Abstract
Wallerian degeneration is an important area of research in modern neuroscience. A large number of genes are differentially regulated in the various stages of Wallerian degeneration, especially during the early response. In this study, we analyzed gene expression in early Wallerian degeneration of the distal nerve stump at 0, 0.5, 1, 6, 12 and 24 hours after rat sciatic nerve injury using gene chip microarrays. We screened for differentially-expressed genes and gene expression patterns. We examined the data for Gene Ontology, and explored the Kyoto Encyclopedia of Genes and Genomes Pathway. This allowed us to identify key regulatory factors and recurrent network motifs. We identified 1 546 differentially-expressed genes and 21 distinct patterns of gene expression in early Wallerian degeneration, and an enrichment of genes associated with the immune response, acute inflammation, apoptosis, cell adhesion, ion transport and the extracellular matrix. Kyoto Encyclopedia of Genes and Genomes pathway analysis revealed components involved in the Jak-STAT, ErbB, transforming growth factor-β, T cell receptor and calcium signaling pathways. Key factors included interleukin-6, interleukin-1, integrin, c-sarcoma, carcinoembryonic antigen-related cell adhesion molecules, chemokine (C-C motif) ligand, matrix metalloproteinase, BH3 interacting domain death agonist, baculoviral IAP repeat-containing 3 and Rac. The data were validated with real-time quantitative PCR. This study provides a global view of gene expression profiles in early Wallerian degeneration of the rat sciatic nerve. Our findings provide insight into the molecular mechanisms underlying early Wallerian degeneration, and the regulation of nerve degeneration and regeneration.
Keywords: Wallerian degeneration, sciatic nerve, microarrays, expression profiling, rats, neural regeneration
Abbreviations
WD, Wallerian degeneration; KEGG, Kyoto Encyclopedia of Genes and Genomes; GO, Genev Ontology
INTRODUCTION
Wallerian degeneration (WD) occurs both in the peripheral nervous system and the central nervous system. Peripheral nerve injuries are quite common in clinical practice, and many studies have been conducted on the underlying molecular mechanisms. WD in the peripheral nervous system occurs following various traumatic injuries, such as nerve fiber transection or crush, and is also known as anterograde degeneration, orthograde degeneration and secondary degeneration. In WD, axonal injury induces a series of responses in the distal nerve segment[1,2]. WD can also be caused by inflammatory or autoimmune responses, in addition to traumatic injuries.
A sequence of events unfolds during WD in the distal stump of an axotomized nerve. WD begins with the disintegration and degeneration of the axoplasm and axolemma, which is complete within 24 hours. Axonal breakdown is mediated by calcium influx and involves the activation of axonal proteases. An important aspect of this response in peripheral nerves is the recruitment and activation of macrophages that leads to the phagocytosis and clearance of myelin and axonal debris. The rapid clearance of myelin, which contains axonal growth inhibitors, appears to contribute to axonal regeneration after peripheral nerve injury[3,4,5,6,7,8,9,10]. WD is currently the focus of much research in neuroscience. Although the mechanisms underlying WD have been studied extensively, the molecular changes in the distal stump during early WD, such as patterns of differential gene expression and the molecular networks involved, are unclear[11,12,13,14,15,16,17,18].
We report here on gene expression patterns in early WD of the rat distal nerve stump at 0, 0.5, 1, 6, 12 and 24 hours after sciatic nerve injury using microarray and bioinformatic analysis. The data were validated with real-time quantitative PCR. We analyzed the data using Series Test Cluster of Gene Ontology (STC-GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG). Our results will help us better understand gene expression profiles and patterns, as well as the signaling pathways and key regulatory networks involved in early WD.
RESULTS
Gene expression profiling and regulatory patterns during early WD after sciatic nerve injury
Gene Ontology (GO) analysis of differentially-expressed genes
To screen for differentially-expressed genes during WD after sciatic nerve injury, we examined the expression of genes at different time points, compared with the 0-hour time point, thereby obtaining 1 546 differentially-expressed genes and 21 significant patterns of gene expression in early WD (Figure 1). The data summarizes all possible expression patterns and statistical judgments based on P < 0.05/N as the standard.
Figure 1.
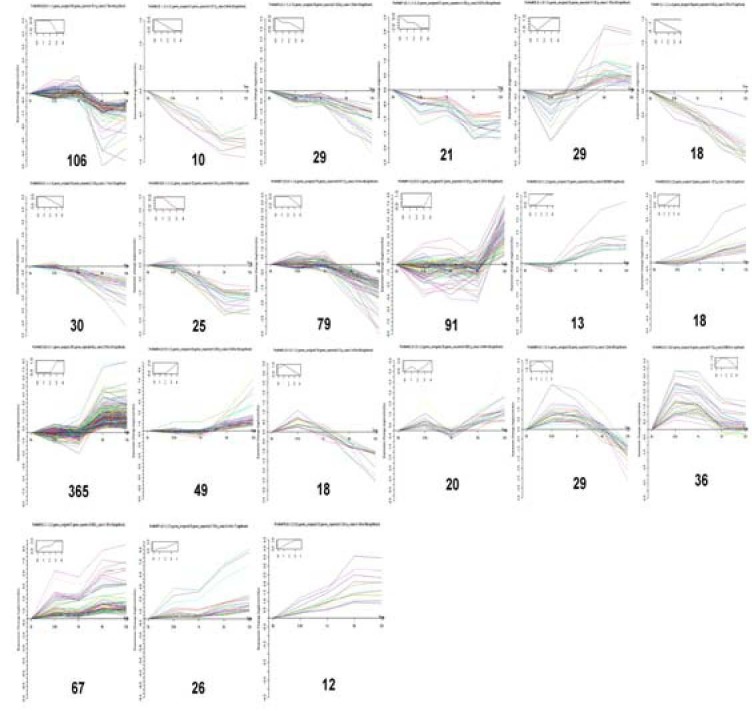
There are 1 546 differentially- expressed genes and 21 types of significant differentially-expressed gene patterns in early Wallerian degeneration of distal sciatic nerve stumps in rats.
