Abstract
Streptococcus pneumoniae has proteins that are attached to its surface by binding to phosphorylcholine of teichoic and lipoteichoic acids. These proteins are known as choline-binding proteins (CBPs). CBPs are an interesting alternative for the development of a cost-effective vaccine, and PspA (pneumococcal surface protein A) is believed to be the most important protective component among the different CBPs. We sought to use CBPs eluted from pneumococci as an experimental vaccine. Since PspA shows variability between isolates, we constructed strains producing different PspAs. We used the nonencapsulated Rx1 strain, which produces PspA from clade 2 (PspA2), to generate a pspA-knockout strain (Rx1 ΔpspA) and strains expressing PspA from clade 1 (Rx1 pspA1) and clade 4 (Rx1 pspA4). We grew Rx1, Rx1 ΔpspA, Rx1 pspA1, and Rx1 pspA4 in Todd-Hewitt medium containing 0.5% yeast extract and washed cells in 2% choline chloride (CC). SDS-PAGE analysis of the proteins recovered by a CC wash showed few bands, and the CBPs PspA and PspC (pneumococcal surface protein C) were identified by mass spectrometry analysis. Subcutaneous immunization of mice with these full-length native proteins without adjuvant led to significantly higher rates of survival than immunization with diluent after an intranasal lethal challenge with two pneumococcal strains and also after a colonization challenge with one strain. Importantly, immunization with recombinant PspA4 (rPspA4) without adjuvant did not elicit significant protection.
INTRODUCTION
Streptococcus pneumoniae causes several diseases, including otitis media, bacteremia, pneumonia, and meningitis. The capsular polysaccharide (PS) is an important virulence factor of pneumococci, and it is classified into more than 90 serotypes. The currently used vaccines are based on the induction of antibodies against PS, providing serotype-specific protection against invasive disease. The widespread use of the 7-valent conjugate vaccine, licensed in 2000, led to a marked reduction in the incidence of disease caused by vaccine serotypes, but there was also an increase in the incidence of disease caused by nonvaccine serotypes (1, 2), a phenomenon known as serotype replacement. More recently, 10- and 13-valent conjugate vaccines have been licensed, but the problem of serotype replacement will probably persist. The development of new vaccines against pneumococcal infections is thus a priority, and the two major requirements for such vaccines for the developing world are (i) that the vaccine be highly efficacious and protective against virtually all pneumococci and (ii) that the vaccine be able to be produced at a cost low enough that it can be made available to children in the poorest countries (3).
Among the proteins exposed on the surface of pneumococci that could be used as vaccine antigens are the choline-binding proteins (CBPs) (4–7). CBPs have a biologically active module and a choline-binding module that anchors these proteins noncovalently to the phosphorylcholine of teichoic and lipoteichoic acids. The number of CBPs varies in different strains (but is approximately 15), and some of these proteins are highly variable (5, 8). The genome of the TIGR4 strain has genes encoding the CBPs CbpI (choline-binding protein I), PspA (pneumococcal surface protein A), CbpC (choline-binding protein C), CbpJ (choline-binding protein J), CbpG (choline-binding protein G), CbpF (choline-binding protein F), Pce (phosphorylcholine esterase), LytB (autolysin B), LytC (autolysin C), LytA (autolysin A), PcpA (pneumococcal choline-binding protein A), PspC (pneumococcal surface protein C), and CbpD (choline-binding protein D). The TIGR4 genome also contains two open reading frames that have the choline-binding domains, but the proteins are truncated or degenerated (8). Some of the CBPs do not have signal secretion sequences. However, proteins found on the surface of pneumococci and other Gram-positive organisms can lack standard signal sequences (9, 10). The genome of the R6 strain does not have CbpI and CbpJ (8). Some of the CBPs, including PspA, PspC, and PcpA, have been described to be important virulence factors (7, 11–13), and PspA has been shown to be the major protein among the CBPs (14, 15).
PspA is produced by virtually all pneumococci and shows variability among different strains. PspA interferes with the host-pathogen interaction through the inhibition of the deposition of complement on the bacterial surface (16, 17), by complement-independent inhibition of phagocytosis (18), and also by protecting pneumococci from killing by apolactoferrin (19). It has recently been proposed that PspA prevents the binding of C-reactive protein to phosphorylcholine, avoiding C3 deposition through the classical pathway (15). PspA is composed of an N-terminal α-helical region exposed on the bacterial surface, followed by a proline-rich region and the C-terminal region with the choline-binding domain (20). Hollingshead and collaborators have proposed a classification based on the amino acid divergence of the most variable part of the α-helical region located just before the proline-rich region, the clade-defining region (CDR) (21). Family 1 comprises clades 1 and 2, and family 2 comprises clades 3, 4, and 5. Family 3, which is rarely isolated, comprises clade 6. Since there is less cross-reactivity between families than within families, it has been proposed that a broad-coverage vaccine should be composed of at least one PspA of each of the two major families (22–24). Previous studies have shown that both the α-helical and proline-rich domains of PspA are able to elicit protection (24, 25), but all phase I vaccine trials with the PspA antigen have been conducted using only the α-helical domain (11). Although a mixture of α-helical domains should be protective against most PspAs (22), recent evidence suggests that the proline-rich domain of PspA is even more cross-protective (25). Vaccine immunogens should thus probably contain both of these domains. It has also been shown that immunization with more than one pneumococcal protein generally elicits better protection than immunization with either domain alone (26–28). Finally, it has been shown that the use of full-length native PspA elicits much more protection than use of the recombinant α-helical domain of PspA (14), but there is at present no way to produce a full-length PspA by a recombinant process because the choline-binding domain appears to be toxic to Escherichia coli.
One way to deal with all of these issues is to produce full-length PspA from nonencapsulated pneumococci and release the PspA either by growing them in medium containing ethanolamine (ET) in place of choline or by using standard bacterial broth for growth and eluting PspA with 2% choline chloride (CC) (14). Both procedures have the advantage that they would be expected to also elute some of the other CBPs, many of which have been shown to be virulence factors for pneumococci that are capable of eliciting measurable protection against colonization and/or invasive disease in mice (12, 29). The purpose of these studies is to confirm some of the earlier work and to do the studies on a somewhat larger scale to determine whether it is possible and desirable to attempt to use one of these approaches to produce a pneumococcal vaccine.
