Abstract
We describe and characterize a method for insertional mutagenesis of the yeast pathogen Candida glabrata using the bacterial transposon Tn7. Tn7 was used to mutagenize a C. glabrata genomic fosmid library. Pools of random Tn7 insertions in individual fosmids were recovered by transformation into Escherichia coli. Subsequently, these were introduced by recombination into the C. glabrata genome. We found that C. glabrata genomic fragments carrying a Tn7 insertion could integrate into the genome by nonhomologous recombination, by single crossover (generating a duplication of the insertionally mutagenized locus), and by double crossover, yielding an allele replacement. We were able to generate a highly representative set of ∼104 allele replacements in C. glabrata, and an initial characterization of these shows that a wide diversity of genes were targeted in the mutagenesis. Because the identity of disrupted genes for any mutant of interest can be rapidly identified, this method should be of general utility in functional genomic characterization of this important yeast pathogen. In addition, the method might be broadly applicable to mutational analysis of other organisms.
Candida species, primarily Candida albicans and Candida glabrata, are important human pathogens, responsible for 7% of all hospital-acquired blood stream infections (Schaberg et al. 1991). Even with available antifungal therapies, the associated mortality for Candida bloodstream infections is high (up to 30% in cancer patients). We are usingC. glabrata as a model to explore the molecular details of the host–pathogen interaction. In frequency of isolation, C. glabrata is second only to C. albicans and is responsible for 15%–20% of both mucosal (Schuman et al. 1998; Vazquez et al. 1999) and systemic (Pfaller et al. 1999, 2001) candidiasis; in spite of this epidemiological similarity, C. glabrata is phylogenetically distant from C. albicans, more highly related, for example, to Saccharomyces cerevisiae than to C. albicans (Barns et al. 1991). What strategies for host colonization are shared by C. glabrata and C. albicans remain to be determined; no genes essential for virulence have yet been described in C. glabrata. Studies primarily in C. albicans have identified multiple factors important in the pathogenesis of Candida species (for review, see Calderone and Fonzi 2001), including the ability to adhere to host tissue, the ability to grow in hyphal and yeast form (for C. albicans), the capacity to switch between different cellular phenotypes, and the ability to acquire iron in vivo. Like C. albicans, C. glabrata is able to adhere specifically to host tissue, recognizing host carbohydrate (Cormack et al. 1999). On the other hand, C. glabrata does not make hyphae, a feature of prime importance in the pathogenesis of C. albicans; rather, it grows solely in the yeast form, making pseudohyphae under conditions of nitrogen starvation (Csank and Haynes 2000).
Because C. glabrata is haploid, the tools of classical genetics can be applied, and mutants defective in various aspects of virulence can be isolated and characterized. An efficient genetic analysis depends on a method of random insertional mutagenesis, and we considered various available options. In other species, numerous approaches have been taken, including the use of bacterial transposons such as Tn3 (Seifert et al. 1986; Ross-Macdonald et al. 1999), Tn7 (Biery et al. 2000), and the Drosophila melanogaster transposon Mariner (Gueiros-Filho and Beverley 1997). Tn3, in particular, has been used to advantage in mutagenesis of S. cerevisiae (Ross-Macdonald et al. 1999). In that efficient and highly random method, fragments of the S. cerevisiae genome are first mutagenized in Escherichia coli; those insertion mutations are then introduced into the S. cerevisiae genome by homologous recombination.
For C. glabrata, the options were somewhat limited, in part because there are no known natural transposons in C. glabrata (like the Ty elements of S. cerevisiae). In an earlier study, we exploited nonhomologous recombination in C. glabrata to make insertion mutants and to analyze these for effects on adherence to epithelial cells (Cormack and Falkow 1999; Cormack et al. 1999). We found that the insertions were distributed more or less randomly in many different genes; however, a close analysis of the sites of insertion for 50 mutants showed that the majority (48/50) were in noncoding regions of the genome. If one were able to analyze a very large number of mutants, this bias might not be an important factor. However, for screens in which only modest numbers of mutants (20,000–30,000) are analyzed, the bias against insertions in coding regions would result in a mutational sampling of only a fraction of the genome.
As an alternative to the problematic nonhomologous-recombination-based method, we describe in this paper a novel mutagenesis approach similar in principle to the Tn3 method described above (Ross-Macdonald et al. 1999), but which exploits recent studies of in vitro transposition by the bacterial transposon Tn7. This method is of some general interest because the generation of mutants requires only two steps: in vitro mutagenesis by Tn7 followed by homologous recombination into the target genome (here C. glabrata). In theory, therefore, our method can easily be applied to any organism with efficient homologous recombination. We describe modifications to Tn7 to allow its use in C. glabrataand to facilitate the recovery of DNA flanking insertion sites for mutants of interest. We demonstrate that this method can be used in the efficient generation of thousands of randomly distributed insertion mutants, possessing an array of phenotypes.
RESULTS
Principle of Mutagenesis and Construction of a Minitransposon With Two Yeast Selectable Markers and a Conditional Origin of Replication for E. coli
We devised a two-step strategy for random insertional mutagenesis in C. glabrata. In this method, a collection of fosmids containing large (∼40-kb) C. glabrata genomic inserts (in essence, a representation of the genome) is first insertionally mutagenized by Tn7 in vitro to create pools of mutants in E. coli. This is followed by introduction of the pool of mutants (at this point carried on fosmids in E. coli) into C. glabrata by homologous recombination. In theory, this should result in an allele replacement, in which individual genes have been replaced by insertionally mutagenized alleles.
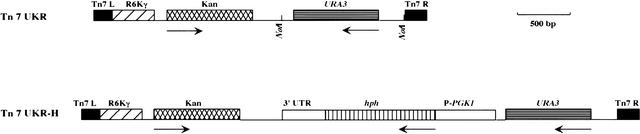
We used a Tn7-based in vitro mutagenesis to mutagenize a set of 100 fosmids (∼0.25× genome coverage). This in vitro mutagenesis system allowed efficient transposition with minimal target-site specificity (Biery et al. 2000). The mini-Tn7 transposon previously described (mini-Tn7 SpeI–KmR–NotI; Biery et al. 2000), carries a kanamycin-resistance cassette flanked by the Tn7 left- and right-end sequences, respectively. As shown in Figure 1, we constructed two different derivatives of this transposon. To generate the first one, Tn7 URA3 · KmR · R6Kγ (Tn7 UKR), we introduced into the mini-Tn7 a yeast selectable marker, the URA3 gene from S. cerevisiae, as well as the conditional origin of replication R6Kγ, which requires the Π protein (the product of the pir gene) for replication (Kolter et al. 1978). The presence of this origin facilitates subsequent recovery of DNA flanking the insertion site in mutants of interest. The resulting transposon can be maintained only in an E. coli strain (BW23473) expressing the pir gene (Metcalf et al. 1996). The second transposon, Tn7 URA3 · hph · KmR · R6Kγ (Tn7 UKR-H), contains, in addition, the hph gene from Klebsiella pneumoniae, which confers resistance to hygromycin B (Gritz and Davies 1983).
Figure 1.
Maps of mini-Tn7 derivatives used. Tn7 UKR (URA3 Kan · R6Kγ ori) is a 3.108-kb derivative of mini-Tn7 SpeI-KmR-NotI containing the conditional origin of replication R6Kγ and the Saccharomyces cerevisiae URA3 gene. Relevant restriction sites are shown. Tn7 UKR-H is a 5.118-kb element derived from Tn7 UKR that contains the Klebsiella pneumoniae hph gene (which confers resistance to hygromycin B) driven by the PGK1 promoter from S. cerevisiae. The arrows underneath each map indicate the direction of transcription.
