Abstract
The corticotropin releasing factors receptor-1 and receptor-2 (CRF1R and CRF2R) are therapeutic targets for treating neurological diseases. Antagonists targeting CRF1R have been developed for the potential treatment of anxiety disorders and alcohol addiction. It has been found that antagonists targeting CRF1R always show high selectivity, although CRF1R and CRF2R share a very high rate of sequence identity. This has inspired us to study the origin of the selectivity of the antagonists. We have therefore built a homology model for CRF2R and carried out unbiased molecular dynamics and well-tempered metadynamics simulations for systems with the antagonist CP-376395 in CRF1R or CRF2R to address this issue. We found that the side chain of Tyr6.63 forms a hydrogen bond with the residue remote from the binding pocket, which allows Tyr6.63 to adopt different conformations in the two receptors and results in the presence or absence of a bottleneck controlling the antagonist binding to or dissociation from the receptors. The rotameric switch of the side chain of Tyr3566.63 allows the breaking down of the bottleneck and is a perquisite for the dissociation of CP-376395 from CRF1R.
The corticotropin-releasing factor (CRF) receptor-1 (CRF1R) and CRF receptor-2 (CRF2R) are family B G-protein-coupled receptors (GPCRs) composed of seven transmembrane helices (TM1 –TM7) linked by three intracellular loops (ICL1 –ICL3) and three extracellular loops (ECL1 –ECL3)1. CRF1R and CRF2R belong to the subfamily of CRF receptors and have been identified to be widely distributed throughout the central nervous system and periphery nervous system and act as key regulators of the hypothalamus-pituitary-adrenal axis2,3,4. It is believed that a well-balanced opposing action between CRF1R and CRF2R is responsible for the initiation of and the recovery from an elicited stress response and that a failed adaptation of the two receptors could lead to neuropathology, including anxiety and depression. Recent studies have revealed that CRF1R and CRF2R are involved in stress-associated anxiety and depression-like behavior in a more complicated way4,5,6. Selectively blocking of CRF1R or CRF2R with an antagonist is an effective way to treat the neuropathology. Efforts have been made to develop antagonists with high selectivity towards CRF1R or CRF2R. Antagonists targeting CRF1R were among the first allosteric GPCR ligands to be evaluated clinically for treating depression and anxiety related disorders7.
In a GPCR subfamily, residues in the ligand binding pocket of the GPCRs are highly conserved, which can lead to the side effects posed by off-target effects8. It is interesting to note that sequence conservation in the subfamily of CRF receptors is even higher than in most of the other GPCR subfamilies. CRF1R and CRF2R show very high sequence conservation on the helices TM5 and TM6 and the residues that directly interact with the antagonists are identical. However, the antagonist CP-376395 in the crystal structure of CRF1R shows a 1000 fold lower affinity towards CRF2R than towards CRF1R9. It has been shown that residues along the ligand binding/dissociation pathway of a target can affect the efficacy of a drug through influencing the binding kinetics of the drug towards its target10. Therefore, we assume that residues remote from the binding pocket play a role for the selectivity of the antagonist CP-376395.
To study the selectivity of the antagonist CP-376395 towards the receptors CRF1R and the role of the remote residues in the selectivity, we built a homology model of CRF2R with CRF1R as the template and carried out unbiased molecular dynamics simulations and well-tempered metadynamics simulations for both CRF1R and CRF2R with CP-376395 binding to them. The dissociation of CP-376395 from CRF1R or CRF2R was observed in the well-temped metadynamics simulations. We found that the hydrogen bond between His2283.40 and Tyr3566.63 in CRF1R, which is absent in CRF2R, plays a pivotal role in controlling the difference of the binding of CP-376395 towards CRF1R and CRF2R (Throughout this paper, the superscript on a residue represents the Wootten generic residue numbering11).
Results and Discussion
Homology modeling of CRF2R
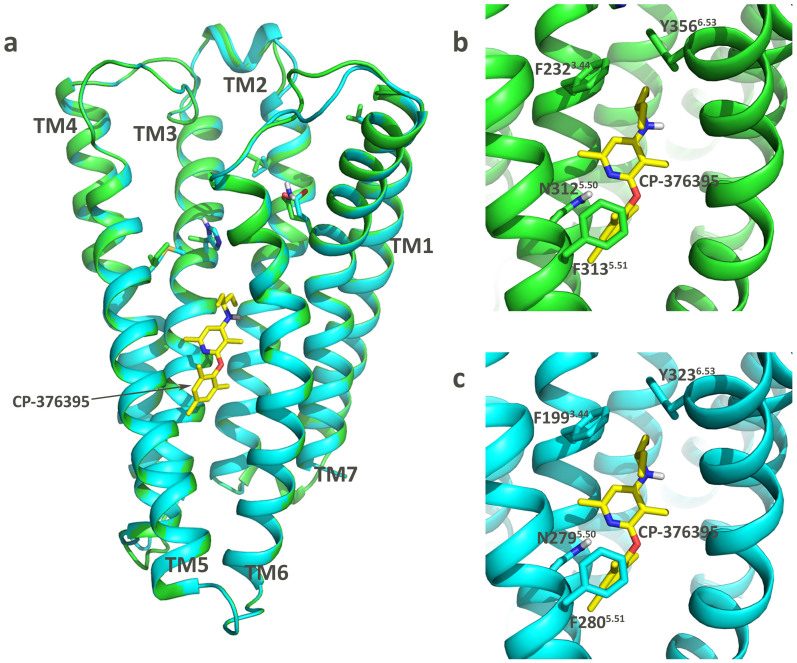
CRF1R and CRF2R belong to the same family and share a high sequence identity. The identity rate is 73% if only the transmembrane parts of the receptors are considered. The sequence alignment of CRF2R to CRF1R is shown in Figure S1. We can see that the most conserved residues match each other (Table S1). A Richardson plot of the modeled CRF2R structure indicates that 98% of the residues are located in the allowed regions, reflecting that the structure is geometrically reasonable (Figure S2)12. The root mean square deviation (RMSD) between the crystal structure of CRF1R and the modeled structure of CRF2R is 0.01 Å (Figure 1).
Figure 1. The crystal structure of CRF1R and the modeled structure of CRF2R.
