Abstract
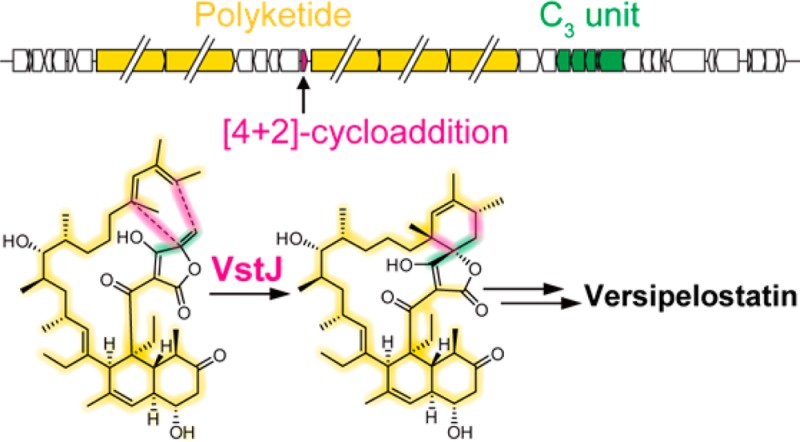
Versipelostatin (VST) is an unusual 17-membered macrocyclic polyketide product that contains a spirotetronate skeleton. In this study, the entire VST biosynthetic gene cluster (vst) spanning 108 kb from Streptomyces versipellis 4083-SVS6 was identified by heterologous expression using a bacterial artificial chromosome vector. Here, we demonstrate that an enzyme, VstJ, catalyzes the stereoselective [4+2]-cycloaddition between the conjugated diene and the exocyclic olefin of a newly identified tetronate-containing intermediate to form the spirotetronate skeleton during VST biosynthesis.
Polyketide compounds constitute a large group of natural products that are important drug resources due to their highly diversified structures. One of the important factors in constructing numerous varieties of carbon skeletons is the presence of a high number of tailoring enzyme cassettes in their biosynthesis that are specified for each polyketide scaffold.1 Spirotetronate moiety (tetronic acid spiro-linked to a cyclohexene ring) is one of the tailored skeletons that is found only in polyketide natural products, including chlorothricin (CHL),2 kijanimicin (KIJ),3 abyssomicin C (ABY),4 quartromicin (QMN),5 lobophorin (LOB),6 and tetrocarcin A (TCA) (Figure 1a).7 Compounds possessing the spirotetronate moiety exhibit broad biological activities.8 Among them, versipelostatin (VST, 1) is a spirotetronate-containing 17-membered macrocyclic polyketide produced by Streptomyces versipellis 4083-SVS6.9−12 VST has been shown to function as a selective GRP78/Bip molecular chaperone down-regulator.13 It has been proposed that the spirotetronate moiety in VST is formed via an intramolecular [4+2]-cycloaddition reaction between a conjugated diene and an exocyclic olefin of the corresponding biosynthetic precursor (Figure 1b). However, whether the proposed [4+2]-cycloaddition involves a specific enzyme has remained elusive, and such an enzyme, if it actually exists, has never been documented.8
Figure 1.
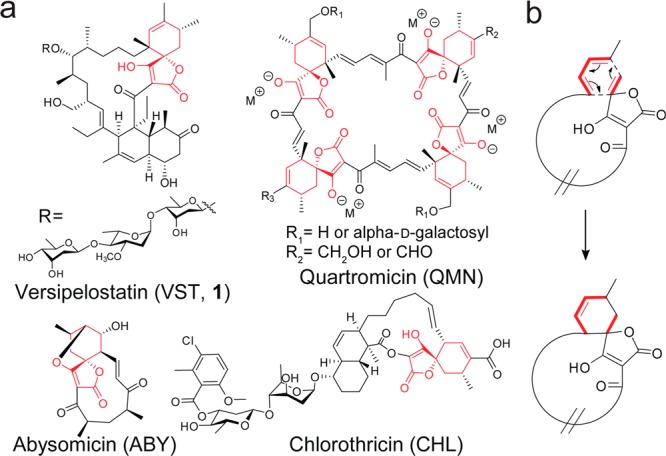
(a) Structures of spirotetronate-containing natural products. Spirotetronate moieties are shown in red. (b) Proposed [4+2]-cycloaddition reaction to form spirotetronate moiety.