We tested the ability of these minitransposons to undergo transposition in vitro using different-sized target DNAs and found no difference with the parent mini-Tn7 (data not shown). We used each transposon for mutagenesis of individual fosmids and cloned and characterized 40 transposition products by digestion with restriction enzymes (data not shown). In no transposition products did we detect a gross rearrangement of fosmid DNA. In addition, the majority (32/40) of transposition products had a single insertion of the transposon in the fosmid with no other rearrangement of the fosmid DNA. In 8 out of the 40 products, there were 2 Tn7 insertions, but these were invariably in two different restriction fragments. This result is not unexpected, because over short distances the phenomenon of transposon immunity prevents the insertion of a second copy of the transposon next to the site of a transposon already present. This effect drops off over distance, so that one might expect two insertions in the same fosmid but separated by >10 kb (DeBoy and Craig 1996).
Mutagenesis of our fosmids was efficient: In a mutagenesis of 200 fosmids, we recovered a minimum of 200 and up to tens of thousands of transformants per fosmid, where each transformant represents an independent insertion in the fosmid. For each fosmid, the transformants were pooled, generating a library of insertions throughout a fosmid-borne 40-kb genomic fragment.
Transformation of Linear Genomic Fragments Into C. glabrata Results in Three Classes of Transformants
Before introducing a pool of Tn7-mutagenized fosmid fragments into the C. glabrata genome, we wanted to characterize in detail the in vivo fate of individual mutagenized genomic fragments, derived from one mutagenized fosmid, and to optimize conditions for recovery of homologous recombinants after transformation of mutagenized fosmids into C. glabrata.Transposon Tn7-UKR (from pIC6) was used to mutagenize one fosmid (fosmid 1). Mutagenized fosmid DNA was linearized with EcoRI, transformed into C. glabrata strain BG14 (ura3Δ), and selection was made for Ura+ transformants.
Ura+ colonies were then patched on a plate lacking uracil and printed onto plates containing 5-fluoroorotic acid (5-FOA). Upon examination of Ura+ transformants obtained, it was clear that there were three different classes of transformants based on the phenotype displayed on 5-FOA plates: Class 1 transformants were unstable Ura+ 5-FOAR colonies; Class 2 transformants were partially stable, Ura+ 5-FOAS transformants in which a patch on 5-FOA plates gave rise to resistant papillae after incubation at 30°C for 48 h; and Class 3 transformants were the expected Ura+ 5-FOAS stable colonies. Each class is the result of a different fate of the transforming linear DNA fragment, as described below.
Three Classes of Transformants Represent Three Different Fates of the Transforming DNA
We hypothesized that Class 1 or unstable transformants (U) corresponded to linear genomic fragments (containing a Tn7 insertion), which happened to contain an autonomous replication sequence (ARS), and which were able to recircularize after transformation and remain as episomes or plasmids. These plasmids would be able to replicate, but because they would generally not contain a CEN sequence, they would be lost at a high frequency, resulting in a mixed colony containing Ura+ 5-FOAS cells (containing the unstable plasmid), as well as Ura− 5-FOAR cells, which have lost the plasmid. If this hypothesis were true, then it should be possible to isolate this freely replicating plasmid from genomic DNA preps from unstable transformants, and functionally identify it by its ability to transform the E. coli strain expressing the pir gene. We prepared genomic DNA from four individual Class 1 transformants, and used it directly to transform the E. coli strain BW23473. As expected, undigested genomic DNA from each unstable transformant produced thousands of transformants in this E. coli host, but not in an E. coli strain not expressing the pir gene (data not shown). Furthermore, plasmid DNA isolated from E. coli strain BW23473 could be transformed back into C. glabrata at the high frequency (>105/μg of DNA) typical of plasmid transformations in C. glabrata (Cormack and Falkow 1999).
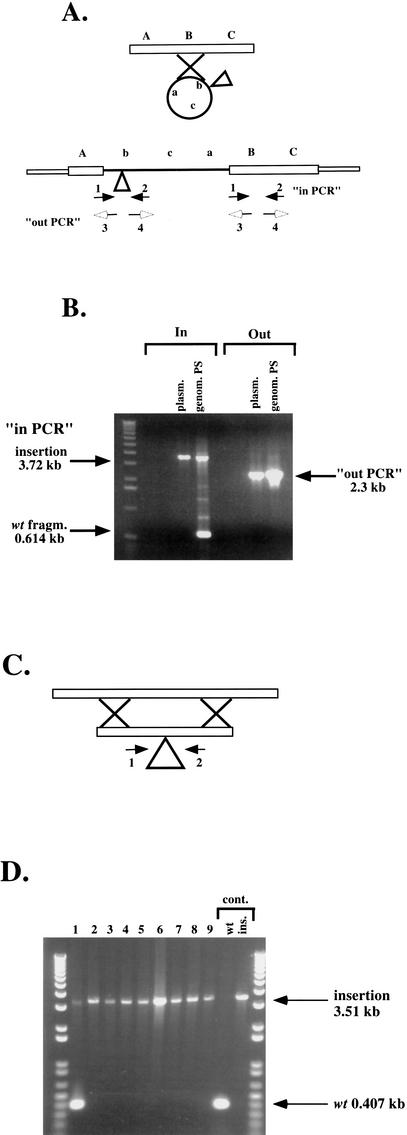
We hypothesized that Class 2 or partially stable (PS) transformants were events in which the incoming, linear DNA recircularized before, or coincident with, the homologous recombination event. This would lead to a partial duplication of this particular locus, in which an insertionally mutagenized allele and the wild-type allele are present as tandem direct repeats (Fig. 2A). Because of the direct repeat, it is possible that at some low frequency, the duplication could resolve by homologous recombination between the direct repeats, yielding Ura− 5-FOAR papillae on an otherwise 5-FOAS patch. To test this hypothesis, we first generated a defined insertion in a single C. glabrata sequence. DNA from the mutagenized fosmid was digested with EcoRI, recircularized, and used to transform E. coli strain BW23473 (pir+) to kanamycin resistance. This generated a smaller, conditional replicon, designated pIC14, containing a single Tn7 insertion and 8 kb of the flanking genomic C. glabrata sequence, which was sequenced using primers that anneal to each end of the Tn7 and are oriented outward. We then designed primers in both directions from the flanking genomic sequence obtained, toward and away from the insertion (Fig. 2A). pIC14 was linearized with EcoRI, transformed into BG14 (ura3Δ), and selection was made for Ura+ transformants. If the homologous recombination resulted in a duplication of this region as illustrated in Figure 2A, then by using the pair of primers whose direction of annealing is away from the insertion (“out-PCR”), it should be possible to amplify a fragment containing the duplication, but not if there was no duplication, because the two primers are oriented away from each other. Furthermore, by using the pair of primers oriented toward the insertion (“in-PCR”), two regions should be amplified from a duplication event: a wild-type small fragment of 614 bp and a larger fragment corresponding to the wild type plus the Tn7 UKR insertion (3720 bp).
Figure 2.
Two of the pathways of homologous recombination after transformation of linearized mutant fosmids. (A) Class 2 (partially stable, PS) transformants: A linearized genomic fragment containing a transposon insertion can recircularize before, or at the time of, homologous recombination, generating a duplication of the region as indicated. Primers 1 and 2 are oriented toward, and primers 3 and 4 oriented away from the Tn7 insertion. (B) Result of PCR amplification of genomic DNA from one Class 2 transformant. (genom.PS lane) Template DNA for PCR was genomic DNA; (plasm. lane) template DNA for PCR was pIC14. (C) Class 3 (stable, S) transformants: allele replacement. A linearized genomic fragment containing a transposon insertion recombines by double crossover and replaces the genomic locus. (D) Result of PCR amplification of genomic DNA from nine Class 3 (S) transformants. (Lane 1) Genomic DNA from a nonhomologous recombinant was used for the PCR and bands for both the wild type and Tn7-disrupted copies are amplified. (Lanes 2–9) Results for homologous recombinants. Controls: (wt) Cg14 genomic DNA was used as template for the PCR; (ins.) pIC14 was used as template.