MATERIALS AND METHODS
Ethical statement.
Immunizations and challenge experiments were performed in mice. Experimental protocols were approved by the Instituto Butantan Animal Care and Use Committee under license numbers CEUAIB 457/08 and 1011/13. During all experiments, animals were housed in cages within ventilated cabinets and supplied with food and water ad libitum.
Pneumococcal strains.
S. pneumoniae strains Rx1 (nonencapsulated, PspA clade 2), St435/96 (serotype 1, PspA clade 1), St255/00 (serotype 14, PspA clade 4), D39 (Rx1 parental strain, serotype 2, PspA clade 2), A66.1 (serotype 3, PspA clade 2), ATCC 6303 (serotype 3, PspA clade 5), 0603 (serotype 6B, PspA clade 1), 245/00 (serotype 14, PspA clade 1), and 3JYP2670 (serotype 3, PspA clade 4) were grown and stocked as previously described (30).
Construction of Rx1 ΔpspA, Rx1 pspA1, and Rx1 pspA4.
The Janus cassette (31) was used to knock out pspA from Rx1. This cassette contains a kanamycin (Kn) resistance gene (kan) and a dominant gene (rpsL) that confers susceptibility to streptomycin (Sm). The region 1,000 bp upstream of pspA (with an XbaI site at the 3′ end), the region 1,000 bp downstream of pspA (with a BamHI site at the 5′ end), and the Janus cassette (with an XbaI site at the 5′ end and a BamHI site at the 3′ end) were amplified by PCR using the primers shown in Table S1 in the supplemental material and Platinum Taq DNA polymerase high fidelity (Invitrogen, Carlsbad, CA). The fragments were digested with the restriction enzymes and ligated, and the fragment containing the upstream region of pspA, the Janus cassette, and the downstream region of pspA was amplified by PCR and cloned into pGEM-T Easy (Promega, Madison, WI), generating pGEM-TE pspA-Janus-pspA. An Rx1 clone naturally resistant to Sm was selected by plating 108 CFU of an Rx1 culture at the exponential phase of growth on blood agar plates containing streptomycin (300 μg/ml). An Sm-resistant Rx1 clone was transformed with pGEM-TE pspA-Janus-pspA digested with NotI. Kn-resistant (200 μg/ml), Sm-sensitive clones were analyzed for the correct deletion of pspA (Rx1 ΔpspA). Rx1 ΔpspA was then transformed with fragments amplified by use of a PCR mixture containing pspA from St435/96 (pspA1) or St255/00 (pspA4) (30) with the 1,000-bp upstream and 1,000-bp downstream regions using the primers shown in Table S1 in the supplemental material. Kn-sensitive and Sm-resistant clones were analyzed for the correct recombination, and the generated strains were Rx1 pspA1 and Rx1 pspA4.
Western blotting.
Cellular protein extracts from bacteria were analyzed for PspA production through Western blotting using anti-recombinant PspA4 (anti-rPspA4; a recombinant protein that includes the mature N-terminal α-helical region plus the proline-rich region of a PspA protein from clade 4) mouse serum (30). The membranes were incubated with anti-mouse IgG conjugated with peroxidase (Sigma-Aldrich, St. Louis, MO), and detection was performed with ECL Prime Western blotting reagent (GE Healthcare, United Kingdom).
Recovery of CBPs from pneumococci grown in CDM-ET.
Strain Rx1 pspA4 was adapted for growth without choline in chemically defined medium (CDM; Gibco-Life Technologies, Carlsbad, CA) containing ethanolamine (CDM-ET) (32). Adaptation was performed as described previously (14). Briefly, cultures grown in Todd-Hewitt medium containing 0.5% yeast extract (THY) were subcultured in CDM containing 0.02% CC and 0.02% ET. The concentration of CC was decreased stepwise from 0.02% to 0.0000002%. Supernatants of overnight cultures were concentrated using an Amicon Ultra centrifugal filter unit with a 10-kDa cutoff (Millipore, Billerica, MA) and dialyzed in phosphate-buffered saline (PBS). The protein concentration was determined by the Bradford method (protein assay; Bio-Rad, Hercules, CA), and recovered proteins were analyzed by SDS-PAGE with Coomassie blue staining.
Elution of CBPs from pneumococci grown in THY.
Pneumococcal strains Rx1, Rx1 ΔpspA, Rx1 pspA1, and Rx1 pspA4 were grown in THY to an optical density at 600 nm (OD600) of 0.4. The cultures were centrifuged, and the cell pellets were washed in PBS. CBPs were eluted through the incubation of the cells for 20 min at room temperature in PBS–2% CC (14). The protein concentration was determined by the Bradford method, and eluted proteins were analyzed by SDS-PAGE with silver staining.
In-gel trypsin digestion and MS analysis by liquid chromatography (LC)-MS/MS.
The protein bands were excised, and in-gel trypsin digestion was done as described by Hanna et al. (33). An aliquot (4.5 μl) of the resulting peptide mixture was injected into a trap column (180 μm [inside diameter {i.d.}] by 20 mm) packed with C18 chromatographic medium (Waters, Milford, MA) for desalting with 100% solvent A (0.1% formic acid) at 5 μl/min for 3 min. Peptides were then eluted onto an analytical C18 column (75 μm [i.d.] by 100 mm) (Waters) using a 20-min gradient at a flow rate of 600 nl/min, where solvent A was 0.1% formic acid and solvent B was 0.1% formic acid in acetonitrile. The gradient was 0 to 80% acetonitrile in 0.1% formic acid over 20 min. A quadrupole time of flight Ultima mass spectrometer (Waters) was used to acquire the spectra. The spray voltage was set at 3.4 kV, and the instrument was operated in data-dependent mode, in which one full mass spectrometric (MS) scan was acquired in the m/z range of 200 to 2,000, followed by MS/MS acquisition using collision-induced dissociation of the three most intense ions from the MS scan. A dynamic peak exclusion was applied to avoid the same m/z being selected for the next 120 s. The resulting fragment spectra were processed using ProteinLynx software (Waters), and pkl* files were created and searched, using the MASCOT search engine (Matrix Science, United Kingdom), against the NCBI nr nucleotide database (downloaded on 5 May 2013). The search was restricted to the Bacteria (Eubacteria) taxonomy with a parent and a fragment tolerance of ±1.2 and ±0.6 Da, respectively. The iodoacetamide derivative of cysteine was specified as a fixed modification, whereas oxidation of methionine and deamidation of asparagine and glutamine were specified as variable modifications.