[4+2]-Cycloaddition is a major reaction used to construct C–C bonds in organic synthesis. However, only a small number of enzymes that catalyze [4+2]-cycloadditions like the Diels–Alder reaction have been identified in nature. These include macrophomate synthase (MPS),14,15 solanopyranone synthase (SPS),16,17 lovastatin synthase (LovB),18,19 and SpnF20 in spinosyn biosynthesis. Although several biosynthetic gene clusters for spirotetronate-containing natural products have been reported,21−26 no candidate for an enzyme that may catalyze the proposed [4+2]-cycloaddition was identified. This situation prompted us to seek such enzymes in the biosynthetic gene clusters for spirotetronates. Here, we report the isolation of the biosynthetic gene cluster of VST and demonstrate that a small enzyme, designated VstJ, which consists of only 142 amino acid residues, catalyzes the stereoselective [4+2]-cycloaddition for the formation of the spirotetronate structure in VST biosynthesis.
To locate the whole gene cluster for VST, we first performed genome scanning of a draft genome sequence of the VST-producing S. versipellis 4083-SVS6. Draft genome sequencing yielded 11.4 Mb of DNA sequence divided into 19 scaffolds. According to the early biosynthetic studies on VST and by analogy to the biosynthesis of other tetronic acids (e.g., Rk-68227 and CHL21), we postulated that the VST aglycon may be biosynthesized through a type I polyketide synthase (PKS), and the ChlD1, ChlD2, and RkD homologues are likely responsible for the formation of the tetronic acid structure. Using these gene sequences as queries, we retrieved a putative biosynthetic gene cluster for VST across two scaffolds of the draft genome sequence spanning over 108 kb. To further identify the whole sequence of the VST biosynthetic gene cluster, we constructed a bacterial artificial chromosome (BAC) library for the S. versipellis 4083-SVS6 genome using a BAC vector, pKU503D (Table S1),28 and screened the library for BAC clones that contained the ChlD1, ChlD2, and RkD homologous genes (Table S2). A positive BAC clone was isolated and subsequently integrated into the genome of Streptomyces albus J1074 using the phiC31 integration site. The resultant transformant S. albus::pKU503DverP10N24 was cultured for heterologous gene expression under conditions identical to those used for S. versipellis 4083-SVS6. The heterologous production of VST by the S. albus::pKU503DverP10N24 transformant was confirmed using LC/MS (Figure 2a). The titer of VST in the transformant was approximately 21.0 mg L–1 broth, which was 14 times higher than that of the wild-type 4083-SVS6 strain (1.5 mg L–1). This result indicated that pKU503DverP10N24 contains all of the vst genes required for VST biosynthesis.
Figure 2.
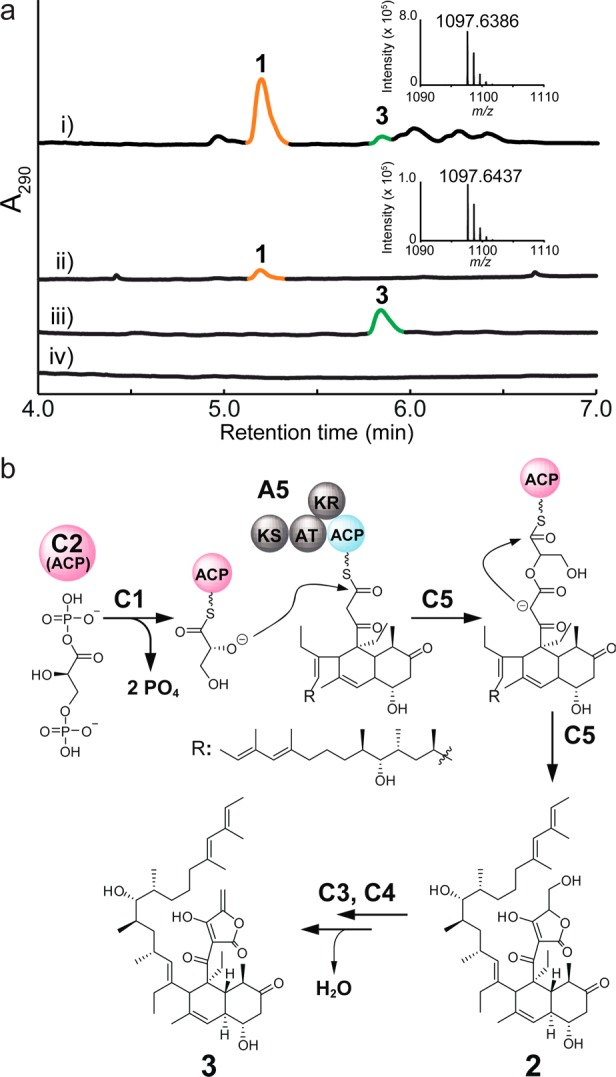
Comparative HPLC analysis of metabolites. (a) HPLC analysis of culture extracts from (i) S. albus J1074::pKU503DverP10N24, (ii) S. versipellis 4083 SVS6 (parent strain), (iii) S. albus J1074::pKU503DverP10N24ΔvstJ, and (iv) S. albus J1074::pKU503D (negative control). VST (1) was detected at 5.18 min. MS spectra of 1 (m/z 1097.6386 [M–H]− in (i) and m/z 1097.6437 [M–H]− in (ii); calculated for [M (C61H94O17)–H]− of VST: 1097.6413) were inserted. S. albus J1074::pKU503DverP10N24ΔvstJ did not produce 1 but gave unknown metabolite (3), which was also detected as a minor product in the extract from S. albus J1074::pKU503DverP10N24. The metabolite was later identified as 3. (b) Proposed biosynthesis of 3.
