Abstract
Applications of microarray technologies to mouse embryology/genetics have been limited, due to the nonavailability of microarrays containing large numbers of embryonic genes and the gap between microgram quantities of RNA required by typical microarray methods and the miniscule amounts of tissue available to researchers. To overcome these problems, we have developed a microarray platform containing in situ-synthesized 60-mer oligonucleotide probes representing approximately 22,000 unique mouse transcripts, assembled primarily from sequences of stem cell and embryo cDNA libraries. We have optimized RNA labeling protocols and experimental designs to use as little as 2 ng total RNA reliably and reproducibly. At least 98% of the probes contained in the microarray correspond to clones in our publicly available collections, making cDNAs readily available for further experimentation on genes of interest. These characteristics, combined with the ability to profile very small samples, make this system a resource for stem cell and embryogenomics research.
[Supplemental material is available online at www.genome.org and at the NIA Mouse cDNA Project Web site, http://lgsun.grc.nia.nih.gov/cDNA/cDNA.html.]
In the past few years, the technology available for microarray-based expression profiling platforms has changed dramatically, from the mechanically deposited cDNA (Schena et al. 1995) and photolithographic short oligo-based (Pease et al. 1994; Lipshutz et al. 1999) systems reported in the early 1990s, to flexible, automated oligo-based systems that only require information as input (Singh-Gasson et al. 1999; Hughes et al. 2001). The newer microarray technologies offer rapid, easy creation of microarrays tailored to specific needs and areas of study.
Although eliminating the need for purified cDNAs makes microarray design and construction faster, more flexible, and more accessible to researchers, these new technologies also present a potential problem for some: Downstream validation of microarray results and characterization of differential transcripts requires a corresponding collection of cDNA clones. This is particularly problematic for novel and/or uncharacterized transcripts from specialized cDNA clone collections, which may not be easily obtainable or publicly accessible.
Gene content and cDNA clone availability requirements are especially exigent to satisfy the growing interest in expression profiling of both stem cell populations (Phillips et al. 2000; Billia et al. 2001; Terskikh et al. 2001; Steidl et al. 2002; Testa et al. 2002) and embryos in early developmental stages (Ko et al. 2000; Lee et al. 2000; Tanaka et al. 2000; Hwang et al. 2001; Stanton and Green 2002). As a step toward relevant gene content for microarray platforms, we described a sequence-verified mouse cDNA clone set representing up to 15,000 unique transcripts (Kargul et al. 2001) derived primarily from preimplantation embryos. This clone set was assembled into a cDNA microarray system that is adapted to the study of early differentiation events (Tanaka et al. 2000). Since the publication of the National Institute on Aging (NIA) 15K mouse cDNA clone set, we have added to our collections many new cDNA libraries made from a variety of newborn tissues, cultured stem cell lines, and purified stem cells. These new libraries have added at least 7400 additional unique transcripts (the NIA 7.4K mouse cDNA clone set), approximately 4000 of which are without high similarity to sequences in GenBank (VanBuren et al. 2002). The expanded library set can support a microarray/clone set combination for studying stem cells, early development, and the connections between them.
Here, we have merged the publicly accessible NIA 15K and 7.4K cDNA clone set sequences and designed an in situ-synthesized 60-mer oligonucleotide probe microarray system, tailored to the expression profiling of early developmental and stem cell tissues, and manufactured using Agilent Technologies' ink-jet based process (Hughes et al. 2001). We have further adapted the system to such studies by developing and verifying labeling protocols for very small tissue samples. We show that the platform gives reproducible, sensitive results, even with low sample inputs, so that it can be used to identify target transcripts with roles in early development, pluripotentiality, and aging-related conditions.
RESULTS AND DISCUSSION
Microarray Design and Annotation
A collection of EST sequences representing 22,927 unique gene clusters was the primary source of input for oligo probe design. The collection was queried for the presence of genes of specific interest to our group, known genes likely to be involved in developmental and stem cell biology, and genes of broad interest. In 397 cases where these genes were not represented, GenBank records for the transcripts were included in the oligo design sequence pool, resulting in a total of 23,324 sequences. Unique 60-mer probes were designed for 21,939 transcripts, with 20,986 designed from 3′ sequences, 556 from 5′ sequences, and 397 from GenBank records.
Probes were annotated by a hierarchical, iterative BLAST-based algorithm, which first compared oligo probe sequences against the NCBI RefSeq and nonredundant (nr) databases (http://www.ncbi.nlm.nih.gov) to identify perfect matches to the sense strand of mRNA entries, followed by searches of the parent clone sequence used to design the oligo against the same databases for a match to an mRNA entry of at least 90% identity with 80% overlap. In cases where no such matches were found, the probe was annotated as “unknown.” Probes for 6711 transcripts were annotated by exact matches of oligo sequence to RefSeq or nr database entries, and 5760 were positively identified by strong parent sequence matches. A group of 1458 were designated unknown but similar to known genes, and the remaining 8009 showed no significant similarity to known sequences. A complete listing of the microarray's annotated gene content can be found at the NIA Mouse cDNA project Web site (http://lgsun.grc.nia.nih.gov/cDNA/cDNA.html), along with information on cDNA clones linked to 98% of the probes.
Experimental Design and Statistical Significance Testing
To evaluate the performance of the system, we generated expression profile data for embryonic day 12.5 (E 12.5) mouse embryos and placentas, and compared this data set to cDNA microarray (Tanaka et al. 2000) and quantitative real-time reverse-transcription polymerase chain reaction (Q-PCR) data. Oligo microarray experiments were designed to match the previously published E12.5 embryo–placenta comparison (Tanaka et al. 2000) as closely as possible. Three separate litters of mice were collected at E12.5, and placentas and embryos were pooled within each litter for RNA extraction. Each RNA sample was used to synthesize two complementary RNA (cRNA) “targets,” each labeled with Cyanine-3 (C3) or Cyanine-5 (C5), and the targets for each litter were “dye-swapped,” or hybridized to produce one microarray with the polarity embryo(C3):placenta(C5) and one with embryo(C5):placenta(C3). Inclusion of multiple litters (biological replicates) allowed the assessment of variation in expression of each gene from litter to litter under the same experimental conditions to be incorporated into statistical significance tests, while dye-swapping allowed identification and correction of probe-specific dye-biases, as well as a measurement of variability between targets and hybridizations from the same RNA sample (technical replicates). This approach allows us to calculate error distributions for biological factors separate from technical ones. We found that error contributed from biological variability was much greater than that contributed by technical factors, despite the pooling of embryos within litters (data not shown), emphasizing the need to include multiple, distinct biological samples for each condition or tissue in a microarray experiment. The same RNA samples were used for Q-PCR validation.
Comparisons made here between 60-mer oligo microarray or Q-PCR data and published data are by definition retrospective, and it was not possible to use the same set of RNA samples. However, we have been careful to reproduce experimental conditions as faithfully as possible, and the combination of tissue pooling and replication of measurements across different pools used to generate all three data sets is designed to minimize the effects of “biological noise,” or random variations in gene expression between individuals and litters. For these reasons, the comparisons presented here should be valid as part of a functional “road test” comparing our results from our previous microarray system to the data presented here.