The protein structures are shown in the cartoon mode and the antagonist CP-376395 is shown in the stick mode. The structures of CRF1R and CRF2R are colored in green and cyan, respectively. The antagonist CP-376395 is colored in yellow. (a) Alignment of the crystal structure of CRF1R and the modeled structure of CRF2R; (b) Key residues in the antagonist binding pocket of CRF1R; (c) Key residues in the antagonist binding pocket of CRF2R.
Comparison of the structures of CRF1R and CRF2R
The residues in the antagonist binding pocket of CRF1R are very similar to those in the corresponding pocket of CRF2R, and, in particular, the residues in the first shell around CP-376395 are conserved (Figure 1). The trimethyl-mesitylene motif on CP-376395 forms a T-shaped π-π stacking interaction with the residue Phe3135.51 in CRF1R or with the residue Phe2805.51 in CRF2R. The nitrogen atom on the dimethylpyridine group of the antagonist forms a hydrogen bond with the side chain of the residue Asn3125.50 in CRF1R (Figure 1b) or with the residue Asn2795.50 in CRF2R (Figure 1c)1. It has been suggested that residues Phe2323.44 and Tyr3566.63 in CRF1R work as a bottleneck for the binding of CP-376395 to the binding pocket (Figure 1b). Residues Phe1993.44 and Tyr3236.63 in CRF2R are located in the corresponding region of the bottleneck residues in CRF1R to restrict the binding of CP-376395 to CRF2R (Figure 1c). Although the residues directly interacting with the antagonist CP-376395 as described above are conserved in these two receptors, CP-376395 is highly selective towards CRF1R, with the Ki values 12 nM towards CRF1R and >10000 nM towards CRF2R9. Some residues along the suggested antagonist binding pathways are different (Figure S3). Such differences lead us to assume that the residues remote from the binding pocket likely affect the binding kinetics to control the selectivity of CP-376395 towards CRF1R. Therefore, we performed unbiased molecular dynamics simulations and well-tempered metadynamics simulations to explain the differences.
MD simulations
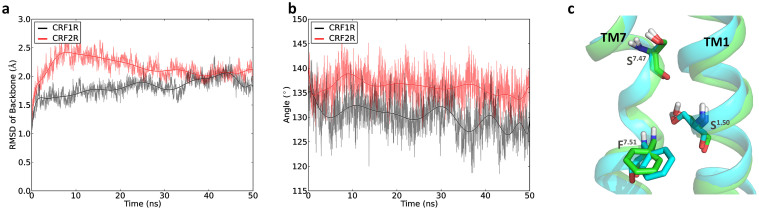
As we can see from the RMSD curves in Figure 2a, CRF1R is rather stable with the RMSD value less than 2.0 Å during the whole simulation. The RMSD value of CRF2R is always larger than that of CRF1R but is still less than 2.5 Å during the whole simulation. We attribute the slightly large RMSD value of CRF2R to the relaxation of the modeled structure. To evaluate the flexibility of the residues in CRF1R and CRF2R, the root mean square fluctuation (RMSF) values of the two receptors are calculated. The RMSF values indicate that in CRF1R and CRF2R, TM1-TM7 are much more stable than the loops connecting these helices. The RMSF values of the two receptors calculated from the MD simulation trajectories were found to follow the same trend of those calculated from the B-factors of the x-ray crystallography structure of CRF1R (Figure S4).
Figure 2.
(a) RMSD values of the backbone atoms with respect to the first snapshots in the simulations; (b) Evolution of the tilt angle of TM7 with the pivot point at Ser7.47 in the simulations. (c) Structural representation of the key residues Ser1.50, Ser7.47 and Phe7.51 which control the kink of TM7 on CRF1R and CRF2R. The structures of CRF1R and CRF2R are colored in green and cyan, respectively. The key residues are shown as sticks.
The kink of TM7
Helices TM1-TM7 form a bundle for the binding of the antagonist and for the signal transmission in the GPCRs. A sharp kink of TM7 has been observed in the crystal structure of CRF1R1. We found that this kink is preserved during the MD simulation and that the kink angle is changing gradually from 135° to 130° with the pivot point at Ser3827.47 (Figure 2b and 2c). An analysis of the structure of CRF1R revealed that the kink is stabilized by the conserved residue Ser1301.50 which forms hydrogen bonds with the backbones of Ser3827.47 and Phe3867.51. The two hydrogen bonds were preserved during the MD simulation (Figure S5).
The structure of CRF2R was built based on the crystal structure of CRF1R. Therefore, the sharp kink on TM7 and the two hydrogen bonds between the corresponding residues are kept in the modeled CRF2R structure (Figure 2b and 2c). The hydrogen bond between Ser1271.50 and Phe3537.51 is also preserved during the whole MD simulation, but the hydrogen bond between Ser1271.50 and Ser3497.47 broke in the beginning of the MD simulation (Figure S5). As a result, a smaller kink angle of TM7 was observed in CRF2R (Figure 2b and 2c). In family A GPCRs, such a kink has also been observed on TM7 with the residue Pro7.50 as the pivot point13,14.
The hydrogen bond between His2.50 and Glu3.50
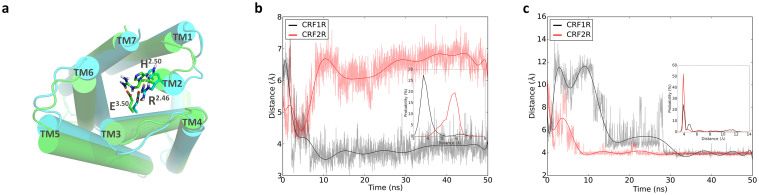
Family B GPCRs lack the sequence motifs to form the conserved ionic lock connecting TM3 and TM6 as found in the inactive conformation of family A GPCRs1. In the family B GPCRs, a hydrogen bond between the residues His2.50 and Glu3.50 is believed to be involved in the receptor activation (Figure 3a)15. In CRF1R, the distance between Nδ of His1842.50 and Cδ of Glu2383.50 is about 4 Å after 8ns of the simulation and a hydrogen bond is formed between the side chains of His1842.50 and Glu2383.50 at the beginning of the simulation and is preserved in the simulation (Figure 3b). This is in line with the observation obtained by Bai. et al16. It can also be seen from Figure 3b that the distances between Nδ of His1842.50 and Cδ of Glu2383.50 are predominantly around 4 Å. In contrast, the hydrogen bond between His2.50 and Glu3.50 is not stable in the simulation of CRF2R. The Nδ of His1522.50 and Cδ of Glu2053.50 became closer in about 5 ns to 7 ns but was separated thereafter with most of the distances distributed around 6 Å as indicated in Figure 3b. The ionic lock between TM3 and TM6 in family A receptors is supposed to interconvert between the two states corresponding to the formation and breaking of the ionic lock17,18,19,20. Our simulation results also revealed that the potentially important hydrogen bond between His2.50 and Glu3.50 can interconvert between the formation and breaking of the hydrogen bond in family B GPCRs. Additionally, it is interesting for us to observe that in CRF2R, Glu2053.50 formed an ionic lock with Arg1482.46 after 10 ns of the simulation and this ionic lock is preserved thereafter. The corresponding ionic lock in CRF1R was formed between Glu2383.50 and Arg1802.46 after 28 ns of the simulation and is preserved thereafter (Figure 3c). The free energy landscapes obtained from the metadynamics simulations clearly indicate that the locked state has a free energy of 5 kcal/mol lower than the unlocked state (Figure S6). This reflects that the system prefers to stay in the locked state. Thus, our results are in agreement with the observations from the long MD simulations18,21.
