Abstract
Smooth pufferfish of the family Tetraodontidae have the smallest vertebrate genomes yet measured. They have a haploid genome size of ∼400 million bp (Mb), which is almost eight times smaller than the human genome. Given that spiny pufferfish from the sister family Diodontidae and a fish from the outgroup Molidae have genomes twice as large as smooth puffers, it appears that the genome size of smooth puffers has contracted in the last 50–70 million years since their divergence from the spiny puffers. Here we use renaturation kinetics to compare the repetitive nature of the smooth and spiny puffer genomes. We also estimate the rates of small (<400 bp) insertions and deletions in smooth and spiny puffers using defunct non-LTR retrotransposons. We find a significantly greater abundance of a transposon-like repetitive DNA class in spiny puffers relative to smooth puffers, in addition to nearly identical indel rates. We comment on the role that large insertions may play in the evolution of genome size in these two groups.
[The sequence data from this study have been submitted to GenBank under accession nos. AY212336–AY212504.]
Early surveys of nuclear DNA content in eukaryotes were baffling. Genome size differences were enormous, with protozoans varying 5800-fold, arthropods 250-fold, fish 350-fold, algae 5000-fold, and angiosperms 1000-fold (for review, see Gregory 2001), but there appeared to be very little correlation between genome size and organismic complexity (the “C-value paradox”). Ultracentrifugation and DNA renaturation kinetics ultimately demonstrated that much of the variation in genome size was not due to differences in the number of genes, but rather in the amount of previously unknown noncoding DNA (Britten and Kohne 1968). Although some of this DNA encodes regulatory information (rather than proteins), much of it was found to consist of presumably nonfunctional simple repeats of varying lengths and random sequence (John and Miklos 1988).
Understanding the significance of this noncoding DNA requires elucidation of the processes responsible for its maintenance. Comparative study of genome size evolution within taxonomic groups is a powerful tool in discovery of these regulatory processes. In this context, the pufferfish are particularly well suited for studies of the process of genome size evolution. Smooth pufferfish (family Tetraodontidae) have the smallest vertebrate genomes measured to date, with a haploid genome size of ∼400 million bp (Mb; Hinegardner and Rosen 1972). Pufferfish have approximately the same complement of genes as other vertebrates (Brenner et al. 1993), thus this curiously small size has presumably resulted from a loss of repetitive or other nonfunctional, noncoding DNA. Spiny pufferfish of the sister family Diodontidae have genomes that are roughly twice as large, ∼800 Mb (Hinegardner and Rosen 1972; Brainerd et al. 2001). Mola mola(Molidae), a member of the closest outgroup to these two families of pufferfish, also has a genome size of ∼800 Mb (Brainerd et al. 2001). Therefore, the difference in genome size between tetraodontid and diodontid puffers presumably has resulted from a reduction of genome size in smooth tetraodontid puffers relative to their spiny diodontid cousins during the 50–70 million years since their divergence (Tyler 1980; Brainerd et al. 2001; Tyler and Santini 2002). Table 1 lists genome size estimates for multiple species within these groups. Finally, because the genomes of two smooth pufferfish (Fugu rubripes and Tetraodon nigroviridis) have been nearly completely sequenced, they offer a tremendous opportunity to study genome size evolution by providing an extremely detailed glimpse of the coding and noncoding sequence of these organisms (Roest Crollius et al. 2000; Aparicio et al. 2002).
Table 1.
Genome Size Variation in Pufferfish
From Pizon et al. 1984.
From Ojima and Yamamoto 1990.
From Brainerd et al. 2002.
From Brenner et al. 1993.
From Hinegardner and Rosen 1972.
From Vinogradov 1998.
From Roest Crollius et al. 2000.
From Tyler 1980; Tyler and Santini 2002.
Here we use DNA renaturation kinetics to quantitatively describe and compare the abundance of repetitive DNA in the differently sized genomes of spiny and smooth pufferfish. Spiny pufferfish are genetically uncharacterized, and the construction of Cot curves using renaturation kinetics (Britten and Kohne 1968) has allowed us to build the first portrait of the abundance of different classes of repetitive DNAs relative to single-copy sequence in this unstudied genome. Analyses of renaturation kinetics are based on the principle that when a solution of sheared denatured DNA is kept at a temperature that permits renaturation, the rate-limiting step of the reaction is the collision of complementary fragments. Because the probability of collision of complementary fragments is proportional to the square of the concentration of those fragments in solution, highly repetitive DNA sequences renature first, followed by repetitive sequences of lower copy number, and then finally the single-copy sequences corresponding to genes and other unique sequences. Measurement of the amount of double-stranded DNA in a renaturing solution over time permits the construction of a Cot (concentration × time) curve, which is used to quantify different repetitive classes.
To complement our Cot-based description of the pattern of genome size change in these fish, we test the hypothesis that a change in the processes of insertion and deletion is responsible for genome size differences in puffers. We estimate the genome-wide incidence of small-scale insertions and deletions using nonfunctional retroelements after the method of Petrov et al. (1996). Many non-LTR retroelement transposition events result in a 5′ truncation of the newly inserted element, rendering it incapable of further transposition. Such elements are said to be “dead on arrival” (DOA). Previous analyses indicate that mutations affecting these DOA elements after truncation are effectively neutral not only in terms of their effect on the elements' coding capacity, but also in terms of their effects on genome size (Petrov and Hartl 1998) and the propensity of the local sequence to participate in ectopic recombination (Blumenstiel et al. 2002). They are therefore ideal for estimation of the neutral mutation rate. Phylogenetic analysis of multiple retrotransposon sequences is used to separate mutations that occurred during the active, nonneutral phase of the elements' lives from those mutations occurring during their nonautonomous DOA phase. The former type of mutation is expected to be found more than once in a densely sampled data set and therefore map to the internal branches of an element tree. Unselected insertions, deletions, and point substitutions are expected to map to the terminal branches of a phylogenetic tree, which permits estimates of DNA loss or gain relative to the nucleotide substitution rate (Petrov et al. 1996; Petrov and Hartl 1997).