Immunization of mice.
Five- to 7-week-old female specific-pathogen-free BALB/c and C57BL/6 mice were obtained from the Medical School of the University of São Paulo (FM-USP, São Paulo, Brazil). Proteins were given through the subcutaneous route in saline. Groups of 6 animals were given three doses of 1 μg of proteins without adjuvant at 14-day intervals. Six animals injected only with saline were used as controls. Proteins were eluted from the pneumococci through incubation with 2% CC and were not dialyzed prior to injection. Mice were immunized with 1 μg rPspA4 without adjuvant for comparison of efficacy.
Measurement of antibodies by ELISA.
Three weeks after the last immunization, mice were individually bled from the retroorbital plexus. Enzyme-linked immunosorbent assay (ELISA) was carried out as described previously in plates coated with 1 μg/ml rPspA1 or rPspA4 (recombinant proteins that include the mature N-terminal α-helical region plus the proline-rich region of a PspA protein from clade 1 or 4, respectively) (30). Goat anti-mouse IgG conjugated with horseradish peroxidase was used as secondary antibody, and the titer was defined as the highest dilution with an A492 of ≥0.1.
Antibody binding to whole pneumococci by flow cytometry.
Assays for antibody binding to whole pneumococci were performed with 1% pooled serum as previously described (16). Bacteria were analyzed using a FACSCanto flow cytometer (BD Biosciences, Franklin Lakes, NJ). Ten thousand gated events were acquired and analyzed in fluorescence intensity histograms using FlowJo (version 7.6.1) software.
Intranasal lethal aspiration and colonization challenges.
S. pneumoniae A66.1, ATCC 6303, and 0603 were grown in THY until the OD600 was 0.4, aliquoted, and kept frozen at −80°C. At 3 weeks after the last immunization, 3 × 105 CFU (ATCC 6303) or 106 CFU (A66.1) was inoculated in 50 μl into one nostril of BALB/c mice anesthetized through the intraperitoneal (i.p.) route with 200 μl of a 0.2% xylazine–1.0% ketamine mixture. Survival was monitored for 10 days. Animals were monitored twice daily, and lethargic mice were euthanized through CO2 narcosis. At 3 weeks after the last immunization, 5 × 106 CFU (strain 0603) was inoculated in 10 μl into the two nostrils of C57BL/6 mice anesthetized i.p. with 200 μl of a 0.2% xylazine–0.5% ketamine mixture. After 5 days, the animals were euthanized through CO2 narcosis, and 200 μl of saline was instilled inside the trachea and washed out through the nostrils twice. The total number of CFU in each sample was determined through plating of diluted samples on blood agar plates containing gentamicin (4 μg/ml) by consideration of the recovered volume. BALB/c mice were used for the intranasal lethal challenge and C57BL/6 mice were used for the nasopharyngeal colonization challenge for comparison with published data from studies that used these mouse lineages for each model (30, 34, 35).
Statistical analysis.
The Fisher exact test was used to compare survival levels between groups, and one-way analysis of variance (ANOVA) with Tukey's multicomparison test was used to compare antibody titers and the number of CFU recovered in the colonization challenge (GraphPad Prism [version 5] software).
RESULTS
Construction of Rx1 ΔpspA, Rx1 pspA1, and Rx1 pspA4.
An Sm-resistant Rx1 clone was transformed with the plasmid containing the Janus cassette, generating Rx1 ΔpspA. Transformation of Rx1 ΔpspA with pspA plus the flanking regions of a PspA1-producing strain (St435/96) and of a PspA4-producing strain (St255/00) generated Rx1 pspA1 and Rx1 pspA4, respectively. The correct insertion was confirmed in all strains through PCR with primers annealing at the flanking regions and also with internal primers. Figure 1 shows the results of analysis of the production of PspA through Western blotting. As expected, PspAs with somewhat different molecular masses were observed for Rx1, Rx1 pspA1, and Rx1 pspA4, whereas no product was detected for Rx1 ΔpspA.
FIG 1.
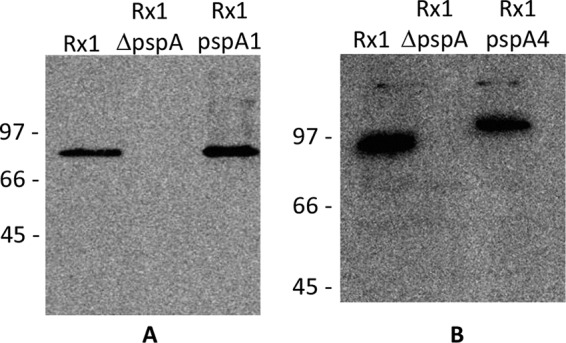
Analysis of PspA expression. Total extracts of pneumococcal strains Rx1, Rx1 ΔpspA, and Rx1 pspA1 (A) or Rx1 pspA4 (B) were analyzed for the production of PspA using anti-rPspA4 serum. Numbers to the left of the gels are molecular masses (in kilodaltons).
Recovery of CBPs from pneumococci grown in CDM-ET.