Figure 3.
(a) The final structures of the two receptors from the unbiased MD simulations viewed from the intracellular side. Residues His2.50, Arg2.46, and Glu3.50 are shown in the stick mode; (b) Evolution of the distances between Nδ of His2.50 and Cδ of Glu3.50 in the two receptors with the distributions shown in the middle panel; (c) Evolution of the distances between Cζ of Arg2.46 and Cδ of Glu3.50 in the two receptors with the distributions shown in the middle panel.
Ligand fluctuations in the unbiased MD simulations
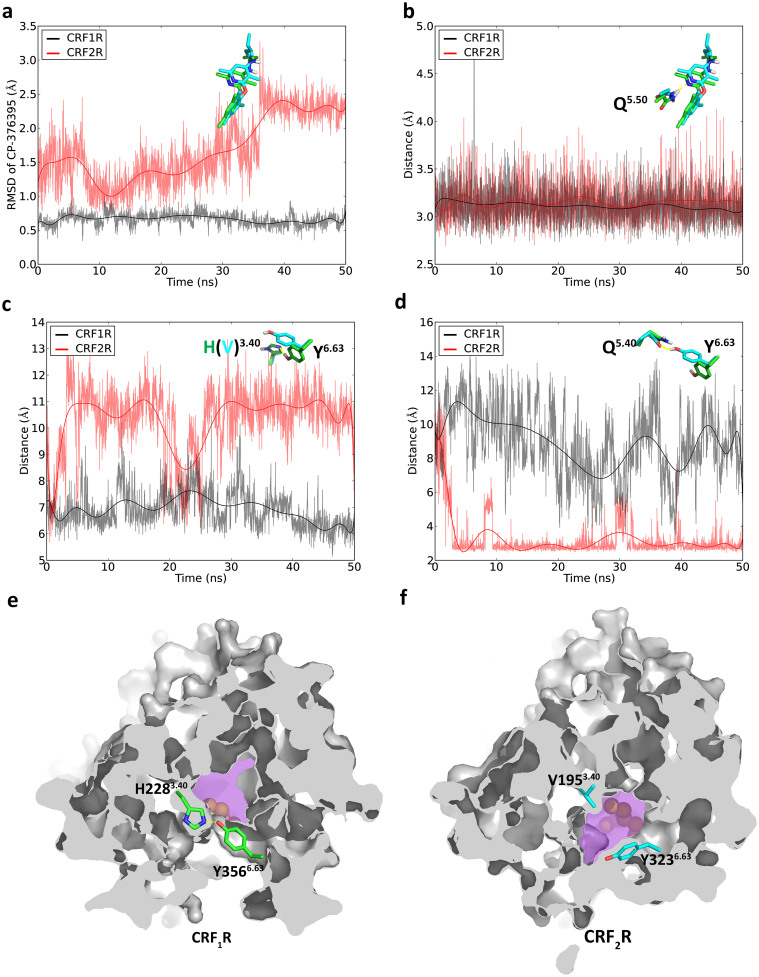
CP-376395 binds to an unexpected site located in the cytoplasmic half of the receptor CRF1R, which is about 18 Å away from the putative agonist binding site of the receptor and is about 13–23 Å away from the corresponding small ligand binding site of family A GPCRs16. Consistent with the observation of Bai et al., small RMSD values (~0.5 Å) of the ligand were obtained from our simulation of CRF1R. In contrast, CP-376395 shows RMSD values in the binding pocket of CRF2R larger than in that of CRF1R as indicated in Figure 4a. The conserved residue Asn5.50 forms an essential hydrogen bond with the nitrogen on the pyridine ring of CP-376395 and mutation of this residue to Ala results in a complete loss of ligand binding1. This key hydrogen bond is preserved during the simulations of both CRF1R and CRF2R and stabilizes the aryloxy moiety of CP-376395 (Figure 4b). Thus, the larger RMSD value of CP-376395 in CRF2R is mainly contributed by the fluctuation of the exocyclic alkylamino group.
Figure 4.
(a) The RMSD values of the ligand with respect to the first snapshots of the simulations; (b) The distance between the side chain of Asn5.50 and the nitrogen on the pyridine ring of CP-376395; (c) The distance between the backbone carbon atoms on Tyr6.63 and His2283.40 (CRF1R) or between the backbone carbon atoms on Tyr6.63 and Val2283.40 (CRF2R); (d) The distance between the side chain oxygen atoms on Tyr6.63 and Gln5.50; (e) Cross-section view of CRF1R with the bottleneck formed; (f) Cross-section view of CRF2R without the bottleneck. In (e) and (f), the antagonist binding pockets are colored in magenta.
