Abstract
We constructed two-dimensional representations of profiles of gene conservation across different genomes using the genome of Escherichia coli as a model. These profiles permit both the visualization at the genome level of different traits in the organism studied and, at the same time, reveal features related to the genomes analyzed (such as defective genomes or genomes that lack a particular system). Conserved genes are not uniformly distributed along the E. coli genome but tend to cluster together. The study of gene distribution patterns across genomes is important for the understanding of how sets of genes seem to be dependent on each other, probably having some functional link. This provides additional evidence that can be used for the elucidation of the function of unannotated genes. Clustering these patterns produces families of genes which can be arranged in a hierarchy of closeness. In this way, functions can be defined at different levels of generality depending on the level of the hierarchy that is studied. The combined study of conservation and phenotypic traits opens up the possibility of defining phenotype/genotype associations, and ultimately inferring the gene or genes responsible for a particular trait.
The number of fully sequenced genomes has opened new possibilities for comparative studies on a genomic scale. Functional annotation of previously uncharacterized genes is now possible by using the information contained in the databases (Pellegrini et al. 1999). Phylogenetic studies involving complete genomes can be carried out (Fitz-Gibbon and House 1999; Tekaia et al. 1999; Lin and Gerstein 2000). Also, horizontal transfer can be detected with higher accuracy than ever, and it seems to be a more frequent event than imagined, even across large phylogenetic distances (Lawrence 1999). At the beginning of this work, and setting aside the eukaryotic genomes, there were 60 bacteria and 13 archaea quoted in the EBI databases (http://www.ebi.ac.uk/genomes/index.html). Comparative genome analysis has been used in different ways to gain information in one of the most important issues of bioinformatics: assigning function to genes. Under the assumption that proteins that are homologs in a number of fully sequenced organisms are likely to be functionally linked, some authors (Gaasterland and Regan 1998; Pellegrini et al. 1999) have used protein phylogenetic profiles (the patterns of presence/absence of genes across genomes) to infer these functional links for proteins without a known function. Combined information that includes not only presence/absence but also the similarity level has been used (Dopazo et al. 2001). In a comparative analysis of the draft genome of Streptococcus pneumoniae type 19F strain (Dopazo et al. 2001), it became quickly apparent that Streptococcus pyogenes, an important human pathogen, is probably capable of natural genetic transformation as previously suspected, as it shares with S. pneumoniae, Lactococcus lactis, and Bacillus subtilis many genes involved in the development of competence for transformation, such as the cglA-D cluster (Pozzi et al. 1996).
In this manuscript, we show how the study of profiles of genome conservation permits a rapid and precise insight into the relative selective pressures (and consequently their relative importance in the survival of the cells) that different genes and operons or putative operons undergo. Clustering of the patterns of presence/absence of genes produces families of genes called clusters of orthologous genes (COGs) (Tatusov et al. 1997), which can be arranged in a hierarchy of closeness. This can be useful in defining common functions from a more general perspective in the highest levels of the hierarchy to more detail in the lowest levels. The combined study of conservation and phenotypic traits opens up the possibility of defining phenotype/genotype associations, and ultimately inferring the gene or genes responsible for a particular trait.