Rx1 pspA4 was adapted for growth in chemically defined medium containing ethanolamine (CDM-ET) in place of choline through several culture passages with decreasing concentrations of choline chloride (CC). We grew the adapted Rx1 pspA4 strain overnight in CDM-ET and collected the supernatant for the recovery of CBPs. The supernatant was concentrated, and the proteins were analyzed by SDS-PAGE. Several bands could be observed in the gel stained with Coomassie blue (Fig. 2A). LC-MS/MS analysis of different bands showed the presence of cytosolic proteins and also of proteins described to be surface exposed in the supernatant of CDM-ET, but no CBPs were identified. Even though PspA could be detected through Western blotting (not shown), CBPs were not the major proteins recovered from the supernatant of Rx1 pspA4 grown overnight in CDM-ET. This result could have been due to the long culture period, which could have resulted in cell death and the release of cytoplasmic content to the culture supernatant.
FIG 2.
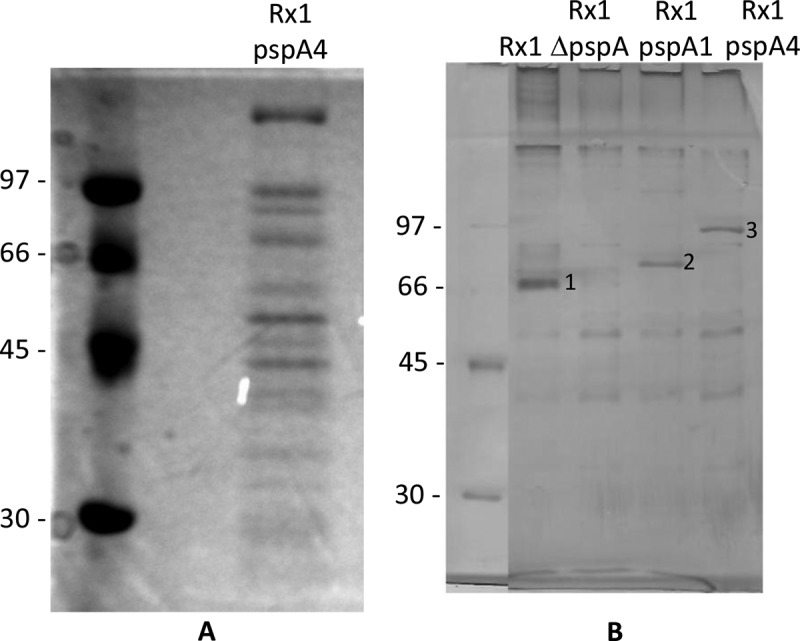
Recovery of pneumococcal proteins. (A) Proteins were recovered from the supernatant of an Rx1 pspA4 culture grown overnight in CDM-ET. The supernatant was concentrated and analyzed by SDS-PAGE with Coomassie blue staining. (B) Proteins were eluted through a 2% CC wash of Rx1, Rx1 ΔpspA, Rx1 pspA1, and Rx1 pspA4 cultures grown to exponential phase in THY. Bacteria were incubated in 2% CC, and the eluted proteins were analyzed by SDS-PAGE with silver staining. Bands indicated by the numbers 1 to 3 were submitted to LC-MS/MS analysis, and the results are shown in Table S2 in the supplemental material. Numbers to the left of the gels are molecular masses (in kilodaltons).
Recovery of CBPs from pneumococci grown in THY.
Rx1, Rx1 ΔpspA, Rx1 pspA1, and Rx1 pspA4 were then grown in THY, and CBPs were recovered through incubation in 2% CC. The recovered proteins were examined by SDS-PAGE, and the gel was silver stained (Fig. 2B). Few bands were observed, and there was retention of proteins in the stacking gel, indicating the presence of aggregates. The major bands in the CC washes of Rx1, Rx1 pspA1, and Rx1 pspA4 were analyzed by LC-MS/MS (see Table S2 in the supplemental material). The Rx1 pspA1 CC wash band showed a hit with PspA, while the Rx1 pspA4 CC wash band showed hits with PspA and PspC. The Rx1 CC wash band showed only one peptide with a hit with the flavoprotein NrdI. Western blot analysis showed the recognition of bands in the protein samples obtained from strains Rx1, Rx1 pspA1, and Rx1 pspA4 by anti-rPspA4 serum, but a considerable amount of the protein recovered from Rx1 was retained in the stacking gel (see Fig. S1 in the supplemental material). It is possible that a large part of the CBPs from Rx1 was retained as aggregates in the stacking gel. We have attempted to perform MS analysis with the proteins from the stacking gel, but no CBPs were recognized. This result could be due to problems with in-gel trypsin digestion due to protein aggregation.
Immunization with CBPs recovered from pneumococci grown in THY.
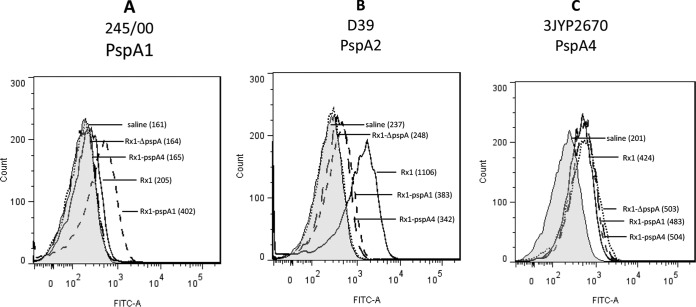
The proteins recovered from the cultures grown in THY through incubation in 2% CC were used to immunize naive BALB/c mice subcutaneously without the use of any adjuvant (6 animals/group). Sera from animals immunized with the CBPs recovered by CC washes of Rx1, Rx1 pspA1, and Rx1 pspA4 showed the presence of antibodies recognizing both rPspA1 and rPspA4 (Fig. 3). Sera were also used to analyze the binding onto the surface of intact pneumococci by flow cytometry (Fig. 4). There was higher binding to strain 245/00 (PspA1) by serum from animals immunized with CBPs from Rx1 pspA1 (Fig. 4A). Higher binding to strain D39 (PspA2) was observed with serum from animals immunized with CBPs from Rx1 (PspA2) (Fig. 4B). For strain 3YJP3670 (PspA4), binding to sera obtained from animals immunized with CBPs from Rx1, Rx1 ΔpspA, Rx1 pspA1, and Rx1 pspA4 was observed (Fig. 4C). The reason for its greater reactivity with the immune sera is not known, but it could be due to the fact that since the type 3 capsule is not covalently attached to the cell body (36), all of it may not have remained on the bacterial surface after the washing steps, allowing antibody to normally poorly exposed surface proteins to provide significant binding.