The data summarizes all possible expression trends and statistical judgments, with P < 0.05/N as the criterion.
Functional classification by GO is an internationally standardized classification system for gene function offering a dynamic, updated and controlled vocabulary employing strictly defined concepts to comprehensively describe the properties of genes and their products in any organism. GO encompasses three domains: molecular function, cellular component and biological process[19,20,21]. GO analysis was conducted using gene expression patterns in a series of experiments, followed by significant and separate analyses of different gene expression trends in early WD. Quantitative changes in selected enriched GO biological processes were present and found to alter the expression of genes involved in these processes. Based on the GO database, the regulated genes were distributed into functional categories; these categories included genes with putative functions in the innate immune response, activation of the acute inflammatory response, promotion of chemokine production, Ras signal transduction, ion transport, nerve growth factor processing, regulation of gene-specific transcription, regulation of gene expression, promotion of axonogenesis, cytokine production, cytokinesis, neurological processing, neural tube development, regulation of cell differentiation and apoptosis (Figure 2).
Figure 2.
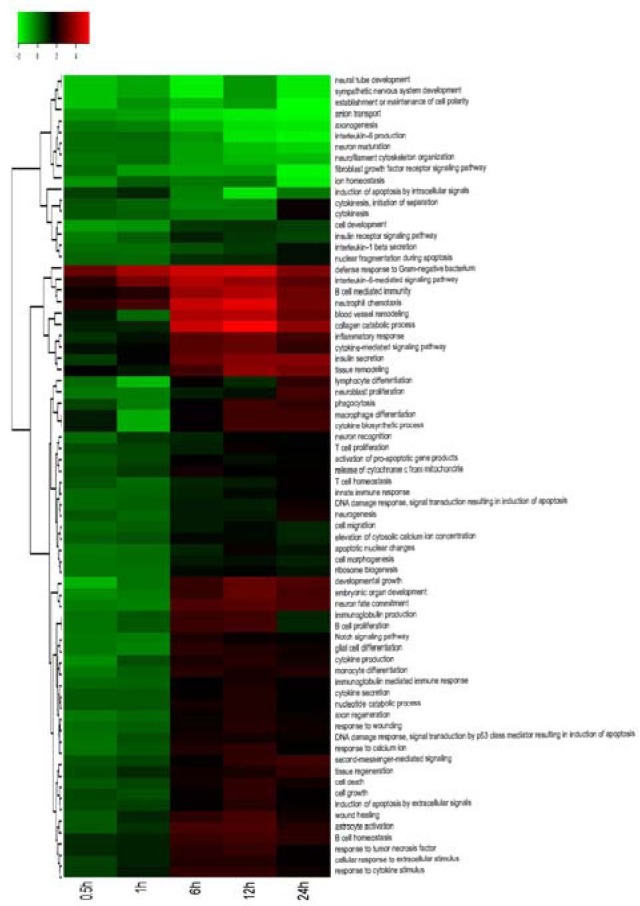
Hierarchical cluster analysis showing partition clustering of genes most highly expressed in the distal nerve stumps after sciatic nerve injury.
Gene Ontology analysis of distal sciatic nerve stumps of rats at 0.5, 1, 6, 12 and 24 hours post-surgery.
Fisher exact test and chi-square test were applied to the differentially-expressed genes, and significance analysis was performed with the target genes by screening for P < 0.05. The heat map shows the intensity of expression as a function of color, with higher expression in red and lower expression in green or black.
KEGG Pathway analysis of differentially-expressed genes during WD
Based on the GO database, Fisher's Exact Test and Chi Square tests were applied to the differentially-expressed genes, significance analysis was performed with the pathways involving target genes, and significant pathways were obtained by screening for P < 0.05. The KEGG Pathway database comprises information on networks of molecular interactions for numerous organisms, permitting functional classification. Pathway-based analysis provides insight into biological functions and interactions of genes. Based on a comparison against the GO database using BLAST with an E value cutoff of 10-5, 1 546 genes had significant matches in the database and were assigned to 70 KEGG pathways in early WD. KEGG pathway analysis identified numerous pathways, including those relating to B-cell receptor signaling, janus kinase and signal transducer and activator of transcription (Jak-STAT) signaling, apoptosis, cytokine-cytokine receptor interactions, toll-like receptor signaling, tight junctions, neuroactive ligand-receptor interactions, axon guidance, Wnt signaling, p53 signaling, T-cell receptor signaling, leukocyte transendothelial migration, vascular endothelial growth factor signaling, adherens junctions, cell adhesion molecules, ErbB signaling, gap junctions, transforming growth factor-β signaling, mitogen-activated protein kinase signaling, the extracellular matrix-receptor interactions, actin cytoskeleton regulation, calcium signaling and the cell cycle (Figure 3).