Genomic DNA was prepared from two Class 2 (PS) transformants and was used as template for PCR reactions using the two pairs of primers from the sequences flanking the insertion. Figure 2B shows the results obtained with one PS transformant. As is clear from the picture, the primer pair oriented toward the insertion amplified the two fragments of the size expected of a duplication. When the primers directed away from the insertion were used, a product of the size expected was amplified. This result indicates that, in fact, a duplication occurred after transformation of this fragment of DNA containing the Tn7 UKR insertion either before or during homologous recombination. We also gel-purified high-molecular-weight genomic DNA from this and one other PS transformant and obtained the same results with “in” and “out” primer pairs, indicating that in PS class transformants, the transforming DNA underwent a recombination pathway that led to a duplication of the region in the genome. Unlike in the case of Class 1 (unstable) transformants, we were unable to transform the E. coli permissive host BW23473 to kanamycin resistance with genomic DNA purified from the two Class 2 transformants (data not shown). This is again consistent with our hypothesis that these transformants represent tandem duplications of the Tn7-disrupted genomic locus.
Class 3 or stable (S) transformants represent allele replacements in which the insertion of Tn7 UKR at that particular locus replaces the wild-type region, resulting in a stable Ura+ FOAS colony (Fig. 2C). We made genomic DNA of 30 stable transformants derived from pIC14 transformation and used the PCR primers directed toward the insertion to analyze them. Figure 2D shows nine transformants of which eight show amplification only of the large 3.51-kb band corresponding to the Tn7-disrupted allele. One transformant shows amplification of both the wild-type band as well as the band corresponding to the insertion, indicating that this transformant had undergone nonhomologous recombination and contained the Tn7 UKR insertion somewhere else in the genome. These transformants are expected to be stable Ura+ FOAS colonies as well. Of the stable transformants, 27/30 or 90% were homologous allele replacements and 3/30 or 10% were nonhomologous recombinants. Taken together, these results show that after transformation of a linear genomic DNA, at least three different events can happen. Only the stable Class 3 transformant is useful for a mutagenesis study, because these colonies represent simple allele replacements in 90% of cases, and therefore are expected to result in a large proportion of null mutants.
In our analysis, the ratio at which each of these classes appeared depended on the particular fosmid, as well as on the enzymes used to digest the mutagenized fosmid before transformation into C. glabrata; but for 96 different mutagenized fosmids, it ranged in our hands from 12% to 58% for Class 1 (US), 8% to 37% for Class 2 (PS), and from 29% to 75% for Class 3 (S) transformants.
Lastly, the classification of transformants as sensitive or partially sensitive depended on there being no background of untransformed Ura− cells carried along with the Ura+ colony when it is picked to be analyzed. This elimination of background untransformed cells was done either by streak-purifying the colonies, or (when using the Tn7 UKR-H, which carries the hygromycin gene) by replica-printing the transformants twice consecutively on hygromycin-containing plates (see Methods) to kill the untransformed background.
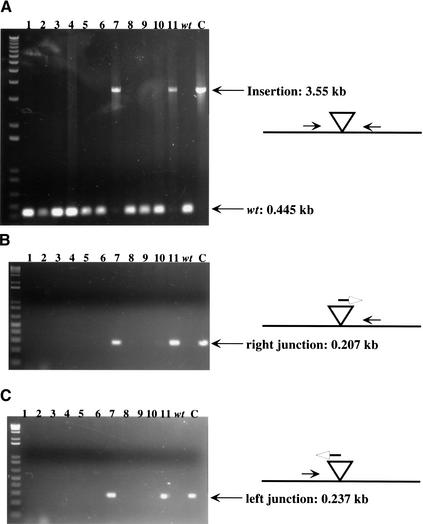
The Majority of 12 Individual Insertions in a Pool Are Represented in a Small Pool of C. glabrata Stable Transformants
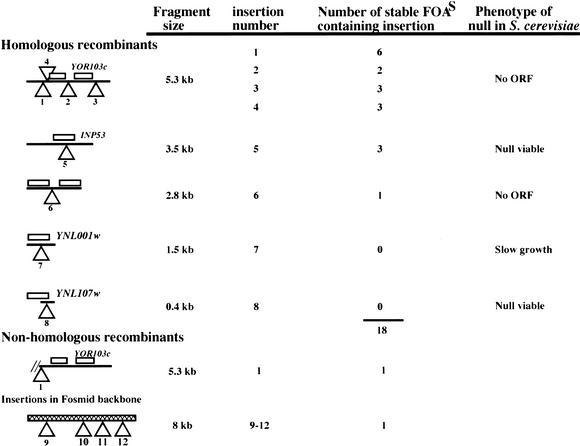
We next wanted to know whether in a small pool of insertions in one fosmid, there would be homologous recombination and replacement of each insertion after transformation into C. glabrata and screening for stable transformants. This was an important parameter to understand because we worried that a single insertion might recombine at much greater than average efficiency and “poison” the pool of C. glabrata transformants derived from a pool of insertions in E. coli. We made a pool of 12 Tn7 UKR insertions in one fosmid, of which 4 were in the fosmid vector, and the remaining 8 were distributed in the five different-sized EcoRI fosmid fragments shown schematically in Table 1. The target site of each of these 8 insertions was sequenced using the Tn7 end primers facing outward, and pairs of primers were designed from each flanking sequence toward the insertion. We made DNA of the pool of 12 insertions from E. coli and digested it with EcoRI and used it to transform BG14 (ura3Δ), selecting for Ura+. Genomic DNA from 20 stable Class 3 (S) FOAS transformants was prepared and used as template for PCR analysis. We used three pairs of primers for each of the insertions in the pool and used these to PCR genomic DNA from each of the 20 FOAS transformants. The first pair is oriented toward the insertion, and the other two amplify each junction of the Tn7 insertion. Figure 3 shows only one example using 11 FOAS DNAs and the three primer pairs specific for one of the eight insertions. In this case, 2 of the 11 genomic DNAs used (numbers 7 and 11) did not give amplification of a wild-type 0.445-kb fragment using primers 708 and 711 (corresponding to insertion 5 in Table 1) toward the insertion; instead, a 3.553-kb band was amplified, corresponding in size to the insertion of the Tn7 UKR at that site. Furthermore, only these same two genomic DNAs showed the expected 0.207-kb and 0.237-kb fragments amplified with primers from the left and right junctions, respectively, of that particular insertion. These results indicate that in these two transformants, a replacement event took place, and also that insertion 5 was not present in any of the 20 other transformants as a nonhomologous recombinant or a double insertion. The results of the complete set of PCR reactions for the 20 genomic DNAs from PS transformants are summarized in Table 1. The first four insertions fell in the largest EcoRI fragment; none of these happened to disrupt any ORF within this fragment, and all four were found among the 20 transformants analyzed; insertion number 1 was the most commonly found (6/20 FOAS genomic DNAs). The four insertions in this fragment accounted for two-thirds (14/20) of the transformants analyzed, not unexpectedly because they accounted for one-half of the total number of insertions in the pooled DNAs, which can recombine by homologous recombination (4/8). Insertion number 5 in the second largest EcoRI fragment is at amino acid 456 of the C. glabrata INP53 homolog and was found three times in the 20 DNAs monitored (disruption of the ortholog of this gene in S. cerevisiae results in no phenotype). Insertion number 6 in the 2.8-kb EcoRI fragment was found once among the genomic DNAs; insertion number 7, in a 1.5-kb EcoRI fragment, which disrupts the YNL001w ortholog, was not found in any of the 20 genomic DNAs tested. Null mutations in this ORF result in slow growth in S. cerevisiae, and it is expected that mutations that confer a slow growth will be underrepresented in our method because in the initial transformation, these colonies would probably be avoided. We also did not find any transformants carrying insertion 8 in the smallest EcoRI fragment (400 bp), which disrupts YNL107w. It may be that this insertion results in a slow growth or inviability phenotype and therefore could not be recovered. Alternatively, 400 bp of homology is too low to give efficient homologous recombination.
