SUMMARY
Proper regulation of chromatin structure is necessary for the maintenance of cell type-specific gene expression patterns. The embryonic stem cell (ESC) expression pattern governs self-renewal and pluripotency. Here, we present an RNAi screen in mouse ESCs of 1008 loci encoding chromatin proteins. We identified 68 proteins that exhibit diverse phenotypes upon knockdown (KD), including seven subunits of the Tip60-p400 complex. Phenotypic analyses revealed that Tip60-p400 is necessary to maintain characteristic features of ESCs. We show that p400 localization to the promoters of both silent and active genes is dependent upon histone H3 lysine 4 trimethylation (H3K4me3). Furthermore, the Tip60-p400 KD gene expression profile is enriched for developmental regulators and significantly overlaps with that of the transcription factor Nanog. Depletion of Nanog reduces p400 binding to target promoters without affecting H3K4me3 levels. Together, these data indicate that Tip60-p400 integrates signals from Nanog and H3K4me3 to regulate gene expression in ESCs.
INTRODUCTION
ESCs are derived from the inner cell mass (ICM) of the blasto-cyst-stage embryo and are defined by two properties: self-renewal, the ability to proliferate without a change in phenotype, and pluripotency, the ability to differentiate into any cell in the organism (Niwa, 2007). Because of these properties, ESCs are regarded as a potential source of material for stem cell therapies. A molecular understanding of the factors necessary for ESC self-renewal and pluripotency is essential for moving toward the implementation of such therapies.
Transcription factors (TFs) and chromatin regulatory proteins play a major role in regulation of self-renewal by maintaining an ESC-specific gene expression pattern (Niwa, 2007; Spivakov and Fisher, 2007). Several TFs like Oct4, Sox2, Foxd3, and Stat3 are required for pluripotency and self-renewal. Another TF, Nanog, is necessary for robust self-renewal, as Nanog mutant ESCs show an increased frequency of differentiated cells in the population (Chambers et al., 2007). In contrast, the role of chromatin regulation in these processes is not as well understood. In ESCs, Polycomb group (PcG) proteins directly repress a large number of genes induced during development, including TFs that have the potential to promote differentiation (Boyer et al., 2006). Like Nanog mutant ESCs, PcG mutant ESCs continue to proliferate in an undifferentiated state with an increased frequency of differentiated cells, indicating that PcG-mediated repression is not essential for ESC self-renewal (Morin-Kensicki et al., 2001; Pasini et al., 2007).
Many developmental genes that are silent in ESCs are marked by histone modifications associated with both transcriptional activation and repression (Bernstein et al., 2006). PcG proteins direct trimethylation of histone H3 at lysine 27 (H3K27me3), a repressive mark, near the transcription start sites (TSS) of their targets (Bernstein et al., 2006; Boyer et al., 2006). Histone H3 lysine 4 trimethylation (H3K4me3), a mark generally associated with expression, is also present at the TSS of most of these genes (Bernstein et al., 2006). It is thought that the presence of both activating and repressive chromatin marks keeps these developmental regulators silent in ESCs, but poised for activation should they receive the appropriate cues.
In addition to PcG proteins, a small set of other chromatin regulatory proteins has been implicated in ESC self-renewal or pluripotency. ESCs mutant for Mbd3, a component of the NuRD corepressor complex, can proliferate normally in an undifferentiated state but are impaired in their ability to differentiate, indicating a defect in pluripotency (Kaji et al., 2006). Mutation of genes encoding subunits of the Swi/Snf family of ATP-dependent chromatin remodeling complexes, Brg1, Baf47/Snf5, and Baf155, results in peri-implantation lethality (Bultman et al., 2000; Guidi et al., 2001; Kim et al., 2001; Klochendler-Yeivin et al., 2000; Roberts et al., 2000), consistent with a defect in self-renewal, pluripotency or derivation of the ICM. Embryos lacking Tip60 and Trrap, two components of the Tip60-p400 his-tone acetyltransferase (HAT) and nucleosome remodeling complex, also die before implantation (Gorrini et al., 2007; Herceg et al., 2001), suggesting a role for this complex in early development. In sum, these data suggest roles for chromatin regulatory complexes in ESC self-renewal, pluripotency and differentiation. However, a direct examination of ESCs lacking individual chromatin regulators is necessary to identify and understand their functions.
Here, we describe a large-scale RNAi screen in ESCs, in which we examine the roles of 1008 loci encoding chromatin proteins in ESCs. We report 68 genes with KD phenotypes, most of which were not previously known to function in ESCs. In depth analysis of one group of hits encoding the Tip60-p400 complex reveals that ESCs lacking this complex continue to express pluripotency TFs but lack most of the defining characteristics of ESCs. We find that Tip60-p400 is required for repression of a large group of genes expressed during differentiation. Binding of p400 to its targets requires both H3K4me3 and the pluripotency TF Nanog. These data broaden the landscape of factors regulating ESC identity and define an important role for Tip60-p400 in the Nanog transcriptional network.
RESULTS
RNAi Screening in ESCs
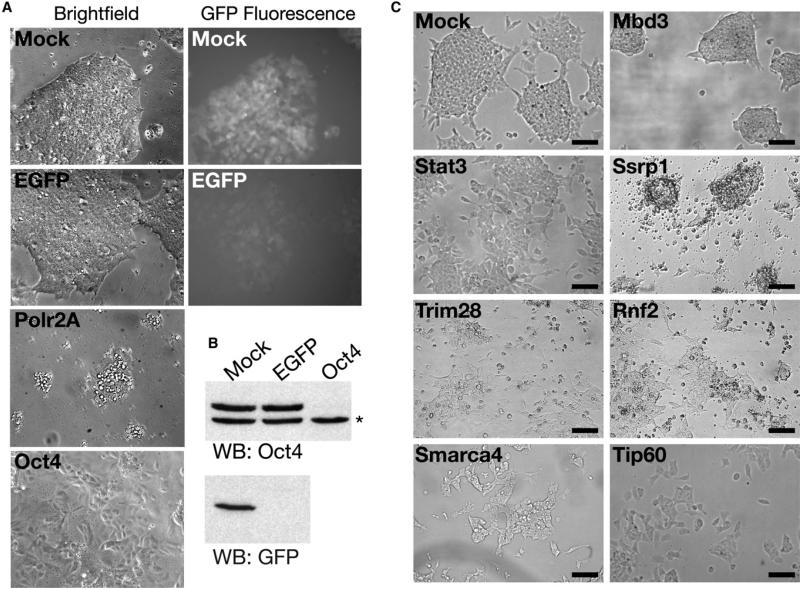
To identify chromatin proteins important in ESCs, we carried out an RNAi screen for proteins necessary for normal ESC growth. We employed esiRNAs, which exhibit minimal off-target silencing (Kittler et al., 2007; Yang et al., 2002). We typically observed esiRNA-mediated KD in > 90% of ESCs (Figure 1A and 1B). Oct4 KD resulted in apparent differentiation of nearly all cells (Figure 1A), consistent with previous reports (Hough et al., 2006; Velkey and O'Shea, 2003). Furthermore, KD of Polr2A, the large subunit of RNA Polymerase II, resulted in extensive cell death (Figure 1A). Therefore, esiRNAs are highly effective in RNAi-mediated gene silencing in ESCs.
Figure 1. Examples of Knockdown Phenotypes in ESCs.
(A) esiRNAs silence effectively in ESCs. Brightfield and GFP fluorescence images of ESCs bearing an EGFP transgene transfected with indicated esiRNAs. Images were obtained after 4 days of KD, except for Polr2A, which was imaged after 2 days.
(B) Western blotting of indicated proteins in ESCs knocked down as in (A). The asterisk indicates a background band in the Oct4 blot, which serves as a loading control.
(C) Examples of phenotypes from the esiRNA screen. ESCs were mock-transfected or transfected with the indicated esiRNAs and imaged after 4 days. Scale bars equal 140 μm.