The exocyclic alkylamino group is located adjacent to the residues Phe3.44 and Tyr6.63 of CRF1R (Figure 1b) and CRF2R (Figure 1c), which have been suggested to be the bottleneck for the binding of the antagonist to CRF1R1. The residue Tyr3566.63 is located within the hydrogen bond distance of His2283.40 in CRF1R. However, the residue Val1953.40 of CRF2R is corresponding to His2283.40 of CRF1R and lacks the hydrogen bond donor or acceptor atoms on its side chain to form a hydrogen bond with Tyr3236.63. In our simulations, the distance between the side chain oxygen atom of Tyr3566.63 and Cα of His2283.40 of CRF1R was preserved during the MD simulation, reflecting that the hydrogen bond formed between the side chains of Tyr3566.63 and His2283.40 was stable (Figure 4d). In contrast, the distance between the side chain of Tyr3236.63 and Cα of Val1953.40 in CRF2R became much larger in the first couple of nanoseconds and remained unchanged in the following simulation with a restoration of the initial value between 20 ns and 25 ns. The increase of the distance alters the conformation of the side chain of Tyr3236.63 in CRF2R (Figure 4d and Figure S7) and this conformational change is not taking place randomly (Figure S8). Interestingly, the rotameric change of this residue generates a hydrogen bond between the side chains of Tyr3236.63 and Gln2696.44. This hydrogen bond was preserved until the end of the 50 ns MD simulation to stabilize the rotameric change of Tyr3236.63 (Figure 4c and Figure S7). Such rotameric switch results in the breaking down of the bottleneck (Figure 4f), while such a bottleneck is preserved during the simulation of CRF1R (Figure 4e). We thus assume that such a difference controls the CP-376395 to dissociate from the antagonist binding pocket in CRF1R and CRF2R.
Metadynamics simulations
In our well-tempered metadynamics simulations, CP-376395 left the antagonist binding pocket in CRF1R and CRF2R, and explored the binding pathways to exit the receptors through the helices bundle (Supporting videos). The free energy surfaces (FESs) for CP-376395 leaving the antagonist binding pockets of CRF1R and CRF2R are displayed in Figure 5 and Figure 6, respectively.
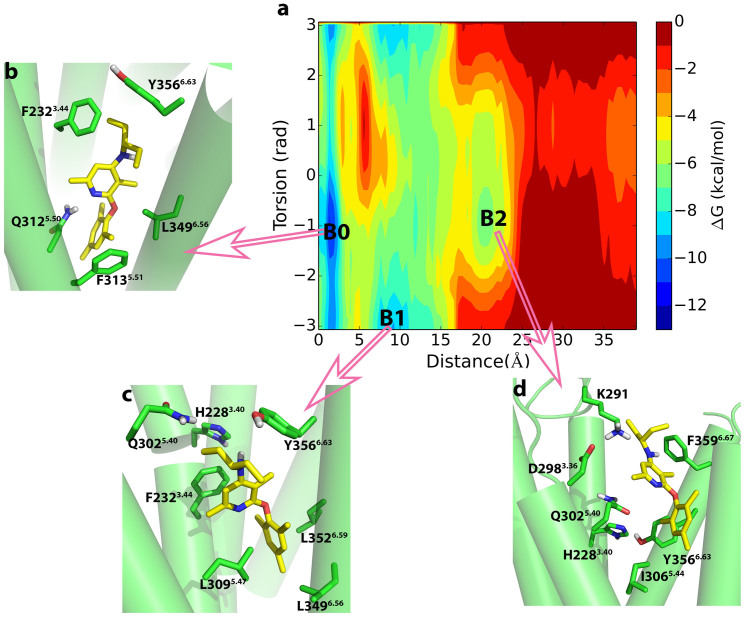
Figure 5. Metastable states in the dissociation of CP-376395 from CRF1R.
(a) The binding free energy surface for the dissociation of CP-376395 from CRF1R as a function of the Z-component of the vector connecting the nitrogen on the dimethylpyridine group of CP-376395 and CY on the Asn3125.50 and χ1 torsional angle of Tyr3566.63.The three main energy basins B0-B2 found in the metadynamics simulation are highlighted in b–d, respectively; (b)–(d) Structural characterization of the metastable states B0–B2.
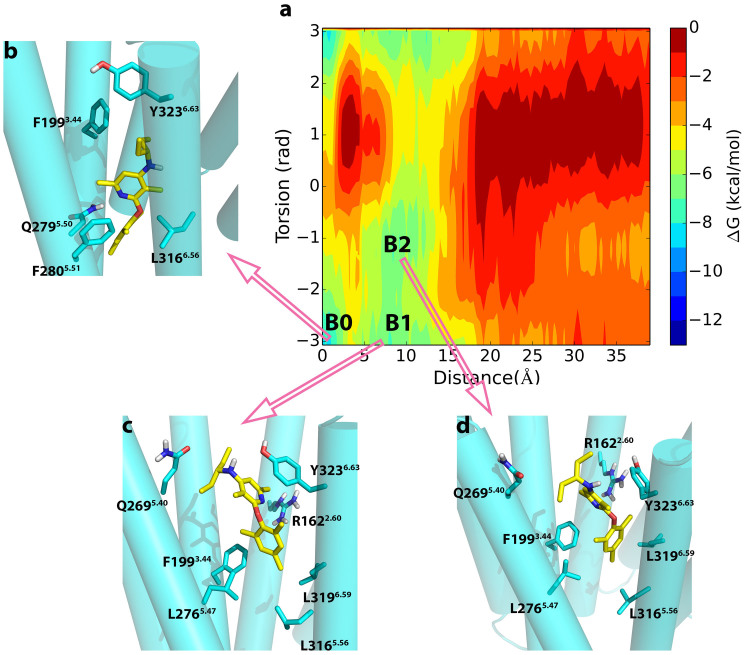
Figure 6. Metastable states in the dissociation of CP-376395 from CRF2R.
(a) The binding free energy surface for the dissociation of CP-376395 from CRF2R as a function of the Z-component of the vector connecting the nitrogen on the dimethylpyridine group of CP-376395 and Cγ on the Asn2795.50 and χ1 torsional angle of Tyr3236.63. The three main energy basins B0–B2 found in the metadynamics simulation are highlighted in b–d, respectively; (b)–(d), Structural characterization of the metastable states B0–B2.
Metastable poses of CP-376395 in CRF1R
For CP-376395 in CRF1R, we observed three energy minima (basin B0, basin B1, and basin B2) as displayed in the FES (Figure 5a). The deepest free energy minimum in the FES is depicted as basin B0 in Figure 5b and corresponds to the conformation that CP-376395 adopts in the x-ray crystallography structure. This conformation corresponds to the energetically most favorable pose for the binding of CP-376395 in the antagonist binding pocket of CRF1R. The aryloxy moiety of CP-376395 is holding tightly in the binding pocket by strongly hydrogen-bonding to Asn3125.50 and by hydrophobic interactions with Leu3165.54, Ile3195.57, Thr3456.42, Leu3486.45, and Leu349. The exocyclic alkylamino group keeps interacting with Gly3536.50, Phe2323.44, Leu3095.47, and Tyr3566.63. The mesityl group on CP-376395 forms a T-shaped π-π stacking interaction with Phe3135.51.