Table 1.
Diagram of Position and Distribution of Insertions in a Pool of 20 Class 3 (S) Candida glabrata Transformants
Transforming pool consisted of an equimolar mix of 12 fragments: 8 different insertions in five Eco RI C. glabrata genomic fragments and 4 different insertions in the pBAC fosmid backbone. Homologous recombinants: 18 stable allele replacements derived by homologous recombination of insertions 1–8. Nonhomologous recombinants: two stable insertions derived by nonhomologous recombination.
Figure 3.
PCR amplification for 11 Class 3 transformants using three primer pairs. (A) Genomic primers flanking the Tn7 insertion site and oriented toward the insertion. (B) One genomic primer combined with a Tn7R primer. (C) Genomic primer combined with the Tn7L primer. Note that the three positive transformants for B and C are the only transformants positive for the longer PCR product in A corresponding to the Tn7-disrupted band. Control lanes: (wt) BG14 genomic DNA, (C) pIC14 DNA used as template for PCR.
From the data in Table 1, we can conclude that many different insertions in a pool can be recovered among a small number of recombinants in C. glabrata; for small fragments, there might be a correlation between the size of the fragment carrying the Tn7 and the likelihood of its integrating by homologous recombination. In all, of the 20 genomic DNAs from stable FOAS transformants, 18 of them were caused by homologous recombination involving six of the eight insertions in the genomic DNA; the remaining two transformants were caused by nonhomologous recombination; in one case, insertion 9 in the vector backbone recombined by nonhomologous recombination and in one case insertion 1 in the largest EcoRI fragment recombined by nonhomologous recombination. The initial pool of 12 E. coli insertions contained 4 (or one third of the pool) insertions in the vector, and these can integrate only by nonhomologous recombination. Consistent with nonhomologous recombination being less efficient than homologous recombination, only 1 of 20 transformants was caused by recombination of a vector-derived fragment (as opposed to the 7 expected if homologous and nonhomologous recombination happened at equal frequency). It is clear from this analysis, as well as from the earlier analysis of a single fragment, that ∼10% of the stable transformants derived from this method are caused by nonhomologous recombination. The corollary is that ∼90% of the stable FOAS transformants are simple allele replacements.
Most Insertions in a Large Pool Transform C. glabrata at Equal Efficiency
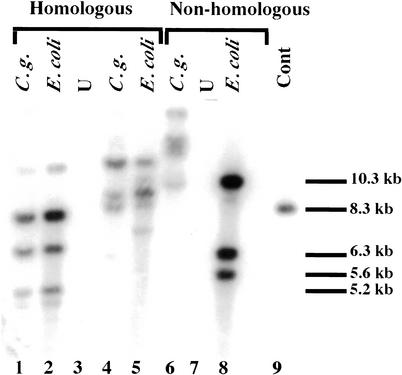
We wanted to further verify for a large pool of Tn7insertions in a fosmid that individual insertions in the pool would give rise to transformants in C. glabrata at approximately the same frequency that they were represented in the E. coli pool. If all fragments in a pool recombined at equal frequency, this would be true. The exception to this would be insertions, which disrupt essential genes (which cannot be recovered at all in our mutagenesis). We mutagenized in vitro two fosmids and pooled ∼150 E. coli individual transformants for each fosmid. DNA from these two pools was digested with BcgI plus MfeI (for neither one of which recognition sites are present in Tn7) and used to transform BG14 (ura3Δ) to Ura+. MfeI recognition sites are present approximately every 3000 bp in the genome; BcgI sites are present on average every 10 kb. The BcgI sites in pBAC-BcgI immediately flank the cloning site for the genomic fragment and therefore digestion with BcgI releases the mutagenized genomic DNA fragments from any vector sequences, leaving genomic fragments with ends precisely complementary to the corresponding genomic locus. Genomic DNA from pools of 35 or 44 stable Class 3 (FOAS) transformants, as well as DNA from the E. coli pool that generated them, was analyzed by Southern blot using a labeled URA3 probe. Figure 4 shows the results of this experiment: The first two lanes correspond to the first C. glabrata pool and the E. coli pool, respectively; all of the bands from the bacterial pool are shared with the yeast pool, indicating that most of the individual insertions are indeed represented in C. glabrata transformants. Because the relative intensities of the individual bands in the E. coli pool and the C. glabrata pools are the same, this shows that the recombination of individual fragments carrying Tn7 occurs at approximately equal frequencies for most of the insertions in the pool, consistent with what we had found for the smaller pool of 12 insertions. The same result was found for the second fosmid (Fig. 4, lanes 4,5). Stripping the nylon membrane and reprobing with vector sequences showed that there were no vector sequences in the C. glabrata pool as expected (data not shown). In a control experiment shown in lanes 6–8, fosmid vector pBAC was mutagenized with Tn7 UKR-H, and a pool of >1000 insertions in pBAC was generated. DNA from the pool was digested with BcgI and MfeI and used to transform BG14 (ura3Δ) to Ura+ by nonhomologous recombination of the vector fragments containing Tn7 UKR-H insertions. Genomic DNA from a pool of five C. glabrata stable transformants was analyzed by Southern blot. When a labeled URA3 probe was used, none of the bands in the E. coli pool (Fig. 4, lane 8) were also present in the yeast pool (Fig. 4, lane 6), showing that all of the C. glabrata transformants were nonhomologous recombinants.
Figure 4.
Southern analysis of Candida glabrata transformants derived from two mutagenized fosmids. The probes are the URA3 gene from Tn7 UKR (lanes 1–9). All DNAs were digested with BcgI and BamHI prior to agarose gel electrophoresis. (Lane 1) C.g., genomic DNA from a pool of 35 Ura+ transformants derived from transformation with mutagenized fosmid 1. (Lane 2) E. coli, genomic DNA from 150 pooled Tn7 insertions in fosmid 1. (Lane 3) U, DNA from unmutagenized fosmid 1. (Lane 4) C.g., genomic DNA from a pool of 44 Ura+ transformants derived from transformation with mutagenized fosmid 2. (Lane 5) E. coli, genomic DNA from 150 pooled Tn7 insertions in fosmid 2. (Lane 6) C.g., genomic DNA from a pool of 5 Ura+ transformants derived from transformation with Tn7-mutagenized pBAC-Bcg3. (Lane 7) DNA from unmutagenized fosmid 2. (Lane 8) E. coli, genomic DNA from >1000 pooled Tn7 insertions in plasmid pBAC-Bcg3. (Lane 9) Cont, control pIC31 digested with PstI.
Thus, all (or the majority) of the linear fragments of genomic DNA derived from a mutagenized fosmid containing Tn7 insertions can readily undergo homologous recombination generating allele replacements, unlike fragments with no homology to C. glabrata, which are integrated, at lower frequency, by nonhomologous recombination.
Spectrum of Mutations Isolated
We used 96 mutagenized fosmid DNAs to mutagenize ∼25% of the C. glabrata genome. DNA from each individual pool (corresponding to a single fosmid) was prepared, digested with either BcgI or MfeI, and transformed into C. glabrata. For each fosmid, 96 Class 3 transformants were identified, generating in all 9216 mutants.
To assess the diversity of generated mutations, we screened for four phenotypes: amino acid auxotrophy, sensitivity or resistance to the triazole antifungal fluconazole, and inability to grow at 37°C, 40°C, or 42°C (a phenotype associated with clinical isolates of S. cerevisiae). We isolated 52 amino acid auxotrophs (from 20 fosmids—see Methods) from this pool of mutants. In terms of fluconazole sensitivity, the parental strain is able to grow well at a concentration of 8 mg/L, is partially growth-inhibited at 16 mg/L, and cannot grow at 32 mg/L (data not shown). We identified 38 insertions (∼12 loci) that rendered the cells resistant to fluconazole (64 mg/L) and 14 insertions (2 loci) that rendered the cells sensitive to fluconazole (4 mg/L). Three mutants (2 loci) were unable to grow at 37°C, whereas 29 additional mutants (18 loci) were unable to grow at 40°C.