Next, we produced esiRNAs directed against most known or predicted chromatin structural and regulatory proteins as well as proteins with motifs common to chromatin regulators (Table S1 available online), and transfected them individually into mouse ESCs. Each KD was scored for defects in viability and alterations in cell or colony morphology in two different ESC lines, each tested twice. We resynthesized esiRNAs for genes exhibiting a phenotype in one or both ESC lines in the primary screen and repeated the KDs in both ESC lines. Of 1008 targets screened, 68 exhibited reproducible phenotypes in both ESC lines (Table 1). To validate the hits we retested 20 with a second, independent esiRNA. All 20 reproduced the original phenotype (Table 1). Fur thermore, the same KD phenotype was observed with individual components of several known multi-subunit complexes. These data strongly suggest that we identified few false positives. Although several of the 68 genes exhibit early embryonic lethality in knockout mice (Table 1), most of these genes had no previously known roles in ESCs, demonstrating the utility of the screen.
Table 1.
Targets with RNAi Phenotypes
| Gene Name | Colony Morphology Phenotype | Viability Defecta | Knockout Phenotypeb | Protein Complex | Known or Predicted Function |
|---|---|---|---|---|---|
| Rnf2c | slightly flattened | 3 | embryonic lethality | Prc1 | histone ubiquitylation |
| Mettl5 | slightly flattened | 2 | DNA, RNA, or ribosomal protein methylase | ||
| Setd8c | none | 4 | histone H4K20 methyltransferase | ||
| Brcal | vertical growth | 1 | peri-implantation lethality | E3 ubiquitin ligase | |
| Sp1 | vertical growth | 0 | embryonic lethality | transcription factor | |
| Ighmbp2 | none | 4 | transcription regulation | ||
| Mbd3c | vertical growth | 0 | peri-implantation lethality | NuRD | histone deacetylation, nucleosome remodeling |
| Sin3ac | none | 4 | embryonic lethality | Sin3 HDAC | scaffold for histone deacetylases |
| Arid4a | slightly flattened | 2 | peri-natal lethality | RB-containing | recruitment of HDAC |
| Cbx3c | slightly flattened | 3 | |||
| Brd4 | none | 3 | peri-implantation lethality | P-TEFb, RFC | transcription initiation, mitosis |
| Chaf1b | none | 3 | CAF-I | chromatin assembly | |
| Tpr | none | 3 | Nuclear pore complex | ||
| Tcp1 | none | 4 | T-complex | protein chaperone, chromosome binding | |
| Rif1 | none | 3 | Rap1 | telomere length regulation | |
| Zcchc4 | slightly flattened | 0 | DNA binding | ||
| Stat3 | slightly flattened | 1 | embryonic lethality | transcription factor | |
| Ints4 | none | 3 | Integrator | RNA 3′ end processing | |
| Trim28c | slightly flattened | 3 | embryonic lethality | transcriptional corepressor | |
| Cdc5l | none | 3 | transcription factor | ||
| Ssrp1c | none | 4 | peri-implantation lethality | FACT | transcriptional elongation |
| Supt16h | none | 4 | FACT | transcriptional elongation | |
| Ruvbl1c | flattened | 3 | Tip60-p400, SRCAP, Ino80 | chromatin remodeling | |
| Ruvbl2c | flattened | 2 | Tip60-p400, SRCAP, Ino80 | chromatin remodeling | |
| Dmap1c | flattened | 1 | Tip60-p400, SRCAP | chromatin remodeling | |
| Ep400c (p400) | flattened | 2 | Tip60-p400 | chromatin remodeling | |
| Htatipc (Tip60) | flattened | 1 | Tip60-p400 | chromatin remodeling | |
| Trrapc | flattened | 1 | preimplantation lethality | Tip60-p400 | chromatin remodeling |
| Yeats4c | flattened | 2 | Tip60-p400, SRCAP | chromatin remodeling | |
| H2afz | none | 1 | embryonic lethality | chromatin structure | |
| Smarcc1 | slightly flattened | 0 | peri-implantation lethality | BRG1 | nucleosome remodeling |
| Smarca4c | slightly flattened | 0 | embryonic lethality | BRG1 | nucleosome remodeling |
| Arid1a | slightly flattened | 0 | BRG1 | nucleosome remodeling | |
| Smc1a | slightly flattened | 1 | cohesin | chromosome cohesion | |
| Smc2 | none | 4 | condensin I, condensin II | Mitotic chromosome condensation | |
| Smc4c | none | 4 | condensin I, condensin II | Mitotic chromosome condensation | |
| Gmnn | none | 4 | preimplantation lethality | DNA replication | |
| Mcm7 | none | 3 | Mcm2-7 helicase | DNA replication | |
| Mcm2c | none | 3 | Mcm2-7 helicase | DNA replication | |
| Mcm6 | none | 3 | Mcm2-7 helicase | DNA replication | |
| Mcm4 | none | 3 | preimplantation lethality | Mcm2-7 helicase | DNA replication |
| Eif4a3c | none | 4 | exon junction complex | NMD, ribosome biogenesis | |
| Ddx54 | none | 4 | Ribosome biogenesis | ||
| Ddx56 | none | 2 | Ribosome biogenesis | ||
| Ddx18c | none | 4 | Ribosome biogenesis | ||
| Ddx47c | none | 3 | Ribosome biogenesis | ||
| Utp3 | slightly flattened | 2 | processome, ribosome biogenesis | ||
| Snrpd1 | none | 4 | core spliceosome | RNA splicing | |
| Snrpd2 | none | 4 | core spliceosome | RNA splicing | |
| Snrpd3 | none | 4 | core spliceosome | RNA splicing | |
| Snrpe | none | 4 | core spliceosome | RNA splicing | |
| Snrpfc | none | 4 | core spliceosome | RNA splicing | |
| Snrpg | none | 4 | core spliceosome | RNA splicing | |
| Snrpn | none | 4 | post-natal size alteration | core spliceosome | RNA splicing |
| Lsm2c | none | 3 | core spliceosome | RNA splicing | |
| Lsm6d | none | 4 | core spliceosome | RNA splicing | |
| Lsm7 | none | 3 | core spliceosome | RNA splicing | |
| Dhx15 | none | 3 | RNA splicing | ||
| Ascc3l1 | none | 4 | U5 snRNP | RNA splicing | |
| Skiv2l2 | none | 3 | spliceosome | RNA splicing | |
| Ddx41 | none | 3 | spliceosome | RNA splicing | |
| Dhx37 | none | 3 | |||
| Dhx8 | none | 4 | RNA splicing | ||
| Ddx39 | none | 4 | spliceosome | RNA splicing | |
| Aqr | none | 4 | spliceosome | RNA splicing | |
| Sf3b1 | none | 4 | preimplantation lethality | spliceosome | RNA splicing |
| Phf5a | none | 4 | spliceosome | RNA splicing | |
| Ddx19a | none | 4 | mRNA export |
Numerical scale of cell division/viability defects: 0, no effect; 1, minor; 2, moderate; 3, moderate to severe; 4, severe.
References in Table S2.
Phenotype validated with second esiRNA (20 of 20 tested).
esiRNAs target Lsm6 and LOC635563 (predicted Lsm6 homolog).
We observed three classes of KD phenotypes: death, alterations in ESC morphology, or alterations in colony morphology (Table 1). The most prevalent KD phenotype was cell death, common among targets that are important for fundamental cellular processes, such as transcriptional elongation, chromo-some condensation, DNA replication, splicing, ribosome bio-genesis, and mRNA export. Depletion of several chromatin regulatory proteins, including histone modifying enzymes, nucleosome remodeling factors, and chromatin assembly proteins also reduced ESC viability (Figure 1C). In addition, KD of 19 genes within this class of targets caused alterations in ESC morphology (Figure 1C), which can indicate a defect in self-renewal (Chambers et al., 2003; Kaji et al., 2006; Mitsui et al., 2003; Niwa et al., 1998, 2000). For example, depletion of the pluripotency TF Stat3, resulted in flattened and elongated cells that grew in a monolayer (Figure 1C). Finally, a small group of KDs, Mbd3, Brca1, and Sp1, resulted in ESC colonies that grew well in the vertical direction, but did not expand well horizontally (Figure 1C). Mbd3−/− ESCs are defective in differentiation (Kaji et al., 2006), suggesting a link between the vertical colony growth and the inability to respond to differentiation cues. Determining how the targets that are important for normal cell or colony morphology function in ESCs should provide new insights into mechanisms that regulate pluripotency and self-renewal.