Data from the 60-mer oligo microarrays were processed using both Rosetta Resolver (a popular software package that uses a combination of proprietary error modeling algorithms and conventional P-value calculations to determine statistical significance) and analysis of variance–false discovery rate (ANOVA-FDR) statistics (a more specialized statistical method designed to minimize false-positive rates; see Methods). To evaluate both the appropriateness of the confidence thresholds employed and the quality of the data set, we analyzed results from pairs of self-against-self control hybridizations, with the polarity of one microarray reversed in each pair to mimic the dye-swapping used in experimental comparisons. ANOVA-FDR identified only six transcripts for embryo and 13 for placenta, using FDR < 0.05 and 2.0 ≤ log(mean intensity) ≤ 5.4, suggesting that the false-positive rate under this analysis is less than 0.06%. Resolver showed a higher false-positive rate in self-against-self experiments, with 288 transcripts for embryo (1.3%) and 461 (2.1%) for placenta (P < 0.05; 2.0 ≤ log(mean intensity) ≤ 5.4). True false-positive rates are likely to be lower, because these control analyses contained data from two replicate microarrays, whereas the experimental data set contained six replicate microarrays.
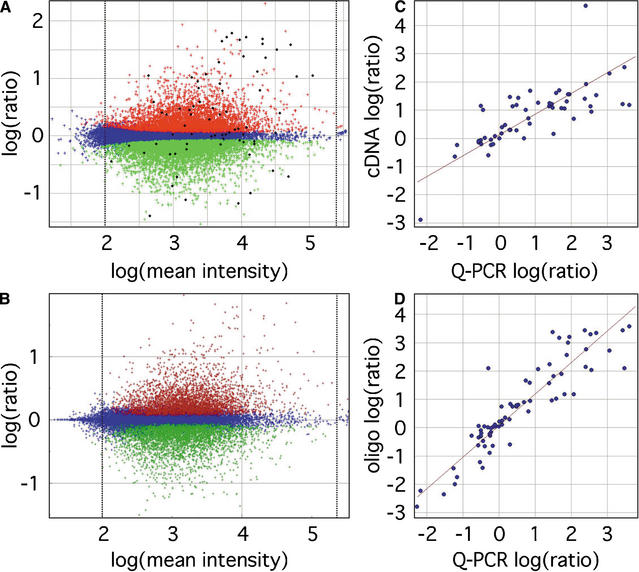
When the same parameters were applied to the experimental data, ANOVA-FDR identified a set of 9389 transcripts that were significant, with 4406 upregulated and 4983 downregulated in placenta compared to embryo. Resolver identified 12,247 transcripts (P < 0.05; 2.0 ≤ log(mean intensity) ≤ 5.4), with 6136 upregulated, 6111 downregulated, and 96.9% overlapping with the ANOVA-FDR set. Whereas ANOVA-FDR controlled false-positive rates more effectively, analysis using more conventional statistical methods gave satisfactory false-positive rates of 2.1% or less, and both methods identify highly similar sets of significant genes. It appears that ANOVA-FDR provides a more conservative analysis of differentially expressed transcripts, and the choice of which package to use depends on confidence level requirements for downstream analysis of differential genes. For purposes of this discussion, the results from the ANOVA-FDR analysis will be used.
Comparison of Q-PCR and Microarray Data
Oligo microarray data were validated by Q-PCR for a set of 71 transcripts, 37 of which were selected from a list of placenta-specific transcripts identified by cDNA microarrays (Table 1; Tanaka et al. 2000), with the remainder being chosen to create a representative sample covering intensity and fold-change ranges (Fig. 1A). There was a strong correlation (0.91) between log(ratio) values determined by Q-PCR and 60-mer microarrays for these 71 transcripts (Fig. 1D; Supplemental Table 1, available online at www.genome.org), and although 60-mer microarrays appeared to underestimate expression differences relative to Q-PCR (slope = 0.53), this effect was consistent and likely to be a result of the kinetic differences between PCR and hybridization reactions. The single significant outlier in this comparison was a homolog of melanoma-associated antigen 10 (MAGE-10), which has high sequence similarity to multiple sites in the genome (as discussed below). The correlation between Q-PCR and cDNA microarray data for 56 transcripts common to the cDNA microarray and the Q-PCR validation gene set (Fig. 1C; Suppl. Table 1) was weaker, at 0.74, with a slope of 0.59. These relationships are consistent with the idea that oligo probes and PCR primers tend to be more specific than cDNA probes, so that the former produce expression data that are more indicative of individual transcript levels, and the latter are more indicative of average expression levels for related transcripts.
Table 1.
Comparison of cDNA Array, 60-mer Oligo Array, and Q-PCR Relative Expression Results for Placenta-Specific Transcripts
| Putative ID | cDNA array | 60-mer oligo array | Q-PCR | |||||
| Clone name | Ratio | Log(R) | Clone name | Ratio | Log(R) | Ratio | Log(R) | |
| Endocrine-related proteins | ||||||||
| secretin (Sct) | H3001A02 | 199.13 | 2.30 | H3001A02 | 20.94 | 1.32 | 1133.23 | 3.05 |
| placental lactogen II (Csh2) | H3046B04 | 329.47 | 2.52 | J0076G09 | 11.98 | 1.08 | <2965.51 | 3.47 |
| calcyclin (S100a6) | H3087H09 | 35.44 | 1.55 | J0721D01 | 41.17 | 1.61 | 89.74 | 1.95 |
| prolactin-like protein A (Pripa) | H3007C10 | 8.73 | 0.94 | C0102C12 | 9.71 | 0.99 | >366.27 | 2.56 |
| prolactin-like protein M (Pripm) | H3018B06 | 15.87 | 1.20 | C0179D05 | 58.47 | 1.77 | 2680.11 | 3.43 |
| proliferin-related protein (PRP) homolog | H3021D07 | 15.04 | 1.18 | C0191E03 | 59.68 | 1.78 | >4083.91 | 3.61 |
| cathepsin L (Ctsl) | H3028F04 | 3.30 | 0.52 | G0111D11 | 6.58 | 0.82 | 7.09 | 0.85 |
| similar to human nuclear VCP-like protein NVLp.2 | H3076C01 | 13.35 | 1.13 | H3076C01 | 1.23 | 0.09 | ||
| mammary-derived growth inhibitor (Fabp3) | H3104D07 | 47.27 | 1.67 | H3104D07 | 3.05 | 0.48 | 7.34 | 0.87 |
| similar to human HEM45 (Isg20) | H3108A01 | 27.43 | 1.44 | H3108A01 | 1.35 | 0.13 | ||
| Epithelial proteins | ||||||||
| EndoA cytokeratin | H3031C01 | 35.46 | 1.55 | H3031C01 | 31.5 | 1.5 | 81.77 | 1.91 |
| EndoB cytokeratin | H3104F03 | 19.74 | 1.30 | H3104F03 | 20.38 | 1.31 | 75.67 | 1.88 |
| band 3-related protein (Slc4a2) | H3137D09 | 26.46 | 1.42 | H3137D09 | 2.61 | 0.42 | 3.50 | 0.54 |