A similar rate of DNA loss between these two families of puffers, combined with an underrepresentation of middle-repetitive DNA in smooth puffers, lead us to invoke a reduced rate of large insertions as a probable cause of the genome size reduction in smooth puffers. We speculate on the role that transposable elements may have played in this genome size reduction, and evaluate the importance of large insertions versus small deletions as determinants of genome size.
RESULTS
Renaturation Kinetics
Cot curves for Fugu rubripes, a smooth puffer, and Diodon hystrix, a spiny puffer, are presented in Figure 1. The curves were analyzed with the least-squares regression program of Pearson et al. (1977). Two-component curves were fitted to the data of each fish and were used to compute the descriptive statistics for the repetitive and single-copy fractions of the two genomes (Table 2). The most obvious difference between the two curves is in the percentage of double-stranded DNA present initially in the renaturation reaction. For F. rubripes and D. hystrix, 19% and 7%, respectively, of the genomic DNA is already present in double-stranded form at the earliest ECot value (ECot represents the product of a correction factor for the salt concentration of the buffer solution (E), the molar concentration of nucleotides (Co), and the time of renaturation in seconds (t; Britten et al. 1974).
Figure 1.
Cot curves for a tetraodontid (smooth) puffer, Fugu rubripes, and a diodontid (spiny) puffer, Diodon hystrix. Filled circles indicate F. rubripes data points, and open circles represent D. hystrix data points. Open arrows mark the Cot1/2 values of the D. hystrix repetitive (left) and single-copy (right) classes, at which half of those components are estimated to have renatured. The filled arrow indicates the Cot1/2 value of the single-copy component of the F. rubripes genome. The Cot1/2 value of the F. rubripes repetitive component is estimated to be −3.3, and does not appear on this graph.
Table 2.
Repetitive Classes in Fugu rubripes and Diodon hystrix Genomic DNA
| Species | Class | Fraction (%) | Frequency (copies/genome) | Complexitya (bp) | Log cot1/2b (M sec) |
| F. rubripes | Foldback | 18 | |||
| Repetitive | 3 | 9.0 × 105 | 140 | −3.3 | |
| Single copy | 79 | 1 | 3.8 × 108 | 2.6 | |
| D. hystrix | Foldback | 7 | |||
| Repetitive | 22 | 150 | 6.4 × 106 | 0.8 | |
| Single copy | 71 | 1 | 9.6 × 108 | 3.0 |
The estimated length of the longest nonrepeating sequence in the class.
The value at which half of the DNA belonging to the class has renatured.
The nature of DNA that has already renatured by the earliest assay point in a Cot curve has been interpreted in various manners by different authors. This DNA is sometimes assumed to be the highly repetitive component of the genome, which renatures too quickly for observation (Krajewski 1989; Verneau et al. 1991). Other authors assert this component to be “foldback DNA,” made up of a mixture of repetitive and nonrepetitive fragments that share the property of being able to form stem–loop structures upon themselves very quickly (Britten et al. 1974; Jack and Hardman 1980; Pizon et al. 1984). To distinguish between these two possibilities, we constructed a partial curve for Escherichia coli with our protocol (data not shown). This curve yielded starting values of renatured DNA around 7%. Because E. coli is known to not contain satellite DNA or other highly repetitive sequences (Britten and Kohne 1968), it is not possible in this case that the already renatured DNA is a purely repetitive class. Therefore, in this analysis we do not consider the already renatured DNA in the puffer curves to be a purely highly repetitive class. The double-stranded DNA present at the earliest ECot values in the pufferfish curves is likely a mixture of fragments from different repetitive classes that are either self-complementary, resistant to the initial denaturation step, or otherwise refractory to digestion with S1 nuclease.
The total repetitive component of the smooth F. rubripes genome based on renaturation analysis is 3%. This value is lower than but comparable to partial library-based analyses of the repetitive content of sequenced smooth pufferfish genomes, which place the repetitive component at <10% of the genome (Elgar et al. 1999; Roest Crollius et al. 2000). In contrast, the repetitive fraction of the spiny D. hystrix genome is ∼22%, indicating a greater abundance of satellite DNA or transposable elements in the spiny pufferfish family. Computation of the repetitive frequency of this fraction from kinetic rate constants (see Methods) indicates that middle repetitive rather than highly repetitive DNA constitutes most of this fraction, because the average repeat occurs ∼150 times in the Diodon genome. The repetitive frequency of the F. rubripes repetitive fraction is higher (9 × 105 copies/genome), and may indicate that most of this fraction is composed of satellite DNA.
Single-copy sequences were estimated to comprise 79% and 71% of the F. rubripes and D. hystrix genomes, respectively (Table 2). The repetitive frequency of these components was assumed to be 1 copy/genome. The complexity of these components, defined as the estimated length of the longest nonrepeating sequence in the class, was, as expected, at least two orders of magnitude greater than the complexity of the repetitive components in both curves.