Tip60-p400 Is Necessary for Maintenance of ESC Identity
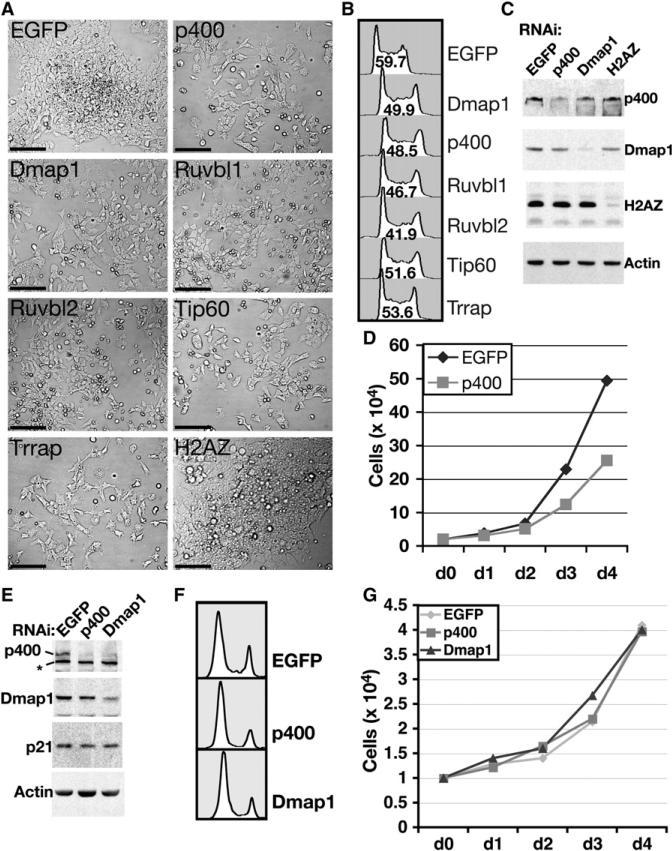
We characterized the group of hits with the most prominent cell morphology KD phenotype. Seven components of the Tip60-p400 HAT and nucleosome remodeling complex, which functions in transcriptional regulation and DNA repair (Sapountzi et al., 2006; Squatrito et al., 2006), exhibited the same unique KD phenotype. Upon depletion of any of these factors, cells exhibited a flattened and elongated morphology, reduced cell-cell contact and growth in a monolayer (Figures 2A and S1). This contrasts with the normal ESC growth pattern, in which small spherical cells grow in dense, three-dimensional colonies.
Figure 2. Phenotypes of ESCs and MEFs Depleted of Tip60-p400 Subunits.
(A) Brightfield images of ESCs depleted of six Tip60-p400 subunits, as well as H2AZ or EGFP as controls. Scale bars equal 120 μm.
(B) Cell cycle profiles of ESCs depleted of the indicated proteins. The percentage of cells in S-phase is indicated within each plot. Shown is one of two experiments with similar results.
(C) Western blotting of indicated proteins from ESCs transfected with the indicated esiRNAs.
(D) Growth curve of ESCs treated with indicated esiRNAs. Shown is one of two experiments with similar results.
(E) Western blotting of indicated proteins from MEFs transfected with the indicated esiRNAs. The asterisk indicates a background band.
(F) Cell cycle profiles of MEFs depleted of the indicated proteins.
(G) Growth curve of MEFs treated with indicated esiRNAs. Shown is one of two experiments with similar results.
In human fibroblasts, KD of p400 results in upregulation of the CDK inhibitor p21, which causes G1 arrest and premature senescence (Chan et al., 2005). In contrast, ESCs depleted of any of several Tip60-p400 subunits continued to cycle, with a reduction in S-phase cells (Figure 2B), an increase in the G2 population, a reduction in proliferation rate (Figure 2D), and no detectable upregulation of p21 (Figure S2). The effect of Tip60-p400 depletion on proliferation was unique to ESCs, as the doubling time, cell morphology and p21 levels of KD primary mouse embryo fibroblasts (MEFs) were similar to controls, despite a modest effect on their cell cycle (Figures 2E–2G).
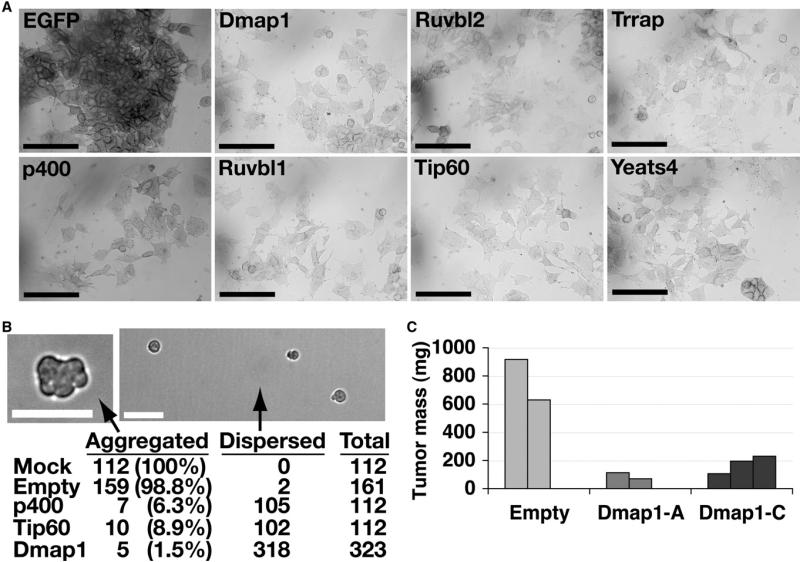
In light of the unusual morphology of Tip60-p400 KD ESCs, we assayed whether other characteristics of ESCs were altered upon KD. Alkaline phosphatase (AP) activity is high in undifferentiated ESCs and becomes weaker upon differentiation (Martin and Evans, 1975). ESCs depleted of Tip60-p400 subunits exhibited weak AP staining relative to controls (Figure 3A). Another feature of ESCs is the ability to aggregate in suspension to form embryoid bodies (EBs) (Martin and Evans, 1975). To test whether Tip60-p400 is important for this process, we placed mock, Tip60, or p400 KD ESCs in hanging drops and scored whether they aggregated within 24 hr (Figure 3B). Control cells formed EBs in almost every drop, while most Tip60-p400 KD ESCs remained dispersed, indicating that Tip60-p400 KD ESCs generally failed to aggregate to form EBs. In addition, ESCs infected with lentiviruses expressing shRNAs specific for the Dmap1 subunit of the Tip60-p400 complex also failed to aggregate, while ESCs infected with a control virus aggregated normally (Figure 3B). Consistent with a defect in aggregation, the few Tip60-p400 KD EBs present after 4 days were much smaller than controls (Figure S3).
Figure 3. Tip60-p400 Is Required for Maintenance of ESC Identity.
(A) AP staining of ESCs transfected for four days with the indicated esiRNAs. Scale bars equal 100 μm.
(B) EB formation. Mock transfected, p400 KD, or Tip60 KD ESCs were aggregated in hanging drops. Alternatively, ESCs infected with an empty vector lentivirus or lentivirus expressing the Dmap1-A shRNA were aggregated. (Top) Brightfield images of empty vector (left) or Dmap1-A shRNA (right) ESCs. Scale bars equal 40 μm. (Bottom) Percentage of hanging drops in which ESCs aggregated or remained dispersed after 24 hr.
(C) Tumor mass of teratomas formed from ESCs infected with lentiviruses expressing Dmap1 shRNAs or empty vector lentiviruses 21 days after injection into nude mice.
A defining feature of ESCs is pluripotency. To assess pluripotency of Tip60-p400 KD ESCs, we infected ESCs with Dmap1 shRNA lentiviruses and tested whether these ESCs could form normal teratomas. While the shRNAs silenced Dmap1 effectively, there was a strong selection against Dmap1 silencing, as ESCs infected with these viruses regained Dmap1 expression after moderate passaging in culture (Figure S4). As with the cultured ESCs, teratomas from Dmap1 shRNA-expressing ESCs exhibited reactivation of Dmap1 expression (Figure S4). Despite this reactivation, Dmap1 shRNA-expressing ESCs formed teratomas that were much smaller (Figure 3C) and less complex than controls, with limited representation of cell types from all three germ layers (Figure S4). These data indicate that Dmap1 expression is necessary for robust proliferation or differentiation of ESCs in teratomas, suggesting a partial defect in pluripotency.