| similar to tazarotene-induced gene 2 (TIG2) | H3137E09 | 83.09 | 1.92 | H3137E09 | 20.5 | 1.31 | 255.90 | 2.41 |
| basic transcription element binding protein 2 (Bteb2) | H3102C04 | 2.91 | 0.46 | H3102C04 | 1.43 | 0.15 | ||
| Oxidative-, stress-, and aging-related proteins | ||||||||
| 25 kDa mammalian stress protein | H3082D09 | 17.57 | 1.24 | H3082D09 | 12.52 | 1.1 | 25.06 | 1.40 |
| carbonic anhydrase IV (Car4) | H3095H12 | 50055.96 | 4.70 | H3095H12 | 10.31 | 1.01 | 249.37 | 2.40 |
| plasma glutathione peroxidase 3 (Gpx3) | H3125A09 | 40.67 | 1.61 | L0067D11 | 3.1 | 0.49 | 38.54 | 1.59 |
| Cell adhesion- or invasion-related proteins | ||||||||
| Ctla-2-beta, proregion of cysteine protease | H3022D05 | 25.00 | 1.40 | H3022D05 | 53.89 | 1.73 | 240.48 | 2.38 |
| fat facets homolog (Fam)/Usp9x | H3149H05 | 1.44 | 0.16 | L0272F03 | 2.27 | 0.36 | 1.20 | 0.08 |
| annexin XI (Anx11) | H3014B09 | 11.77 | 1.07 | H3014B09 | 8.9 | 0.95 | 53.87 | 1.73 |
| transglutaminase (Tgase) | H3137C06 | 13.29 | 1.12 | H3137C06 | 6.08 | 0.78 | 24.62 | 1.39 |
| ligatin (Lgtn) | H3138F09 | 2.96 | 0.47 | L0236F05 | 1.57 | 0.2 | 1.48 | 0.17 |
| Homeobox and transcription-related proteins | ||||||||
| eHAND (Hand1) | H3020C07 | 4.93 | 0.69 | H3020C07 | 3.88 | 0.59 | 116.49 | 2.07 |
| placenta and embryonic expression gene (Pem) | H3027G05 | 33.28 | 1.52 | G0109E12 | 41.43 | 1.62 | 499.07 | 2.70 |
| placenta-specific homeobox 1 (Psx1) | H3120B02 | 11.88 | 1.07 | L0042E08 | 43.08 | 1.63 | 30.52 | 1.48 |
| cellular repressor of E1A-stimulated genes (Creg) | H3037D10 | 49.63 | 1.70 | J0034A07 | 34.25 | 1.53 | 47.21 | 1.67 |
| melanocyte-specific gene (Msg1) | H3076B01 | 16.42 | 1.22 | H3076B01 | 3.03 | 0.48 | 29.21 | 1.47 |
| transcriptional repressor SIN3B (Sin3B) | H3129D02 | 18.00 | 1.26 | H3129D02 | 11.76 | 1.07 | 12.23 | 1.09 |
| similar to rat Frag1 | H3036E12 | 7.66 | 0.88 | J0029C02 | 0.97 | −0.01 | ||
| Tbx1 | H3051H03 | 2.47 | 0.39 | H3108B10 | 1.86 | 0.27 | 6.57 | 0.82 |
| similar to rat Hbp1 | H3054G12 | 2045.88 | 3.31 | K0136F09 | 1.26 | 0.1 | 1.24 | 0.09 |
| Others expressed in placenta | ||||||||
| LPS-binding protein (Lbp) | H3086G08 | 14.88 | 1.17 | H3086G08 | 2.33 | 0.37 | >3.12 | 0.49 |
| pyruvate carboxylase (Pcx) | H3137A09 | 26.42 | 1.42 | H3137A09 | 1.08 | 0.03 | ||
| Newly defined genes | ||||||||
| protein kinase inhibitor p58 | H3094A04 | 10.09 | 1.00 | H3094A04 | 6.74 | 0.83 | 5.60 | 0.75 |
| similar to human unknown protein IT1 | H3137F09 | 33.60 | 1.53 | L0233B10 | 0.88 | −0.05 | ||
| uroporphyrinogen III synthase (Uros) | H3137F11 | 19.87 | 1.30 | H3137F11 | 1.06 | 0.02 | ||
| Unknown transcripts | ||||||||
| H3005D09 | 11.24 | 1.05 | H3005D09 | 3.73 | 0.57 | 65.10 | 1.81 | |
| H3009D03 | 14.64 | 1.17 | H3009D03 | 39.45 | 1.6 | |||
| H3016D10 | 13.44 | 1.13 | C0172H02 | 50.19 | 1.7 | 341.71 | 2.53 | |
| H3046B06 | 12.66 | 1.10 | J0076H03 | 0.98 | −0.009 | |||
| H3049B11 | 13.32 | 1.12 | H3049B11 | 2.27 | 0.36 | 2.41 | 0.38 | |
| H3050F11 | 7116.94 | 3.85 | H3050F11 | 1.25 | 0.1 | |||
| H3087H10 | 17.02 | 1.23 | H3087H10 | 1.14 | 0.06 | |||
| H3101H01 | 16.32 | 1.21 | H3101H01 | 0.97 | −0.01 | |||
| H3137A10 | 11.82 | 1.07 | H3137A10 | 1.22 | 0.09 | |||
| H3137C08 | 28.54 | 1.46 | L0231F10 | 0.45 | −0.34 | 2.18 | 0.34 | |
| H3137E08 | 95.10 | 1.98 | H3137E08 | 0.93 | −0.03 | |||
| H3137F10 | 10.24 | 1.01 | L0233B11 | 0.58 | −0.24 | 0.37 | −0.44 | |
cDNA microarray data for a set of transcripts previously reported as placenta-specific (modified here from Tanaka et al. 2000) were compared to oligo microarray and Q-PCR expression data. Only those 49 transcripts from the original list which are represented on the NIA 15K cDNA microarray by a sequence-verified clone (the clone set has been sequence-verified since the publication of Tanaka et al. 2000), and on the 60-mer oligo microarray by a BLAST-verified oligo probe, are shown. Of the 71 transcripts validated by Q-PCR, 35 overlap with this list, and this Q-PCR data is included here for comparison. See Supplementary Table 1 for Q-PCR data on all 71 transcripts. Expression ratios are expressed as placenta:embryo, and nonsignificant results are italicized.
Figure 1.
Differential gene set identification and Q-PCR validation. (A) ANOVA-FDR analysis of oligo microarrays identified a set of 9389 transcripts significant at FDR < 0.05, with 4406 upregulated and 4983 downregulated in placenta, indicated by red and green points, respectively. Blue points indicate nonsignificant probes. Average log(mean intensity) values below 2.0 or greater than 5.4, indicated by the dotted lines, were discarded. (B) Analysis of the same data set using Resolver 3.0 software identified a similar set of transcripts (12,247 total, 6136 upregulated and 6111 downregulated in placenta) containing 96.9% of the transcripts identified in A. A set of 71 transcripts, indicated by the black points in A, was chosen as a representative sample for validation by Q-PCR, and log(ratio) values were compared against those from cDNA arrays for 56 transcripts held in common (C), for a correlation coefficient of 0.74 and slope of 0.60. (D) Q-PCR results more closely matched oligo microarray results (using data from all 71 Q-PCR transcripts), with a correlation coefficient and slope of 0.91 and 0.55, respectively. See Supplemental Table 1 for a complete listing of data used in microarray/Q-PCR comparisons.