In the metadynamics simulation, CP-376395 moved along the helix bundle to the extracellular side half of the receptor CRF1R and reached a position sufficiently close for interacting directly with the bottleneck residues Tyr3566.63 and Phe2323.44 (Figure 5c). The mesityl group of CP-376395 is placed in a hydrophobic cavity surrounded by Leu3095.47, Leu3486.55, Leu3496.56, Leu3526.59, and Gly3536.60. The dimethylpyridine group of the antagonist is surrounded by the residues Ile3065.44, Gln3025.40, and Tyr3566.63. Met3055.43 also interacts with the dimethylpyridine group. The pyridine ring of CP-376395 forms a face to face π-π stacking interaction with the benzene ring of Phe2323.44. This π-π stacking interaction can stabilize the ligand in the binding site corresponding to the basin B1. In this basin, the torsional angle χ1 of the residue Tyr3566.63 is around π (or - π), while χ1 of Tyr3566.63 is about -1 in basin B0. Such a difference indicates that a rotameric change of Tyr3566.63 is required for CP-376395 moving from the site corresponding to the basin B0 to that corresponding to the basin B1.
Once all the metastable states in the helix bundle were filled, the antagonist CP-376395 crossed the helix bundle to reach the extracellular vestibule of CRF1R to form a metastable vestibule-bound state, where a local minimum (basin B2) was found as displayed in Figure 5d. The vestibule-bound state is the first step for CP-376395 to enter the antagonist binding site. In this local minimum, the antagonist CP-376395 is stabilized by the hydrophobic interactions with the residues Ile3065.44, Phe3606.67, and Tyr3566.63, and an additional amount of interaction energy can be gained from the relative closeness of the residues Asp2983.36 and Lys291.
Metastable poses of CP-376395 in CRF2R
We observe three energy minima (basin B0, basin B1 and basin B2) during the dissociation of the antagonist CP-376395 from CRF2R as displayed in Figure 6a. Basin B0 represents the energetically most stable site. This basin approximately corresponds to the crystallized conformation of CP-376395 in CRF1R (Figure 6b). At the basin, the hydrophobic interactions between CRF2R and CP-376395, which have been observed in the unbiased MD simulation, are kept. The dimethylpyridine group adopts a conformation with the nitrogen atom on the ring hydrogen-bonded to Asn2795.50, which allows CP-376395 to hold tightly with the antagonist binding pocket. The hydrophobic interactions originating from Leu2835.54, Ile2865.57, Leu3155.55, and Leu3165.56 further stabilize CP-376395 in this region. The exocyclic alkylamino group keeps interacting with Gly3136.50, Phe1993.44, Leu2765.47, and Tyr3236.63.
With the action of metadynamics, the antagonist CP-376395 moved along the helix bundle and reached another minimum displayed as basin B1 (Figure 6c). The traverse from basin B0 to basin B1 results in a breaking of the hydrogen bond between the nitrogen on the dimethylpyridine group of CP-376395 and the side chain of Asn2795.50. This hydrogen bond was found to be preserved during our 50 ns of the unbiased MD simulation, reflecting the inability of the unbiased MD in traversing energy barriers to sample the conformation states at different energy minima. Here, the dimethylpyridine group of CP-376395 resides in the pocket surrounded by Phe1913.36, Val1953.40, Ile3216.61, and Met3246.64. The mesityl moiety on CP-376395 is in a highly hydrophobic cage defined by Leu3165.56, Leu3196.59, Phe1993.44, and Leu2765.47. It is interesting to observe the π-π interaction between Tyr3236.63 and the pyridine ring on CP-376395 to stabilize the pose of CP-376395 in CRF2R.
We observed another metastable state (basin B2) residing at about 10 Å away from the residue Asn2795.50 (Figure 6d). As shown in Figure 6a, a large region surrounding basins B1 and B2 is energetically favorable, with the torsional angle ranging from −1 to −π and the distance kept at about 10 Å. This means that the torsional angle can change between 1 and −π with a very low energy barrier. Interestingly, the π-π interactions between Tyr3236.63 and the pyridine ring on CP-376395 are kept in this energetically favorable region. On the other hand, the hydrogen bond between Tyr3236.63 and Asn2695.40 is broken in basins B1 and B2. We thus suggest that the π-π interactions between Tyr3236.63 and the pyridine ring on CP-376395 compensate for the energy required for the breaking of the hydrogen bond between Tyr3236.63 and Asn2695.40.
Comparison of the ligand dissociation pathways and energy profiles
Three energy minima were identified for the dissociation of CP-376395 from both CRF1R and CRF2R as displayed in Figure 5 and Figure 6, respectively. As depicted in the figures, the basin B0 corresponds to the region located about 1–2 Å away from the key residue Asn5.50 with the torsional angle χ1 of the residue Tyr6.63 stabilized at −1 in CRF1R or −π (or π) in CRF2R. With CP-376395 moving from the basin B0 to the basin B1, the torsional angle χ1 of the residue Tyr3566.63 in CRF1R switches from −1 to −π (or π) while the corresponding angle in CRF2R stays at −1 (Figure S). Additionally, Phe3.44 displays no significant conformational change in neither the unbiased MD simulations nor in the metadynamics MD simulations of CRF1R or CRF2R. These observations strongly support our suggestion that the conformational change of Tyr3566.63 plays a pivotal role for CP-376395 binding to or dissociation from the antagonist binding site of CRF1R. With the formation of the hydrogen bond between the side chains of the residues Tyr3236.63 and Gln2696.44 in CRF2R, the crucial role of Tyr6.63 which controls the antagonist binding to or to dissociation from the binding site in CRF1R does not occur in CRF2R.
Bai et al. explored the dissociation pathway of CP-376395 from CRF1R by using random acceleration molecular dynamics simulations16. They found that breaking of the hydrogen bond between CP-376395 and Asn3125.50 results in the first energy barrier for the dissociation. This observation was confirmed from our study. Besides, we found that breaking of the hydrogen bond formed between Tyr3566.63 and His2283.40 also contributes to this barrier. The barrier is about 5 kcal/mol from our simulation, while it is 9.9 kcal/mol from Bai's work. Bai and coworkers also found the second energy barrier for the dissociation of CP-376395 along the pathway they detected because there exist two hydrogen bonds formed by CP-376395 with His2283.40 and Gln3025.40 in the pathway. However, these two hydrogen bonds were not observed along the CP-376395 dissociation pathway in our study. We found that CP-376395 forms π- π stacking interactions with Tyr3566.63, which was not described by Bai. et al. The π-π stacking interactions, together with the interactions between CP-376395 and the remaining residues, form the second energy barrier of about 4 kcal/mol. This barrier is much lower than the one obtained by Bai et al., which is 11.4 kcal/mol.