Deletion/Insertion Profile
We used genomic DNA from four species of smooth pufferfish and four species of spiny pufferfish to estimate insertion and deletion rates (see Methods for species and sample sources). We PCR-amplified a conserved region of the reverse transcriptase from non-LTR retrotransposons. Between 15 and 25 sequences 300–600 bp in length were generated from cloned PCR-amplified mixtures from one of each species of fish. The final smooth pufferfish alignment contained 73 sequences of the previously described Maui non-LTR retrotransposon (Poulter et al. 1999) and was 706 bp in length. The final spiny pufferfish alignment contained 96 sequences of an undescribed non-LTR retrotransposon and was 485 bp in length. This undescribed element is more similar to Maui than any other non-LTR element identified in F. rubripes according to BLAST analysis (Altschul et al. 1997), but exhibits only 25%–30% amino acid identity, and therefore cannot be assumed to be orthologous. It was not possible to amplify any non-LTR element sequences from spiny puffer genomic DNA using Maui-specific primers, and degenerate primers failed to amplify any spiny elements with greater homology to Maui.
Terminal branch mutations occurring in the sequences were determined through phylogenetic analysis of aligned sequences for each group of puffers. Maximum parsimony and neighbor-joining analyses yielded very similar topologies and branch lengths for each data set, with most differences in topology occurring in deep internal branches. The single most parsimonious tree for the spiny puffer alignment and a 50% majority rule consensus of the six most parsimonious trees for the smooth puffer alignment were used for calculation of the mutation rate. Mutations that mapped to terminal branches of each tree were considered to have occurred during the neutral phase of an element's evolution, and were used in subsequent analyses of mutation profiles.
Virtually all insertions and deletions mapped to the terminal branches of the smooth and spiny puffer trees, but a small number of shared deletions were observed in both data sets. These deletions were in each case shared between two elements amplified from the same species, and therefore likely represent alleles of a common locus rather than separate insertions from a functional element containing a deletion. Sequences did not group strictly according to species in the trees, indicating that multiple active element lineages were possibly present in the ancestors of each family. The average number of terminal branch substitutions per base pair in the smooth and spiny alignments was 0.016 and 0.021, respectively. Assuming a time-uniform nucleotide substitution rate on the order of 10−9 per base pair per generation and a 1.5-yr generation time indicates that on average the sequences in these alignments have been neutrally evolving for 25 to 35 million years. We are confident, then, that our mutation rate estimates reflect the period of time during which the change in genome size may have taken place in the tetraodontid lineage, rather than a time period preceding the divergence of the two pufferfish families.
The distribution of insertion and deletion sizes was similar between the two groups of fish (Fig. 2). The number of indel events in each sequence is expected to be positively correlated with the number of point substitutions after a period of neutral evolution. This is the case for both the smooth and the spiny pufferfish data sets, as seen in Figure 3 (Spearman's rs = 0.501, p < 0.0001 for spiny; rs = 0.367, p < 0.002 for smooth).
Figure 2.
Frequency histogram of deletion sizes for tetraodontid (smooth) and diodontid (spiny) puffers. Represented are 31 deletions from the tetraodontid alignment and 48 deletions from the diodontid alignment.
Figure 3.
Number of terminal indels in individual sequences plotted against the number of terminal nucleotide substitutions in each sequence for the tetraodontid (A) and diodontid (B) data sets. Lines are linear regressions with the intercept fixed at 0. A significant correlation is detected in both plots using Spearman's rank correlation statistic.
Neutral evolution in the terminal branches of the smooth and spiny puffer trees is further supported by analysis of the distribution of point substitutions among codon positions. Terminal branch substitutions are approximately equally distributed among the three codon positions (χ2 = 2.98, p < 0.22 for spiny; χ2 = 5.61, p < 0.061 for smooth). Third-position substitutions are highly overrepresented on the internal branches, however, where purifying selection favors synonymous mutations (χ2 = 16.13, p < 0.001 for spiny; χ2 = 29.42, p < 0.001 for smooth).
Estimates of deletion and insertion profiles for these two pufferfish groups are very similar (Table 3). Deletion and insertion rates scaled according to terminal nucleotide substitutions (tns) were estimated with a maximum likelihood technique that assumes these mutations are Poisson processes (Blumenstiel et al. 2002). The 95% confidence intervals were estimated by performing bootstrap resampling of the data to re-estimate the parameter (Blumenstiel et al. 2002). Tests for significance of rate differences were carried out by comparing 1000 bootstrap estimates of the parameters and determining the proportion of comparisons in which one parameter estimate was larger than the other estimate (Blumenstiel et al. 2002).
Table 3.
Deletion and Insertion Profile in Tetraodontid (Smooth) and Diodontid (Spiny) Puffers
| Tetraodontidae | Diodontidae | Significance | |
| Ratio of deletions to point substitutions | 0.04 (0.026–0.063) | 0.06 (0.044–0.079)a | NSb |
| Ratio of insertions to point substitutions | 0.03 (0.017–0.049) | 0.03 (0.018–0.043) | NS |
| Average deletion size (bp) | 19.8 | 19.1 | NS |
| Average insertion size (bp) | 2.7 | 2.6 | NS |
| Deletion bias (base pairs lost/substitution)c | 0.7 | 1.1 | NS |
Numbers in parentheses are 95% confidence intervals estimated by bootstrapping.
Not statistically significant.
Defined as (deletion rate) × (average deletion size) − (insertion rate) × (average insertion size).
Neither the scaled deletion rate nor the scaled insertion rate is significantly different between smooth and spiny puffers (deletions: p < 0.095; insertions: p < 0.569). In spiny puffers the deletion rate is significantly larger than the insertion rate (0.06 deletions/tns vs. 0.03 insertions/tns; p < 0.001). In smooth puffers the insertion and deletion rates are not significantly different (0.04 deletions/tns vs. 0.03 insertions/tns; p < 0.142).