Global Quantitative Comparison of Oligo and cDNA Microarray Results
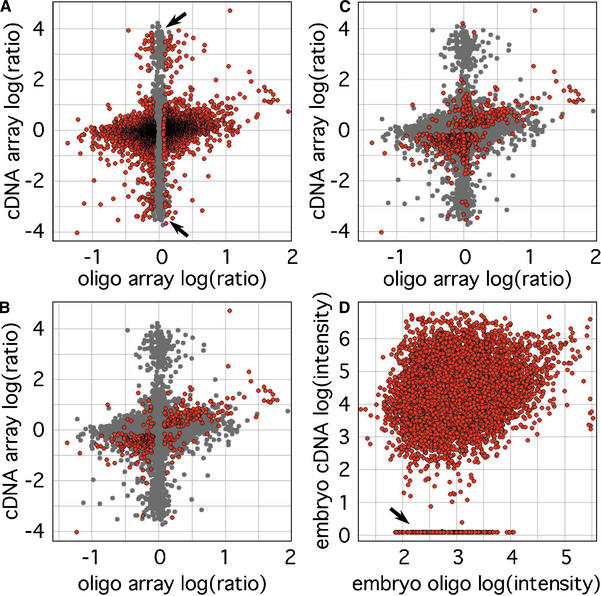
A common set of 11,938 transcripts was represented by probes on both microarray designs, and there was a very weak correlation (0.16) of log(ratio) data for this unrestricted set (Fig. 2A), resulting mainly from a group of probes that are low-intensity outliers on cDNA microarrays, with values at or below background in one or both tissues, but significant intensities on 60-mer microarrays (Fig. 2D). Most of these outliers are nonsignificant in both data sets, but many are significant in the oligo microarray data set only, and serve as examples of the increased sensitivity and reproducibility of 60-mer oligo probes (Fig. 2A,B). Restriction of the data set to only those 545 transcripts that were previously reported as significant by cDNA microarrays (Tanaka et al. 2000) and present on both arrays showed an improved correlation of 0.52. The fact that this correlation is better than that for a 5200-probe set common to both microarrays and significant only in 60-mer oligo microarrays (0.27, Fig. 2B) suggests that most of the discrepancy involved probes that were not significant in the cDNA data set (such as the cDNA low-intensity outliers). Further restriction of the probe set to 336 sequences with significant differential expression on both platforms (Fig. 2C) removed data for probes that are significant in cDNA but not in oligo microarray data, and improved the correlation coefficient and slope to 0.67 and 0.51, respectively. These comparisons demonstrate that the degree of quantitative agreement between the two data sets is directly related to the statistical confidence threshold used—generally, the better the reproducibility in a pair of measurements, the better the correlation between them.
Figure 2.
Scatter plots comparing log expression ratios in mouse E12.5 embryo and placenta measured by 60-mer oligo and cDNA microarrays. Each marker represents averaged results from dye-swapped duplicate microarrays using three biological replicates. (A) Probes showing significant differential expression by ANOVA-FDR analysis of oligo microarray measurements are indicated in red (n = 5200, FDR < 0.05, correlation coefficient [cc] = 0.27, slope = 0.30). (B) Significant transcripts identified by cDNA array show better agreement (n = 545, P < 0.05, cc = 0.52, slope = 0.32), and (C) the intersection of these two sets are even more closely matched (n = 336, cc = 0.67, slope = 0.51). (D) A number of transcripts were measured at background intensity by cDNA arrays, but had substantial intensities on oligo microarrays, as indicated by the arrow. This group was the major component of two sets of log(ratio) outliers, indicated by the arrows in A.
Although detailed comparisons with other cDNA-based platforms (such as two-channel fluorescent glass cDNA arrays) will require additional experiments, the work presented here does shed some light on the general differences and similarities between cDNA- and oligo-based microarrays. Our discussion of many of the issues explored here, such as probe sequence length, position, and composition as they relate to probe specificity, applies to comparisons with two-channel cDNA systems as well.
Comparison of Placenta-Specific Genes
We previously identified a set of transcripts that were more abundant in placenta compared to embryo (Tanaka et al. 2000), many of which are independently established as placenta-specific and/or important in placental development (Hamilton and Millis 1990; Hashido et al. 1991; Cross et al. 1994; Rinkenberger et al. 1997; Chun et al. 1999; Linzer and Fisher 1999; Tanaka et al. 2000). To assess the utility of the oligo microarray in a practical context (i.e., are the same genes identified?), we compared expression ratios determined by cDNA and 60-mer oligo microarrays for these transcripts (Table 1). Of 47 transcripts with significant expression differences of at least 20%, 45 (96%) were positively correlated, with 31 (66%) showing a placenta:embryo ratio greater than 2.0 in the oligo system. Q-PCR measurements of a subset of these transcripts (see above) showed strong agreement with oligo microarray ratios, and in 6 of 8 cases with large quantitative discrepancies between cDNA and oligo microarray measurements where Q-PCR data are available (Table 1: Csh2, Fabp3, Slc4a2, Car4, Gpx3, Hbp1, H3137C08, H3137F10), PCR-based ratios were in better agreement with the oligo microarray.
Comparison of Detection Sensitivity
General properties of 60-mer oligo microarrays such as the signal dynamic range and lower limits of detection have been reported (Hughes et al. 2001), but more practically relevant measures of performance are partially dependent on probe content and experimental protocol. One of the most striking differences between the oligo and cDNA microarray data sets is the number of transcripts identified as more abundant in placenta by this experiment. Without considering differences in differential transcript identification rates, we should expect the larger microarray to detect more significant transcripts, and this was indeed the case—whereas 289 were identified by the cDNA system as more abundant in placenta, 4406 were identified by the 60-mer oligo microarray system, with 1754 of those being present only on the larger microarray. However, such detection rate differences are highly significant, as there are 2491 transcripts common to both platforms that were measured as significantly upregulated in placenta by the 60-mer oligo microarray system only. The overall result was that many transcripts known to be more abundant in placenta, such as Prlpc (Dai et al. 1998), AP-2 gamma/Tcfap2c, adenosine deaminase (Shi et al. 1997; Shi and Kellems 1998), Tpbpa (Lescisin et al. 1988), Keratin 19 (Morrish et al. 1996), adrenomedullin (Yotsumoto et al. 1998), and Rex1/Zfp42 (Rogers et al. 1991), were not present in or not identified by the cDNA platform but showed statistically significant placenta:embryo ratios greater than 4.8 using the 60-mer oligo micro arrays (Table 2). Therefore, the 60-mer oligo microarray system detected most of the placenta-specific transcripts identified using cDNA microarrays, as well as many transcripts that were not identified.
Table 2.
Examples of Additional Known Placenta-Specific Transcripts Identified Using 60-mer Oligo Arrays
| Putative ID | Probe source sequence ID | Ratio | Present on cDNA array | Rank |
| Genes with known roles in placenta | ||||
| prolactin-like protein C (Prlpc) | C0229F08-3 | 204.8 | no | 1 |
| Dtprp | H3015H06-3 | 86.9 | yes | 2 |
| proliferin (Plf) | K02245.1 | 68.4 | yes | 3 |
| trophoblast-specific protein (Tpbp) | NM_009411 | 69.1 | no | 4 |
| adenosine deaminase (Ada) | C0609E09-3 | 47.1 | no | 9 |
| Keratin 19 | H3022G09-3 | 21.5 | yes | 28 |
| AP-2 transcription factor (Tcfap2c) | C0152H01-3 | 7.5 | no | 54 |
| Rex-1 (Zfp42) | M28382 | 8.2 | no | 100 |
| adrenomedullin (Adm) | C0102F09-3 | 6.2 | no | 157 |
| Top-ranked newly identified placenta-specific genes | ||||
| similar to proliferin | C0128F02-3 | 36.1 | no | 15 |
| TBP-associated factor II Q | L0287F02-3 | 35.9 | no | 16 |
| serine protease inhibitor 12 | C0141H10-3 | 33.0 | no | 17 |
| system A amino acid transporter 3 | C0180A05-3 | 30.4 | yes | 20 |
| voltage-dependent Ca++ channel | C0129G10-3 | 27.0 | no | 22 |
| inhibin beta-B subunit | C0329D09-3 | 25.5 | no | 24 |
| unknown | K0537F01-3 | 23.0 | no | 26 |
| C/EBP-beta | NM_009883.1 | 18.4 | no | 30 |
| GM2 ganglioside activator protein | H3035D04-3 | 16.5 | yes | 31 |
| unknown | C0228C02-3 | 16.4 | no | 32 |
| unknown | H3009H12-3 | 16.1 | yes | 33 |
| unknown | C0144F05-3 | 15.6 | no | 34 |
| PPAR-gamma | U01664.1 | 15.3 | no | 35 |
| cubilin | C0026H06-3 | 15.2 | yes | 36 |
| unknown | C0118H11-3 | 15.2 | yes | 37 |
| unknown | K0522G10-3 | 14.9 | no | 38 |
| unknown | H3002F03-3 | 14.4 | yes | 39 |
| homolog of testin 1/2 precursor | C0189H10-3 | 14.1 | no | 40 |
| similar to transglutaminase | K0209G07-3 | 13.9 | no | 41 |
| unknown | C0147A07-3 | 13.3 | no | 42 |
| CD9 antigen | H3139C03-3 | 13.0 | yes | 44 |
| unknown | C0157D02-3 | 13.0 | yes | 45 |
| zinc finger protein 42 | J0029E06-3 | 12.8 | yes | 46 |
| unknown | G0110D05-3 | 12.8 | yes | 47 |
Transcripts identified as significant by ANOVA-FDR were ranked by fold-change in expression.