RESULTS AND METHODS
Genome Profiles
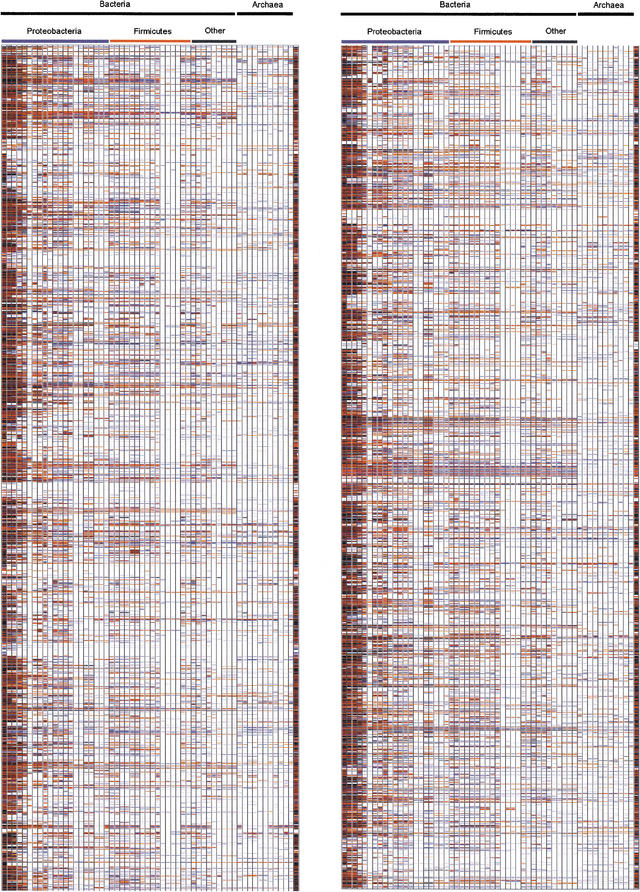
E. coli is a model organism for biochemical and biological studies as it is one of the best characterized prokaryotes. We compared the 4358 proteins encoded by the genome of E. coli K-12 by aligning the sequence of each E. coli protein to the proteins from 57 other fully sequenced genomes (listed at the Web site of The European Bioinformatics Institute http://www.ebi.ac.uk/proteomes/index.html). To quickly visualize which genes of E. coli (Blattner et al. 1997) have homologs in other organisms and which other genes display a more restricted distribution in other bacterial genomes, a graphical representation, the Genomic Conservation Profile, was used. It presents a full-genome view of the degree of conservation of all proteins within the set of publicly available complete (or nearly finished) prokaryotic genomes. A two-dimensional plot is obtained where each row corresponds to an E. coli gene, arranged by its position in the chromosome, and each column corresponds to a prokaryotic genome, arranged in decreasing taxonomic proximity to E. coli. Table 1 shows the prokaryote and archaea species used in this work. The plot is a color-coded representation of the e values of BLAST (Altschul el al. 1997) hits of the E. coli genes in each of the genomes analyzed. Because we are using directly the e values instead of a presence/absence discrete value, like in Pellegrini et al. (1999) approach, we are not using any statistical criterion to decide if the homologous gene exists or not. A low e value can be obtained because of a poor conservation along the complete gene or, alternatively, could be because of a high conservation in a small fragment or a domain. Our approach does not differentiate between these situations and both cases are considered just a low conservation. Nevertheless, the interesting information in the plot is the change in the conservation (as measured by the e value) across genomes that diverged at different times in the evolutionary scale. For the sake of the clarity in the representation, genes with e values >10−2 were not considered as hits. In the picture obtained, it is easy to distinguish patterns of conservation across taxonomic groups (Fig. 1; see also additional information at http://bioinfo.cnio.es/data/GenomeProfile/; or http://www.ebi.ac.uk/proteome/ECOLI/ and follow the link “Genome Conservation Profile for Escherichia coli K-12”). Defective genomes, such as Buchnera, micoplasmas, and chlamydias, can rapidly be identified in Figure 1 because of the unusually low number of homologs they display with respect to E. coli genes. It is also evident that the patterns of conservation of genes across genomes are not randomly distributed along the E. coli genome. Some stretches are poorly conserved beyond enterobacteria or the γ subdivision of proteobacteria. On the other hand, other stretches are highly conserved, with homologs even among the archaea. Gene order is extensively conserved between closely related bacterial species, and archaeal genomes are likely to behave similarly to bacterial genomes (Tamames 2001). Even across very distant species, remnants of gene order conservation exist in the form of highly conserved clusters of genes. This suggests the existence of selective processes that maintain the organization of these regions (Tamames 2001).
Table 1.