Average deletion and insertion sizes were also computed for both groups, and no significant difference between smooth and spiny puffers was found for either parameter (Wilcoxon test, deletions: p < 0.122; insertions: p < 0.94). The average deletion size is significantly larger than the average insertion size for both groups, however (Wilcoxon test, smooth: p < 0.001; spiny: p < 0.004). The average deletion size in smooth and spiny puffers, respectively, is 19.8 and 19.1 bp. The average insertion size in smooth and spiny puffers, respectively, is 2.7 and 2.6 bp. Given that the smooth and spiny pufferfish alignments differ in length, the possibility exists that the shorter spiny pufferfish alignment might bias the average insertion and deletion size estimates downward as a result of the alignment's smaller detection window. We sequenced 30% more spiny than smooth elements (96 vs.73) to provide some protection against this bias. Furthermore, we note that there were no deletions in the smooth puffer alignment large enough to escape capture in the smaller spiny puffer window (Fig. 4). Truncation of the observed deletion spectra above 40 bp yields average deletion size estimates of 8.96 bp in smooth puffers and 7.96 bp in spiny puffers, which are not significantly different (Wilcoxon test, p < 0.2122). We conclude that the small-scale insertion/deletion profiles for these two families of pufferfish are remarkably similar.
Figure 4.
Plot of insertion and deletion size in the tetraodontid (A) and diodontid (B) data sets. Points above the abscissa are insertions. Points below the abscissa are deletions. The abscissa indicates the midpoint of insertions and deletions on alignment. The tetraodontid alignment is 480 bp in length, and the diodontid alignment is 720 bp in length.
DISCUSSION
Spiny and smooth pufferfish have different genome size, and show variation in the proportions of DNA in different repetitive classes. Cot curve analysis has permitted us to efficiently characterize the repetitive nature of the undescribed genome of the spiny pufferfish D. hystrix, and compare it to the thoroughly sequenced genome of the smooth pufferfish F. rubripes. Our complementary estimation of the small-scale deletion/insertion profile in these puffer families has yielded new information on a mutational process thought to affect genome size (Petrov 2002). When the different repetitive composition of these genomes is considered together with the similar small-scale deletion and insertion rates of these two families of pufferfish, a decline in the rate of large-scale insertions is implicated as a probable cause of the genome size reduction in the tetraodontid (smooth) puffer lineage.
Renaturation Data
Analysis of the Cot curves produced for smooth and spiny pufferfish reveal that the genomes of fish in these two families differ not only in size, but also repetitive content (Fig. 1, Table 2).
We detected a significant amount of double-stranded “foldback” DNA present at the start of the renaturation reactions for both F. rubripes and D. hystrix. Although the foldback components of these pufferfish curves must be considered impure in terms of their repetitive composition (see Results), it is nevertheless likely that they contain the bulk of the highly repetitive satellite DNA found in these genomes. The centromeric heterochromatin of the smooth puffers F. rubripes and T. nigroviridis has been found to be largely composed of 118-bp tandem repeats (Brenner et al. 1993; Roest Crollius et al. 2000; Fischer et al. 2000). These tandem repeats possess some degree of self-complementarity, and analysis with the Mfold software program (SantaLucia 1998; SantaLucia et al. 1999) indicates they are capable of forming stem–loop structures at the temperatures and salt concentrations used in this renaturation experiment. Assuming there are similar tandem repeats in D. hystrix, these satellite sequences may contribute heavily to the foldback components observed in the curves. Although the proportion of foldback DNA differs between the two puffer genomes, the absolute amount in base pairs is very similar owing to the difference in genome size (∼56 Mb in spiny vs. ∼76 Mb in smooth).
The quantity of highly repetitive and middle repetitive DNA in the F. rubripes genome was too low to resolve the repetitive component into these two fractions with our experimental protocol. As the average repetitive frequency of this component is on the order of 105, however, this indicates that satellite DNA is the primary component of this fraction. Pizon et al. (1984) constructed a Cot curve for another species of smooth puffer, Arothron diadematus, and recorded values of 5% and 7%, respectively, for the proportions of highly repetitive and middle repetitive DNA classes in the genome of this fish. The smaller amount of repetitive DNA we detected in F. rubripes may be explained in part by the fact that F. rubripes has a slightly smaller genome than A. diadematus, with a haploid genome size of 0.42 pg of DNA/cell vs. 0.45 pg of DNA/cell for A. diadematus (Pizon et al. 1984; Brenner et al. 1993). The discrepancy may also be caused in part by the different protocols used to obtain each estimate. Some of the sequences belonging to our foldback component, for example, may be represented in the repetitive components of A. diadematus. Both estimates place the smooth puffer genome in stark contrast to the repetitive nature of other well-characterized vertebrate genomes in which, for example, ∼45% of the human genome is composed of transposable elements (International Human Genome Sequencing Consortium 2001).
Our renaturation data for D. hystrix indicate that ∼22% of its genome is composed of repetitive DNA, which is almost seven times the proportion of repetitive DNA we detected in F. rubripes. As with F. rubripes, highly repetitive and middle repetitive DNA classes were not resolvable. However, the repetitive frequency of the repetitive component of the genome is ∼150, indicating an abundance of middle repetitive sequences such as transposable elements relative to highly repetitive sequences such as satellite DNA. As with F. rubripes, this highly repetitive satellite DNA is likely represented in the foldback component of the D. hystrix Cot curve.