Figure 3.
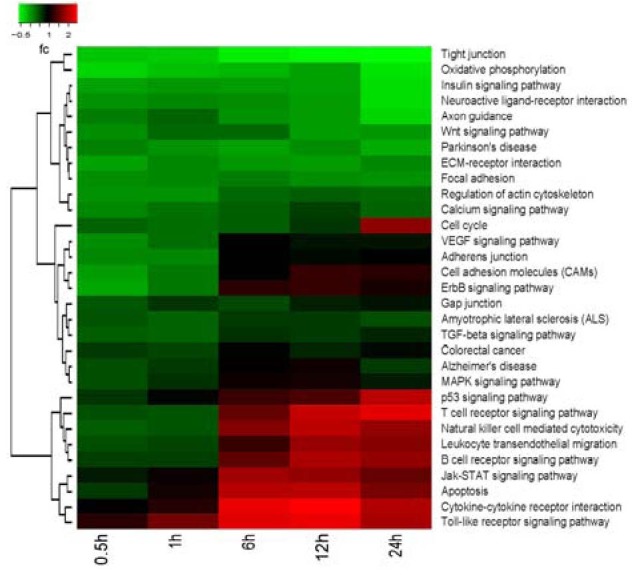
Hierarchical cluster analysis showing partition clustering of genes most highly expressed in the distal nerve stumps after sciatic nerve injury. Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of distal sciatic nerve stumps of rats at 0.5, 1, 6, 12 and 24 hours post-surgery.
Based on the KEGG database, Fisher exact test and Chi square test were applied to the differential genes, significance analysis was performed with the pathways involving target genes, and the significant pathways were obtained by screening for P < 0.05.
The heat map shows the intensity of expression as a function of color, with higher expression in red and lower expression in green or black.
Key network analysis of differentially-expressed genes during WD
External stimuli affect cellular behavior, as reflected in protein interactions and gene expression kinetics, and we infer from this the presence of dynamic gene regulatory networks. These networks are computed based on fold-changes in gene expression and gene interactions in pathways. The relationships among the gene expression data were inferred using a Continuous Time Recurrent Neural Network (CTRNN) as an abstract, dynamic model for gene regulatory networks mediating the cellular decision to migrate upon an external stimulus. The model describes the mutual influence of genes and their stimulus responses as dynamic elements, regardless of how such interactions or stimuli are realized in concrete biological terms. Using a genetic algorithm, we estimated the model parameters. A heat map and dendrogram showed partition clustering of genes most highly expressed in the distal nerve stumps after sciatic nerve injury.
Key networks were constructed from active experiments. The gene expression data was used as the input for the generation of biological networks, using the analyze network algorithm with default settings. This is a variant of the shortest paths algorithm with main parameters of relative enrichment and relative saturation of networks with canonical pathways. Genes encoding signal transducers or factors involved in processes such as cell death, the immune response, transport and transcriptional regulation showed injury-specific gene expression. The expression of numerous genes is required for regeneration[22,23,24]. In this study, we used CTRNN to dynamically model GO processes for gene regulatory networks. A number of key networks were identified, including those comprising interleukin-6, interleukin-1, integrin (ITG), c-sarcoma, carcinoembryonic antigen-related cell adhesion molecules, chemokine (C-C motif) ligand, matrix metalloproteinase, BH3 interacting domain death agonist (BID), baculoviral IAP repeat-containing 3 (BIRC3) and Rac (a subfamily of the Rho family of GTPases) (Figure 4). These key networks cover the most important pathways and play important roles in regulating gene expression profiles through mainly biological GO processes during WD of the sciatic nerve. The data obtained here are consistent with published data and support the conclusion that genetic programs of regenerating and developing peripheral nerves exhibit relevant differences. The data revealed that signaling molecules significantly regulated in the lesioned distal stump after sciatic nerve injury were steadily up and down-regulated. All of these molecules were significantly regulated after sciatic nerve injury and are known to modulate signaling pathways composing the key networks[15,25,26,27,28]. The data also suggest that the functional group of transcriptional regulators identified by the gene expressional changes may be an essential prerequisite for nerve degeneration and/or regeneration/development[29,30,31].
Figure 4.