Complete Genomes Used in the Work Arranged as They Appear in the Figures
| Kingdom | Taxonomical adscription | Species | ||
| Rank 1 | Rank 2 | Rank 3 | ||
| Bacteria | Proteobacteria | Gamma | Enterobacteria | Escherichia coli K12 Escherichia coli O157 Salmonella typhimurium LT2 Salmonella typhi Yersinia pestis |
| Vibronaceae | Vibrio cholerae | |||
| Enterobacteria | Buchnera aphidicola | |||
| Pasteurellaceae | Haemophilus influenzae Pasteurella multicocida | |||
| Pseudomonadaceae | Pseudomonas aeruginosa | |||
| Xantomonas | Xilella fastidiosa | |||
| Alpha | Rhizobiaceae | Rhizobium meliloti Rhizobium loti Agrobacterium tumefaciens | ||
| Caulobacter | Caulobacter crescentus | |||
| Rickettsiales | Rickettsia prowazekii Rickettsia conorii | |||
| Beta | Neisseriaceae | Neisseria meningitidis MC58 Neisseria meningitidis Z2491 | ||
| Epsilon | Helicobacter | Helicobacter pylori J99 Helicobacter pylori | ||
| Campylobacter | Campylobacter jejuni | |||
| Firmicutes | Bacillus/Clostridium | Bacillus/Staphylococcus | Bacillus subtilis Bacillus halodurans Staphylococcus aureus Staphylococcus aureus Mu50 Listeria innocua Listeria monocytogenes | |
| Streptococcaceae | Streptococcus pyogenes Streptococcus pneumoniae Lactococcus lactis | |||
| Clostridia | Clostridium acetobitylicum | |||
| Mollicutes | Ureaplasma parvum Mycoplasma pulmonis Mycoplasma pneumoniae Mycoplasma genitalum | |||
| Actinobacteria | Actinobacteridae | Mycobacterium tuberculosis Mycobacterium leprae | ||
| Thermus | Deinococcus | Deinococcus radiodurans | ||
| Cyanobacteria | Chroococcales | Synechocystis | Synechocystis sp | |
| Aquificales | Aquificaceae | Aquifex | Aquifex aeolicus | |
| Thermotogales | Thermotoga | Thermotoga maritima | ||
| Spirochaetales | Spirochaetaceae | Borrellia | Borrellia burgdorferi | |
| Treponema | Treponema pallidum | |||
| Chlamydiales | Chlamydiaceae | Chlamydia pneumoniae Chlamydia muridarum Chlamydia trachomatis | ||
| Archaea | Euryarchaeota | Halobacteriales | Halobacteriaceae | Halobacterium sp |
| Thermococcales | Thermococcaceae | Pyrococcus horikoshii Pyrococcus abysii | ||
| Thermoplasmales | Thermoplasmaceae | Thermoplasma volcanicum Thermoplasma acidophilum | ||
| Archaeoglobales | Archaeoglobaceae | Archaeglobus fulgidus | ||
| Methanococcales | Methanococcaceae | Methanococcus janascii | ||
| Methanobacteriales | Methanobacteriaceae | Methanobacteria thermoautotrophicum | ||
| Crenarchaeota | Desulfurococcales | Desulfurococcaceae | Aeropyrum pernix | |
| Sulfolobales | Sulfolobaceae | Sulfolobus solfataricus Sulfolobus tokodaii | ||
References for the genomes can be found in the Web page of genomes at EBI (http://www.ebi.ac.uk/genomes/index.html)
Figure 1.
Genome profile of the Escherichia coli K12 genome against the genomes listed in Table 1. The last genome on the right part of the figure is E. coli O157 again to serve as reference. The Genome Conservation Profile was obtained comparing each protein encoded by the genome of E. coli K12 to each of the databases of all proteins for each organism listed in Table 1. Comparisons were done using the NCBI's BLAST2 for a fast identification of homologous sequences. For each protein of the E. coli K12 genome, we obtain a vector whose components can be either the absence of hit, or the e value of the best hit obtained for a given gene. These values are color coded, and the genomes are arranged following the NCBI taxonomy. This is the picture we obtained. Color codes range from black e = 0.0 to pale blue e = 10−10. The darker the color, the more homologous the genes are with respect to the E. coli counterparts. A full color figure can be seen in the additional information Web page http://bioinfo.cnio.es/data/GenomeProfile/.
The arrangement of the genomes in taxonomic groups is very important for any analysis of variability. It has long been recognized that species cannot be treated as independent observations (Felsenstein 1985), as groups of species within phylogenetic lineages share common attributes, such as sequence. The profile of genome conservation provides a convenient representation of the relative conservation of genes and clusters of genes across the different taxonomic lineages of prokaryotes. Figure 1 shows how the conserved genes tend to cluster together along the E. coli genome, and how they maintain a correlated degree of conservation across genomes, which is probably because of the fact that they are playing some common role in the cell.
Patterns of Distribution of Genes Across Genomes
Although different organisms may have developed distinct solutions to their physiological requirements through evolution, genes involved in the same pathway are expected to show a similar degree of conservation in genomes having this particular pathway (Pellegrini et al. 1999). There are two considerations that must be taken into account when studying the genes distribution patterns across genomes. One of them is the fact that many genes will show a pattern of distribution among genomes corresponding to differences in the time of separation of such genomes as independent species, that is, the pattern of the phylogeny. This is not informative itself for defining distribution patterns of genes among genomes. Its importance comes, precisely, from the fact that these genes define the global phylogenetic pattern of the species and, consequently, can be used to reconstruct the phylogeny of the genomes. The second consideration to be taken into account is that, when many genes are compared, it is expected that some of them will show similar patterns by chance.
