Abstract
Background
Microalgal oil is a promising alternative feedstock for biodiesel fuel (BDF). Mixotrophic cultivation with glycerol, the primary byproduct of BDF production, may be used to optimize BDF production. This strategy would reduce costs through glycerol recycling and improve lipid productivity and biomass productivity by overcoming the growth retardation caused by decreased light penetration in high-density culture.
Results
Overexpression of the endogenous glycerol kinase (GK) gene in an oleaginous marine diatom, Fistulifera solaris JPCC DA0580, accelerates glycerol metabolism and improves lipid and biomass productivities. Two candidates were selected from a collection of 90 G418-resistant clones, based on growth and confirmation of genome integration. GK gene expression was higher in the selected clones (GK1_7 and GK2_16) than in the wild-type culture. The GK2_16 clone achieved a 12% increase in lipid productivity.
Conclusion
We have demonstrated the potential of metabolic engineering in oleaginous microalgae to improve lipid productivity. Metabolic engineering techniques can be used to optimize BDF production.
Electronic supplementary material
The online version of this article (doi:10.1186/s13068-014-0184-9) contains supplementary material, which is available to authorized users.
Keywords: Oleaginous microalgae, Fistulifera solaris, Mixotrophic cultivation, Glycerol kinase, Biodiesel fuel, Metabolic engineering
Introduction
The global energy crisis and climate change have aroused considerable interest in alternative fuels. Microalgae are widely accepted as one of the most promising biodiesel fuel (BDF) feedstocks because of their high photoautotrophic biomass productivity and neutral lipid content [1,2]. Various oleaginous microalgal strains, including Nannochloropsis sp., Botryococcus braunii, and Chlorella sorokiniana, are promising candidates for BDF production [2-4]. A transformable [5] oleaginous marine pennate diatom, Fistulifera solaris JPCC DA0580, was the strongest producer of triacylglycerol (TAG) in a collection of 1,393 strains of domestic marine microalgae [6,7]. The lipid content of F. solaris is comparable to that of Nannochloropsis sp. [2,8]. The fatty acid composition of F. solaris is also similar to that of Nannochloropsis sp., with high lipid quality for BDF production, as demonstrated by cetane number, kinematic viscosity, iodine value, and oxidative stability [9]. A high density cultivation technique and outdoor cultivation have been used for F. solaris [8,10]. The whole genome of F. solaris has been sequenced, and the chloroplast genome has been published [11]. This knowledge has empowered further study to enhance BDF productivity and quality in F. solaris. However, industrial application of microalgal BDF is hampered by a combination of high cost and low energy yield. One strategy to overcome these problems is to utilize the byproducts of BDF production.
Recycling of glycerol, the major byproduct of BDF production, may help reduce the cost of BDF [12,13]. Indeed, production of BDF from 1 kg of microalgal TAG releases approximately 0.1 kg of crude glycerol [12]; such yields could decrease the global price of glycerol. Mixotrophic cultivation of various microalgae enhances biomass productivity as well as the production of corresponding valuable bioproducts such as eicosapentaenoic acid (EPA) [14], lipids for BDF [4,15], and pigments [14,16]. Overexpression of glycerol kinase (GK) in the transformed yeast Saccharomyces cerevisiae improves lipid productivity [17], since the first reaction of glycerol metabolism occurs when GK converts glycerol to glycerol-3-phosphate (G3P), a precursor for TAG biosynthesis through the Kennedy pathway [18]. Overexpression of the GK gene leads to accumulation of intracellular G3P, which might then stimulate TAG synthesis and accumulation. Genetic engineering of GK to improve glycerol metabolism and lipid productivity has not been performed in microalgae.
Metabolic engineering to enhance microalgal BDF production has only been achieved in model microalgae such as the green alga Chlamydomonas reinhardtii [19] and the diatoms Thalassiosira pseudonana [20] and Phaeodactylum tricornutum [21,22]. However, C. reinhardtii, T. pseudonana, and P. tricornutum are not ideal for BDF production because their lipid content is moderate [23]. In the oleaginous microalgae, stable transformation has only been established for Nannochloropsis sp. [2,24] and F. solaris [5], neither of which has been metabolically engineered.
In this study, F. solaris was transformed with a vector containing the endogenous GK gene. Glycerol utility, biomass productivity, and lipid productivity were higher than in the wild-type strain. Overexpression of the GK gene in the marine diatom F. solaris may have great utility in commercial BDF production.