Implications for the drug design
The binding free energy for CP-376395 towards CRF1R, averaged over those from the first and repeated runs (−11.42 and −11.27 kcal/mol, respectively), is −11.35 kcal/mol, while that for CP-376395 towards CRF2R, averaged over those from the two simulations (−8.18 kcal/mol and −8.03 kcal/mol, respectively) is −8.10 kcal/mol (Figures S9 and S10). The binding free energy for the CP-376395 towards CRF1R is in agreement with the experimental value (−10.87 kcal/mol) while that for the CP-376395 towards CRF2R is a little larger than the experimental result (weaker than −6.86 kcal/mol)9. The decrease of the antagonist binding affinity from CRF1R to CRF2R is in agreement with the experimental results.
Mutation of His2283.40 has also been performed to evaluate the effect of His2283.40 on the binding of the antagonist NBI27914, which shares a similar scaffold to CP-376395, towards CRF1R22. Compared to the 1000-fold higher binding affinity of CP-376395 towards CRF1R than towards CRF2R, the His288Val mutation only leads to a 40-fold lower binding affinity of NBI27914 towards CRF1R. Such a difference likely comes from two aspects. One aspect is that the exocyclic alkylamino group in NBI27914 is bigger than that in CP-376395, which results in the hydrogen bond between His2283.40 and Tyr3566.63 being less stable in the binding of NBI27914 to CRF1R. Another aspect is that the decrease of the binding affinity of CP-376395 to CRF2R is likely contributed partially by the difference in the rotameric properties of Tyr6.63 in CRF1R and CRF2R.
Recent studies have indicated that the binding kinetics of a ligand towards its target could be one of the most crucial factors for sustainable drug efficacy, and in some cases, even more important than the binding affinity in determining the drug efficacy23,24. The recently published crystal structure of the smoothened receptor by Stevens's group revealed that there are multiple distinct binding sites for the ligand in the helix bundle of the receptor25,26. Both smoothened receptor and CRFR family receptors possess deep ligand binding cavities. This allows us to suggest that the sites along the pathway of an antagonist binding to the CRFR family of receptors, especially those corresponding the basins and saddle points we discovered in our metadynamics simulations, are important in the design of new drug candidates with attenuated side effects and chemoresistance.
Conclusion
In this work, we have carried out homology modeling to build the structure of CRF2R with the crystal structure of CRF1R as the template. Based on the crystal structure of CRF1R and the homology model of CRF2R, we performed unbiased MD simulations as well as well-tempered metadynamics simulations to investigate the origin of the selectivity of the antagonist CP-376395 towards the two receptors. From the unbiased MD simulations, we found that in CRF1R the oxygen atom on Tyr3566.63 forms a hydrogen bond with the side chain of His2283.40 which allows the formation of a bottleneck consisting of the residues Phe2323.44 and Tyr3566.63, while in CRF2R, the side chain oxygen on Tyr3236.63 is hydrogen bonded to the side chain of Gln2696.44, leading to a lack of such a bottleneck. The existence of the bottleneck in CRF1R and its absence in CRF2R provide an explanation for the origin of the high selectivity of the antagonist CP-376395 towards CRF1R. The metadynamics simulations provided an even stronger support for that explanation. The rotameric switch of the side chain of Tyr3566.63 results in the breaking down of the bottleneck in CRF1R and is a prerequisite for the dissociation of CP-376395 from CRF1R, but it is not required for the dissociation of CP-376395 from CRF2R as indicated by the FES. Thus, our studies provide important structural information in explaining the origin of the high selectivity of CP-376395 towards CRF1R.
Methods
Homology modeling of CRF2R
The crystal structure of CRF1R (PDB entry ID: 4K5Y)1 was used as the template for the homology modeling of the structure of CRF2R. ClustalW2 was used for the sequence alignments and manual adjustments were carried out to guarantee no gaps in the secondary structures27. Prime 3.5 was used to build the model of CRF2R28,29.
Unbiased molecular dynamics simulations
System preparation. Two systems, one containing the crystal structure of CRF1R and the other the modeled structure of CRF2R, were built for the simulations. A POPC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine) bilayer with the surface area of 75 Å × 75 Å on the X-Y plane was constructed using VMD13. For each system, the receptor was first embedded into the POPC bilayer using our in-house program according to the orientations provided by the OPM database30. The antagonist CP-376395 was placed in the antagonist binding pocket. A box of 75 × 75 × 100 Å3 with water molecules was then used to solvate the protein. Lipid molecules within 0.85 Å of the heavy atoms on the protein structure and water molecules in the bilayer were removed. Thereafter, sodium and chloride ions were added to produce the neutral system of 0.15 M NaCl. The resulting systems are summarized in Table 1.
Table 1. Systems prepared for the MD simulations.
| System ID | Protein | POPC | Na+ | Cl− | water |
|---|---|---|---|---|---|
| A | CRF1R | 103 | 51 | 60 | 11218 |
| B | CRF2R | 103 | 51 | 53 | 11232 |
Simulation details. MD simulations were performed using Gromacs 4.6.531,32 with the CHARMM36 parameters for the proteins, lipids, and ions and the TIP3P model for water. Force field parameters for the ligand molecule were generated with the CHARMM General Force Field (version 2b8) interface (version 0.9.7.1 beta)33 and were listed in Appendix 1. Three steps were used to equilibrate each system. In the first step, the system was subject to a 50000-step energy minimization with 1000.0 kJ/mol/nm as the force threshold. Then, the system was relaxed by an MD simulation of 100 ps with 1 fs as the time step using the NVT ensemble. In the last step, the system underwent an NPT MD simulation for 1 ns with the time step of 2 fs for equilibration.