In summary, ESCs depleted of Tip60-p400 complex have defects in three distinguishing features of ESCs: AP activity, EB formation and teratoma formation. These defects, combined with the altered cellular and colony morphology, cell cycle and growth rate upon Tip60-p400 KD indicate that this complex is necessary for maintenance of normal ESC identity. Consistent with these results, mouse embryos homozygous for deletion of Trrap or Htatip (Tip60), which encode subunits of the Tip60-p400 complex, die before implantation (Gorrini et al., 2007; Herceg et al., 2001) and Trrap mutant ESCs exhibit phenotypes comparable to those of Tip60-p400 KD ESCs (Z. Herceg, personal communication).
Tip60-p400 Represses Transcription of Genes Induced during Development
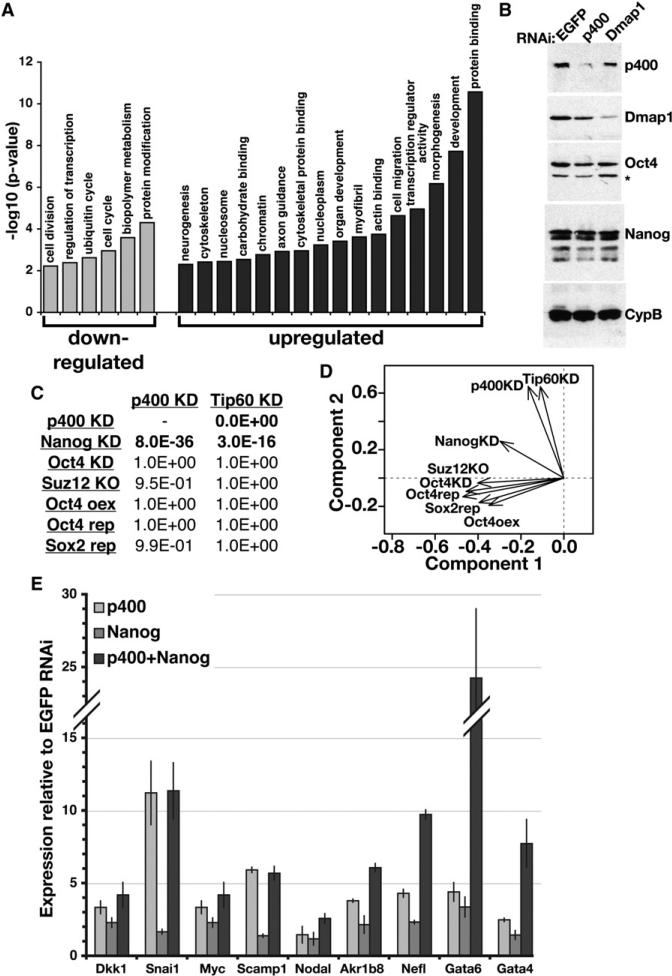
To gain insight into the transcriptional role of the Tip60-p400 complex in maintenance of ESC identity, we examined changes in ESC gene expression upon KD of Tip60 or p400. 802 genes were differentially expressed in both Tip60 and p400 KD ESCs, including 128 downregulated and 674 upregulated genes (Tables S3 and S4), suggesting that Tip60-p400 acts as a transcriptional repressor at most of its targets. RT-qPCR for ESCs depleted of different Tip60-p400 subunits showed similar effects on expression of several genes tested (Figure S5), arguing that the whole Tip60-p400 complex is necessary to repress transcription.
Genes misregulated in Tip60-p400 KD ESCs were significantly enriched for a number of functional categories (Figure 4A, Table S5). Downregulated genes were enriched for cell-cycle regulators, metabolic genes and genes required for cell division, which could be either a cause or consequence of the cell cycle phenotype observed upon KD of Tip60-p400 subunits (Figure 2B). In contrast, upregulated genes were enriched for several categories relating to differentiation and embryonic development (Figure 4A).
Figure 4. Overlapping Functions of Tip60-p400 and Nanog Revealed by Expression Profiling.
(A) The significance of enrichment of a subset of Gene Ontology (GO) terms common among genes misregulated in p400 KD ESCs. Similar categories were enriched in the Tip60 KD dataset.
(B) Protein levels of Tip60-p400 subunits (p400, Dmap1), TFs (Oct4, Nanog), and a loading control (cyclophilin B, CypB) were determined by Western blotting. The asterisk indicates a background band.
(C) Comparison of ESC expression profiles upon KD, knockout (KO), inducible repression (rep) or overexpression (oex) of the indicated factors (Loh et al., 2006; Masui et al., 2007; Matoba et al., 2006; Pasini et al., 2007). p-values (one-sided Fisher's exact test) indicating statistical significance of overlap with Tip60 or p400 KD are shown.
(D) Principal component analysis of gene expression profiles from (C).
(E) RT-qPCR measuring mRNA levels of Nanogand Tip60-p400-target genes (indicated below) in ESCs depleted of p400, Nanog, or both, relative to control (EGFP)-depleted ESCs. Data are represented as mean ± SD.
Although p400 KD ESCs lack many features of normal, pluripotent ESCs and express a large number of developmental genes, we did not observe a significant decrease in mRNA levels of ESC markers Rex1/Zfp42, Sox2, Utf1, Esg1/Dppa5, Stat3, or Cripto1/Tdgf1 (Tables S3 and S4) or in protein levels of Oct4 and Nanog (Figure 4B). Therefore, Tip60-p400 depleted ESCs simultaneously express differentiation and pluripotency genes. In principle, these data could be explained by the presence of a small percentage of differentiated cells in an otherwise normal ESC population. However, there was no obvious heterogeneity within the Tip60-p400 knockdown population, which uniformly exhibited a flattened and elongated cell morphology. In addition, there was no notable cell-to-cell variation in levels of Oct4 for both the mock and Tip60-p400 KD populations (data not shown). This suggests that Tip60-p400 depleted ESCs self-renew in an intermediate state, lacking most of the defining features of ESCs (Figures 2, 3, S1, S3, and S4), but fail to fully differentiate. Several possibilities can account for this result. First, developmental cues present during differentiation may be required for downregulation of pluripotency genes. Second, Tip60-p400 may be required to repress both differentiation genes in ESCs and ESC-specific genes upon differentiation. Third, Tip60-p400 may function downstream of ESC-specific TFs, such that Tip60-p400 depletion perturbs ESC identity without affecting expression of known self-renewal and pluripotency master regulators. We explored the last possibility further.
Tip60-p400 and Nanog Regulate Overlapping Sets of Genes
To better understand the function of Tip60-p400 in regulation of the ESC transcription program, we compared the Tip60 and p400 KD expression profiles to those of factors known to function in ESCs. There was significant overlap between the sets of genes misregulated in Tip60-p400 KD and Nanog KD, but not between those in Tip60-p400 KD and Oct4 KD, Suz12 knockout, Oct4 overexpression or repression, or Sox2 repression (Figure 4C). We used principal component analysis to graphically project the correlations between the Tip60-p400 KD expression data and these other expression profiles (Figure 4D; Table S6). The Tip60 and p400 KD expression profiles were again distinct from those of Oct4, Sox2 and Suz12. In contrast, the Nanog KD expression profile overlapped partially with both the Oct4/Sox2/Suz12 group and the Tip60-p400 group, indicating that Tip60-p400 depletion induces changes in expression comparable to Nanog depletion for many genes that are unaffected in the other data sets.