We studied the patterns of distribution of genes across genomes taking into account the considerations above mentioned. We clustered the genes on the basis of their similarity in the patterns of distribution across genomes using a hierarchical clustering method called self-organizing tree algorithm (SOTA) (Dopazo and Carazo 1997; Herrero et al. 2001). This method arranges in a hierarchy of closeness the clusters of genes that display a similar distribution across genomes. Clusters of codistributed genes are found by using a permutation test that minimizes the number of false positives present in them (Herrero et al. 2001). The SOTA algorithm is based on the paradigm of self-organization used by SOMs (Kohonen 1997) but implemented on a growing binary-tree structure. SOTA is a divisive method that starts the classification with a binary topology composed of a root node with two leaves. The self-organizing process arranges the data (in this case the vectors of patterns of presence/absence of each gene in the analyzed genomes) into two clusters. After reaching convergence at this level, the network is inspected. If the level of variability in one or more terminal nodes is over a given threshold, the tree grows by expanding these terminal nodes. Two new descendants are generated from the most heterogeneous node that becomes internal and does not receive direct updates from the data in future steps. The growth of the network is directed by the resource value that is defined as the mean value of the distances between a node and the vectors of patterns of presence/absence associated with it (Fritzke 1994; Dopazo and Carazo 1997). SOTA structures grow from the root of the tree toward the leaves, producing a representation of the data from lower to higher resolution. If a threshold of resource value has been fixed, once all terminal nodes fall below such a threshold, the expansion of the binary topology is stopped. This allows the generation of a hierarchical cluster structure at the desired level of resolution (Herrero et al. 2001 for details).
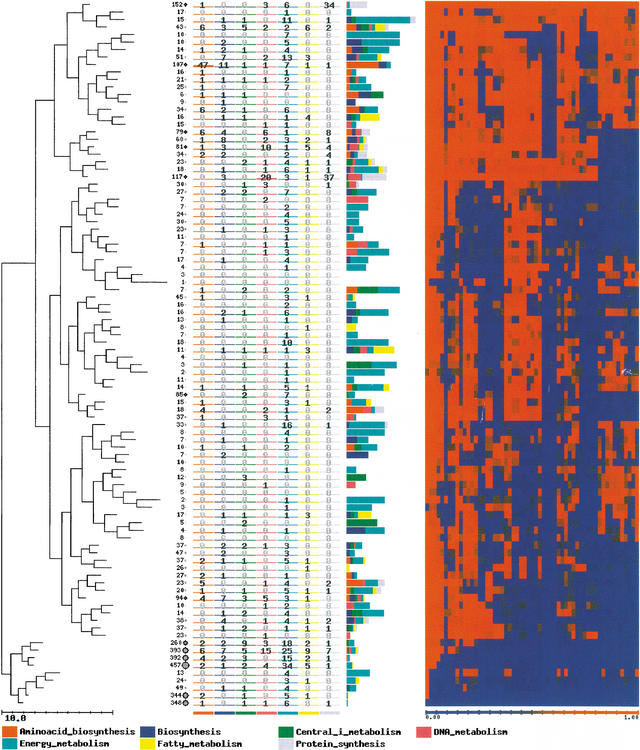
Clustering of the patterns of presence/absence of genes produces families of genes called COGs by some authors (Tatusov et al. 1997). Because we cannot distinguish between orthologous and paralogous genes, especially over large evolutionary distances, we will term them just clusters of homologous genes (CHGs). Figure 2 shows the CHGs obtained upon the application of SOTA to the E. coli genes. Obtaining CHGs in this way has two advantages with respect to the classical way of obtaining them. First, we obtain CHGs with the desired P value (that is, with a predetermined rate of false positives) and, in addition, we obtain a hierarchical relationship among the different CHGs. A look at Figure 2 shows that the genes are unequally distributed among CHGs. In the bottom of the figure, there are clusters with several hundred genes; these are primarily genes unique to E. coli which, in addition, are poorly annotated. For example, in the case of the two CHGs in the bottom of the figure, with 348 and 344 genes, respectively, only 10 and 9, respectively, have annotation for function. On the other hand, at the top of Figure 2, there is a CHG that is highly conserved across all the genomes, with 152 genes, whose function (as can be inferred from the annotated genes) is probably related mainly to protein synthesis. The ninth cluster is another interesting case. It is composed of 107 genes, many of them annotated as being involved in biosynthesis and biosynthesis of amino acids. The genes are quite conserved (even among archaea) except in two groups of bacteria: mycoplasmas and chlamydias. The reduction of the genome size of Mycoplasma pneumoniae, as well as other mycoplasmas (Mycoplasma genitalium, Ureaplasma urealyticum), during its reductive evolution from ancestral bacteria can be explained by the loss of complete anabolic (e.g., no amino-acid synthesis) and metabolic pathways. Therefore, M. pneumoniae, as other mycoplasmas, depends in nature on an obligate parasitic lifestyle that requires the provision of exogenous essential metabolites (Himmelreich et al. 1996). The case of chlamydias is quite similar (Kalman et al. 1999). Some of the genes of the cluster, and many other CHGs in the vicinity, do not have homologs in another genome: Buchnera. Again, this is an analogous case of an intracellular parasitic bacteria that lives in aphids species (Shigenobu et al. 2000). In the Buchnera genome there are genes for the biosynthesis of amino acids essential for the hosts, which explains why the genes in the CHG previously mentioned are in this genome.