Results and discussion
Identification of glycerol kinase genes in Fistulifera solaris
Two endogenous GK genes in F. solaris, designated GK1 [DDBJ/EMBL/GenBank:LC009547 (Gene ID: g11728)] and GK2 [DDBJ/EMBL/GenBank:LC009548 (Gene ID: g14921)], were identified in the draft genome sequence. The nucleotide and amino acid sequences of GK1 and GK2 exhibited 92% nucleotide sequence similarity and 93% amino acid sequence similarity. Both genes are identical in length (1827 bp) and are relatively longer than the glycerol kinase genes (1728 bp and 1737 bp) previously identified in the model diatom Thalassiosira pseudonana [GenBank IDs: XP_002296792 and XP_002295903]. GK1 in F. solaris shares 58% sequence identity and GK2 shares 57% sequence identity with the GK [XP_002296792] of T. pseudonana. The amino acid sequences of GK1 and GK2 in F. solaris and the GKs in T. pseudonana contain an FGGY carbohydrate kinase domain, characteristic of glycerol kinases [25].
Selection of candidate clones based on cell growth
After transformation with pSP-GK1 and pSP-GK2, 48 GK1 clones (GK1_1 to GK1_48) and 42 GK2 clones (GK2_1 to GK2_42) were obtained from an agar plate containing the antibiotic reagent G418. These clones were cultured in 96-well microtiter plates, and the final cell density was evaluated by determining the optical density to estimate the effect of genetic transformation on cell growth. The final cell densities of the transformants varied from 1% to 102% of the mean density of the control strains (n = 5), which were transformed with the empty pSP-NPT/H4 vector (Figure 1). The variation of transformant cell density may be derived from different loci and copy numbers of the integrated transgenic fragments [5,26,27]. The final cell densities of 10 pSP-GK1 and pSP-GK2 transformants, including GK2_16, GK2_40, GK2_41, GK1_7, GK2_24, GK2_38, GK2_39, GK1_16, GK1_24, and GK1_14 (Figure 1, black columns), were not significantly varied as compared with the mean ± SD (OD = 0.157 ± 0.015) of the control strains (n = 5), and were selected for subsequent experiments.
Figure 1.
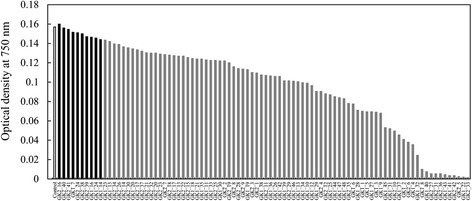
Final cell densities of the transformants (optical density at 750 nm). The glycerol kinase (GK) transformants were generated with pSP-GK1/fcpB or pSP-GK2/fcpB. The white bar indicates the cell density of the negative control transformed with pSP-NPT/H4 (n = 5). The black bar indicates 10 clones with cell densities similar to that of the negative control.
Confirmation of the integrated GK genes from the transformants
The presence of the integrated genes in the 10 selected clones was verified by PCR (Figure 2). A 795-bp fragment encoding the nptII gene region was amplified from the genomic DNA of G418-resistant clones. All 10 clones, as well as the negative control with the empty pSP-NPT/H4 vector, showed the presence of the nptII gene, while no amplified product was obtained from the wild type. On the other hand, the combined fragment of the nptII gene and the GK1 or GK2 gene region (2622 bp) was amplified from only three of the clones (GK1_7, GK2_16, and GK2_39). The wild type and the negative control showed no nptII-GK product. These results suggest that partial gene integration of the vector may occur during transformation in F. solaris. The GK2_39 clone showed a decrease in growth rate (Additional file 1) and was eliminated from subsequent experiments.
Figure 2.
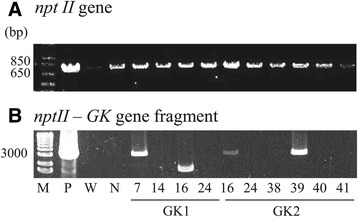
Heterologous gene detection in Fistulifera solaris transformed with pSP-GK1/fcpB or pSP-GK2/fcpB. The nptII (A) and nptII-glycerol kinase (GK) gene fragments (B) were PCR-amplified. M, 1-kb marker; P, pSP-GK1/fcpB as the positive control; W, wild type; N, negative control transformed with pSP-NPT/H4; GK1, transformed with pSP-GK1/fcpB; GK2, transformed with pSP-GK2/fcpB.