The single-copy components of the F. rubripes and D. hystrix genomes do not differ very much in proportion, but do vary in terms of absolute size when the difference in genome size is considered. Our renaturation data indicate that single-copy sequences comprise ∼316 Mb (79%) of the ∼400 Mb F. rubripes genome, and 568 Mb (71%) of the ∼800 Mb D. hystrix genome. The contraction of this component in smooth puffers has probably come mainly at the expense of noncoding intergenic spaces and introns. Even the smooth puffer genome is not at the minimum viable size for vertebrates, however. It is estimated that only 108 Mb of the F. rubripes genome is genic sequence (Aparicio et al. 2002), which leaves nearly 200 Mb of single-copy sequence to which no function has been assigned.
Deletion/Insertion Profile
All previous measurements of small insertions and deletions in other organisms have revealed an overall bias toward DNA loss (Graur et al. 1989; Petrov and Hartl 1998; Petrov et al. 2000; Robertson 2000; Bensasson et al. 2001). This bias is observed whether the sequence data are derived from nuclear pseudogenes, nuclear mitochondrial pseudogenes, or defunct non-LTR elements as in the case of our data (for review, see Petrov 2002). This bias derives from a higher rate of deletions than insertions, as well as a larger average deletion size than insertion size. Pufferfish of the families Tetraodontidae and Diodontidae exhibit a similar bias toward DNA loss, generated by deletions that are larger and more frequent than insertions.
Higher rates of DNA loss through small (⪝400 bp) deletions have been observed to be negatively correlated with genome size in arthropods (Petrov et al. 2000; Bensasson et al. 2001). Laupala crickets and Podisma grasshoppers, which have genomes roughly 10 times and 100 times, respectively, the size of Drosophila melanogaster, have respective rates of DNA loss 13 times and 75 times slower than the rate of loss in Drosophila (for review, see Petrov 2002).
We did not detect a significant difference in the insertion or deletion profiles between the Tetraodontidae and Diodontidae families, despite a twofold difference in genome size. This similarity in mutational profiles should not necessarily be surprising, however, as Petrov and Hartl (1998) failed to detect such a difference between two Drosophila species with a similar difference in genome size. Either the small-scale deletion bias is not an important determinant of genome size variation among arthropods or vertebrates with only twofold differences in genome size, or perhaps the present method is insufficient to detect important variations in this parameter.
Dasilva et al. (2002) recently reported an independent estimation of the small-scale deletion/insertion profile in the smooth puffer T. nigroviridis from pseudogene data. Their data indicate a smaller deletion bias than do our observations from DOA retrotransposons (0.56 vs. 0.71 bp lost per nucleotide substitution; Table 3), but are concordant with our results in that they found a deletion bias greater than that detected in mammals (Graur et al. 1989). From analysis of 66 copies of the Trapeze pseudogene, Dasilva et al. (2002) found an average deletion size of 7.7 bp and an average insertion size of 1.5 bp. These values are approximately half the size of the averages we computed from mutations observed in DOA retrotransposon sequences (Table 3). One cause for the discrepancy may be selection acting on mutations in the T. nigroviridis pseudogenes (Dasilva et al. 2002). Dasilva et al. (2002) found a higher incidence of substitutions, insertions, and deletions in the exons than in the introns of the pseudogene sequences, and they suggest that there may be selection pressure to inhibit the pseudogenes' capacity to form functional mRNA or proteins. In any case, the discrepancy between the pseudogene estimate and our estimate of deletion bias in smooth puffers does not affect conclusions deriving from the relative values of rates we measured in smooth and spiny puffers.
Mechanisms of Genome Size Evolution in Pufferfish
Our complementary analyses of repetitive pattern and mutational process in pufferfish genomes have permitted us to build a dynamic understanding of genome evolution in this taxonomic group. To extend these observations it is important to establish whether these pufferfish genomes are in flux or equilibrium (Petrov 2002). If tetraodontid and diodontid pufferfish genomes are presently at equilibrium with regard to size, then a deletion bias at the length scale observable in this experiment (<400 bp) must be counteracted by an insertion bias at a larger scale (else the genomes would be quickly reduced to their minimum viable size). It is not clear whether the tetraodontid genomes are still contracting, but it seems likely that the diodontid genomes are at an equilibrium size, given that the four measurements of genome size for fish in this family are highly similar to each other as well as to the genome size of the outgroup taxon M. mola (Table 1). Given this equilibrium, we may therefore infer that there is an unobserved input of DNA at larger length scales into the diodontid genome to compensate for the observed small-scale deletion bias.
The difference in genome size between the puffer families implies that an ancestral equilibrium was disturbed in the smooth tetraodontid lineage following its divergence from the spiny diodontids. A lack of variation between the pufferfish families in the insertion/deletion profile at the length scale observable in this experiment indicates that the disturbance in equilibrium resulted from a change in the insertion/deletion profile in the tetraodontid lineage at a larger scale. In other words, the rate or size of large deletions relative to large insertions may have increased, or the rate or size of large insertions relative to large deletions may have decreased. Petrov (2002) has speculated on the nature of large-scale insertions and deletions, and points out that there are fundamental limits to the size of deletions that are tolerated by the genome. Because all deletions have two breakpoints, a larger deletion creates a greater chance that one or both of the breakpoints will occur in a region that is under natural selection, or that the deletion wholly removes a functional sequence. Such deletions will probably be selectively deleterious, and because they will be eliminated from the population they will have no effect on genome size. Insertions have no such constraints. If an insertion occurs between two individual nucleotides in a stretch of noncoding DNA, it should make no difference whether the insertion is 4 nucleotides in length or 4000. The insertion will likely be selectively neutral in either case, and will be fixed or lost from the population with the same probability as other neutral mutations. A reduction in the rate of large insertions in the tetraodontid lineage, rather than an increase in large deletions, may therefore explain that family's genome size contraction.