Figure 2.
Clusters of homologous genes (CHGs) obtained upon the application of Self Organizing Tree Algorithm (SOTA) to the patterns of conservation of Escherichia coli genes after being transformed to presence/absence values. The value of variability used for the clustering process was of five. This means that patterns of conservation that appear in each cluster can have up to 10% of differences. The number in each leaf of the tree represents the number of genes in each cluster. Each column represents the number of genes annotated as members of one of the following TIGRFam families (Peterson et al. 2001): amino-acid biosynthesis; biosynthesis of cofactors, prosthetic groups, and carriers; central and intermediate metabolism; DNA metabolism; energy metabolism; fatty-acids metabolism and protein synthesis. Labels corresponding to the TIGR families are represented for the different clusters. They indicate the proportion of genes labeled with the different terms in each cluster. Each column in the heat map (right) represents one of the genomes in the same order as they appear in Table 1. A full color figure can be seen in the additional information Web page http://bioinfo.cnio.es/data/GenomeProfile/.
The conservation profile can be built from the opposite side, that is taking Methanococcus jannaschii, an archaea, as reference genome (see additional information at http://bioinfo.cnio.es/data/GenomeProfile/). In this case, proteins labeled as “binding” and “enzyme” are among the most conserved ones.
Different Conservation of Biological Processes (GO) Along the Species Spectra
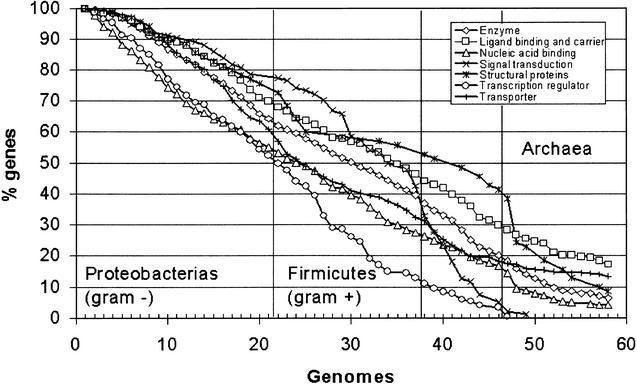
It has previously been shown that patterns of conservation of genes across species are not randomly distributed along the E. coli genome (see Fig. 1). In functional terms, this conservation must be related to some extent to the role of the proteins in the cell. An important aspect to be studied is the conservation of proteins that carry out different biological processes. Gene Ontology (GO) (Ashburner et al. 2000) is a useful and widely accepted collection of definitions for molecular function, biological processes, and cellular components. Figure 3 shows the proportion of genes labeled with different cellular processes that are conserved across a number of genomes. The GO annotations were selected from the EBI's GOA project (http://www.ebi.ac.uk/GOA/index.html). While some of the classes display a nearly constant decrease in the degree of conservation across increasing phylogenetic distances, others display abrupt changes. For example, enzymes, ligand-binding and carrier, and transporters display quite a uniform trend of reduction in the number of conserved proteins that can be considered a reflection of the underlying phylogenies. This suggests that these proteins follow a molecular clock (Zuckerlandl and Pauling 1965) and, generally speaking, constitute a good choice for phylogenetic studies. Interestingly, other proteins display a pattern of conservation more related to the phenotypic characteristics that differentiate distinct taxa of organisms. In the case of transporters, it can be observed that the number of conserved proteins remains nearly constant for all the archaea. This core of 72 genes includes many highly conserved essential genes such as MALK_ECOLI, which is one of the five proteins essential to the active binding protein-dependent transport system for maltose and maltodextrin; DPPF_ECOLI, which is part of the binding-protein–dependent transport system for dipeptides (Abouhamad and Manson 1994), etc. Many of them belong to the family of ABC transporters (see additional information).