GK expression levels in GK1_7 and GK2_16 clones were examined by quantitative reverse transcription PCR (qRT-PCR), with housekeeping genes ribosomal protein subunit (rps) and ubiquitin (Ub) as controls (Figure 3). The rps and Ub genes showed similar expression levels in the transformants and wild-type strains. In contrast, large differences were observed in GK1 and GK2 gene expression. GK1 expression in the GK1_7 clone increased by 4.3-fold in comparison to that observed in the wild type (Figure 3A) and GK2 expression in the GK2_16 clone increased by 3.4-fold (Figure 3B). These results indicate that GK gene expression was enhanced in the GK1_7 and GK2_16 transformants. Moreover, expression of the GK2 gene in the GK1_7 clone and of the GK1 gene in the GK2_16 clone changed negligibly, suggesting that specific overexpression of GK1 and GK2 had been achieved. However, based on the comparison of the cycle threshold (Ct) values, the expression levels of GK (Ct = 22.5) seemed to be lower than those of the two housekeeping genes rps (Ct = 20.6) and Ub (Ct = 21.6). Further enhancement of the GK gene expression would be attained in the future through the optimization of the promoter region.
Figure 3.
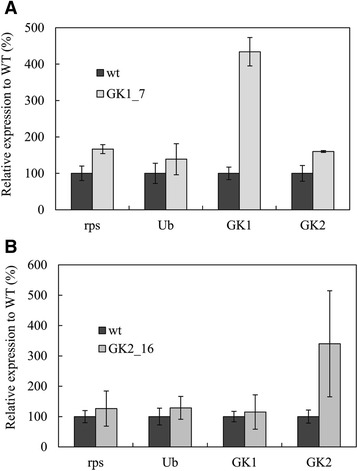
Relative mRNA abundance in the wild-type and glycerol kinase ( GK ) gene transformants. Gene expression was evaluated in the GK1_7 (A) and GK2_16 (B) transformants, with housekeeping genes rps and Ub as controls.
Evaluation of lipid productivity in transformants
Prior to exploring glycerol utilization by transformants, we investigated the optimum glycerol concentration for mixotrophic growth in wild-type F. solaris (Additional file 2). The highest cell density was obtained when the initial glycerol concentration was 50 mM, similar to reports in other microalgae such as the marine diatom P. tricornutum (100 mM) and the green alga Chlorella vulgaris (110 mM) [14,15]. Growth was inhibited at more than 250 mM glycerol, perhaps because of the increasing viscosity of the culture media. Based on these results, we chose to use 50 mM glycerol for mixotrophic growth.
To investigate lipid productivity, the final cell concentration, dry cell weight, lipid content, and lipid yield were evaluated in the transformants and wild-type strains. Table 1 summarizes the data for transformants and wild-type cultures in the absence of glycerol. The GK1_7 clones had a significantly lower lipid content in comparison to the wild type and GK2_16, leading to a slightly lower lipid yield in GK1_7, although the final cell concentration and dry cell weight of this strain were increased. In comparison to the wild type, GK2_16 showed a slightly increased dry cell weight and lipid content, thus increasing the final lipid yield. The GK2_16 results suggest that, even under photoautotrophic culture conditions, GK overexpression may also affect glycerol utilization and result in variable cell growth and lipid production.
Table 1.
Biomass and lipid productivities in transformants cultivated in f/2 medium
| Clone | Final cell density | Dry cell weight | Lipid content | Lipid yield |
|---|---|---|---|---|
| (× 10 7 cells/mL) | (mg/L) | (% dry w/w) | (mg/L) | |
| Wild type | 1.03 ± 0.04 | 407 ± 12 | 36.2 ± 0.8 | 148 ± 7 |
| GK1_7 | 1.12 ± 0.03 | 426 ± 11 | 31.7 ± 3.5 | 135 ± 15 |
| GK2_16 | 1.03 ± 0.08 | 426 ± 49 | 40.9 ± 1.3 | 152 ± 13 |
Table 2 summarizes the data of the wild type and the transformants cultured with 50 mM glycerol. The increases in final cell concentration, dry cell weight, lipid content, and lipid yield were compared in wild-type cultures with and without glycerol (Table 1). This result suggests that it is possible to improve BDF production by mixotrophic cultivation of F. solaris. As expected, increases in final cell concentration (GK1_7: 108%, GK2_16: 116%) and dry cell weight (GK1_7: 103%, GK2_16: 110%) were observed in both transformants; however, in comparison to GK1_7 cultured without glycerol (Table 1), the addition of glycerol did not significantly affect all tested parameters, suggesting that the GK1 gene may not be fully functional. The GK1 gene in GK1_7 may be integrated into unexpected loci in the genome, thus affecting the lipid content in this strain. On the other hand, all of the tested parameters significantly increased in the GK2_16 clone versus the same clone cultured without glycerol, and a 112% increase in lipid yield was observed in comparison to that observed in the wild type. These results suggest that GK2 is fully functional in this transformant. Remaining glycerol concentrations in the filtered culture media of the GK2_16 clone were measured by HPLC. Glycerol consumption was markedly increased to 2.4 mM (221.0 mg/L) in the GK2_16 clone in comparison to the wild type (1.7 mM, 156.6 mg/L) cultivated in f/2 medium containing 7.5 mM glycerol. Overexpression of the GK2 gene resulted in a 1.4-fold increase in glycerol utilization, which is the most likely explanation for the increase in biomass and lipid productivity.