It is possible to calculate whether a difference in large insertions alone would be sufficient to create the difference in genome size observed between diodontid and tetraodontid puffers. If one assumes that no large insertions or deletions >400 bp are occurring, the amount of DNA lost (L) from small-scale deletion bias over a given time period can be calculated:
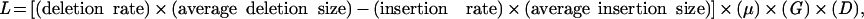 |
where insertion and deletion rates are scaled according to nucleotide substitutions, μ is the nucleotide substitution rate, G is the haploid genome size in base pairs, and D is the number of generations since divergence. Assuming a conservatively low nucleotide substitution rate of 10−9 per base pair per generation, a diploid genome size of 800 Mb, and 40 million generations since the divergence of the families (assuming a 1.5-yr generation time), one can calculate that the amount of DNA lost is estimated to be on the order of 350 Mb. This is nearly sufficient to account for the difference in size between the smooth and spiny puffer genomes. As a result, the contraction of smooth puffer genome size could be caused by a severe reduction of large insertions in this lineage compared with the spiny puffers. Indeed, our renaturation data indicate that some portion of these missing large insertions may result from reduced transposable element activity, as middle repetitive sequences are rare in F. rubripes but comprise roughly a fifth of the D. hystrix genome.
Small Deletions Versus Large Insertions as Determinants of Genome Size
The small size of introns and intergenic spaces in the F. rubripes genome has long been thought to have been generated by a higher rate of DNA loss in this fish compared with humans (Brenner et al. 1993; Aparicio et al. 2002). This hypothesis is supported by our study. Although Graur et al. (1989) measured a small-scale deletion rate in primates and rodents very similar to what we measured in pufferfish, they detected a much smaller average deletion size. The average deletion size in mammals is only 3.2 bp, which is nearly six times smaller than the 19-bp average deletion size seen in puffers. The insertional profile (size and rate) is similar in mammals and pufferfish. Thus, the small-scale deletion bias seems to explain at least part of the nearly eightfold difference in genome size between smooth tetraodontid puffers and humans.
Indeed, the small-scale deletion biases we measured in smooth and spiny pufferfish accord well with an observed inverse correlation between this parameter and genome size in a wide variety of organisms (Petrov 2002), and support a hypothesis of a causal relationship between these factors. For example, the rate of small-scale DNA loss (base pairs per 1 substitution) in smooth and spiny puffers is slower than the rate in Drosophila melanogaster (Petrov and Hartl 1998) and Caenorhabditis elegans (Robertson 2000), each of which have smaller genomes than smooth and spiny pufferfish. Similarly, Laupala crickets and Podisma grasshoppers have larger genomes than pufferfish, and exhibit a slower rate of small-scale DNA loss (Petrov et al. 2000; Bensasson et al. 2001).
Although the small-scale deletion bias may offer explanatory power with regard to many-fold differences in genome size between these distant animal taxa, it is not capable of explaining the twofold difference in genome size between diodontid (spiny) and tetraodontid (smooth) pufferfish. A substantial amount of DNA has been lost in smooth pufferfish relative to their spiny cousins since their divergence 50–70 million years ago, without any significant change in the rate of small-scale DNA loss. Indeed, the rate of deletion in the larger genome of spiny puffers is actually slightly larger than in smooth puffers, although not significantly so (Table 3). Because the average-sized spiny puffer genome appears to be in equilibrium, one might argue that it is primarily a recent contraction of genome size in smooth puffers that is responsible for their curiously small genome size. To evolve the smallest vertebrate genome yet measured, it was necessary that tetraodontid puffers not only experience a high rate of DNA loss, but also a low rate of large DNA insertions.
Small deletions have been given much attention as a possible determinant of eukaryotic genome size. However, given that the minuscule size of the smooth puffer genome may have been achieved through a change in the rate of large insertions, it may be argued that this side of the mutational spectrum should receive more attention. Indeed, the observation that few eukaryotic genomes are even close to their presumed minimum viable size implies that large-scale insertions must constantly buoy them up against a seemingly ubiquitous small-scale deletion bias. Systematic exploration of both increases and decreases in transposable element activity over time at various taxonomic scales should do much to clarify the role these pervasive characters have played in eukaryotic genome evolution.
METHODS
Renaturation Analysis
Somatic tissue samples from F. rubripes and D. hystrix were collected in Japan and Puerto Rico, respectively. Genomic DNA was obtained from somatic tissue of F. rubripes and D. hystrix using a phenol/chloroform extraction after the method of Sambrook et al. (1989). Samples were treated with RNase and then verified through a UV/vis spectrophotometer to have an OD260/280 ratio of at least 1.80 and an OD260/230 ratio of at least 2.30. High-molecular-weight DNA was diluted with water to a concentration of <500 μg/mL and then sonicated for four periods of 30 sec to fragment it. DNA was checked on 1.5% agarose gels to ensure that no fragments larger than 1 kb were present and that the average fragment size was ∼500 bp.
Sheared DNA was precipitated and then resuspended in a solution of 2× SSC to a concentration of 0.25 μg/μL. Purity was rechecked on a spectrophotometer. Resuspended DNA was aliquoted in 10-μL volumes to 200-μL PCR tubes for denaturation and renaturation in a thermalcycler. Aliquots were denatured through boiling for 9 min. The temperature in the thermalcycler was then dropped to 55°C to permit renaturation. Aliquots were removed from the thermalcycler at predetermined time points in groups of three replicates and immediately frozen in liquid nitrogen to prevent further renaturation. Frozen aliquots were then thawed at room temperature after a 10-μL volume of 1.5 U/μL S1 nuclease solution was added to each tube. Samples were incubated at 37°C for 80 min to permit nuclease digestion of single-stranded sequences. Following incubation, nuclease activity was halted by adding 20 μL of stop buffer (1 M Tris at pH 9.0, 0.1 M EDTA). The remaining double-stranded DNA in solution was labeled with Hoechst 33258 bisbenzamide dye and quantified with a fluorescence spectrophotometer (with excitation at 356 nm and emission at 456 nm).