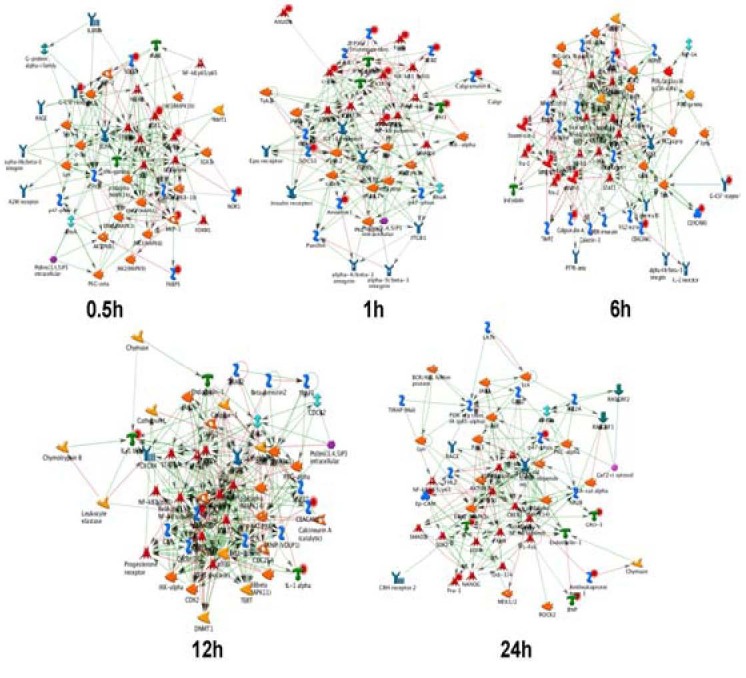
Key network analysis of distal sciatic nerve stumps of rats at 0.5, 1, 6, 12 and 24 hours post-surgery.
The gene expression data were used as the input file for the generation of biological networks using the analyze network algorithm with default settings.
The networks are prioritized based on the number of fragments of canonical pathways. Thick cyan lines indicate the fragments of canonical pathways.
Up-regulated genes are marked with red circles; down-regulated genes with blue circles. The checker board pattern indicates mixed expression for the gene between files or between multiple tags for the same gene.
Real-time quantitative PCR assay
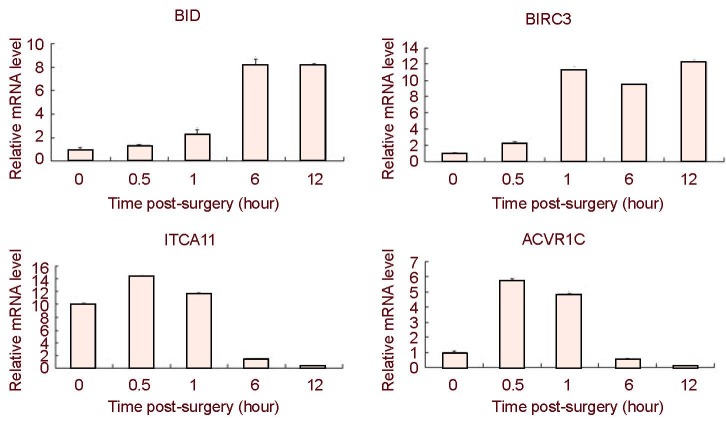
Although the reliability of our array data is demonstrated by a number of genes which have been previously described in response to peripheral nerve injury [such as c-jun (Herdegen and Zimmerman 1994), cyclin D1 (Kim et al. 2000), GADD45 (Befort et al. 2003), phospholipase A2 (Edstrm et al. 1996), PMP22 (Spreyer et al. 1991) and the p75NTR receptor (Heumann et al. 1987)], we have performed additional validation experiments for a number of genes, the expression or regulation of which has not been described in this context. We carried out real-time quantitative PCR for the following genes: BID, BIRC3, ITGA11, and ACVR1C. Real-time quantitative PCR reactions were run in triplicate. Levels of mRNAs were normalized against GAPDH. Expression levels for each time point were compared to the 0-hour time point and statistically analyzed (Figure 5). The real-time quantitative PCR results are consistent with the gene expression patterns identified by microarray analysis.
Figure 5.
Real-time quantitative reverse transcription-PCR analysis of distal sciatic nerve stumps of rats at 0.5, 1, 6 and 12 hours post-surgery.
The relative expression values of each mRNA were calculated using comparative Ct and were normalized using GAPDH for each data point.
The average of three independent experiments is shown as mean ± SEM.
DISCUSSION
WD is a process that occurs before nerve regeneration and can be described as a cleaning or clearing process that prepares the distal stump for reinnervation. The macrophage response required to clear myelin from the distal portion of damaged peripheral nerves is well underway in the first week[32,33,34], and myelin and axonal debris are cleared within 7–14 days. Macrophages in degenerating peripheral nerves are mainly of hematogenous origin[35,36]. Understanding the factors that regulate the rapid macrophage responses in the peripheral nervous system during WD and comparing the differences in the expression of these molecules in the lesioned peripheral nervous system may provide insight into the reasons for slow WD in the peripheral nervous system.
In this study, we performed validation experiments for a number of genes; the expression and regulation of a number of these has not been described previously. The BID gene is a pro-apoptotic member of the Bcl-2 protein family responsible for apoptotic signaling. The expression of BID is up-regulated by the tumor suppressor p53, a protein transcription factor, and participates in p53-mediated apoptosis, as part of the cell response to stress. BIRC3 is a member of the Inhibitor of Apoptosis family, and inhibits apoptosis by interfering with the activation of caspases. It inhibits apoptosis induced by serum deprivation, but does not significantly affect apoptosis mediated by reactive oxygen species. ITGA11 plays a role in cell signaling and cellular shape regulation, motility, and the cell cycle. Integrins work alongside other proteins, such as cadherins, immunoglobulin superfamily cell adhesion molecules, selectins and syndecans to mediate cell-cell and cell-matrix interactions and signaling. ACVR1C is a dimeric growth and differentiation factor that belongs to the transforming growth factor beta superfamily. ACVR1 is a structurally related receptor that mediates BMP signaling.