Given the overlap between the Tip60-p400 and Nanog KD expression profiles, we examined whether these factors function in the same pathway or in different pathways to repress developmental genes. ESCs were depleted of p400, Nanog, or both and mRNA levels were analyzed. For some genes (Dkk1, Snai1, Myc, Scamp1), simultaneous KD of both p400 and Nanog increased expression to levels similar to p400 KD alone (Figure 4E). In contrast, two master regulators of primitive endoderm differentiation, Gata4 and Gata6 (Fujikura et al., 2002), were more severely affected in the double KD than in either single KD. A few genes (Akr1b8, Nefl, Nodal) exhibited intermediate effects. These data suggest that Nanog and Tip60-p400 function together to regulate a common set of genes.
p400 Localization Strongly Correlates with H3K4me3 at Both Active and Silent Genes in ESCs
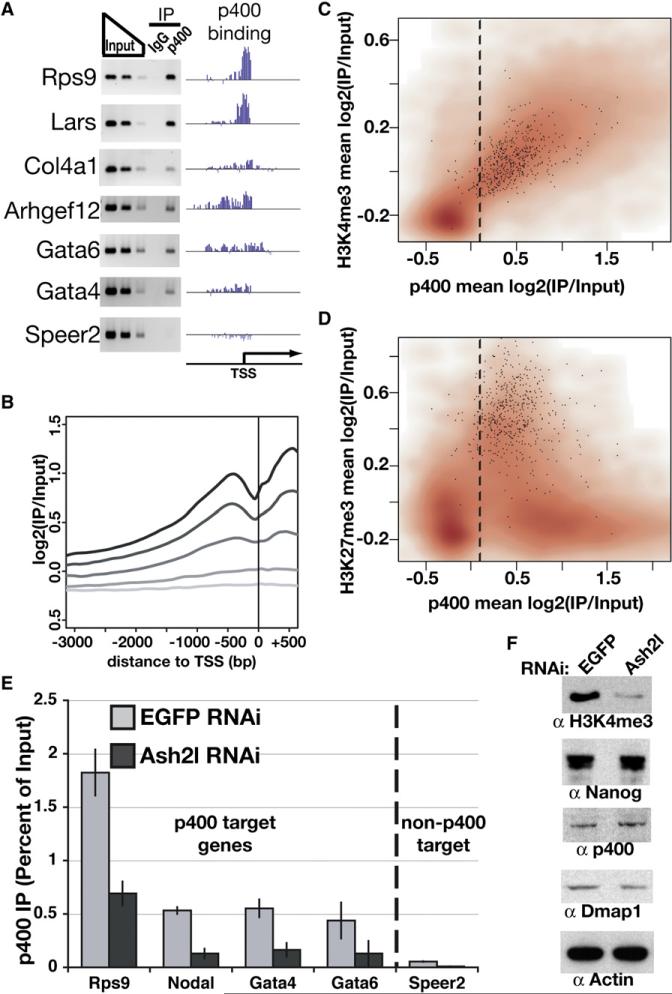
We next performed p400 chromatin immunoprecipitation and hybridization to promoter tiling arrays to determine which genes misregulated upon Tip60-p400 KD were bound by this complex in ESCs (Table S7). We observed high confidence binding of p400 at 55% of all promoter regions (Figure S6). These results were validated by PCR for several targets (Figure 5A).
Figure 5. Genome-Wide Location Analysis of p400.
(A) PCR verification of p400 ChIP at genes with varying enrichment of p400. “p400 binding” plots p400 enrichment (y axis) for each probe (x axis) tiling the promoters indicated.
(B) p400 enrichment was plotted relative to the TSS. Genes were binned into quintiles based on expression level in ESCs, ranging from highly expressed genes (black) to nonexpressed genes (lightest gray).
(C and D) Density scatterplots of each gene promoter average for p400 enrichment versus H3K4me3 (Maherali et al., 2007) (C) or H3K27me3 (Maherali et al., 2007) (D) enrichment. Direct targets of Polycomb repressive complex 2 (PRC2) (Boyer et al., 2006) are indicated with black dots. The dashed line separates p400-enriched from nonenriched genes, as in Figure S6.
(E) ChIP in Ash2l or EGFP KD ESCs for p400 measured by qPCR and expressed as a percentage of input. The dashed line separates p400 target genes (according to genome-wide location data and confirmed for several genes in Figure 5A) from a gene without p400 enrichment (Speer2), included as a control. IgG control qPCRs were near zero (Figure S12). Data are represented as mean ± SD.
(F) Western blotting for the proteins indicated in EGFP and Ash2l KD ESCs.
Since p400 was enriched at most promoters, we examined whether p400 occupancy correlated with gene expression or abundant histone modifications. There was a positive correlation between p400 enrichment and gene expression (r = 0.64); the most highly expressed genes showed the greatest enrichment for p400 (Figure 5B). Despite the fact that p400 binds 55% of all promoters in ESCs, we also observed a significant overlap between p400-bound promoters and genes whose expression is affected by p400 KD (Figure S7).
While the magnitude of p400 enrichment at promoters generally correlated with the level of gene expression, PcG targets were a notable exception (Figure S8). PcG targets are unusual among silent genes in ESCs, in that they have chromatin modifications associated with silent (H3K27me3) and active (H3K4me3) genes (Bernstein et al., 2006). There was a strong positive correlation between H3K4me3 and p400 levels at promoters (r = 0.77) (Figure 5C). Furthermore, most PcG targets are enriched for p400 (Figure 5D). These data suggest that p400 binds to promoters marked by H3K4me3, including both highly expressed genes and targets of PcG repression.
H3K4me3 and Nanog Independently Promote p400 Binding
The strong correlation between H3K4me3 and p400 localization raised the possibility that this mark may be necessary for p400 to bind its target promoters. To test this possibility, we made use of the fact that mammalian SET1-like complexes (which catalyze H3K4me3) all contain several common subunits, including Ash2l and Wdr5 (Dou et al., 2006). We therefore tested the effect of Ash2l KD on binding of p400 to its target promoters. Ash2l KD sharply reduced H3K4me3 levels, but had no effect on expression of p400 or Dmap1 (Figure 5F). Binding of p400 to its chromatin targets was significantly reduced in Ash2l KD ESCs (Figure 5E), suggesting that H3K4me3 is necessary for normal levels of chromatin binding by Tip60-p400. Similar effects on p400 binding were also observed upon KD of Wdr5 (data not shown).
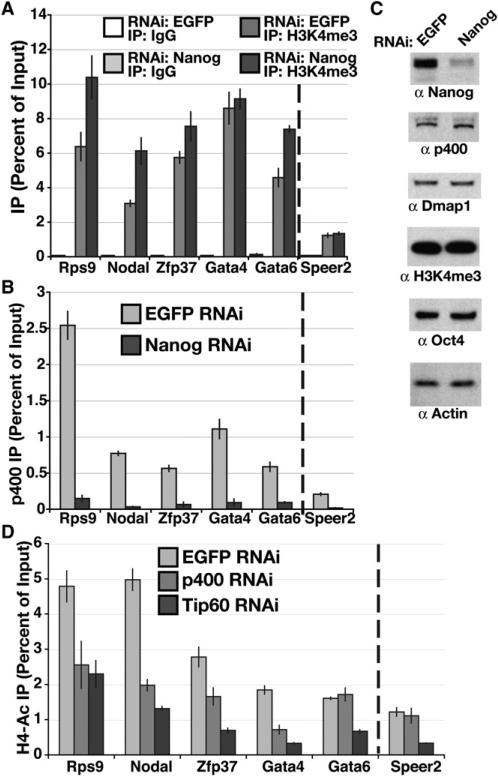
We next examined the basis of the overlap between the Nanog and Tip60-p400 KD expression profiles. We considered the possibility that Nanog regulates binding of Tip60-p400 to chromatin in ESCs, perhaps by regulation of H3K4me3 levels at Tip60-p400 target promoters. To test this possibility, we examined the localization of p400 and H3K4me3 to p400 targets upon Nanog KD. H3K4me3 levels at p400 target promoters ranged from unaffected to slightly increased in Nanog KD ESCs (Figure 6A), indicating that Nanog was not required for deposition of the H3K4me3 mark. In contrast, p400 binding to its targets was significantly reduced in Nanog-depleted ESCs (Figure 6B). Nanog depletion does not significantly affect mRNA levels of components of the Tip60-p400 complex (Loh et al., 2006) or protein levels of p400 and Dmap1 (Figure 6C), suggesting that Nanog is necessary for Tip60-p400 to bind its targets efficiently. Nanog KD ESCs showed no signs of differentiation and had levels of Oct4 protein and AP activity that were indistinguishable from controls (Figure 6C and S9), consistent with the recent finding that Nanog−/− ESCs remain undifferentiated, albeit less robustly (Chambers et al., 2007). These data indicate that the alteration in p400 binding is unlikely to be due to differentiation of ESCs upon Nanog KD. In addition, the differences in KD phenotypes between Nanog and Tip60-p400 strongly suggest that Tip60-p400 has Nanog-independent functions important for maintenance of ESC identity. The reduction in p400 binding upon Nanog KD occurred at p400 targets regardless of whether they were also Nanog targets, suggesting that Nanog does not directly recruit p400 to target promoters. We observed no effect of p400 depletion on Nanog binding or H3K4me3 levels at p400 targets (data not shown). In sum, these data indicate that H3K4me3 is necessary, but not sufficient, for targeting of Tip60-p400. H3K4me3 is not required for Nanog expression (Figure 5F), nor is Nanog required for H3K4me3 deposition (Figures 6A and 6C). Therefore, Nanog and H3K4me3 appear to act independently to promote binding of Tip60-p400 to its targets.
