be identified for a given query SNPs, SNPsnap samples SNPs with replacement from the set matched SNPs. The SNPsnap output consists of a matrix with dimensions equal to the number of query SNPs times the number of matched SNPs requested
For the default parameters, SNPsnap provides two visual and two numeric scores that indicate how well the requested number of SNPs could be retrieved (see Supplementary Material).
3 WEB SERVER
The query set of SNPs should be independent and must be uploaded using rs-numbers or chromosomal coordinates. SNPsnap allows for test of input SNPs’ independence. Besides the number of SNP to be sampled, the investigator must specify the maximum allowed deviation for each of the four properties. The investigator can choose whether matched SNPs may contain the query SNPs. The output consists of a matrix with query SNPs as rows and matched SNPs as columns. The investigator can optionally enable annotation of the SNPs to yield output files that include the genetic properties, nearest genes and genes in loci.
4 EXAMPLE
We used SNPsnap to illustrate how using properly matched SNPs can avoid spurious results. We first retained the top 500 independent SNPs from a simulated GWAS based on random phenotypes with no genetic basis, and retrieved a set of synapse genes (see Supplementary Material). Next, we conducted a Fisher’s exact test, which indicated that the synapse genes were 2.4-fold enriched at the random GWAS loci (P = 0.005). We then used SNPsnap to retrieve 10 000 matched SNPs for each of the 500 simulated GWAS SNPs and computed the fold enrichment for each of the 10 000 SNP sets. From this we calculated an empirical P-value showing no enrichment of synapse genes (P = 0.16), as expected.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank The Genetic Investigation of ANthropometric Traits (GIANT) Consortium height working group for useful discussions.
Funding: T.H.P. is supported by The Danish Council for Independent Research Medical Sciences (FSS) and The Alfred Benzon Foundation. P.T. is supported by The Novo Scholarship Programme. This work was supported by The National Institute of Diabetes and Digestive and Kidney Diseases [2R01DK075787 to J.N.H.].
Conflict of interest: none declared.
REFERENCES
- Abecasis GR, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P, et al. Ensembl 2014. Nucleic Acids Res. 2014;42:D749–D755. doi: 10.1093/nar/gkt1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamazon ER, et al. Chemotherapeutic drug susceptibility associated SNPs are enriched in expression quantitative trait loci. Proc. Natl Acad. Sci. USA. 2010;107:9287–9292. doi: 10.1073/pnas.1001827107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamazon ER, et al. Enrichment of cis-regulatory gene expression SNPs and methylation quantitative trait loci among bipolar disorder susceptibility variants. Mol Psychiatry. 2013;18:340–346. doi: 10.1038/mp.2011.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindorff LA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl Acad. Sci. USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschhorn JN. Genomewide association studies—illuminating biologic pathways. N. Engl. J. Med. 2009;360:1699–1701. doi: 10.1056/NEJMp0808934. [DOI] [PubMed] [Google Scholar]
- Lango Allen H, et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurano MT, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolae DL, et al. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcel S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri S, et al. Accurately assessing the risk of schizophrenia conferred by rare copy-number variation affecting genes with brain function. PLoS Genet. 2010;6:e1001097. doi: 10.1371/journal.pgen.1001097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaub MA, et al. Linking disease associations with regulatory information in the human genome. Genome Res. 2012;22:1748–1759. doi: 10.1101/gr.136127.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, et al. Analysing biological pathways in genome-wide association studies. Nat. Rev. Genet. 2010;11:843–854. doi: 10.1038/nrg2884. [DOI] [PubMed] [Google Scholar]
- Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood A, et al. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat. Genet. 2014;46:1173–1186. doi: 10.1038/ng.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.