In this study, large-scale microarray analysis of WD in the lesioned peripheral nervous system was conducted, which can be used to discover novel genes associated with nerve degeneration and regeneration. WD is a complex process that involves a large number of diverse and overlapping signals, requiring the participation of numerous endogenous signaling molecules. Distinct up-and down-regulation of gene expression after nerve crush indicates that these genes are probably of functional relevance for WD and/or nerve repair. GO and functional analysis demonstrated that genes with similar functions often display similar regulation profiles[37,38]. Of the thousands of genes analyzed using microarrays, numerous differentially-expressed transcripts were identified in early WD. Preferentially up-regulated genes comprised growth factors and cytokines and their receptors, cell adhesion and extracellular matrix molecules, and those involved in axonal growth and plasticity. We compared gene expression patterns in the sciatic nerve during developmental myelination and during remyelination after injury. Pro-inflammatory chemokines and cytokines play an important role in WD after peripheral nerve injury. These pro-inflammatory signals are down-regulated in a timely manner to ensure that the inflammatory response in the injured nerve is limited. The factors that suppress the pro-inflammatory state are not fully understood. In our study, we applied microarray technology using a genome-wide, systems-based approach to explore transcriptional responses that provide evidence of biological processes/pathways impacting WD after peripheral nerve injury. Future studies, for example, by using microarray analysis to compare gene expression patterns in peripheral nerves during WD in a congenital hypomyelinating rat model, will help us understand how the peripheral nervous and immune systems interact to modulate nerve repair, and identify the molecules that drive these responses, such as in the injured peripheral nerve during early WD.
Due to the complexity of the peripheral nervous system, especially in the context of injury, which involves the proliferation, recruitment and differentiation of various cell types, it is difficult to precisely determine the roles of the hundreds of genes that are regulated, or their relative importance in the regenerative repair process. For these reasons, we decided to focus our attention on genes that are differentially regulated in the sciatic nerve distal stump, as opposed to where cell bodies of axotomized neurons are located. Furthermore, analyzing gene expression profiles in the distal nerve stump offers the possibility to study how genes are regulated after sciatic nerve injury in WD[12,39,40,41,42]. Our results revealed distinct patterns of up and down-regulation of genes in the injured distal nerve, which suggests that these genes play a role in nerve degeneration and/or regeneration. Further studies are necessary to determine whether these differentially-expressed genes are also expressed by Schwann cells in vivo, and to clarify their functions during WD after sciatic nerve injury.
MATERIALS AND METHODS
Design
A randomized, controlled, animal experiment.
Time and setting
The experiment was performed at Jiangsu Key Laboratory of Neuroregeneration, Nantong University, China, from October 2009 to March 2011.
Materials
Male Sprague-Dawley rats, weighing 180–200 g, were provided by the Experimental Animal Center of Nantong University. All animal tests were conducted in accordance with NIH Guidelines for the Care and Use of Laboratory Animals.
Methods
Establishment of sciatic nerve injury models
All rats were anesthetized using an injection of complex narcotics (10 mg/kg xylazine, 95 mg/kg ketamine, acepromazine 0.7 mg/kg) and the sciatic nerve was identified and lifted through an incision on the lateral aspect of the mid-thigh of the right hind limb. A 1-cm segment of the right sciatic nerve was excised, and the distal and proximal nerve stumps were separately inserted into nearby inter-muscular spaces to prevent nerve regeneration. After the surgical incisions were closed, the animals were housed in temperature and humidity-controlled cages, maintained under a 12 hour light-dark cycle, and were allowed free access to water and food. All rats randomly divided into six groups, with six animals in each group, to undergo sciatic neurectomy. One group of rats was killed immediately and the other groups were killed at 0.5, 1, 6, 12 or 24 hours post surgery. Distal nerve stumps were obtained from these rats after euthanization.
RNA isolation and Affymetrix microarray scanning
Total RNA was isolated from the distal nerve stumps using Trizol (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's protocol. An Affymetrix Genechip Hybridization Oven 640 and Gene Array Scanner 3000 were used for genechip analysis. The labeling and hybridization were performed at Shanghai Biochip Company, Ltd., China according to the protocols in the Affymetrix GeneChip System. Affymetrix GeneChip scan control software was used for scanning the microarray slides, and an Affymetrix Fluidics Station 450 was used for image analysis.