We determined the DNA sequence surrounding the insertion point for many of the amino acid auxotrophic mutants. Genomic DNA for any mutant of interest was digested either with XbaI or MfeI, and DNA ligase was added. This generated a circular plasmid containing the transposon and the DNA flanking the insertion site, which was recovered by transformation into E. coli selecting for Km resistance. Of 68 sequenced mutants, we determined that 58 (86%) were derived by homologous recombination. For the auxotrophs, the insertion points and identity of the disrupted gene for one representative insertion in each of 11 loci are shown in Table 2. Approximately three-fourths of the insertions recovered from the C. glabrata genome were in the coding regions of genes.
Table 2.
Analysis of Auxotrophic Mutants
| Mutant | Gene | Insertion site |
| 15-04 | ILV1 | aa 268 |
| 31-36 | ARG2 | aa 441 |
| 37-26 | SUL2 | aa 388 |
| 42-37 | LYS14 | aa 755 |
| 47-40 | MET3 | aa 245 |
| 48-14 | LYS12 | aa 348 |
| 53-73 | ARG8 | aa 113 |
| 59-09 | ARG7 | aa 110 |
| 85-62 | LYS9 | aa 116 |
| 92-53 | GDH1 | aa 148 |
| 95-75 | LYS5 | aa 240 |
Shown are the genes disrupted for the 11 different loci disrupted in a total of 43 mutants (derived by homologous recombination) isolated by screening 9216 mutants. The position of the Tn7 insertion for each of the insertions is given. For multiple auxotrophic mutants derived from the same fosmid (and therefore predicted to be mutants in the same gene), at least two insertions were sequenced. In all cases, as expected, the same gene was disrupted but at a different site of insertion.
DISCUSSION
We have mutagenized C. glabrata using a modified Tn7 transposon, derivatized with the S. cerevisiae URA3 gene, the K. pneumonia hph gene, and the E. coli R6Kγ conditional origin. After mutagenesis of large fosmid-borne fragments of the genome in vitro and recovery of these insertion mutants in E. coli, the insertion mutants were introduced by allele replacement into the genome of C. glabrata. We had first tried the simpler approach of mutagenizing C. glabrata chromosomal DNA with Tn7 and directly transforming this pool of DNA into the organism, bypassing the need for the fosmid library or recovery of insertions in E. coli. However, probably because C. glabrata transformation is relatively inefficient and because, as detailed in this paper, there are multiple ways in which a linear DNA can integrate into the genome, direct transformation of mutagenized chromosomal DNA yielded no simple Tn7 insertions in the genome. We therefore used the scheme described in this paper.
The distribution of insertion sites for Tn7 itself in vitro showed essentially no bias at all, and we present evidence here that the full range of mutations generated in vitro can be introduced by homologous recombination into the genome of C. glabrata. The resultant library of mutants is a highly random collection of insertion mutations. This method, although not as random as high-throughput disruption approaches, nonetheless generates, with moderate effort, more highly distributed mutations than, for example, those generated by Ty1 mutagenesis in S. cerevisiae. The Tn3 mutagenesis technique in S. cerevisiae (Ross-Macdonald et al. 1999) probably yields a similar spectrum of mutations as our Tn7 method, although no analysis (similar to that presented here) of fates of Tn3-mutagenized linear fragments has been reported forS. cerevisiae. However, given the low rates of nonhomologous recombination in S. cerevisiae compared with C. glabrata, it is likely that the majority of transformants in S. cerevisiae resulting from transformation with a linear genomic fragment containing a Tn3 insertion are, in fact, allele replacements. The complications in our mutagenesis strategy resulting from the isolation of three classes of C. glabratatransformants were related to recombination parameters, which were specific to C. glabrata. Lastly, the ratio of homologous to nonhomologous recombinants is likely to be species-specific and the utility of Tn7 as a mutagenesis method in other organisms will certainly depend in part on the efficiency of homologous recombination and equally importantly on the ratio of homologous to nonhomologous recombination.
The R6Kγ origin that we engineered into the transposon facilitates recovery of the transposon and associated flanking genomic DNA for any insertion of interest. The R6Kγ origin is silent in any E. coli strain lacking the o-ring proteins; thus, fosmids carrying the Tn7 insertion could be maintained stably in DH5αMCR. When cloning genomic DNA flanking the transposon, we transform the permissive host and easily recover the transposon and associated DNA.
The insertions that we isolated are targeted to a diverse subset of genes in C. glabrata. Among the mutations that we found among the first 9216 generated by this method were 52 amino acid auxotrophs with insertions in 11 genes known from S. cerevisiae to be required for amino acid prototrophy. We also found insertions in two loci that profoundly altered susceptibility to the antifungal fluconazole. The first of these was in CgCDR1, the C. glabrata ortholog of the S. cerevisiae PDR5 gene, which encodes an ABC transporter (Balzi et al. 1994). This gene has previously been shown to be required for resistance to azoles (Miyazaki et al. 1998; Sanglard et al. 1999); the second was caused by a nonhomologous recombination event and has not been characterized further. We identified 38 fluconazole-resistant mutants that will be characterized elsewhere.
Our analysis of small pools of insertion mutants indicated that the mutagenesis method is, indeed, random. Southern analysis of pools of ∼40 insertions in C. glabrata derived from pools of 150 insertions in E. coli showed that all the bands in E. coli were, in fact, present at approximately the same intensity in the pool of C. glabrata mutants. This indicated strongly that most mutants in a pool of insertions in E. coli recombine at approximately equal frequency into the C. glabrata genome. However, because each band on the Southern could in theory correspond to any one of many different insertions in that fragment, it is possible that each band on the Southern in the C. glabrata lane was, in fact, caused by differential recombination (poisoning) by one or a small subset of the many insertions in that fragment present in the E. coli library (each of which contributes to the corresponding band in the E. coli lane). We view this formal possibility as unlikely given the randomness of the insertions that we have analyzed by sequencing. For 21 different loci, we isolated and sequenced multiple insertions giving the same phenotype (as many as 7 insertions for the same locus). For none of these 54 mutants were the insertion sites the same, implying that for a variety of loci, multiple insertions in the same genomic fragment (in the E. coli library) all recombine into the C. glabrata genome. Our characterization of ∼100 initial insertion mutants indicates in other ways that the method is, indeed, random. First, three-fourths of the insertions generated in C. glabrata were in coding regions of the genome, which corresponds well with the ratio of coding to noncoding regions in the S. cerevisiae genome. Second, we identified 11 loci giving tight auxotrophic phenotypes in a mutagenesis that covers 20%–25% of the genome. Extrapolating from S. cerevisiae, in which disruptions of ∼65 genes (taken from the SGD database) show tight auxotrophic phenotypes on media lacking amino acids, we might have expected to identify ∼14 mutants.
The presence of the R6Kγ origin in the Tn7 transposon permits the rapid rescue of a plasmid containing the Tn7 transposon and DNA flanking the insertion site. This plasmid can be used to obtain sequence information for the locus disrupted. Equally importantly, this rescued plasmid can be used (via homologous recombination and allele replacement) to introduce the mutation into a clean background and formally verify that the phenotype in question is the result of the Tn7 insertion. Lastly, although they are not used for analysis here, the C. glabrata mutants are all barcoded with short oligonucleotide tags (see Cormack et al. 1999 and Methods). This will permit parallel analysis of the mutants in future screens.