Table 2.
Biomass and lipid productivities in transformants cultivated with 50 mM glycerol
| Clone | Final cell density | Dry cell weight | Lipid content | Lipid yield |
|---|---|---|---|---|
| (× 10 7 cells/mL) | (mg/L) | (% dry w/w) | (mg/L) | |
| Wild type | 1.06 ± 0.03 | 432 ± 12 | 38.6 ± 2.8 | 167 ± 12 |
| GK1_7 | 1.14 ± 0.07 | 443 ± 30 | 30.3 ± 0.6 | 134 ± 6 |
| GK2_16 | 1.23 ± 0.10 | 477 ± 56 | 39.6 ± 4.3 | 187 ± 11 |
Glycerol can be utilized as the sole carbon source in various pathways including glucose metabolism and CO2 fixation. A GK mutant of Arabidopsis which was not able to utilize glycerol showed no variance in lipid content [28]. This study suggested that GK in essential for glycerol metabolism; however, glycerol metabolism is not directly linked to lipid synthesis. On the other hand, by increasing the G3P level through the enhancement of GK activity by metabolic engineering, TAG accumulation was reported to be induced in higher plants and yeast [17,29]. Thus, we propose to overexpress GK to clarify whether the glycerol metabolism in F. solaris is linked with TAG synthesis.
Conclusion
The lipid productivity of microalgae closely depends on the basic physical laws of photosynthesis. Mixotrophic cultivation could overcome the mutual shading of cells under high cell density photoautotrophic cultivation. In other words, mixotrophic cultivation may reduce the dependence of cell growth on the supply of sunlight for photosynthesis in large-scale cultivation. By enhancing organic carbon assimilation through metabolic engineering, the oleaginous microalgae may become a reliable, renewable feedstock for BDF production.
In this study, the GK gene was overexpressed in the oleaginous microalga F. solaris to accelerate glycerol metabolism. The transformant GK2_16 showed a 12% increase in lipid productivity. To our knowledge, this is the first report of metabolic engineering in oleaginous microalgae that has focused on enhanced lipid productivity. Our results and observations will promote the development of technologies to improve lipid productivity through glycerol recycling. The establishment of metabolic engineering techniques for the oleaginous microalgae has great potential to optimize BDF production. Through further metabolic engineering of this oleaginous microalga, improved BDF quality and productivity can be expected.
Materials and methods
Strain and culture conditions
The marine pennate diatom F. solaris JPCC DA0580 was isolated from the mouths of the Sumiyo and Yakugachi Rivers in Amami-Ohshima, Kagoshima, Japan [7,30]. The cells were cultured as previously reported [5]. Briefly, pre-cultivated cells containing approximately 40% lipid component were inoculated in half-strength Guillard’s ”f” medium (f/2) [31] at an initial density of 1 × 105 cells/mL. The cells were grown at 25°C under continuous, cool-white fluorescent lights (140 μmol/m2/s). The cell density was estimated using a hemocytometer.
For the purpose of positive transformant selection, G418-resistant colonies were cultured with f/2 liquid medium containing 500 μg/mL G418 (Roche Applied Science, Indianapolis, IN), an aminoglycoside antibiotic, in a 96-well plate (Sumilon, Tokyo, Japan) at 25°C under constant illumination of 50 μmol/m2/s. After three weeks, 20 μL of the G418-resistant clone cultures were transferred to a fresh 96-well plate and incubated for another six days. In order to determine the cell density, the optical density (at 750 nm) of each well was measured in a microplate reader (SH-9000, Corona Electric, Ibaraki, Japan). Furthermore, to evaluate the effect of glycerol on cell growth, wild-type and transformant strains were cultured in the presence of 10 mM to 500 mM glycerol.