Bioinformatic analysis
Gene screening and bioinformatic analyses were performed by the Bioinformatics Center at the Key Laboratory of Systems Biology, Shanghai Institute for Biological Sciences. We selected differentially-expressed genes in a logical sequence according to Random Variance Model corrective ANOVA. Differentially-expressed genes were identified by Affymetrix scan at each time point with ratios set for different genes. Using a strategy for clustering the early WD gene expression data, we defined some unique profiles. The expression model profiles were related to the actual or expected number of genes assigned to each model profile. Significant profiles demonstrated a higher probability than expected by Fisher's Exact Test and multiple comparison tests[43,44]. Stc GO analysis was applied to determine the main functions of the differentially-expressed genes according to GO, the key functional classification of NCBI. Pathway analysis was used to determine the significant pathways of the differentially-expressed genes according to KEGG[19,20,45]. Gene regulatory networks were analyzed using a CTRNN. Key regulatory factors and networks were calculated according to gene fold-change expression and gene interaction in pathways[46]. The networks were constructed extempore and unique for the uploaded data, using an analytical network algorithm with default settings for biological networks.
Real-time quantitative PCR assay
The distal nerve stumps of rats killed at 0, 0.5, 1, 6 or 12 hours following injury were dissected and then suspended in RNA Stabilization Reagent (Qiagen Tokyo, Japan) and stored at –80 °C. The total RNA fraction was prepared from stored tissue specimens using an RNeasy Mini Kit (Qiagen), according to the manufacturer's protocol. The cDNA was synthesized using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster, CA, USA), and real-time quantitative PCR was performed using a 7 300 Real-Time PCR System (Applied Biosystems), both according to the manufacturers’ protocols. All reactions were run in triplicate. The cycle threshold (Ct) values were analyzed using the comparative Ct (ΔΔCt) method. Levels of mRNAs were normalized using GAPDH as a reference. The primers used in the real-time quantitative PCR assays are provided in supplementary information online. The relative expression values of each mRNA were calculated using comparative Ct and were normalized using GAPDH mature mRNA for each data point.
Statistical analysis
Microarray data were analyzed using GeneSpring GCOS1.2 software. The data were analyzed statistically using the two-sample (independent group) t-test, and differences were considered statistically significant at P < 0.05. All data were expressed as mean ± SEM and analyzed by one-way analysis of variance and Scheffe's post-hoc test with the SPSS software package. All values of P < 0.05 were considered statistically significant.
Footnotes
Funding: This research was supported by the National Natural Science Foundation of China (Key Program), No. 81130080; the National Natural Science Foundation of China, No. 30870811; Scientific Research Foundation for Returned Scholars, Ministry of Education of China; the Natural Science Foundation of Jiangsu Province, No. BK2010282; A Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions, PAPD.
Conflicts of Interest: None declared
Ethical approval: This study was approved by the Animal Ethics Committee of Nantong University, China.
(Edited by Surguchov A, Lou XY/Song LP)
REFERENCES
- [1].Stoll G, Jander S, Myers RR. Degeneration and regeneration of the PNS: From Augustus Waller's observations to neuroinflammation. J Peripher Nerv Syst. 2002;7:13–27. doi: 10.1046/j.1529-8027.2002.02002.x. [DOI] [PubMed] [Google Scholar]
- [2].Waller A. Experiments on the section of the glossopharyngeal and hypoglossal nerves of the frog, and observations of the alterations produced thereby in the structure of their primitive fibres. Phil Transact Royal Soc London. 1850;140:423–429. [Google Scholar]
- [3].Ambron RT, Walters ET. Priming events and retrograde injury signals: a new perspective on the cellular and molecular biology of nerve regeneration. Mol Neurobiol. 1996;13:61–79. doi: 10.1007/BF02740752. [DOI] [PubMed] [Google Scholar]
- [4].Boivin A, Pineau I, Barrette B, et al. Toll-like receptor signaling is critical for Wallerian degeneration and functional recovery after peripheral nerve injury. J Neurosci. 2007;27:12565–12576. doi: 10.1523/JNEUROSCI.3027-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].De S, Trigueros MA, Kalyvas A, et al. Phospholipase A2 plays an important role in myelin breakdown and phagocytosis during Wallerian degeneration. Mol Cell Neurosci. 2003;24:753–765. doi: 10.1016/s1044-7431(03)00241-0. [DOI] [PubMed] [Google Scholar]