FIG 3.
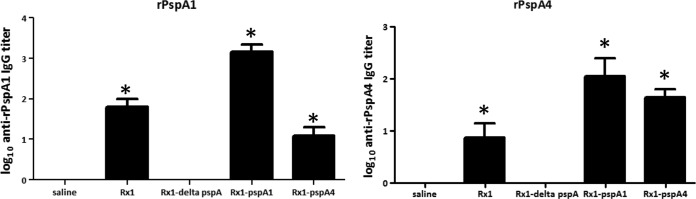
Induction of anti-PspA antibodies. Sera from mice immunized with proteins eluted from Rx1, Rx1 ΔpspA, Rx1 pspA1, and Rx1 pspA4 through a CC wash were analyzed for the induction of anti-rPspA1 (rPspA1) or anti-rPspA4 (rPspA4) antibodies by ELISA. Sera from animals injected only with saline were used as a control. Bars indicate the mean titer plus the standard error. *, statistically significant difference from the results obtained with saline (P < 0.05, one-way ANOVA with Tukey's multicomparison test).
FIG 4.
Binding of antibodies onto intact pneumococci. Sera from mice immunized with proteins eluted from Rx1, Rx1 ΔpspA, Rx1 pspA1, and Rx1 pspA4 through a CC wash were analyzed for binding to pneumococcal strains St255/00 (A), D39 (B), and 3JYP2670 (C) by flow cytometry. Sera from animals injected with saline were used as a control, and the results for the control are shown in the areas filled in gray. The median bacterial fluorescence for each sample is shown in parentheses. FITC-A, fluorescein isothiocyanate A channel.
Challenge of immunized mice intranasally with live pneumococci.
Next, immunized BALB/c mice were intranasally challenged with a lethal dose of A66.1 (PspA2) or ATCC 6303 (PspA5), and survival was analyzed (Table 1). Mice immunized with the CBPs recovered by CC washes of Rx1, Rx1 ΔpspA, Rx1 pspA1, and Rx1 pspA4 showed statistically significantly higher rates of survival after challenge with A66.1 than animals injected with saline. Mice immunized with the CBPs recovered by CC washes of Rx1 ΔpspA and Rx1 pspA4 also showed statistically significantly higher rates of survival after challenge with ATCC 6303 than animals injected with saline.
TABLE 1.
Survival of mice after challenge with A66.1 (PspA2) and ATCC 6303 (PspA5)e
| Immunogen | A66.1 |
ATCC 6303 |
||||
|---|---|---|---|---|---|---|
| No. of animals alive/total no. tested | % survival | Pa | No. of animals alive/total no. tested | % survival | P | |
| Saline | 1/12 | 8 | 0/12 | 0 | ||
| Rx1b | 12/12 | 100 | <0.0001 | 1/12 | 8 | 1.0000 |
| Rx1 ΔpspAb | 10/12 | 83 | 0.0006 | 5/12 | 42 | 0.0373 |
| Rx1 pspA1b | 9/12 | 75 | 0.0028 | 4/12 | 33 | 0.0932 |
| Rx1 pspA4b | 11/12 | 92 | 0.0001 | 7/12 | 58 | 0.0046 |
| rPspA4c,d | 5/12 | 42 | 0.1550 | 1/6 | 17 | 0.3333 |
P values by comparison with the results for the saline group (two-sided Fisher exact test).
Immunization with CBP eluted from the indicated strain with 2% choline chloride.
Immunization with recombinant PspA from clade 4.
The comparison of protection elicited by rPspA4 and a choline chloride eluate of Rx1 pspA4 was significant at P values of 0.027 and 0.15 by the two-sided Fisher exact test for A66.1 and ATCC 6303 challenge, respectively.
Mice were immunized with the indicated immunogens in saline in the absence of any adjuvant. Survival was monitored for 10 days, and the numbers of mice that survived in each group were recorded. Results are pooled data from two independent experiments.
Intranasal challenge of mice in a pneumococcal carriage model.
Proteins recovered from the cultures grown in THY by elution of pneumococci with CC were also used to immunize C57BL/6 mice subcutaneously without adjuvant in order to evaluate protection against a colonization challenge model with strain 0603 (PspA1). Figure 5 shows the numbers of CFU recovered from the nasopharynx of mice 5 days after challenge. Only the group of animals immunized with CBPs from Rx1 pspA1 showed a significantly lower colonization density than animals immunized with saline. In most prior studies, protection against colonization was seen after immunization with recombinant proteins and a mucosal adjuvant related to cholera toxin (28). In the present studies, the reduction in colonization that has been observed was detected with subcutaneous immunization and in the absence of adjuvant.
FIG 5.
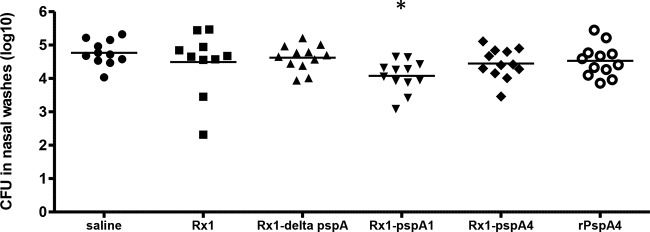
Intranasal colonization challenge. Mice immunized with proteins eluted from Rx1, Rx1 ΔpspA, Rx1 pspA1, and Rx1 pspA4 through a CC wash and rPspA4 were challenged intranasally with strain 0603 (capsule type 6B, PspA clade 1). The density of pneumococcal colonization, expressed as the log10 number of total CFU in upper respiratory tract lavage fluid, was determined for individual mice at day 5 after challenge. Mean values are indicated for each group. *, statistically significant difference from the results for the saline group (P < 0.05, one-way ANOVA with Tukey's multicomparison test). The results shown are pooled data from two independent experiments.