Plasmid construction
A homology search, using BLASTX, was performed with reference to the 19,859 genes from the draft genome sequence of F. solaris to obtain the GK-coding genes [11] (the whole genome is undergoing preparation for publication). The pSP-NPT/H4 vector [5] was used to construct the expression vector for the two predicted GK genes. The nucleotide sequence comprising a promoter region (500 bp) from a fucoxanthin chlorophyll a/c-binding protein B (fcpB) gene in F. solaris and a terminator region (227 bp) from a fucoxanthin chlorophyll a/c-binding protein A (fcpA) gene [27] in P. tricornutum was synthesized (Integrated DNA Technologies, Inc., Coralville, IA, USA) and cloned into the SalI-SphI site of pSP-NPT/H4. The GK1 and GK2 gene fragments, with restriction sites (EcoRI and PstI), were PCR-amplified from F. solaris genomic DNA. These fragments were then inserted into the EcoRI-PstI site within the expression vector pSP-NPT/H4 to construct pSP-GK1 and pSP-GK2 (Additional file 3). Transformants carrying the empty pSP-NPT/H4 vector served as the negative control.
Transformation
The transformation of F. solaris was performed by microparticle bombardment, as described (Biolistic PDS-1000/He Particle Delivery System; Bio-Rad Laboratories, Inc., Hercules, CA) [5]. Tungsten particles (3 mg; 0.6 μm in diameter; Kojundo Chemical Laboratory, Saitama, Japan) were coated with pSP-GK1 or pSP-GK2 (5 μg), according to the manufacturer’s protocols. Microalgal cells were harvested at mid-log phase. Approximately 5 × 107 cells in the agar plate were bombarded with 600 ng plasmid DNA by using an 1,100-psi rupture disk under a negative pressure of 28 in of Hg. After bombardment, the cells were incubated on plates at 25°C under constant illumination (140 μmol/m2/s1) for 24 h and then resuspended in 1 mL f/2 medium. Then, approximately 107 cells were inoculated at 25°C under constant illumination on a 1% f/2 plate containing 500 μg/mL G418. G418-resistant colonies were obtained after three weeks.
PCR and qRT-PCR
Successful integration of the target gene region, including the promoter, GK gene, and terminator, was confirmed by PCR. Selected G418-resistant clones (approximately 4 × 106 cells) were used for direct PCR. The full-length nptII gene (795 bp), GK genes (1.9 kb), and nptII-GK gene fragment (3.2 kb) were PCR-amplified with specific primer sets (Additional file 4) and TaKaRa Taq polymerase (TaKaRa Bio, Otsu, Japan) under the following cycling conditions: 94°C for 1 min, (94°C for 30 s, 60°C for 30 s, and 72°C for 2 min) × 30 cycles, and 72°C for 5 min. Amplification products were analyzed by electrophoresis.
qRT-PCR was performed to measure GK expression in selected transformants. Total RNA was isolated with Plant RNA Isolation Reagent (Invitrogen Corporation) according to manufacturer instructions. The extracts were further purified with the RNeasy Mini kit (Qiagen, Valencia, CA). After purification, the total RNA solution was treated with DNase I (TaKaRa). The cDNA was synthesized by using a PrimeScript II First Strand cDNA Synthesis Kit (TaKaRa) and 1 μg total RNA. The cDNAs were PCR-amplified to determine the transcript levels of GK1 and GK2. The housekeeping genes rps and Ub were used as controls. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used for standardization between samples [32]. Amplified fragments were purified with a QIAquick PCR Purification Kit (Qiagen). After DNA concentrations were measured by e-spect UV-Vis spectrometry (Malcom, Tokyo, Japan), the purified DNA fragments were diluted and used to obtain standard curves. Individual qRT-PCR reactions contained 10 μL Fast SYBR Green Master Mix (Applied Biosystems), 2 μL of 1 ng/μL cDNA, and 1 μL of 10 mM forward and reverse primers. Two-step qRT-PCR amplification (40 cycles of 95°C for 3 s, followed by 60°C for 30 s) was performed on a ViiA 7 Real-Time PCR System (Life Technologies). Based on the standard curves, the quantities of GK1, GK2, rps, and Ub transcripts were calculated relative to the wild-type sample. The data represented an average of three independent biological samples.
Cell lipid content and glycerol concentration in the medium
Lipids were extracted from lyophilized microalgal cells (about 50 mg) with 10 mL n-hexane as described [30]. The crude extracts were dried under argon gas, and the ratio of dried lipid weight to dried cell weight was defined as the lipid content.
To determine glycerol consumption in the wild-type and transformant strains, cells were cultured in f/2 medium containing 7.5 mM glycerol. This low concentration enabled us to detect significant variations in glycerol consumption. The glycerol concentration of the filtered culture supernatants was measured on a Shimadzu Prominence HPLC system (Shimadzu Scientific Instruments, Kyoto, Japan) equipped with an Aminex HPX-87H+ column (300 mm × 7.8 mm, Bio-Rad Laboratories). The column was maintained at 50°C. Glycerol was eluted with 25 mM NH4HCO2 at 1.0 mL/min and detected with an Evaporative Light Scattering Detector System (Shimadzu).