After the equilibration run, each system was simulated for 50 ns using the NPT ensemble with the temperature and pressure set to 310 K and 1 bar, respectively. The Nose-Hoover thermostat and the Parrinello-Rahman pressure coupling were applied during the simulation. The bonds containing hydrogen atoms were constrained with the LINCS algorithm and a time step of 2 fs was used. The cubic periodic boundary conditions were applied. The cut-offs for the electrostatic and van der Waals interactions were set to 12 Å, with the long range electrostatic interactions recovered by the Particle Mesh Ewald summation.
Metadynamics simulations
Theory. Metadynamics34,35,36 has been successfully applied in describing the selectivity of a ligand towards different targets37. We performed metadynamics simulations to detect the residues that are relevant to the selectivity of CP-376395 towards CRF1R and CRF2R. In a metadynamics simulation, an additional history-dependent biased potential VG(S, t) was introduced into the system,
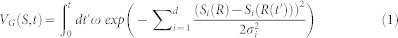 |
where t represents time, S represents the collective variables, ω is the energy rate and σi controls the width of the Gaussian for the ith collective variable. With the evolution of the system, the wells in the FES of the collective variables are filled up with the biased potential VG. The underlying free energy −F(S) is assumed to be estimated from the biased potential once all the wells have been filled after a sufficiently long time,
 |
The correctness of the relationship as shown in equation 2 has proven to be empirical by extensive tests under the assumption that the stochastic dynamics in the collective variable space is memoryless in the absence of the bias. Under the assumption, the error in FES construction has proven to be:
 |
Where D is the intrinsic system diffusion coefficient in the collective variable space, kB is the Boltzmann constant, and T is the temperature of the system38.
In fact, if one is interested in reconstructing the free energy surface from a metadynamics simulation, the simulation should be stopped once the biased potential fulfills the underlying FES in the region of interest. If the simulation does not stop as soon as the system exits from the minima, the biased potential would overfill the minima and push the system to high energy regions with respect to the collective variable space. To solve the problem, a “well-tempered” and “smoothly converging” algorism is introduced by Barducci et al39. In the well-tempered metadynamics, the deposition rate for the biased potential decreases by rescaling the Gaussian height (W) over the simulation time
 |
where VG(S, t) is the biased potential at the current position and current time, τG is the deposition stride, and ΔT is a temperature-like parameter. The underlying free energy is a scaled approximation to the VG(S, t), with
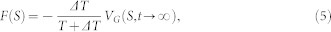 |
With respect to the standard metadynamics, the biased potential decreases as 1/t when the simulation proceeds, which allows to smoothly converge to an approximation of F(S).
Simulation details. We have carried out 50 ns well-tempered metadynamics simulations for the systems of CRF1R and CRF2R with the antagonist CP-376395 in their antagonist binding pockets. For each system, the metadynamics simulations were carried out two times, with the last snapshot from the unbiased MD simulation used as the initial structure for the simulations. The metadynamics simulations were carried out using plumed 2.02 implemented in Gromacs 4.6.5. The collective variables were selected based on the unbiased MD simulations. The residues Phe3.44 and Tyr6.63 have been suggested to be working as the bottleneck for the binding of CP-376395 to its binding pocket1. We have observed a rotational switch of Tyr3236.63 in CRF2R in the unbiased MD simulation. This rotational switch opens the bottleneck that controls the binding of CP-376395 to the antagonist binding pocket. Thus, the χ1 torsional angle of Tyr6.63 was selected to control the opening and closing of the bottleneck and used as the first collective variable. The nitrogen on the dimethylpyridine group of the antagonist forms a hydrogen bond with the side chain of Asn5.50 in CRF1R and CRF2R. This hydrogen bond plays a pivotal role in the binding of CP-376395 to the antagonist binding pockets. In addition, a dissociation pathway along the Z-axis from the antagonist binding pocket to the extracellular side of the receptor CRF1R was proposed base on the random acceleration MD simulations16. We thus select the Z-component of the vector connecting the nitrogen on the dimethylpyridine group of CP-376395 and CY on the Asn5.50 as the second collective variable. For the metadynamics simulations, the biasing potential was added every 250 steps, with the width and height of the Gaussian hills set to 0.05 and 0.3 kJ/mol, respectively, and ΔT = 2700.
Author Contributions
X.S. and X.W. carried out the molecular dynamics simulations. X.S. and Y.T. designed the study and analyzed the data. Y.T. was responsible for the project. X.S., J.C., Y.T., H.Å. and Y.T. contributed to writing and commenting on the manuscript.
Supplementary Material
Supporting information
Dissociation of CP-376395 from CRF1R
Dissociation of CP-376395 from CRF2R
Additional SI
Acknowledgments
This work was supported by the grants from the Swedish National Infrastructure for Computing (SNIC), SNIC2013-26-31 and SNIC2013-1-236. X.S. acknowledges the China Scholarship Council for financial support.