DISCUSSION
A mixture of CBPs containing full-length native PspA as the major antigen is an interesting alternative for a broad-coverage vaccine formulation with a low production cost. We evaluated the recovery of CBPs from the supernatant of cultures grown overnight in CDM-ET and from CC washes of cultures grown to exponential phase in complete medium. MS analysis showed the presence of cytosolic proteins and also of proteins described to be surface exposed in the supernatant of CDM-ET, but no CBPs were identified. Pneumococci grown in chemically defined medium without choline were described to grow in long chains and not to undergo autolysis (14, 37, 38), but we cannot rule out the possibility of cell death and lysis during the overnight growth under our culture conditions. On the other hand, CC washes contained few bands by SDS-PAGE analysis, and the major ones were identified to be PspA and PspC, as expected. Previous work has evaluated the supernatant of cultures grown to exponential phase in the absence of choline and with supplementation with ethanolamine or putrescine, but MS analysis was not performed and only challenge strains of the type 3 capsular type were examined (38, 39).
We thus used CBPs recovered by the CC washes from strains producing PspA2 (Rx1), PspA1 (Rx1 pspA1), and PspA4 (Rx1 pspA4) or lacking pspA (Rx1 ΔpspA) to immunize mice without adjuvant. The use of the nonencapsulated Rx1 strain rules out any influence of polysaccharide contamination in the preparations. Sera from mice immunized with CBPs from Rx1, Rx1 pspA1, and Rx1 pspA4 recognized rPspA1 and rPspA4 by ELISA. When the binding of antibodies to intact pneumococci was analyzed by flow cytometry, there was better binding to bacteria producing PspA from the same clade as the strain used to recover the CBPs. Furthermore, immunization with CBPs from Rx1, Rx1 ΔpspA, Rx1 pspA1, and Rx1 pspA4 elicited protection against a lethal challenge with a PspA2-producing strain, whereas immunization with CBPs from Rx1 ΔpspA and Rx1 pspA4 elicited protection against a lethal challenge with a PspA5-producing strain. Thus, in agreement with our data showing cross-protection through immunization with rPspA4 adjuvanted with alum (30), CBPs recovered from Rx1 pspA4 protected against strains expressing PspA from family 1 (clade 2) and family 2 (clade 5). Interestingly, protection against both challenges was observed in the absence of PspA in the group immunized with the CBPs recovered from Rx1 ΔpspA. It thus seems that the other CBPs can compensate for the absence of PspA in the formulation. It is important to note that we inoculated a fixed amount of protein in all groups and that PspA is probably the major component of the CBPs recovered from the strains producing PspA. These results are in agreement with previous data showing that PspA is the main protective component but other CBPs can contribute to protection (14). It should be noted that recombinant PspA4, which lacks the choline-binding domain, did not elicit protection in the absence of adjuvant, an illustration of the greater vaccine potential of CC eluates containing full-length molecules than the vaccine potential of rPspA.
When subcutaneously immunized mice were challenged with a PspA1-producing strain intranasally, only immunization with CBPs from Rx1 pspA1 elicited protection against colonization, indicating that clade-specific immunization might be required to elicit protection against carriage, whereas broad cross-protection against fatal challenge was observed. The findings of these studies are in contrast to those of earlier studies using rPspA to protect against carriage, where intranasal immunization and a mucosal adjuvant were normally used (28).
CBPs can thus induce protection in the absence of adjuvants, while rPspA is known to require the use of adjuvants, such as alum and the cholera toxin B subunit, to elicit protection against lethal and colonization challenges (27, 28, 30). This finding suggests that the full-length CBPs exhibit more protective efficacy than the recombinant N-terminal portions of CBPs, which invariably require adjuvant to elicit strong protective responses. Importantly, a vaccine formulation based on CBPs could have a low production cost and require few steps to recover the proteins from the pneumococcal culture. Since we observed possible PspA clade-specific protection against colonization, the use of CBPs obtained from strains producing PspA from family 1 and family 2 is still advisable to attain broad protection and avoid the emergence of vaccine escape mutants.
Other vaccine formulations currently being evaluated by several groups include multiple antigens of pneumococci (11). One advantage of this approach is that selective pressure would be on multiple antigens, making the emergence of vaccine escape mutants a rare event. One of the most promising formulations currently in clinical trials is composed of an inactivated nonencapsulated whole-cell vaccine, which was shown to induce protection against nasal colonization with a variety of pneumococcal serotypes in mice (40), as well as against a lethal challenge with a serotype 3 strain in rats (41). The present studies confirm that a formulation based on CBPs is also an interesting alternative for a pneumococcal vaccine based on multiple antigens, since it would comply with the requirements of low cost and induction of protection against different pneumococcal strains and would be focused on surface antigens.
ACKNOWLEDGMENTS
We are grateful to Jorge M. C. Ferreira for the flow cytometry analysis.
This work was supported by CNPq (302070/2011-7 and 472253/2012-3), FAPESP (2011/06853-2), and the Fundação Butantan (Brazil).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/CVI.00692-14.