Figure 6. H3K4me3 and Nanog Are Independently Required for p400 Localization.
(A and B) ChIP in Nanog or EGFP KD ESCs for IgG and H3K4me3 (A) or p400 (B) at genes indicated. The dashed line separates p400 targets from a gene without p400 enrichment, as in Figure 5E. Data are represented as mean ± SD for (A), (B), and (D).
(C) Western blotting for the proteins indicated in EGFP and Nanog KD ESCs.
(D) ChIP in p400, Tip60 or EGFP KD ESCs for tetra-acetylated histone H4 (H4ac) at genes indicated. IgG control qPCRs were near zero (Figure S12).
Tip60-p400 Complex Promotes Histone H4 Acetylation at Both Active and Silent Target Promoters
Two major functions of Tip60-p400, H2AZ incorporation and histone acetylation, are thought to be associated with gene expression (Squatrito et al., 2006), yet expression profiling suggests that this complex functions mainly in transcriptional repression in ESCs. H2AZ KD did not alter ESC morphology (Table 1 and Figure 2A), suggesting that H2AZ deposition is not the essential function of the Tip60-p400 complex in ESCs. Tip60-p400 is also implicated in exchange of a second H2A variant, H2AX (Ikura et al., 2007; Kusch et al., 2004). However, as with H2AZ, H2AX loss does not pheno-copy Tip60-p400 KD (Figure S10). Therefore, we focused on the role of histone acetylation at targets of Tip60-p400-mediated repression. Bulk H4 and H2A acetylation were unaltered upon p400 KD (Figure S11), suggesting that Tip60-p400 may mediate its effects in ESCs through a gene-specific mechanism.
In p400 or Tip60 KD ESCs, we observed a decrease in histone H4 acetylation (H4ac) at the promoters of most p400-bound genes tested (Figure 6D). These included genes normally repressed by Tip60-p400 complex in ESCs (Gata4, Nodal) and a gene normally expressed in ESCs whose expression is minimally affected by Tip60-p400 KD (Rps9). Therefore, Tip60-p400 appears to acetylate histones at target promoters, regardless of whether it functions in repression of those targets. Furthermore, these data indicate that H4ac does not promote gene expression at targets of Tip60-p400-mediated silencing.
DISCUSSION
We report the first large-scale RNAi screen in ESCs, targeting most known or predicted chromatin structural or regulatory proteins. We found 68 genes with reproducible KD phenotypes in two different ESC lines, 22 of which exhibited alterations in cell and/or colony morphology, which can be indicative of a defect in self-renewal. These 22 genes include members of several nucleosome remodeling complexes, including NuRD, Brg1 and Tip60-p400, as well as histone modifying enzymes, transcriptional co-regulators and chromatin binding proteins. The remaining 46 genes include some chromatin regulators, but are enriched for chromatin structural proteins and enzymes with roles in core nuclear processes, such as DNA replication, chromosome condensation and RNA processing. It is noteworthy that several of these 46 genes have been knocked down in somatic cells without causing major defects in viability or proliferation (Baillat et al., 2005; Cortez et al., 2004; Ono et al., 2003), suggesting that ESCs are uniquely sensitive to certain perturbations of chromatin structure. This is consistent with the idea that nuclear architecture, global chromatin structure and chromatin fluidity are distinctly different in ESCs and differentiated cells (Meshorer and Misteli, 2006).
The genes with the most prominent change in ESC morphology upon KD each encode a subunit of the Tip60-p400 complex. ESCs depleted of Tip60-p400 subunits exhibited phenotypes consistent with a defect in maintenance of ESC identity, including failure to grow in colonies, poor expression of AP, failure to aggregate into EBs and defects in teratoma formation. Tip60-p400 depletion has effects on the cell cycle and proliferation rate of ESCs, which might contribute to these phenotypes.
These data represent the first demonstration that Tip60-p400 plays a role in maintenance of ESC identity. Upon KD of Tip60-p400 4% of genes were misregulated, the majority of which were upregulated. Misregulated genes overlapped significantly with genes misregulated upon Nanog KD, suggesting functional overlap of these factors. These data indicate that transcriptional repression of developmental genes is a major function of Tip60-p400 complex in ESCs.
While the most prominent effect of Tip60-p400 KD was upregulation of differentiation genes normally silent in ESCs, we found that p400 binds to more than half of all promoters, with the strongest binding at genes that are actively transcribed. Two major catalytic activities of Tip60-p400, H2AZ deposition and HAT activity, are usually associated with active transcription, which may explain why the genes bound most strongly by p400 are highly transcribed. Why then are most actively transcribed genes only minimally affected by Tip60-p400 depletion? We found that Tip60-p400 promotes H4ac at targets that are normally expressed in ESCs (and are not strongly affected by Tip60-p400 KD), as well as genes that require Tip60-p400 for silencing in ESCs. In light of these data, one explanation is that H4ac might have different functions in different contexts. At active promoters, H4ac is thought to promote transcription. However, this may be one of several redundant marks for gene activation, which would explain why actively transcribed targets of p400 are minimally affected by Tip60-p400 KD. Perhaps H4ac at inactive genes is a nonredundant mark that mediates gene silencing when found in the presence of other silencing marks. Alternatively, a nonhistone protein that mediates repression of transcription may be a more important target for acetylation by Tip60-p400 at silent genes.
p400 localization was found to closely mirror that of histone H3K4me3: they are present at highly overlapping sets of genes and both localize near the TSS (Figures 5B and 5C, (Maherali et al., 2007)). Indeed, we found that p400 binding requires this mark, consistent with in vitro data showing that Ing3, a subunit of the Tip60-p400 complex, binds peptides bearing this modification (Shi et al., 2006). In addition, binding of p400 to its chromatin targets requires Nanog, despite the fact that the H3K4me3 mark remains present upon Nanog KD. Therefore, H3K4me3 is a necessary, but not sufficient, determinant of p400 binding.
Nanog does not appear to recruit p400 by direct binding (Wang et al., 2006 and data not shown). This is not surprising, given that the binding sites for Nanog and p400 at common target genes do not frequently overlap (Loh et al., 2006 and data not shown). Therefore, we propose a model in which binding of Tip60-p400 to target promoters in ESCs is regulated by at least two mechanisms, direct binding to H3K4me3 and an indirect mechanism that depends on Nanog. The Nanog-dependent determinant of p400 binding could be mediated by a number of mechanisms. Nanog could promote expression of a transcription factor that directly recruits Tip60-p400 to specific targets that also contain H3K4me3. Alternatively, Nanog could regulate one or more enzymes that covalently modify and promote stabilization of the complex. However, no matter the mechanism by which Nanog regulates p400 binding, our data show that Tip60-p400 is an important effector of Nanog-mediated transcriptional repression, integrating signals from at least two different pathways to regulate gene expression in ESCs.
While Nanog expression is relatively restricted, Tip60-p400 is broadly expressed.Itwillbeinteresting to investigate how the function of Tip60-p400 in differentiated cells differs from its role in ESCs and how Tip60-p400 chromatin binding and enzymatic activities are regulated during development. Also, human NANOG is among the group of TFs that can direct reprogramming of differentiated cells into induced pluripotent stem (iPS) cells (Yu et al., 2007), revealing thatectopically expressed NANOG can function in differentiated cells. Determining whether NANOG can promote chromatin binding of human TIP60-p400 in differentiated cells, and whether TIP60-p400 is necessary for generation of iPS cells may provide important insights into the events that mediate reprogramming.