Cot curves were constructed by plotting the proportion of DNA remaining in solution after digestion against the log of the ECot value of each sample. ECot values were determined by multiplying a correction factor for nonstandard salt concentrations (E) with the molar concentration of nucleotides (Co) and time permitted for renaturation in seconds (Britten et al. 1974). Data were analyzed with the least-squares reassociation kinetics program of William Pearson (Pearson et al. 1977), which fits a least-squares curve to the data and then calculates a rate constant and complexity value for each component. Cot1/2 values were determined by taking the log of the inverse of the rate constants provided by the program for each component (data not shown). The repetitive frequency of the repetitive components was estimated by assuming the repetitive frequency of the single-copy component was 1 copy/genome, and then dividing the rate constant of each repetitive component by the rate constant of the single-copy component for the same genome.
A partial Cot curve for E. coli strain TOP10 (Invitrogen) was also constructed using the above techniques.
Insertion and Deletion Rate Estimates
Genomic DNA was extracted from somatic tissue samples of four species of tetraodontid (smooth) puffers and four species of diodontid (spiny) puffers using NucleoSpin columns (Clontech). The smooth puffers studied were F. rubripes, T. nigroviridis, Canthigaster valentini, and Canthigaster jactatus. The spiny puffers used were D. hystrix, Diodon holacanthus, Diodon eydouxii, and Chilomycterus schoepfi. Tissue samples from F. rubripes and D. eydouxii were collected in Japan, samples from T. nigroviridisand D. holacanthus were obtained from fish in the aquarium trade, and the remaining samples were obtained from the University of Kansas Museum of Natural History Tissue Collection. The reverse transcriptase region of non-LTR retrotransposons was initially amplified from D. holacanthus and F. rubripes using the degenerate primers DVO144 and DVO145 of Wright et al. (1996) according to the PCR protocol that accompanies their description. The resultant amplifications were cleaned with Qiaquick columns (QIAGEN) and cloned into XL1-Blue cells using a TOPO TA cloning kit (Invitrogen). Random clones were screened with vector primers, and inserts of all sizes were sequenced to prevent bias in insertion/deletion rate estimates. Multiple sequences from each transformation were generated on an ABI 3100 sequencer. All inserts were sequenced in the forward and reverse directions. Ambiguous base calls were discounted from further analysis or resequenced. BLAST analysis (Altschul et al. 1997) confirmed that the sequences from D. holacanthus were an undescribed non-LTR retrotransposon, and indicated that the majority of sequences from F. rubripes were the previously described non-LTR retrotransposon Maui (Poulter et al. 1999). Sequences were aligned with CLUSTAL X (Thompson et al. 1997), with all parameters set to default except for the gap opening penalty (set to 20.0) and the DNA transition rate (set to 0). Alignments were then adjusted by hand in BioEdit (Hall 1999) to minimize the number of insertions and deletions. Adjustments were performed to minimize the number of terminal substitutions when such adjustments did not add to the number of insertions or deletions. Ambiguous indel alignments were discounted from further analysis. Variations in the length of monomeric microsatellite repeats in both alignments were ignored. Both alignments contained a minority of sequences that appeared to be from different element families. These sequences were discarded to prevent bias in mutation rate estimation. Conserved regions from alignable sequences were used to design family-specific primers, as the degenerate DVO144/145 primers only weakly amplified reverse transcriptase sequences from these groups. MAUIF1 (ACCAGATGTGCTGACTGTGG) and MAUIR1 (TTGAGGAACTCCATGGCTAAC) were used to amplify Maui non-LTR reverse transcriptase sequences from smooth puffer samples, and DIODF1 (GTGGACAACAATAGCGCCAC) and DIODR1 (CCTTACAGATGAAATTACGGAGC) were used to amplify the undescribed element from the spiny puffer samples. All PCR reactions were performed under standard conditions with an annealing temperature of 56°C. Products amplified with these family-specific primers were cleaned, cloned, sequenced, and aligned as above. Sequences are deposited in GenBank under the following accession numbers: AY212336–AY212504.
Reverse transcriptase alignments for smooth and spiny puffers were used to build maximum parsimony and neighbor-joining phylogenies in PAUP* 4.0b10 (Swofford 2002). Heuristic searches with TBR branch-swapping were performed, and gaps were ignored. Branch length estimates from parsimony and distance trees were compared and verified to be significantly correlated (Spearman's rs < 0.001). Nucleotide substitutions, insertions, and deletions that mapped to the terminal branches of the most parsimonious tree for spiny puffers and a 50% majority rule consensus of six most parsimonious smooth puffer trees were used in subsequent analyses of mutation rate. These terminal, autapomorphic mutations are assumed to have arisen during the nonautonomous, neutral phase of the retroelements' evolution, and are assumed to represent the true, neutral, genome-wide mutation rate (Petrov et al. 1996). Maximum likelihood estimates of the ratio of insertions to nucleotide substitutions and the ratio of deletions to nucleotide substitutions were made using a method described in Blumenstiel et al. (2002), which accounts for biases resulting from differences in the observed versus the presumed original length of the reverse transcriptase sequences. This method assumes that nucleotide substitution and indel formation are Poisson processes. Bootstrap resampling of the original data was used to generate 1000 replicates of each parameter estimate, from which 95% confidence intervals for the rate estimates were derived (Blumenstiel et al. 2002).