REFERENCES
- 1.Singleton RJ, Hennessy TW, Bulkow LR, Hammitt LL, Zulz T, Hurlburt DA, Butler JC, Rudolph K, Parkinson A. 2007. Invasive pneumococcal disease caused by nonvaccine serotypes among Alaska native children with high levels of 7-valent pneumococcal conjugate vaccine coverage. JAMA 297:1784–1792. doi: 10.1001/jama.297.16.1784. [DOI] [PubMed] [Google Scholar]
- 2.Hicks LA, Harrison LH, Flannery B, Hadler JL, Schaffner W, Craig AS, Jackson D, Thomas A, Beall B, Lynfield R, Reingold A, Farley MM, Whitney CG. 2007. Incidence of pneumococcal disease due to non-pneumococcal conjugate vaccine (PCV7) serotypes in the United States during the era of widespread PCV7 vaccination, 1998-2004. J Infect Dis 196:1346–1354. doi: 10.1086/521626. [DOI] [PubMed] [Google Scholar]
- 3.PATH. 2014, posting date Developing new vaccines against pneumonia and other pneumococcal diseases. PATH, Seattle, WA: http://www.path.org/publications/detail.php?i=1783. [Google Scholar]
- 4.Gamez G, Hammerschmidt S. 2012. Combat pneumococcal infections: adhesins as candidates for protein-based vaccine development. Curr Drug Targets 13:323–337. doi: 10.2174/138945012799424697. [DOI] [PubMed] [Google Scholar]
- 5.Perez-Dorado I, Galan-Bartual S, Hermoso JA. 2012. Pneumococcal surface proteins: when the whole is greater than the sum of its parts. Mol Oral Microbiol 27:221–245. doi: 10.1111/j.2041-1014.2012.00655.x. [DOI] [PubMed] [Google Scholar]
- 6.Gosink KK, Mann ER, Guglielmo C, Tuomanen EI, Masure HR. 2000. Role of novel choline binding proteins in virulence of Streptococcus pneumoniae. Infect Immun 68:5690–5695. doi: 10.1128/IAI.68.10.5690-5695.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosenow C, Ryan P, Weiser JN, Johnson S, Fontan P, Ortqvist A, Masure HR. 1997. Contribution of novel choline-binding proteins to adherence, colonization and immunogenicity of Streptococcus pneumoniae. Mol Microbiol 25:819–829. doi: 10.1111/j.1365-2958.1997.mmi494.x. [DOI] [PubMed] [Google Scholar]
- 8.Hakenbeck R, Madhour A, Denapaite D, Bruckner R. 2009. Versatility of choline metabolism and choline-binding proteins in Streptococcus pneumoniae and commensal streptococci. FEMS Microbiol Rev 33:572–586. doi: 10.1111/j.1574-6976.2009.00172.x. [DOI] [PubMed] [Google Scholar]
- 9.Pribyl T, Moche M, Dreisbach A, Bijlsma JJ, Saleh M, Abdullah MR, Hecker M, van Dijl JM, Becher D, Hammerschmidt S. 2014. Influence of impaired lipoprotein biogenesis on surface and exoproteome of Streptococcus pneumoniae. J Proteome Res 13:650–657. doi: 10.1021/pr400768v. [DOI] [PubMed] [Google Scholar]
- 10.Henderson B, Martin A. 2011. Bacterial virulence in the moonlight: multitasking bacterial moonlighting proteins are virulence determinants in infectious disease. Infect Immun 79:3476–3491. doi: 10.1128/IAI.00179-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miyaji EN, Oliveira ML, Carvalho E, Ho PL. 2013. Serotype-independent pneumococcal vaccines. Cell Mol Life Sci 70:3303–3326. doi: 10.1007/s00018-012-1234-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glover DT, Hollingshead SK, Briles DE. 2008. Streptococcus pneumoniae surface protein PcpA elicits protection against lung infection and fatal sepsis. Infect Immun 76:2767–2776. doi: 10.1128/IAI.01126-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hava DL, Camilli A. 2002. Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol Microbiol 45:1389–1406. doi: 10.1046/j.1365-2958.2002.03106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Briles DE, King JD, Gray MA, McDaniel LS, Swiatlo E, Benton KA. 1996. PspA, a protection-eliciting pneumococcal protein: immunogenicity of isolated native PspA in mice. Vaccine 14:858–867. doi: 10.1016/0264-410X(96)82948-3. [DOI] [PubMed] [Google Scholar]
- 15.Mukerji R, Mirza S, Roche AM, Widener RW, Croney CM, Rhee DK, Weiser JN, Szalai AJ, Briles DE. 2012. Pneumococcal surface protein A inhibits complement deposition on the pneumococcal surface by competing with the binding of C-reactive protein to cell-surface phosphocholine. J Immunol 189:5327–5335. doi: 10.4049/jimmunol.1201967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ren B, Szalai AJ, Hollingshead SK, Briles DE. 2004. Effects of PspA and antibodies to PspA on activation and deposition of complement on the pneumococcal surface. Infect Immun 72:114–122. doi: 10.1128/IAI.72.1.114-122.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tu AH, Fulgham RL, McCrory MA, Briles DE, Szalai AJ. 1999. Pneumococcal surface protein A inhibits complement activation by Streptococcus pneumoniae. Infect Immun 67:4720–4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Genschmer KR, Accavitti-Loper MA, Briles DE. 2013. A modified surface killing assay (MSKA) as a functional in vitro assay for identifying protective antibodies against pneumococcal surface protein A (PspA). Vaccine 32:39–47. doi: 10.1016/j.vaccine.2013.10.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shaper M, Hollingshead SK, Benjamin WH Jr, Briles DE. 2004. PspA protects Streptococcus pneumoniae from killing by apolactoferrin, and antibody to PspA enhances killing of pneumococci by apolactoferrin. Infect Immun 72:5031–5040. doi: 10.1128/IAI.72.9.5031-5040.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yother J, Briles DE. 1992. Structural properties and evolutionary relationships of PspA, a surface protein of Streptococcus pneumoniae, as revealed by sequence analysis. J Bacteriol 174:601–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hollingshead SK, Becker R, Briles DE. 2000. Diversity of PspA: mosaic genes and evidence for past recombination in Streptococcus pneumoniae. Infect Immun 68:5889–5900. doi: 10.1128/IAI.68.10.5889-5900.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nabors GS, Braun PA, Herrmann DJ, Heise ML, Pyle DJ, Gravenstein S, Schilling M, Ferguson LM, Hollingshead SK, Briles DE, Becker RS. 2000. Immunization of healthy adults with a single recombinant pneumococcal surface protein A (PspA) variant stimulates broadly cross-reactive antibodies to heterologous PspA molecules. Vaccine 18:1743–1754. doi: 10.1016/S0264-410X(99)00530-7. [DOI] [PubMed] [Google Scholar]