Here, we have used RNAi to screen for chromatin proteins necessary for normal ESC growth. ESCs hold considerable promise for the treatment of disease. Characterization of pathways important for ESC self-renewal and differentiation will be essential to their possible therapeutic use. Besides Tip60-p400, we have identified a number of proteins that are likely involved in these processes—understanding how these proteins regulate and are regulated by self-renewal, differentiation and reprogramming pathways will be an important next step.
EXPERIMENTAL PROCEDURES
Embryonic Stem Cells
ESC lines used in this study were E14 (Hooper et al., 1987); E14-EGFP, constructed by integration of pEGFP-C1 in E14 cells; Oct4-GiP (provided by A. Smith); E14-H2A-GFP, constructed by integration of pBOS-H2A-GFP (provided by D. Nusinow); and H2AX−/− (Bassing et al., 2002).
Construction of esiRNA Library and RNAi
Most known or predicted chromatin proteins in mouse were identified by searching the NCBI Gene database for genes encoding proteins with known roles in chromatin structure or regulation or for those with domains common among chromatin regulators. Templates for esiRNA production were produced using a two-step PCR procedure (Foley and O'Farrell, 2004). In vitro transcription was performed with T7 RNA polymerase and dicing of double stranded RNA was performed using RNase III. esiRNAs were transfected into ESCs using Lipofectamine 2000. See Supplemental Data for details.
Cell Cycle Analysis
Cell cycle profiling of propidium iodide stained cells was performed as described (Benanti et al., 2002). Fractions of cells in S-phase were quantified using FlowJo software.
Alkaline Phosphatase Staining
ESCs were transfected with esiRNAs for 4 days (except where noted) and stained for AP activity using a kit (Chemicon) according to the manufacturers instructions.
Embryoid Body Generation
EBs were made by placing 10 ESCs in a 20 ml volume and suspending the drops upside down over a bath of PBS. For p400 or Tip60 KD EBs, ESCs were mock transfected or transfected with p400 or Tip60 esiRNA, and after 24 hr were trypsinized and placed in hanging drops. For Dmap1 KD EBs, normal ESCs infected with Dmap1 shRNA lentiviruses (in pSICOR vector; Ventura et al., 2004) or control virus were placed in hanging drops. After 24 hr, each drop was scored for whether the ESCs had aggregated.
Teratomas
ESCs were infected with lentiviruses expressing either no shRNA or one of two shRNAs targeting the Dmap1 subunit of Tip60-p400. Infected cells were purified by FACS and injected into the posterior flanks of nude mice (5 × 106 cells per injection). After 21 days, mice were sacrificed. Tumors were weighed, fixed in 4% paraformaldehyde, sectioned and stained with hematoxylin and eosin.
Expression Profiling
RNA was isolated using Trizol reagent combined with PureLink Micro-to-Midi columns (Invitrogen). Labeled material was hybridized to whole mouse genome 4x44K Ink-jet arrays (Agilent). See Supplemental Data for details.
Chromatin Immunoprecipitation
For chromatin Immunoprecipitation (ChIP), we used 1 or 0.25 mg of formalincrosslinked chromatin for genome-wide location analysis or qPCR, respectively. ChIP was performed essentially as described (Fazzio et al., 2005) with a few modifications (Supplemental Data). Genome-wide location analysis was carried out using Mouse MM8 promoter arrays (NimbleGen; 50 base probes tiling ~every 100 base pairs from 3250 base pairs upstream to 750 base pairs downstream of each TSS). See Supplemental Data for details.
RT-qPCR and ChIP-qPCR
RT-qPCR
Cells were transfected with esiRNAs for 72 hr and total RNA was extracted. Reverse transcription was performed for 1 hr using random priming. RT-qPCR was performed in 25 μl reactions using 0.5 μl of cDNA. qPCR reactions (1× AmpliTaq Gold Buffer, 1.5 mM MgCl2, 0.2 mM dNTPs, 0.2 μM each primer, SYBR green, Taq enzyme) were performed on a Stratagene Mx3000P qPCR system, using primers specific for each gene. Data were normalized to a loading control (Cyclophilin B).
ChIP-qPCR
Dilutions of input and IP were analyzed as above and the percentage of input in the IP sample was calculated from the relative copy numbers. Oligonucleotides used in RT-qPCR and ChIP-qPCR are listed in the Supplemental Data.
Supplementary Material
ACKNOWLEDGMENTS
We thank H. Madhani, J. Benanti, M. Royce-Tolland, K. Worringer, and F. Chu for comments on the manuscript and J. Delrow, D. Nusinow, J. Sharp, J. Benanti, and D. Toczyski for technical advice, F. Alt for the H2AX mutant ESCs, M. Bigos, and V. Stepps for assistance with flow sorting and G. Martin for analysis of teratomas. We thank Z. Herceg for sharing unpublished results. We also thank J.M. Bishop, in whose lab some of these experiments were performed. Expression array data collection and preliminary analyses were performed by R. Barbeau, A. Barczak, W. Xu, and D. Erle at the Sandler Asthma Basic Research (SABRE) Center Functional Genomics Core Facility. This work was supported by NIH grant RO1 GM63671. T.G.F. was a fellow of the Jane Coffin Childs Memorial Fund for Medical Research. J.T.H. is funded by a UCSF NIH Genetics Training Grant.
Footnotes
ACCESSION NUMBERS
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO Series accession number GSE11243.
SUPPLEMENTAL DATA
Supplemental Data include Supplemental Experimental Procedures, Supplemental References, twelve figures, and seven tables and can be found with this article online at http://www.cell.com/cgi/content/full/134/1/162/DC1/.
The authors declare that they have no competing financial interests.
REFERENCES
- Baillat D, Hakimi MA, Naar AM, Shilatifard A, Cooch N, Shiekhattar R. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell. 2005;123:265–276. doi: 10.1016/j.cell.2005.08.019. [DOI] [PubMed] [Google Scholar]
- Bassing CH, Chua KF, Sekiguchi J, Suh H, Whitlow SR, Fleming JC, Monroe BC, Ciccone DN, Yan C, Vlasakova K, et al. Increased ionizing radiation sensitivity and genomic instability in the absence of histone H2AX. Proc. Natl. Acad. Sci. USA. 2002;99:8173–8178. doi: 10.1073/pnas.122228699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benanti JA, Williams DK, Robinson KL, Ozer HL, Galloway DA. Induction of extracellular matrix-remodeling genes by the senescence-associated protein APA-1. Mol. Cell. Biol. 2002;22:7385–7397. doi: 10.1128/MCB.22.21.7385-7397.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Bultman S, Gebuhr T, Yee D, La Mantia C, Nicholson J, Gilliam A, Randazzo F, Metzger D, Chambon P, Crabtree G, Magnuson T. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol. Cell. 2000;6:1287–1295. doi: 10.1016/s1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- Chambers I, Silva J, Colby D, Nichols J, Nijmeijer B, Robertson M, Vrana J, Jones K, Grotewold L, Smith A. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- Chan HM, Narita M, Lowe SW, Livingston DM. The p400 E1A-associated protein is a novel component of the p53–> p21 senescence pathway. Genes Dev. 2005;19:196–201. doi: 10.1101/gad.1280205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez D, Glick G, Elledge SJ. Minichromosome maintenance proteins are direct targets of the ATM and ATR checkpoint kinases. Proc. Natl. Acad. Sci. USA. 2004;101:10078–10083. doi: 10.1073/pnas.0403410101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Y, Milne TA, Ruthenburg AJ, Lee S, Lee JW, Verdine GL, Allis CD, Roeder RG. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat. Struct. Mol. Biol. 2006;13:713–719. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- Fazzio TG, Gelbart ME, Tsukiyama T. Two distinct mechanisms of chromatin interaction by the Isw2 chromatin remodeling complex in vivo. Mol. Cell. Biol. 2005;25:9165–9174. doi: 10.1128/MCB.25.21.9165-9174.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley E, O'Farrell PH. Functional dissection of an innate immune response by a genome-wide RNAi screen. PLoS Biol. 2004;2:E203. doi: 10.1371/journal.pbio.0020203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujikura J, Yamato E, Yonemura S, Hosoda K, Masui S, Nakao K, Miyazaki Ji J, Niwa H. Differentiation of embryonic stem cells is induced by GATA factors. Genes Dev. 2002;16:784–789. doi: 10.1101/gad.968802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorrini C, Squatrito M, Luise C, Syed N, Perna D, Wark L, Martinato F, Sardella D, Verrecchia A, Bennett S, et al. Tip60 is a haplo-insufficient tumour suppressor required for an oncogene-induced DNA damage response. Nature. 2007;448:1063–1067. doi: 10.1038/nature06055. [DOI] [PubMed] [Google Scholar]