This method of mutagenesis is potentially broadly applicable to other organisms, requiring only that the organism be transformable and that transforming DNA integrate via homologous recombination. Its advantages over chemical methods of mutagenesis are obviously in the rapidity of the subsequent analysis of mutants with phenotypes of interest. We would also suggest that it has potential advantages over in vivo methods of transposon mutagenesis depending on any in vivo bias for transposon insertion sites (e.g., the bias to insertion sites upstream of RNA polymerase III promoters or in silent chromatin displayed by the S. cerevisiae Ty family of transposons; Ji et al. 1993; Zou et al. 1996; Kim et al. 1998). In summary, we present the characterization of a novel approach to insertional mutagenesis in the pathogenic yeast C. glabrata. This method yields a randomly distributed set of insertion mutations throughout the genome, which can be screened for phenotypes affecting diverse aspects of metabolism, physiology, or virulence.
METHODS
Strains and Growth Conditions
E. coli strain BW23473 (Δlac-169 robA1 creC510 hsdR514 ΔuidA::pir endA recA1; Metcalf et al. 1996) was used for maintenance of conditional replicons carrying an R6Kγ origin of replication that require expression of the protein Π (the product of the pir gene) for replication. Otherwise, strain DH10 (GIBCO BRL) was used routinely for electroporation of plasmids or fosmids. Barcoded derivatives of C. glabrata strain BG14 (ura3Δ::Tn903NeoR; Cormack and Falkow 1999) were used for all the experiments described. These 96 barcoded strains are identical except that each has a unique oligonucleotide integrated in the genome, which permits tracking of the strain in a mixed pool of barcoded strains (Cormack et al. 1999).
Media
Electrocompetent cells were prepared according to Dower et al. (1988); plasmid and fosmid DNA preps were made using Qiaspin miniprep columns (QIAGEN) according to the manufacturer's instructions. To obtain reasonable yields for fosmid DNA preps that are maintained at a single copy in E. coli, 10-mL cultures were grown in 2× YT at 37°C overnight. The cell pellet was resuspended and lysed in twice the volume recommended for regular minipreps, and then passed over two Qiaspin columns. For growth of fosmid DNAs, media was supplemented with Cm to 10 mg/L and for mutagenized fosmids with Km to 30 mg/L.
Yeast cells were routinely grown on standard S. cerevisiae media (Sherman et al. 1986): YPD (YEP supplemented with 2% final glucose concentration at 30°C). For Ura+ selection of BG14, SD plates supplemented with 0.6% of casamino acids (GIBCO) were used. To score for resistance or sensitivity to 5-fluorotic acid (5-FOA), YNB plates were supplemented with casamino acids (0.6%), uracil (25 mg/mL) and 1.4 g/L of 5-FOA. Scoring of the hygromycin phenotype was done on YPD plates supplemented with a 500 μg/mL final concentration of hygromycin B. The phenotype of the insertion mutants was scored by growth on YPD plates incubated at 30°C, 37°C, and 42°C; by growth on YPD plates supplemented with fluconazole at concentrations of 4 mg/mL, 8 mg/mL, 16 mg/mL, 32 mg/mL, and 64 mg/mL incubated at 30°C; by growth on minimal SD plates; and by growth on minimal SD plates supplemented with 0.6% casamino acids.
C. glabrata Transformation
We used a modification of the LiAc method (Gietz et al. 1992). Briefly, cells were grown to early log phase in YPD, harvested, and washed with an equal volume of sterile water. The cells were then resuspended in 1/100 volume of 100 mM LiAc, and 50-μL aliquots were used for each transformation. To each tube containing 50 μL of cell suspension, 240 μL of 50% PEG (3500) was added, followed by 36 μL of 1 M LiAc, 50 μg of heat-denatured salmon sperm DNA, and the transforming DNA dissolved in 50 μL of sterile water. This mix was incubated at 30°C for 30 min, after which 45 μL of DMSO was added, mixed, and immediately incubated at 42°C for 15 min. Cells were centrifuged, resuspended in H2O, and plated on selective media.
Construction of Fosmid Library
The plasmid pBAC carries the F origin, the cat gene conferring Cm resistance, and a cos site for packaging (Kim et al. 1992). We modified pBAC by introducing two BcgI sites immediately flanking the BamHI site. This was accomplished by first digesting pBAC with NotI and then cloning a double-stranded DNA oligonucleotide obtained by allowing oligonucleotides BCGI (GGCCGCCGAATTATTTGCGGATCCCGAATTAT TTGCGAAGCTTGC) and BCG2 (GGCCGCAAGCTTCG CAAATAATTCGGGATCCGCAAATAATTCGGC) to anneal. The final vector, pBAC-Bcg3, is similar to the original, but the BamHI cloning site is now immediately flanked by BcgI sites (sequence deposited at GenBank accession number). The advantage of this is that once a fragment of the genome has been cloned into the BamHI site, it can be precisely released with BcgI, which cleaves 7 bases downstream of its site (inside the cloned genomic insert). Thus, the genomic DNA ends generated by BcgI digestion are precisely homologous to the genome, with no nucleotides derived from the pBAC vector.
Genomic DNA from strain BG14 was carefully prepared according to Goshorn et al. (1992). The DNA was digested with decreasing amounts of Sau3A and after addition of EDTA (10 mM) to stop the digestion, fractions were analyzed by gel electrophoresis to find the sample in which the average molecular weight of the DNA was well above 40 kb. This sample was treated with shrimp alkaline phosphatase (Epicenter), and the phosphatase was inactivated by heat. To generate the fosmid arms, pBAC-Bcg3 was digested with HindIII and phosphatased with Calf Intestinal Phosphatase (Boehringer Mannheim). The phosphatase was removed by phenol extraction, and the sample was redigested with BamHI to generate one of the fosmid arms (a HindIII–BamHI fragment). The second fosmid arm (EcoRI–BamHI) was generated by digesting the fosmid with EcoRI, followed by treatment with Calf Intestinal Phosphatase. The phosphatase was removed by phenol extraction, and the product was digested with BamHI. These two fosmid arms were ligated to the genomic DNA; the resultant ligation was packaged using XL packaging extract (Stratagene) and used for infection of DH5αMCR cells. Individual CmR transformants were picked and arrayed in 96-well plates. For 700 of these, fosmid DNA was prepared from 10-mL cultures using the REAL prep kit in a 96-well format (Stratagene). The library is highly representative. We picked 5500 fosmids (∼15× genome coverage) and analyzed by PCR how many times particular genes were present. The majority of genes were present between 10 and 15 times (TRP1, 16 times; HIS3, 15 times; YERO19w homolog, 12 times; EPA1, 7 times).
Plasmid Constructions
pBC166.2
pBC166.2 is derived from plasmid pMCB40 (p-oriR6Kγ::miniTn7 SpeI-KmR-NotI). This plasmid contains the conditional origin of replication R6Kγ and a mini-Tn7 element conferring resistance to kanamycin (Biery et al. 2000). We first introduced the URA3 gene of S. cerevisiae into this mini-Tn7. We PCR-amplified the gene with primers 341 and 343 containing NotI sites. The PCR product was digested with NotI and ligated to pMCB40 to generate plasmid pBC166.2.
pIC6
Plasmid pBC166.2 was used as template to PCR-amplify a 398-bp fragment containing the R6Kγ origin of replication, using primers 538 and 539 containing XbaI sites; the PCR product was digested with XbaI and ligated to SpeI-digested pBC166.2 to generate plasmid pIC2. This plasmid contains two R6Kγ origins of replication: the original one in the backbone, and the newly cloned copy within the Tn7 element. To remove the backbone R6Kγ origin, plasmid pIC2 was digested with AatII and PmlI, blunted, ligated, and transformed into the permissive strain BW23473. The plasmid generated, pIC6, carries the element Tn7 UKR (Tn7 URA3 · KmR · R6Kγ). This minitransposon contains within it a yeast-selectable marker (URA3), a kanamycin-resistance cassette, and the conditional origin of replication R6Kγ. The element carries the only origin of replication of the plasmid, and must be maintained in strain BW23473 (Metcalf et al. 1996), which expresses the Π protein from a chromosomal fusion to the uidA gene.