When probes detecting statistically significant expression differences were broken down into defined fold-change ranges, the 60-mer oligo microarray identified more transcripts as statistically significant than did cDNA at all but the highest ratios. Oligo probes were especially sensitive to small changes in expression, with over 56 times as many oligo probes detecting significant expression changes ≤ 1.5-fold, and over 24 times as many for changes ≤ twofold, normalized to the number of probes on each microarray. Larger expression changes showed more moderate sensitivity advantages, with 60-mer oligos detecting over five times more significant changes in the 2- to 5-fold range, and 1.8 times in the 5- to 10-fold range. For expression differences > 10-fold, cDNA probes were more sensitive, detecting 1.5 times as many significant changes.
Much of the past work in expression profiling has concentrated on larger differences in expression, due to their ease of detection and the belief that larger expression changes are more biologically important. However, a report that expression changes in stem cells of less than twofold for the candidate regulator of pluripotency Oct3/4 result in differentiation (Niwa et al. 2000) challenges this view, suggesting that future utility of microarrays in developmental studies may require the ability to measure small changes reliably. Furthermore, many clustering methods analyze patterns which include both small and large expression changes, and are less robust when values are omitted due to poor reproducibility (Eisen et al. 1998). As a result, microarray systems which provide larger numbers of reliable measurements across a wider range of expression changes are more appropriate for the comparison of expression patterns under many different conditions.
Gene Families Versus Individual Transcripts
It is outside the scope of this discussion to examine and characterize all instances of disagreement between 60-mer oligo- and cDNA-based expression measurements individually, but a few examples can illustrate issues that contribute to differences seen for individual genes. For instance, cDNA and 60-mer probes for a MAGE-10 homolog produced anticorrelative results (data not shown). Recent BLASTN searches of the Ensembl mouse genome database (http://www.ensembl.org) revealed that the cDNA clone sequence has high homology to at least three sites in the genome, and the 60-mer oligo probe and Q-PCR primers designed to detect this transcript also match the same sites, but to slightly different degrees. The relative affinities of each potential transcript (if they are all indeed expressed) for the cDNA probe, the oligo probe, or the Q-PCR primers are unknown, but are likely responsible for disparity between measurements made with different systems. Related gene families can also cause disagreement between cDNA and oligo microarray data—the serine protease inhibitor (Spi) gene family is a good example of this. Probes for Spi10 show a 1.4-fold nonsignificant expression difference with cDNA probes, a significant, approximately 27-fold difference with an oligo probe, and a difference of at least 75-fold by Q-PCR. Whereas most of the Spi family transcripts measured by Q-PCR were more abundant in placenta, Spi8 was approximately fivefold more abundant in embryo (data not shown). Again, the contribution of each transcript to the overall signals is unknown, but these and other examples raise two points to consider generically in microarray design: (1) Probe cross-reactivity is very difficult to eliminate completely, albeit somewhat easier when using oligomers, especially in the case of probes for members of closely related gene families; and (2) design algorithms that aim to avoid cross-reactivity are dependent on transcript and genome annotation data, which are improving with time. These considerations are being applied to an improved version of this oligo microarray.
Array Platform Comparison: Conclusions
These comparisons illustrate that there is general agreement between cDNA and oligo microarray platforms at the quantitative (ratio) level, and at the qualitative (differential gene list) level. The 60-mer oligo microarray data were more highly correlated with Q-PCR data for specific transcripts, and they identified several times as many statistically significant, differential genes, compared to cDNA microarray data. Because 60-mer oligo probes are generally more specific than cDNA probes, their increased detection rate is likely due to reduced cross-hybridization, which can mask expression differences in cDNA microarrays. It is important to keep in mind that this was a retrospective comparison, using a fresh set of RNA samples for the oligo microarray and Q-PCR data. Nonetheless, the intrinsic cross-hybridization problem of cDNA microarrays appears to diminish detection of expression differences, making 60-mer oligo probe microarrays the more appropriate system for general use. The importance of obtaining average expression levels of transcript families versus levels of each specific transcript will determine the more appropriate system for a particular use, and the comparisons given can help in making informed decisions. Complete microarray data sets are available at the NIA Mouse cDNA Project Web site (http://lgsun.grc.nia.nih.gov/cDNA/cDNA.html).
Adaptation to Small RNA Samples
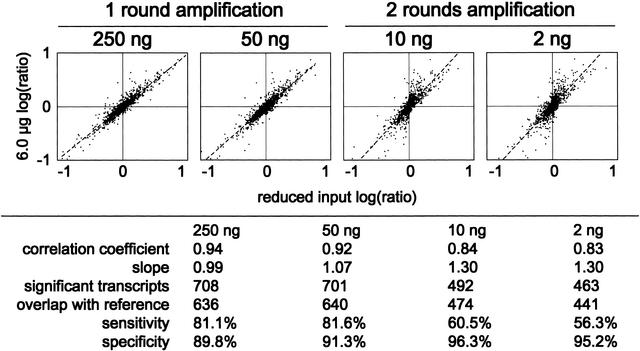
To test the performance of the microarray system with very small RNA samples on the scale of those available from stem cell and embryonic tissues, we prepared linearly amplified cRNA targets labeled with Cyanine-3 and Cyanine-5 dyes from 250, 50, 10, and 2 ng of E12.5 embryo or placenta total RNA. One round of amplification was used for 250 and 50 ng, whereas two successive rounds were used to prepare the 10- and 2-ng targets. Quadruplicate dye-swapped microarrays at each input level were compared to results from targets labeled with the standard 6 μg protocol (Fig. 3). The correlation coefficient for the entire probe set decreased from 0.94 to 0.83 as the input level was decreased from 250 to 2 ng (Fig. 3), with some compression of the log(ratio) distribution for the targets labeled with two rounds of amplification (Fig. 3). Inter- and intra-array error distributions were highly similar and intensity-dependent for singly amplified targets, but when two rounds of amplification were used, interarray error was intensity-independent (data not shown).
Figure 3.
Performance comparison of reduced-input labeling to standard protocol for use with 60-mer oligo microarrays. Decreasing amounts of total RNA from mouse E12.5 embryo and placenta were used to prepare labeled linear-amplified targets, using one round of amplification for 250- and 50-ng and two rounds for 10- and 2-ng inputs. Each spot represents averaged ratios from four dye-swapped microarrays, each containing eight replicates of each probe. The correlation coefficient, slope, number of significant transcripts, overlap with the reference set of 784 significant transcripts, specificity, and sensitivity are shown beneath each panel.