We sequenced and analyzed the insert DNA of pKU503DverP10N24. The length of the inserted DNA spans approximately 120 kb (Table S3 and Figure S1) and contains 52 ORFs, including the RkD, ChlD1, and ChlD2 homologues. There are five polyketide synthase (PKS) genes (vstA1–A5) harboring 13 keto synthase (KS) domains that are responsible for the 13 decarboxylative condensations in the chain elongation process using two carbon units derived from five molecules of malonyl-CoA, seven molecules of methylmalonyl-CoA, and two molecules of ethylmalonyl-CoA as building blocks (Figure S2). Based on a bioinformatics analysis of the vst gene cluster (Table S3, Figures S4–S7), the biosynthesis of VST can be proposed as shown in Figures S2 and S3. A tetronate-containing intermediate (2) is likely biosynthesized through the sequential reactions catalyzed by PKSs (VstA1–A5) and VstC1, VstC2, and VstC5, which are respectively the ChlD1, ChlD2, and RkD homologues essential for the construction of the tetronate structure (Figure S2).
To identify the putative gene that is responsible for the [4+2]-cycloaddition, we compared the vst genes with the biosynthetic gene clusters of CHL,21 TCA,23 and LOB,26 all of which contain a spirotetronate moiety. We hypothesized that the spirotetronate moiety may be synthesized by the actions of a specific enzyme(s) conserved in the biosynthetic pathways of these compounds. Comparing their gene clusters led to a hypothetical vstJ gene (429 bp) adjacent to the PKS vstA3 gene in addition to vstC1, vstC2, and vstC5, which are respectively the ChlD1, ChlD2, and RkD homologous genes in the vst cluster (Figure S8).
To investigate the function of VstJ in VST biosynthesis, we inactivated the vstJ gene in S. albus J1074::pKU503DverP10N24 (Figure S9). The resultant transformant S. albus::pKU503DverP10N24ΔvstJ was cultured, and the metabolites were analyzed using an LC/MS instrument (Figure 2a). LC/MS analysis revealed that the transformant was unable to produce VST (1), strongly suggesting that the vstJ gene is essential for VST biosynthesis. In addition, an unknown metabolite (3) accumulated in the extract prepared from the transformant. This metabolite (12.2 mg) was purified from 4 L of culture broth of the ΔvstJ transformant and identified as the acyclic intermediate 3 with a conjugated diene and an exocyclic olefin by HR/MS and extensive NMR spectral analyses (Table S4, Figures S10–S16). We speculate that 3 is biosynthesized through dehydration of 2 via acetylation. The conversion is catalyzed by the successive actions of VstC3 (an acyltransferase) and VstC4 (a dehydratase) (Figure 2b, Figure S2b).29,30 As a consequence, VstJ may catalyze the proposed stereoselective intramolecular [4+2]-cycloaddition between the terminal conjugated diene and the exocyclic olefin in 3 to yield a product with a spirotetronate skeleton.