Acknowledgments
This study was supported by JST-CREST.
Abbreviations
- BDF
biodiesel fuel
- BLAST
Basic Local Alignment Search Tool
- bp
base pairs
- Ct
cycle threshold
- fcpA
fucoxanthin chlorophyll a/c-binding protein A
- fcpB
Fucoxanthin chlorophyll a/c-binding protein B
- G3P
glycerol-3-phosphate
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GK
glycerol kinase
- HPLC
high performance liquid chromatography
- PCR
polymerase chain reaction
- qRT-PCR
quantitative reverse transcription PCR
- rps
ribosomal protein subunit
- TAG
triacylglycerol
- Ub
ubiquitin
Additional files
Growth curves of the wild-type (solid triangles) and glycerol kinase ( GK )-expressing lines GK1_7 (open circles), GK2_16 (large open squares), and GK2_39 (small open diamonds) during flat flask cultivation in f/2 medium.
Final cell density of the wild-type (gray bar) and glycerol kinase ( GK ) gene transformant (GK2_16; white bar) in the presence of glycerol.
Restriction map of the pSP-GK/fcpB (pSP-GK1/fcpB, pSP-GK2/fcpB) vector for GK overexpression. Abbreviations: H4 promoter, histone H4 gene promoter from Fistulifera solaris; nptII gene, neomycin phosphotransferase gene; Terminator, fucoxanthin chlorophyll a/c-binding protein A gene terminator from Phaeodactylum tricornutum; fcpB promoter, fucoxanthin chlorophyll a/c-binding protein B gene promoter from F. solaris; glycerol kinase gene, glycerol kinase gene (g10050, g13546) from F. solaris.
Primers used for quantitative qRT-PCR and PCR to obtain the glycerol kinase ( GK ) overexpression clone.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
MM, MT, and TT designed the experiments. MMuto and MM conducted the experiments. MMuto, MT, YL, and TY performed data analysis. MMuto, MT, YL, TY, and TT participated in the experimental design and wrote the manuscript. All the authors have approved the manuscript and agree with submission to your esteemed journal.
Contributor Information
Masaki Muto, Email: m_muto@cc.tuat.ac.jp.
Masayoshi Tanaka, Email: m_tanaka@chemeng.titech.ac.jp.
Yue Liang, Email: y_liang@cc.tuat.ac.jp.
Tomoko Yoshino, Email: y-tomoko@cc.tuat.ac.jp.
Mitsufumi Matsumoto, Email: Mitsufumi_Matsumoto@jpower.co.jp.
Tsuyoshi Tanaka, Email: tsuyo@cc.tuat.ac.jp.
References
- 1.Georgianna DR, Mayfield SP. Exploiting diversity and synthetic biology for the production of algal biofuels. Nature. 2012;488:329–335. doi: 10.1038/nature11479. [DOI] [PubMed] [Google Scholar]
- 2.Radakovits R, Jinkerson RE, Fuerstenberg SI, Tae H, Settlage RE, Boore JL, Posewitz MC. Draft genome sequence and genetic transformation of the oleaginous alga Nannochloropsis gaditana. Nat Commun. 2012;3:686. doi: 10.1038/ncomms1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banerjee A, Sharma R, Chisti Y, Banerjee UC. Botryococcus braunii: a renewable source of hydrocarbons and other chemicals. Crit Rev Biotechnol. 2002;22:245–279. doi: 10.1080/07388550290789513. [DOI] [PubMed] [Google Scholar]
- 4.Wan MX, Liu P, Xia JL, Rosenberg JN, Oyler GA, Betenbaugh MJ, Nie ZY, Qiu GZ. The effect of mixotrophy on microalgal growth, lipid content, and expression levels of three pathway genes in Chlorella sorokiniana. Appl Microbiol Biotechnol. 2011;91:835–844. doi: 10.1007/s00253-011-3399-8. [DOI] [PubMed] [Google Scholar]
- 5.Muto M, Fukuda Y, Nemoto M, Yoshino T, Matsunaga T, Tanaka T. Establishment of a genetic transformation system for the marine pennate diatom Fistulifera sp. strain JPCC DA0580 - a high triglyceride producer. Mar Biotechnol. 2013;15:48–55. doi: 10.1007/s10126-012-9457-0. [DOI] [PubMed] [Google Scholar]
- 6.Matsumoto M, Tanaka T, Matsunaga T. Performance of marine diatom Navicula sp. JPCC DA0580 as high lipids producer for biofuel production. J Biosci Bioeng. 2009;108:S42-S42. [Google Scholar]
- 7.Matsumoto M, Mayama S, Nemoto M, Fukuda Y, Muto M, Yoshino T, et al. Morphological and molecular phylogenetic analysis of the high triglyceride-producing marine diatom, Fistulifera solaris sp. nov. (Bacillariophyceae). Phycol Res 2014;62:257–68.