Figure 3.
Proportion Escherichia coli of genes labeled with different GO (Ashburner et al. 2000) biological processes, which are conserved across different genomes. Genomes in the horizontal axis are arranged as in Table 1.
On the other hand, structural proteins change rapidly from proteobacterias (Gram-positive) to firmicutes (Gram-negative) and even more abruptly from eubacteria to archaea, accounting most probably for the phenotypic differences among these groups. Despite the fall in the conservation level, there are 17 structural proteins conserved in some archaea, nine of which are conserved in all the genomes (these belong to different families of ribosomal proteins). Proteins related to signaling processes are among the less conserved. For example, proteins belonging to the signal transduction class are more conserved among proteobacterias, but the conservation falls abruptly across the rest of the bacteria, and none are conserved in archaea. A similar trend, but with a lower basal conservation, can be observed for transcription regulators.
It is interesting to note that defective genomes do not cause, by themselves, a significant reduction in the trends of conservation shown. The changes are observed in the interfaces between phenotypically different taxa (e.g., Proteobacterias to firmicutes, bacteria to archaea, etc.)
Genotype–Phenotype Associations
The availability of all the genes of the genome makes possible the study of differences at gene level that account for the differences at phenotype level.
The rationale of the approach proposed is based on a similar method we have used for the analysis of microarray data (Mateos et al. 2002). Both types of data have in common that a large number of genes account for some properties (in this case phenotypes). Each gene is represented by its pattern of presence/absence along the studied genomes. Some of these patterns are identical or, at least, very similar. Obviously, the phenotypes of the distinct genomes are a consequence of the different distributions of genes. We can try to infer which genes are associated with a set of genomes having a specific phenotype. As a proof of concept, we extracted the genes with a differential distribution between Gram-positive and Gram-negative genomes (essentially represented by proteobacterias and firmicutes, respectively; see Table 1).
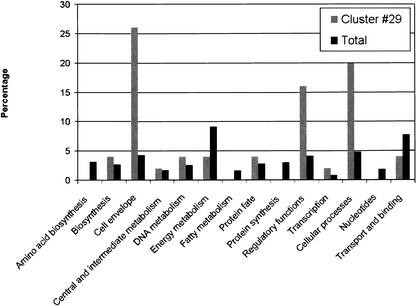
The approach proposed here consists of several steps. In the first step, we used SOTA (Dopazo and Carazo 1997; Herrero et al. 2001), a divisive clustering method that is able to define the number of nonredundant clusters in the data to reduce the gene dataset to the different representative profiles of presence/absence. In the second step, we trained a neural network with the average conservation values of the clusters found, to learn the different phenotypes on the basis of the distribution of genes in the genomes. The level of resolution at which the clusters are used as inputs for the perceptron is found by examination of their informative contents. To find this optimal value, different levels of resolution are tested with a perceptron, and the number of true positives found in each case is used to assess this value (see Mateos et al. 2002 for details). Once the optimal level was found, the magnitudes of the interconnection weights of the perceptron used for the classification provided an idea of the importance of the different clusters of co-occurring genes in the definition of the classes. The method can compress the patterns of co-occurrence in genomes to a level of 31 different clusters. In other words, if the hierarchy of CHGs (see Fig. 2) is climbed up to a level in which we only have 31 different average patterns of co-occurrence, and these average patterns are used to infer whether a genome is Gram-positive or Gram-negative, we still have 100% accuracy in the classification of Gram-positive and 95.45% in the case of Gram-negative. The strongest weights of the perceptron indicate which of these clusters are more important in the definition of the classes (Gram positive and negative). In this case, cluster No. 29 has the strongest weight. Figure 4 shows a comparison of the role of the 50 proteins contained in cluster No. 29 with respect to the role of the rest of proteins. It is clear that roles like cell envelope, regulatory functions, and cellular processes (which include adaptations to atypical conditions, cell adhesion, cell division, chemotaxis and motility, conjugation, detoxification, pathogenesis, and toxin production and resistance: see TIGR assigned role categories at http://www.tigr.org/CMR2/role_id.shtml) are overrepresented in that cluster. All these roles are clearly related to the differences among Gram-positive and Gram-negative bacteria.