References
- Hollenstein K. et al. Structure of class B GPCR corticotropin-releasing factor receptor 1. Nature 499, 438–443 (2013). [DOI] [PubMed] [Google Scholar]
- Vale W., Spiess J., Rivier C. & Rivier J. Characterization of a 41-residue ovine hypothalamic peptide that stimulates secretion of corticotropin and beta-endorphin. Science 213, 1394–1397 (1981). [DOI] [PubMed] [Google Scholar]
- Owens M. J. & Nemeroff C. B. Physiology and pharmacology of corticotropin-releasing factor. Pharmacol. Rev. 43, 425–473 (1991). [PubMed] [Google Scholar]
- Janssen D. & Kozicz T. Is it really a matter of simple dualism? Corticotropin-releasing factor receptors in body and mental health. Front. Endocrinol. 4, 28 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Refojo D. et al. Glutamatergic and dopaminergic neurons mediate anxiogenic and anxiolytic effects of CRHR1. Science 333, 1903–1907 (2011). [DOI] [PubMed] [Google Scholar]
- Giardino W., Mark G., Stenzel-Poore M. & Ryabinin A. Dissociation of corticotropin-releasing factor receptor subtype involvement in sensitivity to locomotor effects of methamphetamine and cocaine. Psychopharmacology 219, 1055–1063 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleck B. A., Hoare S. R. J., Pick R. R., Bradbury M. J. & Grigoriadis D. E. Binding Kinetics Redefine the Antagonist Pharmacology of the Corticotropin-Releasing Factor Type 1 Receptor. J. Pharmacol. Exp. Ther. 341, 518–531 (2012). [DOI] [PubMed] [Google Scholar]
- Magnani F. et al. Electronic Sculpting of Ligand-GPCR Subtype Selectivity: The Case of Angiotensin II. ACS Chem. Biol. 9, 1420–1425 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. L. et al. 2-Aryloxy-4-alkylaminopyridines: Discovery of Novel Corticotropin-Releasing Factor 1 Antagonists. J. Med. Chem. 51, 1385–1392 (2008). [DOI] [PubMed] [Google Scholar]
- Wang J. & Verkhivker G. M. Energy Landscape Theory, Funnels, Specificity, and Optimal Criterion of Biomolecular Binding. Phys. Rev. Lett. 90, 188101 (2003). [DOI] [PubMed] [Google Scholar]
- Wootten D., Simms J., Miller L. J., Christopoulos A. & Sexton P. M. Polar transmembrane interactions drive formation of ligand-specific and signal pathway-biased family B G protein-coupled receptor conformations. Proc. Natl. Acad. Sci. U.S.A. 110, 5211–5216 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell S. C. et al. Structure validation by Cα geometry: φ,Ψ and Cβ deviation. Proteins: Struct. Funct. Bioinf. 50, 437–450 (2003). [DOI] [PubMed] [Google Scholar]
- Rosenbaum D. M., Rasmussen S. G. F. & Kobilka B. K. The structure and function of G-protein-coupled receptors. Nature 459, 356–363 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatakrishnan A. J. et al. Molecular signatures of G-protein-coupled receptors. Nature 494, 185–194 (2013). [DOI] [PubMed] [Google Scholar]
- Hjorth S. A., Orskov C. & Schwartz T. W. Constitutive activity of glucagon receptor mutants. Mol. Endocrinol. 12, 78–86 (1998). [DOI] [PubMed] [Google Scholar]
- Bai Q., Shi D., Zhang Y., Liu H. & Yao X. Exploration of the antagonist CP-376395 escape pathway for the corticotropin-releasing factor receptor 1 by random acceleration molecular dynamics simulations. Mol. Biosyst. 10, 1958–1967 (2014). [DOI] [PubMed] [Google Scholar]
- Dror R. O. et al. Activation mechanism of the beta2-adrenergic receptor. Proc. Natl. Acad. Sci. U. S. A. 108, 18684–18689 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dror R. O. et al. Identification of two distinct inactive conformations of the beta2-adrenergic receptor reconciles structural and biochemical observations. Proc. Natl. Acad. Sci. U. S. A. 106, 4689–4694 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebon G., Warne T. & Tate C. G. Agonist-bound structures of G protein-coupled receptors. Curr. Opin. Struct. Biol. 22, 482–490 (2012). [DOI] [PubMed] [Google Scholar]
- Moukhametzianov R. et al. Two distinct conformations of helix 6 observed in antagonist-bound structures of a β1-adrenergic receptor. Proc. Natl. Acad. Sci. U.S.A. 108, 8228–8232 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya S., Hall S. E., Li H. & Vaidehi N. Ligand-Stabilized Conformational States of Human β(2) Adrenergic Receptor: Insight into G-Protein-Coupled Receptor Activation. Biophys. J. 94, 2027–2042 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liaw C. W., Grigoriadis D. E., Lorang M. T., De Souza E. B. & Maki R. A. Localization of Agonist- and Antagonist-Binding Domains of Human Corticotropin-Releasing Factor Receptors. Mol. Endocrinol. 11, 2048–2053 (1997). [DOI] [PubMed] [Google Scholar]
- Pan A. C., Borhani D. W., Dror R. O. & Shaw D. E. Molecular determinants of drug-receptor binding kinetics. Drug Discov. Today 18, 667–673 (2013). [DOI] [PubMed] [Google Scholar]
- Copeland R. A., Pompliano D. L. & Meek T. D. Drug-target residence time and its implications for lead optimization. Nat. Rev. Drug Discov. 5, 730–739 (2006). [DOI] [PubMed] [Google Scholar]
- Wang C. et al. Structural basis for Smoothened receptor modulation and chemoresistance to anticancer drugs. Nat. Commun. 5, 4355 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C. et al. Structure of the human smoothened receptor bound to an antitumour agent. Nature 497, 338–343 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. D., Higgins D. G. & Gibson T. J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic. Acids. Res. 22, 4673–4680 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson M. P. et al. A hierarchical approach to all-atom protein loop prediction. Proteins: Struct. Funct. Bioinf. 55, 351–367 (2004). [DOI] [PubMed] [Google Scholar]
- Humphrey W., Dalke A. & Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38 (1996). [DOI] [PubMed] [Google Scholar]
- Lomize M. A., Lomize A. L., Pogozheva I. D. & Mosberg H. I. OPM: orientations of proteins in membranes database. Bioinformatics 22, 623–625 (2006). [DOI] [PubMed] [Google Scholar]
- Van Der Spoel D. et al. GROMACS: fast, flexible, and free. J. Comput. Chem. 26, 1701–1718 (2005). [DOI] [PubMed] [Google Scholar]
- Hess B., Kutzner C., van der Spoel D. & Lindahl E. GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. J Chem. Theory. Comput. 4, 435–447 (2008). [DOI] [PubMed] [Google Scholar]
- Vanommeslaeghe K. et al. CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 31, 671–690 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laio A. & Parrinello M. Escaping free-energy minima. Proc. Natl. Acad. Sci. U.S.A. 99, 12562–12566 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonomi M. et al. PLUMED: A portable plugin for free-energy calculations with molecular dynamics. Comput. Phys. Commun. 180, 1961–1972 (2009). [Google Scholar]
- Tribello G. A., Bonomi M., Branduardi D., Camilloni C. & Bussi G. PLUMED 2: New feathers for an old bird. Comput. Phys. Commun. 185, 604–613 (2014). [Google Scholar]
- Limongelli V. et al. Molecular basis of cyclooxygenase enzymes (COXs) selective inhibition. Proc. Natl. Acad. Sci. U.S.A. 107, 5411–5416 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barducci A., Bonomi M. & Parrinello M. Metadynamics. Wiley Interdiscip. Rev. Comput. Mol. Sci. 1, 826–843 (2011). [Google Scholar]
- Barducci A., Bussi G. & Parrinello M. Well-Tempered Metadynamics: A Smoothly Converging and Tunable Free-Energy Method. Phys. Rev. Lett. 100 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information
Dissociation of CP-376395 from CRF1R
Dissociation of CP-376395 from CRF2R
Additional SI