- [6].Girolami EI, Bouhy D, Haber M, et al. Differential expression and potential role of SOCS1 and SOCS3 in Wallerian degeneration in injured peripheral nerve. Exp Neurol. 2010;223(1):173–182. doi: 10.1016/j.expneurol.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Guertin AD, Zhang DP, Mak KS, et al. Microanatomy of axon/glial signaling during Wallerian degeneration. J Neurosci. 2005;25:3478–3487. doi: 10.1523/JNEUROSCI.3766-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Martini R, Fischer S, Lopez-Vales R, et al. Interactions between Schwann cells and macrophages in injury and inherited demyelinating disease. Glia. 2008;56:1566–1577. doi: 10.1002/glia.20766. [DOI] [PubMed] [Google Scholar]
- [9].Tofaris GK, Patterson PH, Jessen KR, et al. Denervated Schwann cells attract macrophages by secretion of leukemia inhibitory factor (LIF) and monocyte chemoattractant protein-1 in a process regulated by interleukin-6 and LIF. J Neurosci. 2002;22:6696–6703. doi: 10.1523/JNEUROSCI.22-15-06696.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Navarro X, Vivo M, Valero-Cabre A. Neural plasticity after peripheral nerve injury and regeneration. Prog Neurobiol. 2007;82:163–201. doi: 10.1016/j.pneurobio.2007.06.005. [DOI] [PubMed] [Google Scholar]
- [11].Feltri ML, Wrabetz L, Behrens A, et al. c-Jun is a negative regulator of myelination. J Cell Biol. 2008;181:625–637. doi: 10.1083/jcb.200803013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kirsch M, Terheggen U, Hofmann HD. Ciliary neurotrophic factor is an early lesion-induced retrograde signal for axotomized facial motoneurons. Mol Cell Neurosci. 2003;24:130–138. doi: 10.1016/s1044-7431(03)00130-1. [DOI] [PubMed] [Google Scholar]
- [13].Lindholm D, Heumann R, Meyer M, et al. Interleukin-1 regulates synthesis of nerve growth factor in non-neuronal cells of rat sciatic nerve. Nature. 1987;330:658–659. doi: 10.1038/330658a0. [DOI] [PubMed] [Google Scholar]
- [14].Perrin FE, Lacroix S, Aviles-Trigueros M, et al. Involvement of monocyte chemoattractant protein-1, macrophage inflammatory protein-1a and interleukin-1b in Wallerian degeneration. Brain. 2005;128:854–866. doi: 10.1093/brain/awh407. [DOI] [PubMed] [Google Scholar]
- [15].Raivich G, Bohatschek M, Da Costa C, et al. The AP-1 transcription factor c-Jun is required for efficient axonal regeneration. Neuron. 2004;43:57–67. doi: 10.1016/j.neuron.2004.06.005. [DOI] [PubMed] [Google Scholar]
- [16].Sendtner M, Gotz R, Holtmann B, et al. Endogenous ciliary neurotrophic factor is a lesion factor for axotomized motoneurons in adult mice. J Neurosci. 1997;17:6999–7006. doi: 10.1523/JNEUROSCI.17-18-06999.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Wiklund P, Ekstrom PA, Edstrom A. Mitogen-activated protein kinase inhibition reveals differences in signalling pathways activated by neurotrophin-3 and other growth-stimulating conditions of adult mouse dorsal root ganglia neurons. J Neurosci Res. 2002;67:62–68. doi: 10.1002/jnr.10073. [DOI] [PubMed] [Google Scholar]
- [18].Zochodne DW, Levy D, Zwiers H, et al. Evidence for nitric oxide and nitric oxide synthase activity in proximal stumps of transected peripheral nerves. Neuroscience. 1999;91:1515–1527. doi: 10.1016/s0306-4522(98)00729-5. [DOI] [PubMed] [Google Scholar]
- [19].Yi M, Horton JD, Cohen JC, et al. Whole Pathway Scope: a comprehensive pathway-based analysis tool for high-throughput data. BMC Bioinformatics. 2006;7:30. doi: 10.1186/1471-2105-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Drăghici S, Khatri P, Tarca AL, et al. A systems biology approach for pathway level analysis. Genome Res. 2007;17:1537–1545. doi: 10.1101/gr.6202607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Zhou S, Yu B, Qian T, et al. Early changes of microRNAs expression in the dorsal root ganglia following rat sciatic nerve transection. Neurosci Lett. 2011;494(2):89–93. doi: 10.1016/j.neulet.2011.02.064. [DOI] [PubMed] [Google Scholar]
- [22].Ghosh A, Greenberg ME. Calcium signaling in neurons: molecular mechanisms and cellular consequences. Science. 1995;268:239–247. doi: 10.1126/science.7716515. [DOI] [PubMed] [Google Scholar]
- [23].Hanz S, Perlson E, Willis D, et al. Axoplasmic importins enable retrograde injury signaling in lesioned nerve. Neuron. 2003;40:1095–1104. doi: 10.1016/s0896-6273(03)00770-0. [DOI] [PubMed] [Google Scholar]
- [24].Kim D, Lee S, Lee SJ. Toll-like receptors in peripheral nerve injury and neuropathic pain. Curr Top Microbiol Immunol. 2009;336:169–186. doi: 10.1007/978-3-642-00549-7_10. [DOI] [PubMed] [Google Scholar]
- [25].Berti Mattera LN, Harwalkar S, Hughes B, et al. Proliferative and morphological effects of endothelins in Schwann cells: roles of p38 mitogen-activated protein kinase and Ca(2+)-independent phospholipase A2. J Neurochem. 2001;79(6):1136–1148. doi: 10.1046/j.1471-4159.2001.00642.x. [DOI] [PubMed] [Google Scholar]