- Guidi CJ, Sands AT, Zambrowicz BP, Turner TK, Demers DA, Webster W, Smith TW, Imbalzano AN, Jones SN. Disruption of Ini1 leads to peri-implantation lethality and tumorigenesis in mice. Mol. Cell. Biol. 2001;21:3598–3603. doi: 10.1128/MCB.21.10.3598-3603.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herceg Z, Hulla W, Gell D, Cuenin C, Lleonart M, Jackson S, Wang ZQ. Disruption of Trrap causes early embryonic lethality and defects in cell cycle progression. Nat. Genet. 2001;29:206–211. doi: 10.1038/ng725. [DOI] [PubMed] [Google Scholar]
- Hooper M, Hardy K, Handyside A, Hunter S, Monk M. HPRT-deficient (Lesch-Nyhan) mouse embryos derived from germline colonization by cultured cells. Nature. 1987;326:292–295. doi: 10.1038/326292a0. [DOI] [PubMed] [Google Scholar]
- Hough SR, Clements I, Welch PJ, Wiederholt KA. Differentiation of mouse embryonic stem cells after RNA interference-mediated silencing of OCT4 and Nanog. Stem Cells. 2006;24:1467–1475. doi: 10.1634/stemcells.2005-0475. [DOI] [PubMed] [Google Scholar]
- Ikura T, Tashiro S, Kakino A, Shima H, Jacob N, Amunugama R, Yoder K, Izumi S, Kuraoka I, Tanaka K, et al. DNA Damage-Dependent Acetylation and Ubiquitination of H2AX Enhances Chromatin Dynamics. Mol. Cell. Biol. 2007;27:7028–7040. doi: 10.1128/MCB.00579-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaji K, Caballero IM, MacLeod R, Nichols J, Wilson VA, Hendrich B. The NuRD component Mbd3 is required for pluripotency of embryonic stem cells. Nat. Cell Biol. 2006;8:285–292. doi: 10.1038/ncb1372. [DOI] [PubMed] [Google Scholar]
- Kim JK, Huh SO, Choi H, Lee KS, Shin D, Lee C, Nam JS, Kim H, Chung H, Lee HW, et al. Srg3, a mouse homolog of yeast SWI3, is essential for early embryogenesis and involved in brain development. Mol. Cell. Biol. 2001;21:7787–7795. doi: 10.1128/MCB.21.22.7787-7795.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittler R, Surendranath V, Heninger AK, Slabicki M, Theis M, Putz G, Franke K, Caldarelli A, Grabner H, Kozak K, et al. Genome-wide resources of endoribonuclease-prepared short interfering RNAs for specific loss-of-function studies. Nat. Methods. 2007;4:337–344. doi: 10.1038/nmeth1025. [DOI] [PubMed] [Google Scholar]
- Klochendler-Yeivin A, Fiette L, Barra J, Muchardt C, Babinet C, Yaniv M. The murine SNF5/INI1 chromatin remodeling factor is essential for embryonic development and tumor suppression. EMBO Rep. 2000;1:500–506. doi: 10.1093/embo-reports/kvd129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusch T, Florens L, Macdonald WH, Swanson SK, Glaser RL, Yates JR, 3rd, Abmayr SM, Washburn MP, Workman JL. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science. 2004;306:2084–2087. doi: 10.1126/science.1103455. [DOI] [PubMed] [Google Scholar]
- Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, Stadtfeld M, Yachechko R, Tchieu J, Jaenisch R, et al. Directly Reprogrammed Fibroblasts Show Global Epigenetic Remodeling and Widespread Tissue Contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Martin GR, Evans MJ. Differentiation of clonal lines of teratocarcinoma cells: formation of embryoid bodies in vitro. Proc. Natl. Acad. Sci. USA. 1975;72:1441–1445. doi: 10.1073/pnas.72.4.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masui S, Nakatake Y, Toyooka Y, Shimosato D, Yagi R, Takahashi K, Okochi H, Okuda A, Matoba R, Sharov AA, et al. Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat. Cell Biol. 2007;9:625–635. doi: 10.1038/ncb1589. [DOI] [PubMed] [Google Scholar]
- Matoba R, Niwa H, Masui S, Ohtsuka S, Carter MG, Sharov AA, Ko MS. Dissecting oct3/4-regulated gene networks in embryonic stem cells by expression profiling. PLoS ONE. 2006;1:e26. doi: 10.1371/journal.pone.0000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshorer E, Misteli T. Chromatin in pluripotent embryonic stem cells and differentiation. Nat. Rev. Mol. Cell Biol. 2006;7:540–546. doi: 10.1038/nrm1938. [DOI] [PubMed] [Google Scholar]
- Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- Morin-Kensicki EM, Faust C, LaMantia C, Magnuson T. Cell and tissue requirements for the gene eed during mouse gastrulation and organogenesis. Genesis. 2001;31:142–146. doi: 10.1002/gene.10017. [DOI] [PubMed] [Google Scholar]
- Niwa H. How is pluripotency determined and maintained? Development. 2007;134:635–646. doi: 10.1242/dev.02787. [DOI] [PubMed] [Google Scholar]
- Niwa H, Burdon T, Chambers I, Smith A. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev. 1998;12:2048–2060. doi: 10.1101/gad.12.13.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- Ono T, Losada A, Hirano M, Myers MP, Neuwald AF, Hirano T. Differential contributions of condensin I and condensin II to mitotic chromosome architecture in vertebrate cells. Cell. 2003;115:109–121. doi: 10.1016/s0092-8674(03)00724-4. [DOI] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K. The polycomb group protein suz12 is required for embryonic stem cell differentiation. Mol. Cell. Biol. 2007;27:3769–3779. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts CW, Galusha SA, McMenamin ME, Fletcher CD, Orkin SH. Haploinsufficiency of Snf5 (integrase interactor 1) predisposes to malignant rhabdoid tumors in mice. Proc. Natl. Acad. Sci. USA. 2000;97:13796–13800. doi: 10.1073/pnas.250492697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapountzi V, Logan IR, Robson CN. Cellular functions of TIP60. Int. J. Biochem. Cell Biol. 2006;38:1496–1509. doi: 10.1016/j.biocel.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, Carney D, Pena P, Lan F, Kaadige MR, et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spivakov M, Fisher AG. Epigenetic signatures of stem-cell identity. Nat. Rev. Genet. 2007;8:263–271. doi: 10.1038/nrg2046. [DOI] [PubMed] [Google Scholar]
- Squatrito M, Gorrini C, Amati B. Tip60 in DNA damage response and growth control: many tricks in one HAT. Trends Cell Biol. 2006;16:433–442. doi: 10.1016/j.tcb.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Velkey JM, O'Shea KS. Oct4 RNA interference induces trophectoderm differentiation in mouse embryonic stem cells. Genesis. 2003;37:18–24. doi: 10.1002/gene.10218. [DOI] [PubMed] [Google Scholar]
- Ventura A, Meissner A, Dillon CP, McManus M, Sharp PA, Van Parijs L, Jaenisch R, Jacks T. Cre-lox-regulated conditional RNA interference from transgenes. Proc. Natl. Acad. Sci. USA. 2004;101:10380–10385. doi: 10.1073/pnas.0403954101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Rao S, Chu J, Shen X, Levasseur DN, Theunissen TW, Orkin SH. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006;444:364–368. doi: 10.1038/nature05284. [DOI] [PubMed] [Google Scholar]
- Yang D, Buchholz F, Huang Z, Goga A, Chen CY, Brodsky FM, Bishop JM. Short RNA duplexes produced by hydrolysis with Escherichia coli RNase III mediate effective RNA interference in mammalian cells. Proc. Natl. Acad. Sci. USA. 2002;99:9942–9947. doi: 10.1073/pnas.152327299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.