pIC31
We then modified pIC6 by cloning the hph gene conferring resistance to hygromycin B (Tn7 UKR-H) into the Tn7 element. To do this, we first destroyed one of the two NotI sites in pIC6 by partially digesting, blunting, and religating. This generated plasmid pIC28, which contains only one NotI site (downstream from URA3). We then PCR-amplified the K. pneumoniae hph gene conferring resistance to hygromycin B (Gritz and Davies 1983) from plasmid pAG26 (Goldstein et al. 1999); the PCR product was cloned in front of the PGK1 gene promoter from S. cerevisiae (P-PGK) in a derivative of pRS316 to generate plasmid pAP358. C. glabrata transformants with this plasmid grow on YPD plates supplemented with 200 μg/mL of hygromycin B. The 2.0-kb hygromycin-resistance cassette from plasmid pAP358 was excised by digesting with SacI and KpnI, blunted, and ligated to pIC28 cut with NotI and blunted, generating plasmid pIC31. This plasmid carries minitransposon Tn7 UKR-H (URA3 · Hph · KmR · R6Kγ), which contains the URA3 selectable marker, the kanamycin-resistance gene, the conditional origin of replication R6Kγ, and the hygromycin-resistance cassette.
pIC5.3::Tn7-UKR
One random insertion of the Tn7 UKR transposon in one arbitrarily chosen fosmid was used to generate a smaller plasmid. The mutagenized fosmid was digested with EcoRI and religated; the resulting plasmid is an 8.4-kb conditional replicon consisting of a 5.3-kb EcoRI C. glabrata genomic fragment with a Tn7 UKR insertion.
In Vitro Mutagenesis
In vitro transposition reactions were performed as described by Biery and Craig (Biery et al. 2000) using purified transposase proteins (TnsA, TnsB, and TnsCA25V purchased from New England Biolabs). For fosmid mutagenesis, we first prepared fosmid templates from 10-mL cultures using the QIAGEN REAL prep, according to the manufacturer's instructions. We used 400 ng of target fosmid DNA (0.12 nM), and 100 ng of donor plasmid carrying the transposon: pIC31 or pIC6 (0.19 and 0.22 nM, respectively). The reaction was stopped by phenol extraction and precipitation, and the DNA was resuspended in 10 μL of TE. Then 1 μL of this mutagenesis mix was used to transform DH10 electrocompetent cells, and transformants were selected on media containing chloramphenicol (10 mg/mL) and kanamycin (50 mg/mL). Only those fosmid molecules into which the mini-Tn7 transposed are able to replicate in this strain, because donor molecules contain only the conditional R6Kγ origin, and unmutagenized fosmid is not resistant to kanamycin. For each fosmid, a minimum of 200 and a maximum of tens of thousands of transformants were pooled and grown in a 100-mL culture to saturation at 30°C. This culture was used to prepare fosmid DNA using the QIAGEN midiprep kit according to the manufacturer's instructions.
Classification of C. glabrata Transformants
To introduce the mutagenized fosmids into C. glabrata, we used DNA from 96 mutagenized fosmids to transform the 96 oligonucleotide-tagged (barcoded) derivatives of BG14 (ura3Δ::Tn903NeoR). First, 1–2 μg of a mutagenized fosmid DNA was digested with either BcgI or with MfeI prior to transformation. The transformation mixes were plated on SD plates lacking uracil, and Ura+ isolates were scored after growth at 30°C for 2–3 d. Ura+ transformants fall in three different classes based on the phenotype displayed on 5-FOA plates. The first class of unstable (U) transformants is composed of colonies that are able to grow both on 5-FOA plates as well as on plates lacking uracil, therefore appearing as Ura+ 5-FOAR colonies. The second class of partially stable (PS) transformants refers to colonies that appear to be Ura+ FOAS; however, upon incubation at 30°C (48–72 h), patches of the transformant engender a small number of 5-FOAR papillae on the 5-FOA plate. The third class of stable (S) transformants consists of colonies that display the expected Ura+ 5-FOAS phenotype without papillation even after prolonged incubation on 5-FOA plates. For any given fosmid tested, the percent of transformants derived from each class of transformant did not change by altering parameters of the transformation protocol; the parameters tested were inclusion or omission of DMSO and varying the time (15 min to 1 h) and temperature (from 37°C or 45°C) of the heat-shock temperature (data not shown).
Killing of Untransformed Background and Abortive Transformants of C. glabrata
C. glabrata was transformed to Ura+ using linearized, in vitro mutagenized fosmids. Transformants were picked and arranged on a plate lacking uracil, allowed to grow overnight at 30°C, and printed onto 5-FOA plates. The majority of Ura+ colonies tested this way were contaminated with untransformed background (Ura− 5-FOAR), which are Ura− and thus 5-FOAR; therefore, most 5-FOAS transformants appeared to give 5-FOAR papillae. To identify transformants that were truly 5-FOAS, it was necessary to eliminate these contaminating cells. This could be easily accomplished by colony-purifying the transformants on plates lacking uracil. However, we decided this was not practical for thousands of transformants. Therefore, a step to screen for hygromycin-resistant transformants was introduced. Transformants patched on plates were replica-plated onto a hygromycin-containing YPD plate, allowed to grow, and printed a second time onto hygromycin plates; patches were subsequently printed onto 5-FOA plates and incubated at 30°C for 48–72 h. After these two consecutive prints on hygromycin plates, all of the untransformed background was killed, because they are sensitive to hygromycin. In control experiments, identical results were obtained by streak purification and replica-printing onto hygromycin plates: that is, the bona fide Ura+ 5-FOAS colonies identified by colony purification were also identified by printing onto hygromycin plates (data not shown).
Consideration of Multiple Mutants Derived From a Single Fosmid
Because the majority of our stable recombinants are derived by homologous recombination, the expectation is that if a fosmid contains a gene that can mutate to a given phenotype, then in a pool of 100 transformants derived from that fosmid, there will likely be multiple mutants, representing independent insertions in the same gene and giving the same phenotype. Given that we have mutagenized only 20%–25% of the genome, most fosmids analyzed here will not physically overlap. Therefore, the number of fosmids from which mutants with a phenotype were derived, rather than the number of mutants themselves, gives a good indication of the number of loci identified. Thus, for auxotrophs, we identified 52 mutants, but these were derived from 20 fosmids (between 1 and 9 auxotroph mutants isolated per fosmid), and we therefore estimate that our mutants identify ∼20 loci. For fluconazole sensitivity, we identified 15 insertions, but all derived from two fosmids. We therefore estimate that our mutants identify only two loci. That this line of reasoning is valid for the majority of cases is indicated by the fact that we have sequenced the insertions for up to three independent auxotrophs derived from the same fosmid; in 5/5 cases, mutants from the same fosmid with the same phenotype represented independent insertions in the same gene; in contrast, auxotrophic mutants derived from different fosmids were all in different genes.
Acknowledgments
We are grateful to Giuseppe Rotondo for helpful discussions during the course of these experiments. We thank A. Goldstein and J. McCusker for plasmids. This work was funded by NIH grants RO1 AI46223 and 2PO1 DK49720 to B.C., by a Searle Scholars award to B.C., and by a UNCF-Pfizer postdoctoral fellowship to A.D.L.P.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
E-MAIL bcormack@jhmi.edu; FAX (410) 502-6718.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.848203. Article published online before print in April 2003.