- 23.Briles DE, Hollingshead SK, Nabors GS, Paton JC, Brooks-Walter A. 2000. The potential for using protein vaccines to protect against otitis media caused by Streptococcus pneumoniae. Vaccine 19(Suppl 1):S87–S95. doi: 10.1016/S0264-410X(00)00285-1. [DOI] [PubMed] [Google Scholar]
- 24.Briles DE, Hollingshead SK, King J, Swift A, Braun PA, Park MK, Ferguson LM, Nahm MH, Nabors GS. 2000. Immunization of humans with recombinant pneumococcal surface protein A (rPspA) elicits antibodies that passively protect mice from fatal infection with Streptococcus pneumoniae bearing heterologous PspA. J Infect Dis 182:1694–1701. doi: 10.1086/317602. [DOI] [PubMed] [Google Scholar]
- 25.Daniels CC, Coan P, King J, Hale J, Benton KA, Briles DE, Hollingshead SK. 2010. The proline-rich region of pneumococcal surface proteins A and C contains surface-accessible epitopes common to all pneumococci and elicits antibody-mediated protection against sepsis. Infect Immun 78:2163–2172. doi: 10.1128/IAI.01199-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paton JC, Briles DE. 2003. Streptococcus pneumoniae vaccines, p 294-310. In Ellis RW, Brodeur BR (ed), New bacterial vaccines. Landes Bioscience, Georgetown, TX. [Google Scholar]
- 27.Briles DE, Hollingshead SK, Paton JC, Ades EW, Novak L, van Ginkel FW, Benjamin WH Jr. 2003. Immunizations with pneumococcal surface protein A and pneumolysin are protective against pneumonia in a murine model of pulmonary infection with Streptococcus pneumoniae. J Infect Dis 188:339–348. doi: 10.1086/376571. [DOI] [PubMed] [Google Scholar]
- 28.Briles DE, Ades E, Paton JC, Sampson JS, Carlone GM, Huebner RC, Virolainen A, Swiatlo E, Hollingshead SK. 2000. Intranasal immunization of mice with a mixture of the pneumococcal proteins PsaA and PspA is highly protective against nasopharyngeal carriage of Streptococcus pneumoniae. Infect Immun 68:796–800. doi: 10.1128/IAI.68.2.796-800.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brooks-Walter A, Briles DE, Hollingshead SK. 1999. The pspC gene of Streptococcus pneumoniae encodes a polymorphic protein, PspC, which elicits cross-reactive antibodies to PspA and provides immunity to pneumococcal bacteremia. Infect Immun 67:6533–6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moreno AT, Oliveira ML, Ferreira DM, Ho PL, Darrieux M, Leite LC, Ferreira JM Jr, Pimenta FC, Andrade AL, Miyaji EN. 2010. Immunization of mice with single PspA fragments induces antibodies capable of mediating complement deposition on different pneumococcal strains and cross-protection. Clin Vaccine Immunol 17:439–446. doi: 10.1128/CVI.00430-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sung CK, Li H, Claverys JP, Morrison DA. 2001. An rpsL cassette, Janus, for gene replacement through negative selection in Streptococcus pneumoniae. Appl Environ Microbiol 67:5190–5196. doi: 10.1128/AEM.67.11.5190-5196.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van de Rijn I, Kessler RE. 1980. Growth characteristics of group A streptococci in a new chemically defined medium. Infect Immun 27:444–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hanna SL, Sherman NE, Kinter MT, Goldberg JB. 2000. Comparison of proteins expressed by Pseudomonas aeruginosa strains representing initial and chronic isolates from a cystic fibrosis patient: an analysis by 2-D gel electrophoresis and capillary column liquid chromatography-tandem mass spectrometry. Microbiology 146(Pt 10):2495–2508. [DOI] [PubMed] [Google Scholar]
- 34.Oliveira ML, Miyaji EN, Ferreira DM, Moreno AT, Ferreira PC, Lima FA, Santos FL, Sakauchi MA, Takata CS, Higashi HG, Raw I, Kubrusly FS, Ho PL. 2010. Combination of pneumococcal surface protein A (PspA) with whole cell pertussis vaccine increases protection against pneumococcal challenge in mice. PLoS One 5:e10863. doi: 10.1371/journal.pone.0010863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ferreira DM, Oliveira ML, Moreno AT, Ho PL, Briles DE, Miyaji EN. 2010. Protection against nasal colonization with Streptococcus pneumoniae by parenteral immunization with a DNA vaccine encoding PspA (pneumococcal surface protein A). Microb Pathog 48:205–213. doi: 10.1016/j.micpath.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 36.Sorensen UB, Henrichsen J, Chen HC, Szu SC. 1990. Covalent linkage between the capsular polysaccharide and the cell wall peptidoglycan of Streptococcus pneumoniae revealed by immunochemical methods. Microb Pathog 8:325–334. doi: 10.1016/0882-4010(90)90091-4. [DOI] [PubMed] [Google Scholar]
- 37.Yother J, White JM. 1994. Novel surface attachment mechanism of the Streptococcus pneumoniae protein PspA. J Bacteriol 176:2976–2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ware D, Watt J, Swiatlo E. 2005. Utilization of putrescine by Streptococcus pneumoniae during growth in choline-limited medium. J Microbiol 43:398–405. [PubMed] [Google Scholar]
- 39.Swiatlo E, Champlin FR, Holman SC, Wilson WW, Watt JM. 2002. Contribution of choline-binding proteins to cell surface properties of Streptococcus pneumoniae. Infect Immun 70:412–415. doi: 10.1128/IAI.70.1.412-415.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Malley R, Morse SC, Leite LC, Areas AP, Ho PL, Kubrusly FS, Almeida IC, Anderson P. 2004. Multiserotype protection of mice against pneumococcal colonization of the nasopharynx and middle ear by killed nonencapsulated cells given intranasally with a nontoxic adjuvant. Infect Immun 72:4290–4292. doi: 10.1128/IAI.72.7.4290-4292.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Malley R, Lipsitch M, Stack A, Saladino R, Fleisher G, Pelton S, Thompson C, Briles D, Anderson P. 2001. Intranasal immunization with killed unencapsulated whole cells prevents colonization and invasive disease by capsulated pneumococci. Infect Immun 69:4870–4873. doi: 10.1128/IAI.69.8.4870-4873.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]