- 8.Satoh A, Ichii K, Matsumoto M, Kubota C, Nemoto M, Tanaka M, Yoshino T, Matsunaga T, Tanaka T. A process design and productivity evaluation for oil production by indoor mass cultivation of a marine diatom, Fistulifera sp. JPCC DA0580. Bioresource Technol. 2013;137:132–138. doi: 10.1016/j.biortech.2013.03.087. [DOI] [PubMed] [Google Scholar]
- 9.Liang Y, Maeda Y, Yoshino T, Matsumoto M, Tanaka T. Profiling of fatty acid methyl esters from the oleaginous diatom Fistulifera sp. strain JPCC DA0580 under nutrition-sufficient and -deficient conditions. J Appl Phychol. 2014;26:2295–2302. doi: 10.1007/s10811-014-0265-y. [DOI] [Google Scholar]
- 10.Sato R, Maeda Y, Yoshino T, Tanaka T, Matsumoto M. Seasonal variation of biomass and oil production of the oleaginous diatom Fistulifera sp. in outdoor vertical bubble column and raceway-type bioreactors. J Biosci Bioeng. 2014;114:720–724. doi: 10.1016/j.jbiosc.2013.11.017. [DOI] [PubMed] [Google Scholar]
- 11.Tanaka T, Fukuda Y, Yoshino T, Maeda Y, Muto M, Matsumoto M, Mayama S, Matsunaga T. High-throughput pyrosequencing of the chloroplast genome of a highly neutral-lipid-producing marine pennate diatom, Fistulifera sp. strain JPCC DA0580. Photosynth Res. 2011;109:223–229. doi: 10.1007/s11120-011-9622-8. [DOI] [PubMed] [Google Scholar]
- 12.Thompson JC, He BB. Characterization of crude glycerol from biodiesel production from multiple feedstocks. Appl Eng Agric. 2006;22:261–265. doi: 10.13031/2013.20272. [DOI] [Google Scholar]
- 13.Johnson DT, Taconi KA. The glycerin glut: options for the value-added conversion of crude glycerol resulting from biodiesel production. Environ Prog. 2007;26:338–348. doi: 10.1002/ep.10225. [DOI] [Google Scholar]
- 14.Ceron Garcia MC, Camacho FG, Miron AS, Sevilla JMF, Chisti Y, Grima EM. Mixotrophic production of marine microalga Phaeodactylum tricornutum on various carbon sources. J Microbiol Biotechn. 2006;16:689–694. [Google Scholar]
- 15.Liang YN, Sarkany N, Cui Y. Biomass and lipid productivities of Chlorella vulgaris under autotrophic, heterotrophic and mixotrophic growth conditions. Biotechnol Lett. 2009;31:1043–1049. doi: 10.1007/s10529-009-9975-7. [DOI] [PubMed] [Google Scholar]
- 16.Kong WB, Song H, Cao YT, Yang H, Hua SF, Xia CG. The characteristics of biomass production, lipid accumulation and chlorophyll biosynthesis of Chlorella vulgaris under mixotrophic cultivation. Afr J Biotechnol. 2011;10:11620–11630. [Google Scholar]
- 17.Yu KO, Jung J, Ramzi AB, Choe SH, Kim SW, Park C, Han SO. Development of a Saccharomyces cerevisiae strain for increasing the accumulation of triacylglycerol as a microbial oil feedstock for biodiesel production using glycerol as a substrate. Biotechnol Bioeng. 2013;110:343–347. doi: 10.1002/bit.24623. [DOI] [PubMed] [Google Scholar]
- 18.Griffiths G, Stobart AK, Stymne S. The acylation of sn-glycerol 3-phosphate and the metabolism of phosphatidate in microsomal preparations from the developing cotyledons of safflower (Carthamus tinctorius L.) seed. Biochem J. 1985;230:379–388. doi: 10.1042/bj2300379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mussgnug JH, Thomas-Hall S, Rupprecht J, Foo A, Klassen V, McDowall A, Schenk PM, Kruse O, Hankamer B. Engineering photosynthetic light capture: impacts on improved solar energy to biomass conversion. Plant Biotechnol J. 2007;5:802–814. doi: 10.1111/j.1467-7652.2007.00285.x. [DOI] [PubMed] [Google Scholar]