Several criteria for gene selection were evaluated across the input level range, to assess the effects of reducing RNA input on the sensitivity and specificity of differential expression detection (Fig. 3). For single-round targets, sensitivity was reduced up to 20%, with 784 significant transcripts detected with a 6-μg input and 640 detected at 50 ng. Specificity was retained better, with 91% of these transcripts also identified at 6-μg input. Two-round amplified targets were less sensitive, but again showed similar specificity, with only 441 significant transcripts detected (56%) and 95% overlap with the 6-μg set.
Clearly, there is a trade-off between performance and reduced RNA input, particularly with sensitivity, and in cases where tissue or cell line RNA is abundant, standard labeling protocols are most appropriate. Experiments using scarce tissues can still identify 50%–80% of differentially expressed transcripts detected using standard labeling inputs. A complete listing of compiled data is found in Supplemental Table 2, and complete raw data sets are available at the NIA Mouse cDNA Project Web site (http://lgsun.grc.nia.nih.gov/cDNA/cDNA.html).
Application of 22K (60-mer) Oligo Microarrays to Mouse Embryogenomics
The 22K 60-mer oligo microarray that we report here has the following unique features: (1) 60-mer oligonucleotide probes, providing more specificity for individual genes and transcripts; (2) freely available clones corresponding to 98% of the probes on the arrays for downstream molecular studies; (3) enriched representation of genes that are relevant for studies of mouse embryogenomics (Ko 2001), particularly in stem cells and early embryos; (4) differential expression detection rates several times higher than those obtainable with cDNA microarrays; and (5) compatibility with reduced amounts of input RNA, allowing the application of microarray technologies to small amounts of mouse embryos, FACS-purified cells, and microdissected tissues.
Although the features listed above describe a system uniquely qualified for expression profiling of mouse embryos, embryonic tissues, and stem cells, making it feasible to profile the expression of developmentally relevant genes in tiny amounts of these tissues, it is also important to note that the comprehensive probe content of this 60-mer oligo microarray platform makes it suitable for general use as a mouse “gene catalog” chip. For example, we have now generated expression profiles of unfertilized mouse oocytes using only 18 cells per hybridization (data not shown). Ongoing improvements in amplification and labeling techniques and the compatibility of this oligo microarray platform with a wide variety of labeling methods (Hughes et al. 2001) should further increase flexibility and value for a wide range of developmental studies.
METHODS
Microarray Design and Fabrication
Sequence data from our entire cDNA clone collection were clustered (Carpenter et al. 2002) and masked for repeat and low-complexity sequence using RepeatMasker (A. Smit and P. Green, unpubl.) and Dust (R. Tatusov and D. Lipman, unpubl.) algorithms, respectively. For each cluster, a representative 3′ sequence was chosen, and in cases where that sequence was shorter than 60 bp or did not satisfy parent-clone similarity or sequence quality criteria, the parental clone 3′ sequence was selected. If parental 3′ sequence failed to meet the above criteria, other 3′ sequences in the cluster were considered, followed by 5′ sequences. For each sequence in this pool, 60-mer oligo probes were evaluated and selected as previously described (Hughes et al. 2001; Shoemaker et al. 2001; van't Veer et al. 2002). Oligonucleotide 60-mer microarrays were manufactured by Agilent Technologies using their ink-jet based SurePrint technology (Hughes et al. 2001), with each probe represented once on each microarray.
RNA Extraction
Mouse embryos were collected from C57BL/6J litters at E12.5, and placentas were dissected away from embryonic tissue. Three to five embryos or placentas were pooled within each litter, and stored at −80°C. Total RNA was extracted and purified using TriZol reagent (Invitrogen) per the manufacturer's protocol, and the quality and quantity of the preparations were assessed using an RNA 6000 Nano Lab-on-a-chip Kit with a 2100-Bioanalyzer system (Agilent Technologies). Aliquots of 12 μg were stored at −80°C for later use in both linear amplification labeling and cDNA synthesis for Q-PCR.
RNA Target Labeling
Amplified cRNA labeled with Cyanine-3 CTP and Cyanine-5 CTP (Perkin-Elmer/NEN Life Sciences) was produced from 6.0-μg aliquots of total RNA using a Fluorescent Linear Amplification Kit (Agilent Technologies) as specified by the manufacturer, except for the following modifications to accommodate total RNA samples: One microliter of 0.3% Triton X-102 (Sigma) was added to each 20-μL cDNA synthesis reaction containing 6.0 μg of total RNA, and the reactions were incubated at 40°C for 240 min. Two rounds of amplification were used for 10-ng and 2-ng targets. First, total RNA was used to synthesize cDNA in a reaction scaled down to a total volume of 4 μL, with half the standard T7-oligo-dT primer concentration and 125 ng/μL of T4gp32 single-stranded DNA-binding protein (United States Biochemical). Linear amplification was performed in a total volume of 16 μL, with half the standard NTP concentration and no labeled CTP. For the second round of amplification, the product of the first reaction was divided in half, and labeled using the manufacturer's standard protocol, with the addition of T4gp32 in the cDNA synthesis reaction. The quality and size distribution of targets were determined by RNA 6000 Nano Lab-on-a-chip Assay (Agilent Technologies), and quantitation was determined using a NanoDrop micro-scale spectrophotometer (NanoDrop).
Array Hybridization, Washing, and Scanning
Fluorescent linear amplified cRNAs used in biological comparisons were hybridized to custom-made in situ synthesized 60-mer oligo microarrays containing 22,575 features including controls (Agilent Technologies), per the manufacturer's instructions. Targets used to optimize reduced-input labeling protocols were hybridized to a 60-mer oligo microarray consisting of eight replicates of approximately 1000 probes that were evenly distributed across the detectable intensity range in previous experiments, with good signal-to-noise (data not shown). Hybridized microarrays were washed according to the manufacturer's protocol and scanned on an Agilent Technologies G2565AA Microarray Scanner System with SureScan technology.
Data Processing and Statistical Analysis
Ratio data were extracted from scanned microarray images using Feature Extraction 5.1.1 software (Agilent Technologies), and dye-normalized, background-subtracted intensity and ratio data were exported to text and GEML-format files. Text output was processed using an application developed in-house to perform ANOVA analysis.
Data were sorted by intensity, and mean error variance was calculated using a sliding window of 1000 probes. Intensity values were filtered to remove values where probe error was greater than two times mean error and relative error was greater than 50%. Surrogate values equal to mean error were inserted for values that were negative or less than probe error. Mean dye-swapped log(ratio) values were calculated, and mixed-model ANOVA (Sokal and Rohlf 1995) was applied, using the following error model:
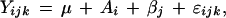 |
1 |
where μ = mean log(ratio), Ai = random effect of biological replication, βj = fixed effect of dye-swapping, and ɛijk = error for biological replication i, dye swap j, and technical replication k. The small numbers of biological replications typical in expression profiling experiments result in a highly variable error variance, and this problem is usually addressed by log-ratio thresholds (Schena et al. 1995) that require subjective decisions about biological significance, or by Bayesian adjustment of error variance (Baldi and Long 2001), which may still underestimate error variance and result in false positive results. We opted for the stronger statistical basis of Bayesian adjustment, with a very conservative error model to reduce false-positives:
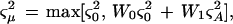 |
2 |
where ςA2 is the probe's biological replication error variance, and ς02 is the mean value of ςA2 for transcripts in the sliding window, not including the highest 5%, which could be outliers. The expression W0ς02 + W1ςA2 is a Bayesian-adjusted error variance (Baldi and Long 2001) with necessary degrees of freedom K = 10. Probes where ‖μ‖/ςμ > 7 were considered outliers and removed. The analysis was repeated until no new outliers were identified, and transcripts with log(mean intensity) values outside of the 2.0–5.4 range were excluded.