Figure 4.
Comparison of the GO terms corresponding to biological processes of the 50 proteins contained in cluster No. 29 with respect to the corresponding ones in the basal distribution of the rest of proteins.
This approach can be very helpful in assisting in the definition of functions for unknown genes. Thus, if the pattern of distribution across genomes of an unknown gene is found to be important in the separation of phenotypic traits, it must be related with the trait. This is especially true if other genes with the same co-occurrence across genomes have annotated roles that reinforce this finding. For example, in cluster No. 29, we have found two proteins of unknown function: YEGN_ECOLI and YEGO_ECOLI. The only annotation both proteins display in the corresponding SWISS-PROT entry is “could be a drug efflux pump (by similarity).” The fact that they appear in this cluster of co-occurrence with many other proteins related to cell envelope and detoxification reinforces their putative role as a drug efflux pump (Pellegrini et al. 1999).
DISCUSSION
There were 60 prokaryotic genomes at the beginning of this work and, by the time of submitting the manuscript, there were 11 more. The possibility of using all this information simultaneously opens up the possibility of studies at genome level. The two-dimensional (2D) representation of the profile of gene conservation along a genome and across genomes constitutes a visualization tool that gives immediate information of features of the genome studied as well as about the other genomes used in the comparison. In addition, quantitative analyses can be made on the basis of the different relative degrees of conservation of the genes and the pattern of distribution of homologs across genomes. Clustering these patterns permits the study, at different levels, of the patterns of distribution as well as the functionality of these genes from different resolution levels. The obvious next step is obtaining the relationship between gene distribution patterns and phenotypic traits, which will point to genes as responsible for these traits.
Future Prospects: Universal Gene Conservation Profile
The profile presented here represents a comparison of one-against-all genomes. A comparison of all-against-all would imply the repetition of this profile for each genome with respect to others. This would give as many profiles as genomes. Each of these profiles would include only genes represented in the genome analyzed. Alternatively, it would be possible to cluster all the families of proteins, using a universal list of proteins that include all the types of proteins represented at least once in any of the genomes. Each type (or family) of proteins must be represented by some sort of average or consensus sequence. The profile of conservation obtained in this way would be unique and universal and would include the information of all possible one-against-all comparisons. Studies of genotype/phenotype correlation would be possible within a common framework. We are currently exploring both representations.
WEB SITE REFERENCES
http://www.ebi.ac.uk/genomes/index.html; Genomes at the EBI.
http://www.tigr.org/CMR2/role_id.shtml; TIGR assigned role categories.
http://www.ebi.ac.uk/GOA/index.html; GO terms for the proteomes analyzed.
http://www.ebi.ac.uk/∼martin/EcoliK12/GenomicProfiles.html; Additional information.
http://bioinfo.cnio.es/data/GenomeProfile/; Additional information.
http://www.ebi.ac.uk/proteomes/index.html; list of proteomes at the EBI.
http://www.ebi.ac.uk/proteome/ECOLI/; E. coli K12 proteome at the EBI.
Acknowledgments
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
E-MAIL jdopazo@cnio.es; FAX +34 912246972.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.678303. Article published online before print in April 2003.