To test our hypothesis, we overexpressed VstJ in Escherichia coli as an N-terminal His8-tagged protein and purified the recombinant VstJ protein to near homogeneity (Figure S17). The recombinant VstJ was incubated with the purified 3, and the reaction mixture was subjected to LC/MS analysis. The analysis revealed the formation of a new product (4) concomitant with consumption of 3 in a time-dependent manner (Figure 3a). The product was purified from the reaction mixture and identified as the cyclized 37-deoxy VST aglycone (4) by HR/MS and extensive NMR spectral analyses (Figure 3b, Table S5, Figures S18–S23). The 13C NMR spectral data indicate that 4 has the same stereochemistry as VST.10 It should be noted that 4 was never detected in the absence of VstJ at 30 °C. These data demonstrate that vstJ is essential for the last step in the stereoselective formation of the spirotetronate skeleton. The Km and kcat of VstJ-catalyzed reaction were determined to be 39 ± 5 μM and 4.4 ± 0.3 min–1 (0.25 ± 0.02 μmol min–1 mg–1), respectively (Figure S17).
Figure 3.
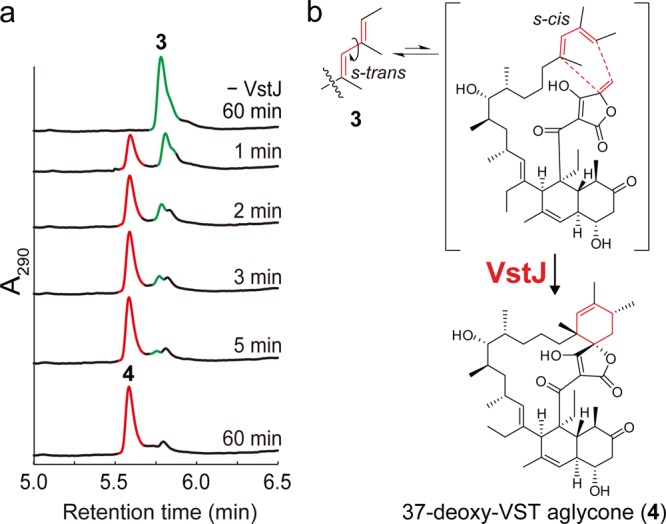
Assay with the recombinant VstJ protein. (a) HPLC analysis of the VstJ reaction mixtures. In the VstJ-catalyzed reaction, 3 was converted into 4 in a time-dependent manner. (b) Proposed VstJ-catalyzed [4+2]-cycloaddition reaction.
The above results clearly establish the catalytic role of VstJ in catalyzing the [4+2]-cycloaddition of 3. The absence of 1 and accumulation of 3 in the ΔvstJ transformant along with the failure to convert 3 to the cyclization product 4 in the absence of VstJ in vitro strongly indicate that this enzyme is indispensable for assembling the spirotetronate skeleton. This observation stands in contrast to the occurrence of enzyme-independent cyclization to afford the [4+2]-cycloaddition products in the biosynthesis of SPS,16,17 LovB,18 and SpnF.20 It is conceivable that the high flexibility of the acyclic substrate 3 may restrict the access of exocyclic olefin and the conjugated diene to each other and thus render spontaneous cyclization difficult. Hence, VstJ may catalyze the [4+2]-cycloaddition by preferentially stabilizing the s-cis conformation of the end-diene and aligning it properly with the exocyclic olefin when compound 3 is bound in the active site, so that 4 can be stereoselectively produced after cyclization. The exocyclic olefin in 3 is conjugated with three carbonyl groups. Any of these carbonyl groups might interact with amino acid residue(s) in the VstJ active site through hydrogen bond(s). Hydrogen-bonding could lower the energy of the lowest unoccupied molecular orbital of the conjugated olefin and drive the [4+2]-cycloaddition. Structural studies on VstJ complexed with 3 or its analogue would provide key insights into the catalytic mechanisms and how VstJ controls the dynamics of binding of the flexible substrate.