REFERENCES
- 1.Balzi E., Wang, M., Leterme, S., Van Dyck, L., and Goffeau, A. 1994. PDR5, a novel yeast multidrug resistance conferring transporter controlled by the transcription regulator PDR1. J. Biol. Chem. 269: 2206-2214. [PubMed] [Google Scholar]
- 2.Barns S.M., Lane, D.J., Sogin, M.L., Bibeau, C., and Weisburg, W.G. 1991. Evolutionary relationships among pathogenic Candida species and relatives. J. Bacteriol. 173: 2250-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biery M.C., Stewart, F.J., Stellwagen, A.E., Raleigh, E.A., and Craig, N.L. 2000. A simple in vitro Tn7-based transposition system with low target site selectivity for genome and gene analysis. Nucleic Acids Res. 28: 1067-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calderone R.A. and Fonzi, W.A. 2001. Virulence factors of Candida albicans. Trends Microbiol. 9: 327-335. [DOI] [PubMed] [Google Scholar]
- 5.Cormack B.P. and Falkow, S. 1999. Efficient homologous and illegitimate recombination in the opportunistic yeast pathogen Candida glabrata. Genetics 151: 979-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cormack B.P., Ghori, N., and Falkow, S. 1999. An adhesin of the yeast pathogen Candida glabrata mediating adherence to human epithelial cells. Science 285: 578-582. [DOI] [PubMed] [Google Scholar]
- 7.Csank C. and Haynes, K. 2000. Candida glabrata displays pseudohyphal growth. FEMS Microbiol. Lett. 189: 115-120. [DOI] [PubMed] [Google Scholar]
- 8.DeBoy R.T. and Craig, N.L. 1996. Tn7 transposition as a probe of cis interactions between widely separated (190 kilobases apart) DNA sites in the Escherichia coli chromosome. J. Bacteriol. 178: 6184-6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dower W.J., Miller, J.F., and Ragsdale, C.W. 1988. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 16: 6127-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gietz D., St. Jean, A., Woods, R.A., and Schiestl, R.H. 1992. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20: 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goldstein A.L., Pan, X., and McCusker, J.H. 1999. Heterologous URA3MX cassettes for gene replacement in Saccharomyces cerevisiae. Yeast 15: 507-511. [DOI] [PubMed] [Google Scholar]
- 12.Goshorn A.K., Grindle, S.M., and Scherer, S. 1992. Gene isolation by complementation in Candida albicans and applications to physical and genetic mapping. Infect. Immun. 60: 876-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gritz L. and Davies, J. 1983. Plasmid-encoded hygromycin B resistance: The sequence of hygromycin B phosphotransferase gene and its expression in Escherichia coli and Saccharomyces cerevisiae. Gene 25: 179-188. [DOI] [PubMed] [Google Scholar]
- 14.Gueiros-Filho F.J. and Beverley, S.M. 1997. Trans-kingdom transposition of the Drosophila element mariner within the protozoan Leishmania. Science 276: 1716-1719. [DOI] [PubMed] [Google Scholar]
- 15.Ji H., Moore, D.P., Blomberg, M.A., Braiterman, L.T., Voytas, D.F., Natsoulis, G., and Boeke, J.D. 1993. Hotspots for unselected Ty1 transposition events on yeast chromosome III are near tRNA genes and LTR sequences. Cell 73: 1007-1018. [DOI] [PubMed] [Google Scholar]
- 16.Kim J.M., Vanguri, S., Boeke, J.D., Gabriel, A., and Voytas, D.F. 1998. Transposable elements and genome organization: A comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence. Genome Res. 8: 464-478. [DOI] [PubMed] [Google Scholar]
- 17.Kim U.J., Shizuya, H., de Jong, P.J., Birren, B., and Simon, M.I. 1992. Stable propagation of cosmid sized human DNA inserts in an F factor based vector. Nucleic Acids Res. 20: 1083-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kolter R., Inuzuka, M., and Helinski, D.R. 1978. Trans-complementation-dependent replication of a low molecular weight origin fragment from plasmid R6K. Cell 15: 1199-1208. [DOI] [PubMed] [Google Scholar]
- 19.Metcalf W.W., Jiang, W., Daniels, L.L., Kim, S.K., Haldimann, A., and Wanner, B.L. 1996. Conditionally replicative and conjugative plasmids carrying lacZ α for cloning, mutagenesis, and allele replacement in bacteria. Plasmid 35: 1-13. [DOI] [PubMed] [Google Scholar]
- 20.Miyazaki H., Miyazaki, Y., Geber, A., Parkinson, T., Hitchcock, C., Falconer, D.J., Ward, D.J., Marsden, K., and Bennett, J.E. 1998. Fluconazole resistance associated with drug efflux and increased transcription of a drug transporter gene, PDH1, in Candida glabrata. Antimicrob. Agents Chemother. 42: 1695-1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pfaller M.A., Messer, S.A., Hollis, R.J., Jones, R.N., Doern, G.V., Brandt, M.E., and Hajjeh, R.A. 1999. Trends in species distribution and susceptibility to fluconazole among blood stream isolates of Candida species in the United States. Diagn. Microbiol. Infect. Dis. 33: 217-222. [DOI] [PubMed] [Google Scholar]
- 22.Pfaller M.A., Diekema, D.J., Jones, R.N., Sader, H.S., Fluit, A.C., Hollis, R.J., and Messer, S.A. 2001. International surveillance of bloodstream infections due to Candida species: Frequency of occurrence and in vitro susceptibilities to fluconazole, ravuconazole, and voriconazole of isolates collected from 1997 through 1999 in the SENTRY Antimicrobial Surveillance Program. J. Clin. Microbiol. 39: 3254-3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ross-Macdonald P., Coelho, P.S., Roemer, T., Agarwal, S., Kumar, A., Jansen, R., Cheung, K.H., Sheehan, A., Symoniatis, D., Umansky, L., et al. 1999. Large-scale analysis of the yeast genome by transposon tagging and gene disruption. Nature 402: 413-418. [DOI] [PubMed] [Google Scholar]
- 24.Sanglard D., Ischer, F., Calabrese, D., Majcherczyk, P.A., and Bille, J. 1999. The ATP binding cassette transporter gene CgCDR1 from Candida glabrata is involved in the resistance of clinical isolates to azole antifungal agents. Antimicrob. Agents Chemother. 43: 2753-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schaberg D.R., Culver, D.H., and Gaynes, R.P. 1991. Major trends in the microbial etiology of nosocomial infection. Am. J. Med. 91: 72S-75S. [DOI] [PubMed] [Google Scholar]
- 26.Schuman P., Sobel, J.D., Ohmit, S.E., Mayer, K.H., Carpenter, C.C., Rompalo, A., Duerr, A., Smith, D.K., Warren, D., and Klein, R.S. 1998. Mucosal candidal colonization and candidiasis in women with or at risk for human immunodeficiency virus infection. HIV Epidemiology Research Study (HERS) Group. Clin. Infect. Dis. 27: 1161-1167. [DOI] [PubMed] [Google Scholar]
- 27.Seifert H.S., Chen, E.Y., So, M., and Heffron, F. 1986. Shuttle mutagenesis: A method of transposon mutagenesis for Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 83: 735-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sherman F., Fink, G.R., and Hicks, J.B., 1986. Methods in yeast genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 29.Vazquez J.A., Sobel, J.D., Peng, G., Steele-Moore, L., Schuman, P., Holloway, W., and Neaton, J.D. 1999. Evolution of vaginal Candida species recovered from human immunodeficiency virus-infected women receiving fluconazole prophylaxis: The emergence of Candida glabrata? Terry Beirn Community Programs for Clinical Research in AIDS (CPCRA). Clin. Infect. Dis. 28: 1025-1031. [DOI] [PubMed] [Google Scholar]
- 30.Zou S., Ke, N., Kim, J.M., and Voytas, D.F. 1996. The Saccharomyces retrotransposon Ty5 integrates preferentially into regions of silent chromatin at the telomeres and mating loci. Genes & Dev. 10: 634-645. [DOI] [PubMed] [Google Scholar]