REFERENCES
- 1.Abouhamad W.N. and Manson, M.D. 1994. The dipeptide permease of Escherichia coli closely resembles other bacterial transport systems and shows growth-phase-dependent expression. Mol. Microbiol. 14: 1077-1092. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S.F., Madden, T.L., Schäffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashburner M., Ball, C.A., Blake, J.A., Botstein, D., Butler, H., Cherry, J.M., Davis, A.P., Dolinski, K., Dwight, S.S., Eppig, J.T., et al. 2000. Gene Ontology: Tool for the unification of biology. Nat. Genet. 25: 25-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blattner F.R., Plunkett, G., III, Bloch, C.A., Perna, N.T., Burland, V., Riley, M., Collado-Vides, J., Glasner, J.D., Rode, C.K., Mayhew, G.F., et al. 1997. The complete genome sequence of Escherichia coli K-12. Science 277: 1453-1474. [DOI] [PubMed] [Google Scholar]
- 5.Dopazo J. and Carazo, J.M. 1997. Phylogenetic reconstruction using a growing neural network that adopts the topology of a phylogenetic tree. J. Mol. Evol. 44: 226-233. [DOI] [PubMed] [Google Scholar]
- 6.Dopazo J., Mendoza, A., Herrero, J., Caldara, F., Humbert, Y., Friedli, L., Guerrier, M., Grand-Schenk, E., Gandin, C., De Francesco, M., et al. 2001. Annotated draft genomic sequence from a Streptococcus pneumoniae type 19F clinical isolate. Microbial Drug Resistance 7: 99-125. [DOI] [PubMed] [Google Scholar]
- 7.Felsenstein J. 1985. Phylogenetics and the comparative method. Am. Naturalist 125: 1-15. [Google Scholar]
- 8.Fitz-Gibbon S.T. and House, C.H. 1999. Whole genome-based phylogenetic analysis of free-living microorganisms. Nucleic Acids Res. 27: 4218-4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fritzke B. 1994. Growing cell structures—A self-organizing network for unsupervised and supervised learning. Neural Networks 7: 1141-1160. [Google Scholar]
- 10.Gaasterland T. and Regan, M.A. 1998. Constructing multigenome views of whole microbial genomes. Microb. Comp. Genomics 3: 177-192. [DOI] [PubMed] [Google Scholar]
- 11.Herrero J., Valencia, A., and Dopazo, J. 2001. A hierarchical unsupervised growing neural network for clustering gene expression patterns. Bioinformatics 17: 126-136. [DOI] [PubMed] [Google Scholar]
- 12.Himmelreich R., Hilbert, H., Plagens, H., Pirkl, E., Li, B.-C., and Herrmann, R. 1996. Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucl. Acids Res. 24: 4420-4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kalman S., Mitchell, W., Marathe, R., Lammel, C., Fan, J., Hyman, R.W., Olinger, L., Grimwood, J., Davis, R.W., and Stephens, R.S. 1999. Comparative genomes of Chlamydia pneumoniae and C. Trachomatis. Nat. Genet. 21: 385-389. [DOI] [PubMed] [Google Scholar]
- 14.Kohonen T., 1997. Self-organizing maps. Springer-Verlag, Berlin, Germany.
- 15.Lawrence J.G. 1999. Gene transfer, speciation, and the evolution of bacterial genomes. Curr. Opin. Microbiol. 2: 519-523. [DOI] [PubMed] [Google Scholar]
- 16.Lin J. and Gerstein, M. 2000. Whole-genome trees based on the occurrence of folds and orthologs: Implications for comparing genomes on different levels. Genome Res. 10: 808-818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mateos A., Herrero, J., Tamames, J., and Dopazo, J. 2002. Supervised neural networks for clustering conditions in DNA array data after reducing noise by clustering gene expression profiles. In Methods for Microarray Data Analysis II (eds. S. Lin & K. Jonson), pp. 91–103. Kluwer Academic Publishers, Boston, MA.
- 18.Pellegrini M., Marcotte, E.M., Thompson, M.J., Eisenberg, D., and Yeates, T.O. 1999. Assigning protein functions by comparative genome analysis: Protein phylogenetic profiles. Proc. Natl. Acad. Sci. 96: 4285-4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peterson J.D., Umayam, L.A., Dickinson, T.M., Hickey, E.K., and White, O. 2001. The comprehensive microbial resource. Nucleic Acids Res. 29: 123-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pozzi G., Masala, L., Iannelli, F., Manganelli, R., Havarstein, L.S., Piccoli, L., Simon, D., and Morrison, D.A. 1996. Competence for genetic transformation in encapsulated strains of Streptococcus pneumoniae: Two allelic variants of the peptide pheromone. J. Bacteriol. 178: 6087-6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shigenobu S., Watanabe, H., Hattori, M., Sakaki, Y., and Ishikawa, H. 2000. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature 407: 81-86. [DOI] [PubMed] [Google Scholar]
- 22.Tekaia F., Lazcano, A., and Dujon, B. 1999. The genomic tree as revealed from whole proteome comparisons. Genome Res. 9: 550-557. [PMC free article] [PubMed] [Google Scholar]
- 23.Tamames, J. 2001. Evolution of gene order conservation in prokaryotes Genome Biology 2: 0020.1–0020.11. [DOI] [PMC free article] [PubMed]
- 24.Tatusov R.L., Koonin, E.V., and Lipman, D.J. 1997. A genomic perspective on protein families. Science 278: 631-637. [DOI] [PubMed] [Google Scholar]
- 25.Zuckerlandl E. and Pauling, L. 1965. Evolutionary divergence and convergence in proteins. In Evolving genes and proteins (eds. V. Bryson & H.J. Vogel), pp. 97–166. Academic Press, New York, NY.