- 20.Trentacoste EM, Shrestha RP, Smith SR, Gle C, Hartmann AC, Hildebrand M, Gerwick WH. Metabolic engineering of lipid catabolism increases microalgal lipid accumulation without compromising growth. Proc Natl Acad Sci U S A. 2013;110:19748–19753. doi: 10.1073/pnas.1309299110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Radakovits R, Eduafo PM, Posewitz MC. Genetic engineering of fatty acid chain length in Phaeodactylum tricornutum. Metab Eng. 2011;13:89–95. doi: 10.1016/j.ymben.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 22.Zaslavskaia LA, Lippmeier JC, Shih C, Ehrhardt D, Grossman AR, Apt KE. Trophic obligate conversion of an photoautotrophic organism through metabolic engineering. Science. 2001;292:2073–2075. doi: 10.1126/science.160015. [DOI] [PubMed] [Google Scholar]
- 23.Demirbas A, Demirbas MF. Importance of algae oil as a source of biodiesel. Energy Convers Manage. 2011;52:163–170. doi: 10.1016/j.enconman.2010.06.055. [DOI] [Google Scholar]
- 24.Kilian O, Benemann CSE, Niyogi KK, Vick B. High-efficiency homologous recombination in the oil-producing alga Nannochloropsis sp. Proc Natl Acad Sci U S A. 2011;108:21265–21269. doi: 10.1073/pnas.1105861108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Armbrust EV, Berges JA, Bowler C, Green BR, Martinez D, Putnam NH, Zhou SG, Allen AE, Apt KE, Bechner M, Brzezinski MA, Chaal BK, Chiovitti A, Davis AK, Demarest MS, Detter JC, Glavina T, Goodstein D, Hadi MZ, Hellsten U, Hildebrand M, Jenkins BD, Jurka J, Kapitonov VV, Kröger N, Lau WW, Lane TW, Larimer FW, Lippmeier JC, Lucas S, et al. The genome of the diatom Thalassiosira pseudonana: Ecology, evolution, and metabolism. Science. 2004;306:79–86. doi: 10.1126/science.1101156. [DOI] [PubMed] [Google Scholar]
- 26.Li YT, Han DX, Hu GR, Sommerfeld M, Hu QA. Inhibition of starch synthesis results in overproduction of lipids in Chlamydomonas reinhardtii. Biotechnol Bioeng. 2010;107:258–268. doi: 10.1002/bit.22807. [DOI] [PubMed] [Google Scholar]
- 27.Apt KE, KrothPancic PG, Grossman AR. Stable nuclear transformation of the diatom Phaeodactylum tricornutum. Mol Gen Genet. 1996;252:572–579. doi: 10.1007/BF02172403. [DOI] [PubMed] [Google Scholar]
- 28.Eastmond PJ. Glycerol-insensitive Arabidopsis mutants: gli1 seedlings lack glycerol kinase, accumulate glycerol and are more resistant to abiotic stress. Plant J. 2004;37:617–625. doi: 10.1111/j.1365-313X.2003.01989.x. [DOI] [PubMed] [Google Scholar]
- 29.Vigeolas H, Geigenberger P. Increased levels of glycerol-3-phosphate lead to a stimulation of flux into triacylglycerol synthesis after supplying glycerol to developing seeds of Brassica napus L. in planta. Planta. 2004;219:827–835. doi: 10.1007/s00425-004-1273-y. [DOI] [PubMed] [Google Scholar]
- 30.Matsumoto M, Sugiyama H, Maeda Y, Sato R, Tanaka T, Matsunaga T. Marine diatom, Navicula sp. strain JPCC DA0580 and marine green alga, Chlorella sp. strain NKG400014 as potential sources for biodiesel production. Appl Biochem Biotech. 2010;161:483–490. doi: 10.1007/s12010-009-8766-x. [DOI] [PubMed] [Google Scholar]
- 31.Guillard RRL, Ryther JH. Studies of marine planktonic diatoms: I. Cyclotella nana Hustedt, and Detonula confervacea (Cleve) Gran. Can J Microbiol. 1962;8:229–239. doi: 10.1139/m62-029. [DOI] [PubMed] [Google Scholar]
- 32.Couso I, Vila M, Rodriguez H, Vargas MA, Leon R. Overexpression of an exogenous phytoene synthase gene in the unicellular alga Chlamydomonas reinhardtii leads to an increase in the content of carotenoids. Biotechnol Progr. 2011;27:54–60. doi: 10.1002/btpr.527. [DOI] [PubMed] [Google Scholar]