Another significant discovery is the finding of VstJ homologues, ChlL, TcaU4, and LobU2, in the biosynthetic gene clusters of CHL, TCA, and LOB, respectively, all of which contain the spirotetronate moiety. This finding prompted us to search further for VstJ homologues in the biosynthetic gene clusters of KIJ22 and ABY,24 both of which contain the spirotetronate moiety. This search retrieved two new open reading frames, Kijcyc and Abycyc, in the biosynthetic gene clusters of KIJ and ABY, respectively (Figure S24). These two DNA segments were not recognized as genes previously. In addition, we identified QmnH in the biosynthetic gene cluster of QMN,25 a more highly symmetrical spirotetronate compound with four spirotetronate moieties. All of the VstJ homologues exhibit about 30% similarity to each other, and some amino acid residues are conserved among them, although these homologues show no sequence similarity to any other characterized proteins (Figure S25). Curiously, QmnH contains two tandem VstJ-like sequences. It is possible that each of the VstJ-like sequences may catalyze the stereoselective formation of two spirotetronate moieties.
In conclusion, we identified the biosynthetic machinery for the 17-membered macrocyclic polyketide VST by heterologous expression using a BAC vector. Most notably, we demonstrated that VstJ enzyme catalyzes the stereoselective [4+2]-cycloaddition to form the spirotetronate skeleton and thus complete the macrocyclic structure during VST biosynthesis. Given that VstJ homologues can be found in the biosynthetic gene clusters of other spirotetronate moiety-containing macrocyclic polyketide compounds, VstJ may prove to be a general enzyme for spirotetronate biosynthesis. Besides SpnF, VstJ represents the second example of a naturally selected carbocyclase whose sole function is to catalyze a [4+2]-cycloaddition reaction. Whether its catalyzed [4+2]-cycloaddition is a concerted Diels–Alder reaction awaits further experiments, which are currently in progress.
Acknowledgments
This work was supported by a grant for “Project focused on developing key technology of discovering and manufacturing drug for next-generation treatment and diagnosis” from METI, Japan (to K.S. and T.K.), and grants from NIH (GM035906 and GM040541) as well as Welch Foundation (F-1501) (to H.-w.L.).
Supporting Information Available
Experimental details and supporting tables and figures. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Hertweck C. Angew. Chem., Int. Ed. 2009, 48, 4688. [DOI] [PubMed] [Google Scholar]
- Kellersc W.; Muntwyle R.; Pache W.; Zahner H. Helv. Chim. Acta 1969, 52, 127. [Google Scholar]
- Waitz J. A.; Horan A. C.; Kalyanpur M.; Lee B. K.; Loebenberg D.; Marquez J. A.; Miller G.; Patel M. G. J. Antibiot. 1981, 34, 1101. [DOI] [PubMed] [Google Scholar]
- Bister B.; Bischoff D.; Strobele M.; Riedlinger J.; Reicke A.; Wolter F.; Bull A. T.; Zahner H.; Fiedler H. P.; Sussmuth R. D. Angew. Chem., Int. Ed. 2004, 43, 2574. [DOI] [PubMed] [Google Scholar]
- Tsunakawa M.; Tenmyo O.; Tomita K.; Naruse N.; Kotake C.; Miyaki T.; Konishi M.; Oki T. J. Antibiot. 1992, 45, 180. [DOI] [PubMed] [Google Scholar]
- Jiang Z. D.; Jensen P. R.; Fenical W. Bioorg. Med. Chem. Lett. 1999, 9, 2003. [DOI] [PubMed] [Google Scholar]
- Tomita F.; Tamaoki T.; Shirahata K.; Kasai M.; Morimoto M.; Ohkubo S.; Mineura K.; Ishii S. J. Antibiot. 1980, 33, 668. [DOI] [PubMed] [Google Scholar]