Although standard tests for statistical significance based on t- or z-distributions will identify significant transcripts even when the null hypothesis is true in all cases, use of the Bonferroni correction (simply multiplying each P-value by the number of transcripts tested) can prove unnecessarily stringent, resulting in the identification of few or no significant transcripts. To more appropriately control the false-positive rate in this analysis, we tested for statistical significance using the False Discovery Rate (FDR) rule:
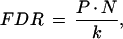 |
3 |
where N is the number of transcripts tested, k is the transcript's rank by decreasing t-value, and P is estimated using the z-distribution (Benjamini and Hochberg 1995).
Real-Time Quantitative RT-PCR
For E12.5 embryo and placenta, 10-μg RNA aliquots were DNAse-treated using a DNA-Free Kit (Ambion), and annealed with random hexamers. cDNA synthesis was performed using SuperScript II (Invitrogen), and cDNA products were diluted to 100 ng total RNA input/μL output. For each selected transcript, the 3′-end sequence of the EST clone used for oligo probe design was loaded into Vector NTI software (Informax), and PCR primer pairs were designed such that both anneal at 60° ± 1°C, the amplicon length is between 75 and 250 bp, and low-complexity sequence was avoided. Primers were tested using a pool of embryo and placenta cDNAs with SYBR Green PCR Master Mix on an ABI 7700 Sequence Detection System (Applied Biosystems). First, each primer pair was run using a matrix of forward and reverse primer concentrations, and threshold cycle measurements were compared with dissociation curves to determine optimal primer concentrations with high amplicon specificity. Second, a 5-log standard curve dilution series was run using each primer pair at optimal concentration, and amplification efficiencies were calculated. Primer sets with suboptimal dissociation curves, or efficiencies outside of the 85%–115% range were discarded, and replacements were designed and tested.
E12.5 embryo and placenta cDNAs were diluted, aliquotted into 96-well plates, and stored at −80°C for later use. Standard curve dilutions, RT− controls, and quintuplicate RT+ samples were included on each plate. The first plate in each batch was used to run a normalizing gene and check for even loading of cDNA. Optimized primer pairs were run on the remaining plates, and dissociation curves for each run were checked for specificity. Unknowns were plotted on the standard curve, normalized to the first plate of the batch, and the expression ratio was calculated for each sample pair.
Animal Experimentation
All experiments were carried out in accordance with guidelines set forth by the NIA, and were reviewed and approved by the Gerontology Research Center's Animal Care and Use Committee, Animal Studies Proposal #220MSK-MI.
WEB SITE REFERENCES
http://lgsun.grc.nia.nih.gov/cDNA/cDNA.html; NIA Mouse cDNA Project Home Page.
http://www.ncbi.nlm.nih.gov; National Center for Biotechnology Information Home Page.
http://www.ensembl.org; Ensembl genome browser home page.
Acknowledgments
We thank Dr. David Schlessinger for critical reading of the manuscript, Dr. Tetsuya Tanaka for assistance with the cDNA data set, and Dr. Glenda Delenstarr for advice and discussion on statistical analysis. We would also like to thank Drs. Ruhikant Meetei, Chang-Yi Cui, Luisa Herrera, and members of the Developmental Genomics and Aging Section for help in selecting the gene content of the microarray. M.G.C. and T.H. were supported by fellowships from the NIGMS PRAT program and The Serono Foundation, respectively.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
E-MAIL kom@grc.nia.nih.gov; FAX (410) 558-8331.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.878903.
REFERENCES
- 1.Baldi P. and Long, A.D. 2001. A Bayesian framework for the analysis of microarray expression data: Regularized t-test and statistical inferences of gene changes. Bioinformatics 17: 509-519. [DOI] [PubMed] [Google Scholar]
- 2.Benjamini Y. and Hochberg, Y. 1995. Controlling the false discovery rate—A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57: 289-300. [Google Scholar]
- 3.Billia F., Barbara, M., McEwen, J., Trevisan, M., and Iscove, N.N. 2001. Resolution of pluripotential intermediates in murine hematopoietic differentiation by global complementary DNA amplification from single cells: Confirmation of assignments by expression profiling of cytokine receptor transcripts. Blood 97: 2257-2268. [DOI] [PubMed] [Google Scholar]
- 4.Carpenter J.E., Christoffels, A., Weinbach, Y., and Hide, W.A. 2002. Assessment of the parallelization approach of d2_cluster for high-performance sequence clustering. J. Comput. Chem. 23: 755-757. [DOI] [PubMed] [Google Scholar]
- 5.Chun J.Y., Han, Y.J., and Ahn, K.Y. 1999. Psx homeobox gene is X-linked and specifically expressed in trophoblast cells of mouse placenta. Dev. Dyn. 216: 257-266. [DOI] [PubMed] [Google Scholar]
- 6.Cross J.C., Werb, Z., and Fisher, S.J. 1994. Implantation and the placenta: Key pieces of the development puzzle. Science 266: 1508-1518. [DOI] [PubMed] [Google Scholar]
- 7.Dai G., Chapman, B.M., Liu, B., Orwig, K.E., Wang, D., White, R.A., Preuett, B., and Soares, M.J. 1998. A new member of the mouse prolactin (PRL)-like protein-C subfamily, PRL-like protein-C α: Structure and expression. Endocrinology 139: 5157-5163. [DOI] [PubMed] [Google Scholar]
- 8.Eisen M.B., Spellman, P.T., Brown, P.O., and Botstein, D. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad.Sci. 95: 14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamilton R.T. and Millis, A.J. 1990. Developmental roles for growth factor-regulated secreted proteins. Curr. Top. Dev. Biol. 24: 193-218. [DOI] [PubMed] [Google Scholar]
- 10.Hashido K., Morita, T., Matsushiro, A., and Nozaki, M. 1991. Gene expression of cytokeratin endo A and endo B during embryogenesis and in adult tissues of mouse. Exp. Cell Res. 192: 203-212. [DOI] [PubMed] [Google Scholar]
- 11.Hughes T.R., Mao, M., Jones, A.R., Burchard, J., Marton, M.J., Shannon, K.W., Lefkowitz, S.M., Ziman, M., Schelter, J.M., Meyer, M.R., et al. 2001. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat. Biotechnol. 19: 342-347. [DOI] [PubMed] [Google Scholar]
- 12.Hwang S.Y., Oh, B., Knowles, B.B., Solter, D., and Lee, J.S. 2001. Expression of genes involved in mammalian meiosis during the transition from egg to embryo. Mol. Reprod. Dev. 59: 144-158. [DOI] [PubMed] [Google Scholar]