- [26].Koehler JA, Moran MF. Regulation of extracellular signal-regulated kinase activity by p120 RasGAP does not involve its pleckstrin homology or calcium-dependent lipid binding domains but does require these domains to regulate cell proliferation. Cell Growth Differ. 2001;12(11):551–561. [PubMed] [Google Scholar]
- [27].Lindwall C, Kanje M. Retrograde axonal transport of JNK signaling molecules influence injury induced nuclear changes in p-c-Jun and ATF3 in adult rat sensory neurons. Mol Cell Neurosci. 2005;29:269–282. doi: 10.1016/j.mcn.2005.03.002. [DOI] [PubMed] [Google Scholar]
- [28].Schwaiger FW, Hager G, Schmitt AB, et al. Peripheral but not central axotomy induces changes in Janus kinases (JAK) and signal transducers and activators of transcription (STAT) Eur J Neurosci. 2000;12:1165–1176. doi: 10.1046/j.1460-9568.2000.00005.x. [DOI] [PubMed] [Google Scholar]
- [29].Lee HK, Seo IA, Park HK, et al. Nidogen is a prosurvival and promigratory factor for adult Schwann cells. J Neurochem. 2007;102:686–698. doi: 10.1111/j.1471-4159.2007.04580.x. [DOI] [PubMed] [Google Scholar]
- [30].Lee N, Neitzel KL, Devlin BK, et al. STAT3 phosphorylation in injured axons before sensory and motor neuron nuclei: potential role for STAT3 as a retrograde signaling transcription factor. J Comp Neurol. 2004;474:535–545. doi: 10.1002/cne.20140. [DOI] [PubMed] [Google Scholar]
- [31].Sheu JY, Kulhanek DJ, Eckenstein FP. Differential patterns of ERK and STAT3 phosphorylation after sciatic nerve transection in the rat. Exp Neurol. 2000;166:392–402. doi: 10.1006/exnr.2000.7508. [DOI] [PubMed] [Google Scholar]
- [32].de Bilbao F, Giannakopoulos P, Srinivasan A, et al. In vivo study of motoneuron death induced by nerve injury in mice deficient in the caspase 1/interleukin-1 beta-converting enzyme. Neuroscience. 2000;98:573–583. doi: 10.1016/s0306-4522(00)00100-7. [DOI] [PubMed] [Google Scholar]
- [33].Herdegen T, Waetzig V. The JNK and p38 signal transduction following axotomy. Restor Neurol Neurosci. 2001;19:29–39. [PubMed] [Google Scholar]
- [34].Kuhn G, Lie A, Wilms S, et al. Coexpression of PMP22 gene with MBP and P0 during de novo myelination and nerve repair. Glia. 1993;8:256–264. doi: 10.1002/glia.440080406. [DOI] [PubMed] [Google Scholar]
- [35].Shubayev VI, Angert M, Dolkas J, et al. TNF alpha induced MMP-9 promotes macrophage recruitment into injured peripheral nerve. Mol Cell Neurosci. 2006;31:407–415. doi: 10.1016/j.mcn.2005.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Sun W, Oppenheim RW. Response of motoneurons to neonatal sciatic nerve axotomy in Baxknockout mice. Mol Cell Neurosci. 2003;24:875–886. doi: 10.1016/s1044-7431(03)00219-7. [DOI] [PubMed] [Google Scholar]
- [37].Ugolini G, Raoul C, Ferri A, et al. Fas/tumor necrosis factor receptor death signaling is required for axotomy-induced death of motoneurons in vivo. J Neurosci. 2003;23:8526–8531. doi: 10.1523/JNEUROSCI.23-24-08526.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Waetzig V, Herdegen T. MEKK1 controls neurite regrowth after experimental injury by balancing ERK1/2 and JNK2 signaling. Mol Cell Neurosci. 2005;30:67–78. doi: 10.1016/j.mcn.2005.06.001. [DOI] [PubMed] [Google Scholar]
- [39].Shadiack AM, Sun Y, Zigmond RE. Nerve growth factor antiserum induces axotomy-like changes in neuropeptide expression in intact sympathetic and sensory neurons. J Neurosci. 2001;21:363–371. doi: 10.1523/JNEUROSCI.21-02-00363.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Yoo S, Nguyen MP, Fukuda M, et al. Plasmalemmal sealing of transected mammalian neurites is a gradual process mediated by Ca(2+)-regulated proteins. J Neurosci Res. 2003;74:541–551. doi: 10.1002/jnr.10771. [DOI] [PubMed] [Google Scholar]
- [41].Bosse F, Hasenpusch-Theil K, Küry P, et al. Gene expression profiling reveals that peripheral nerve regeneration is a consequence of both novel injury-dependent and reactivated developmental processes. J Neurochem. 2006;96:1441–1457. doi: 10.1111/j.1471-4159.2005.03635.x. [DOI] [PubMed] [Google Scholar]
- [42].Makwana M, Raivich G. Molecular mechanisms in successful peripheral regeneration. FEBS J. 2005;272:2628–2638. doi: 10.1111/j.1742-4658.2005.04699.x. [DOI] [PubMed] [Google Scholar]
- [43].Ramoni MF, Sebastiani P, Kohane IS. Cluster analysis of gene expression dynamics. Proc Natl Acad Sci U S A. 2002;99:9121–9126. doi: 10.1073/pnas.132656399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Miller LD, Long PM, Wong L, et al. Optimal gene expression analysis by microarrays. Cancer Cell. 2002;2:353–361. doi: 10.1016/s1535-6108(02)00181-2. [DOI] [PubMed] [Google Scholar]
- [45].Kanehisa M, Goto S, Kawashima S, et al. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Busch H, Camacho-Trullio D, Rogon Z, et al. Gene network dynamics controlling keratinocyte migration. Mol Syst Biol. 2008;4:199. doi: 10.1038/msb.2008.36. [DOI] [PMC free article] [PubMed] [Google Scholar]