- Vieweg L.; Reichau S.; Schobert R.; Leadlay P. F.; Süssmuth R. D. Nat. Prod. Rep. 2014, 31, 1554. [DOI] [PubMed] [Google Scholar]
- Park H. R.; Furihata K.; Hayakawa Y.; Shin-ya K. Tetrahedron Lett. 2002, 43, 6941. [Google Scholar]
- Park H. R.; Chijiwa S.; Furihata K.; Hayakawa Y.; Shin-Ya K. Org. Lett. 2007, 9, 1457. [DOI] [PubMed] [Google Scholar]
- Ueda J.-y.; Chijiwa S.; Takagi M.; Shin-ya K. J. Antibiot. 2008, 61, 752. [DOI] [PubMed] [Google Scholar]
- Zhao P.; Ueda J.-y.; Kozone I.; Chijiwa S.; Takagi M.; Kudo F.; Nishiyama M.; Shin-ya K.; Kuzuyama T. Org. Biomol. Chem. 2009, 7, 1454. [DOI] [PubMed] [Google Scholar]
- Matsuo J.; Tsukumo Y.; Sakurai J.; Tsukahara S.; Park H.-R.; Shin-ya K.; Watanabe T.; Tsuruo T.; Tomida A. Cancer Sci. 2009, 100, 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa H.; Yagi K.; Watanabe K.; Honma M.; Ichihara A. Chem. Commun. 1997, 97. [Google Scholar]
- Ose T.; Watanabe K.; Mie T.; Honma M.; Watanabe H.; Yao M.; Oikawa H.; Tanaka I. Nature 2003, 422, 185. [DOI] [PubMed] [Google Scholar]
- Oikawa H.; Katayama K.; Suzuki Y.; Ichihara A. J. Chem. Soc., Chem. Commun. 1995, 1321. [Google Scholar]
- Kasahara K.; Miyamoto T.; Fujimoto T.; Oguri H.; Tokiwano T.; Oikawa H.; Ebizuka Y.; Fujii I. ChemBioChem 2010, 11, 1245. [DOI] [PubMed] [Google Scholar]
- Auclair K.; Sutherland A.; Kennedy J.; Witter D. J.; Van den Heever J. P.; Hutchinson C. R.; Vederas J. C. J. Am. Chem. Soc. 2000, 122, 11519. [Google Scholar]
- Ma S. M.; Li J. W. H.; Choi J. W.; Zhou H.; Lee K. K. M.; Moorthie V. A.; Xie X.; Kealey J. T.; Da Silva N. A.; Vederas J. C.; Tang Y. Science 2009, 326, 589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. J.; Ruszczycky M. W.; Choi S.-h.; Liu Y.-n.; Liu H.-w. Nature 2011, 473, 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia X. Y.; Tian Z. H.; Shao L.; Qu X. D.; Zhao Q. F.; Tang J.; Tang G. L.; Liu W. Chem. Biol. 2006, 13, 575. [DOI] [PubMed] [Google Scholar]
- Zhang H.; White-Phillip J. A.; Melancon C. E.; Kwon H. J.; Yu W. L.; Liu H.-w. J. Am. Chem. Soc. 2007, 129, 14670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J.; Zhang Y. P.; Huang L. J.; Jia X. Y.; Zhang Q.; Zhang X.; Tang G. L.; Liu W. J. Bacteriol. 2008, 190, 6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottardi E. M.; Krawczyk J. M.; von Suchodoletz H.; Schadt S.; Muhlenweg A.; Uguru G. C.; Pelzer S.; Fiedler H. P.; Bibb M. J.; Stach J. E. M.; Sussmuth R. D. ChemBioChem 2011, 12, 1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H.-Y.; Pan H.-X.; Wu L.-F.; Zhang B.-B.; Chai H.-B.; Liu W.; Tang G.-L. Chem. Biol. 2012, 19, 1313. [DOI] [PubMed] [Google Scholar]
- Li S.; Xiao J.; Zhu Y.; Zhang G.; Yang C.; Zhang H.; Ma L.; Zhang C. Org. Lett. 2013, 15, 1374. [DOI] [PubMed] [Google Scholar]
- Sun Y. H.; Hahn F.; Demydchuk Y.; Chettle J.; Tosin M.; Osada H.; Leadlay P. F. Nat. Chem. Biol. 2010, 6, 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M.; Uchiyama T.; Omura S.; Cane D. E.; Ikeda H. Proc. Natl. Acad. Sci. U.S.A. 2010, 107, 2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanchanabanca C.; Tao W. X.; Hong H.; Liu Y. J.; Hahn F.; Samborskyy M.; Deng Z. X.; Sun Y. H.; Leadlay P. F. Angew. Chem., Int. Ed. 2013, 52, 5785. [DOI] [PubMed] [Google Scholar]
- Wu L.-F.; He H.-Y.; Pan H.-X.; Han L.; Wang R.; Tang G.-L. Org. Lett. 2014, 16, 1578. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