- 13.Kargul G.J., Dudekula, D.B., Qian, Y., Lim, M.K., Jaradat, S.A., Tanaka, T.S., Carter, M.G., and Ko, M.S. 2001. Verification and initial annotation of the NIA mouse 15K cDNA clone set. Nat. Genet. 28: 17-18. [DOI] [PubMed] [Google Scholar]
- 14.Ko M.S. 2001. Embryogenomics: Developmental biology meets genomics. Trends Biotechnol. 19: 511-518. [DOI] [PubMed] [Google Scholar]
- 15.Ko M.S., Kitchen, J.R., Wang, X., Threat, T.A., Hasegawa, A., Sun, T., Grahovac, M.J., Kargul, G.J., Lim, M.K., Cui, Y., et al. 2000. Large-scale cDNA analysis reveals phased gene expression patterns during preimplantation mouse development. Development 127: 1737-1749. [DOI] [PubMed] [Google Scholar]
- 16.Lee K.F., Kwok, K.L., and Yeung, W.S. 2000. Suppression subtractive hybridization identifies genes expressed in oviduct during mouse preimplantation period. Biochem. Biophys. Res. Commun. 277: 680-685. [DOI] [PubMed] [Google Scholar]
- 17.Lescisin K.R., Varmuza, S., and Rossant, J. 1988. Isolation and characterization of a novel trophoblast-specific cDNA in the mouse. Genes & Dev. 2: 1639-1646. [DOI] [PubMed] [Google Scholar]
- 18.Linzer D.I. and Fisher, S.J. 1999. The placenta and the prolactin family of hormones: Regulation of the physiology of pregnancy. Mol. Endocrinol. 13: 837-840. [DOI] [PubMed] [Google Scholar]
- 19.Lipshutz R.J., Fodor, S.P., Gingeras, T.R., and Lockhart, D.J. 1999. High density synthetic oligonucleotide arrays. Nat. Genet. 21: 20-24. [DOI] [PubMed] [Google Scholar]
- 20.Morrish D.W., Linetsky, E., Bhardwaj, D., Li, H., Dakour, J., Marsh, R.G., Paterson, M.C., and Godbout, R. 1996. Identification by subtractive hybridization of a spectrum of novel and unexpected genes associated with in vitro differentiation of human cytotrophoblast cells. Placenta 17: 431-441. [DOI] [PubMed] [Google Scholar]
- 21.Niwa H., Miyazaki, J., and Smith, A.G. 2000. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 24: 372-376. [DOI] [PubMed] [Google Scholar]
- 22.Pease A.C., Solas, D., Sullivan, E.J., Cronin, M.T., Holmes, C.P., and Fodor, S.P. 1994. Light-generated oligonucleotide arrays for rapid DNA sequence analysis. Proc. Natl. Acad. Sci. 91: 5022-5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Phillips R.L., Ernst, R.E., Brunk, B., Ivanova, N., Mahan, M.A., Deanehan, J.K., Moore, K.A., Overton, G.C., and Lemischka, I.R. 2000. The genetic program of hematopoietic stem cells. Science 288: 1635-1640. [DOI] [PubMed] [Google Scholar]
- 24.Rinkenberger J.L., Cross, J.C., and Werb, Z. 1997. Molecular genetics of implantation in the mouse. Dev. Genet. 21: 6-20. [DOI] [PubMed] [Google Scholar]
- 25.Rogers M.B., Hosler, B.A., and Gudas, L.J. 1991. Specific expression of a retinoic acid-regulated, zinc-finger gene, Rex-1, in preimplantation embryos, trophoblast and spermatocytes. Development 113: 815-824. [DOI] [PubMed] [Google Scholar]
- 26.Schena M., Shalon, D., Davis, R.W., and Brown, P.O. 1995. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270: 467-470. [DOI] [PubMed] [Google Scholar]
- 27.Shi D. and Kellems, R.E. 1998. Transcription factor AP-2γ regulates murine adenosine deaminase gene expression during placental development. J. Biol. Chem. 273: 27331-27338. [DOI] [PubMed] [Google Scholar]
- 28.Shi D., Winston, J.H., Blackburn, M.R., Datta, S.K., Hanten, G., and Kellems, R.E. 1997. Diverse genetic regulatory motifs required for murine adenosine deaminase gene expression in the placenta. J. Biol. Chem. 272: 2334-2341. [PubMed] [Google Scholar]
- 29.Shoemaker D.D., Schadt, E.E., Armour, C.D., He, Y.D., Garrett-Engele, P., McDonagh, P.D., Loerch, P.M., Leonardson, A., Lum, P.Y., Cavet, G., et al. 2001. Experimental annotation of the human genome using microarray technology. Nature 409: 922-927. [DOI] [PubMed] [Google Scholar]
- 30.Singh-Gasson S., Green, R.D., Yue, Y., Nelson, C., Blattner, F., Sussman, M.R., and Cerrina, F. 1999. Maskless fabrication of light-directed oligonucleotide microarrays using a digital micromirror array. Nat. Biotechnol. 17: 974-978. [DOI] [PubMed] [Google Scholar]
- 31.Sokal R.R. and Rohlf, F.J., 1995. Biometry: The principles and practice of statistics in biological research. Freeman, New York, NY.
- 32.Stanton J.L. and Green, D.P. 2002. A set of 1542 mouse blastocyst and preblastocyst genes with well-matched human homologs. Mol. Hum. Reprod. 8: 149-166. [DOI] [PubMed] [Google Scholar]
- 33.Steidl U., Kronenwett, R., Rohr, U.P., Fenk, R., Kliszewski, S., Maercker, C., Neubert, P., Aivado, M., Koch, J., Modlich, O., et al. 2002. Gene expression profiling identifies significant differences between the molecular phenotypes of bone marrow-derived and circulating human CD34+ hematopoietic stem cells. Blood 99: 2037-2044. [DOI] [PubMed] [Google Scholar]
- 34.Tanaka T.S., Jaradat, S.A., Lim, M.K., Kargul, G.J., Wang, X., Grahovac, M.J., Pantano, S., Sano, Y., Piao, Y., Nagaraja, R., et al. 2000. Genome-wide expression profiling of mid-gestation placenta and embryo using a 15,000 mouse developmental cDNA microarray. Proc. Natl. Acad. Sci. 97: 9127-9132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Terskikh A.V., Easterday, M.C., Li, L., Hood, L., Kornblum, H.I., Geschwind, D.H., and Weissman, I.L. 2001. From hematopoiesis to neuropoiesis: Evidence of overlapping genetic programs. Proc. Natl. Acad. Sci. 98: 7934-7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Testa U., Torelli, G.F., Riccioni, R., Muta, A.O., Militi, S., Annino, L., Mariani, G., Guarini, A., Chiaretti, S., Ritz, J., et al. 2002. Human acute stem cell leukemia with multilineage differentiation potential via cascade activation of growth factor receptors. Blood 99: 4634-4637. [DOI] [PubMed] [Google Scholar]
- 37.VanBuren V., Piao, Y., Dudekula, D.B., Qian, Y., Carter, M.G., Martin, P.R., Stagg, C.A., Bassey, U.C., Aiba, K., Hamatani, T., et al. 2002. Assembly, verification, and initial annotation of the NIA mouse 7.4K cDNA clone set. Genome Res. 12: 1999-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van't Veer L.J., Dai, H., van de Vijver, M.J., He, Y.D., Hart, A.A., Mao, M., Peterse, H.L., van der Kooy, K., Marton, M.J., Witteveen, A.T., et al. 2002. Gene expression profiling predicts clinical outcome of breast cancer. Nature 415: 530-536. [DOI] [PubMed] [Google Scholar]
- 39.Yotsumoto S., Shimada, T., Cui, C.Y., Nakashima, H., Fujiwara, H., and Ko, M.S. 1998. Expression of adrenomedullin, a hypotensive peptide, in the trophoblast giant cells at the embryo implantation site in mouse. Dev. Biol. 203: 264-275. [DOI] [PubMed] [Google Scholar]