Acknowledgments
We thank Justin Blumenstiel and William Pearson for help with our data analysis. We thank Toshiaki Itami, Christian Landry, Steve Vollmer, and the Museum of Natural History at the University of Kansas for tissue samples. Comments and criticism from Elena Lozovsky, Cristian Castillo-Davis, and two anonymous reviewers improved this paper. This work was supported by grants from the NSF to S.R.P. D.E.N. was supported in part by an NSF Pre-Doctoral Fellowship.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
E-MAIL neafsey@oeb.harvard.edu; FAX (617) 496-5854.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.841703.
REFERENCES
- 1.Altschul S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aparicio S., Chapman, J., Stupka, E., Putnam, N., Chia, J.M., Dehal, P., Christoffels, A., Rash, S., Hoon, S., Smit, A.F., et al. 2002. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science 297: 1301-1310. [DOI] [PubMed] [Google Scholar]
- 3.Bensasson D., Petrov, D.A., Zhang, D.X., Hartl, D.L., and Hewitt, G.M. 2001. Genomic gigantism: DNA loss is slow in mountain grasshoppers. Mol. Biol. Evol. 18: 246-253. [DOI] [PubMed] [Google Scholar]
- 4.Blumenstiel J.P., Hartl, D.L., and Lozovsky, E.R. 2002. Patterns of insertion and deletion in contrasting chromatin domains. Mol. Biol. Evol. 19: 2211-2225. [DOI] [PubMed] [Google Scholar]
- 5.Brainerd E.L., Slutz, S.S., Hall, E.K., and Phillis, R.W. 2001. Patterns of genome size evolution in tetraodontiform fishes. Evol. Int. J. Org. Evol. 55: 2363-2368. [DOI] [PubMed] [Google Scholar]
- 6.Brenner S., Elgar, G., Sandford, R., Macrae, A., Venkatesh, B., and Aparicio, S. 1993. Characterization of the pufferfish (Fugu) genome as a compact model vertebrate genome. Nature 366: 265-268. [DOI] [PubMed] [Google Scholar]
- 7.Britten R.J. and Kohne, D.E. 1968. Repeated sequences in DNA. Hundreds of thousands of copies of DNA sequences have been incorporated into the genomes of higher organisms. Science 161: 529-540. [DOI] [PubMed] [Google Scholar]
- 8.Britten R.J., Graham, D.E., and Neufeld, B.R. 1974. Analysis of repeating DNA sequences by reassociation. Methods Enzymol. 29: 363-418. [DOI] [PubMed] [Google Scholar]
- 9.Dasilva C., Hadji, H., Ozouf-Costaz, C., Nicaud, S., Jaillon, O., Weissenbach, J., and Roest Crollius, H. 2002. Remarkable compartmentalization of transposable elements and pseudogenes in the heterochromatin of the Tetraodon nigroviridis genome. Proc. Natl. Acad. Sci. 99: 13636-13641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elgar G., Clark, M.S., Meek, S., Smith, S., Warner, S., Edwards, Y.J., Bouchireb, N., Cottage, A., Yeo, G.S., Umrania, Y., et al. 1999. Generation and analysis of 25 Mb of genomic DNA from the pufferfish Fugu rubripes by sequence scanning. Genome Res. 9: 960-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fischer C., Ozouf-Costaz, C., Roest Crollius, H., Dasilva, C., Jaillon, O., Bouneau, L., Bonillo, C., Weissenbach, J., and Bernot, A. 2000. Karyotype and chromosome location of characteristic tandem repeats in the pufferfish Tetraodon nigroviridis. Cytogenet Cell Genet. 88: 50-55. [DOI] [PubMed] [Google Scholar]
- 12.Graur D., Shuali, Y., and Li, W.H. 1989. Deletions in processed pseudogenes accumulate faster in rodents than in humans. J. Mol. Evol. 28: 279-285. [DOI] [PubMed] [Google Scholar]
- 13.Gregory T.R. 2001. Coincidence, coevolution, or causation? DNA content, cell size, and the C-value enigma. Biol. Rev. Camb. Philos. Soc. 76: 65-101. [DOI] [PubMed] [Google Scholar]
- 14.Hall T.A. 1999. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acid Symp. Ser. 41: 95-98. [Google Scholar]
- 15.Hinegardner R. and Rosen, D.E. 1972. Cellular DNA content and the evolution of teleostean fishes. Am. Nat. 106: 621-644. [Google Scholar]
- 16.International Human Genome Sequencing Consortium 2001. Initial sequencing and analysis of the human genome. Nature 409: 860-921. [DOI] [PubMed] [Google Scholar]
- 17.Jack P.L. and Hardman, N. 1980. Sequence organization in nuclear deoxyribonucleic acid from Physarum polycephalum. Physical properties of foldback sequences. Biochem. J. 187: 105-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.John B. and Miklos, G.L.C., 1988. The eukaryotic genome in development and evolution, pp. 1–416. Allen and Unwin, London, UK.
- 19.Krajewski C. 1989. Comparative DNA reassociation kinetics of cranes. Biochem. Genet. 27: 131-136. [PubMed] [Google Scholar]
- 20.Ojima Y. and Yamamoto, K. 1990. Cellular DNA contents of fishes determined by flow cytometry. La Kromosomo II 57: 1871-1888. [Google Scholar]
- 21.Pearson W.R., Davidson, E.H., and Britten, R.J. 1977. A program for least squares analysis of reassociation and hybridization data. Nucleic Acids Res. 4: 1727-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petrov D.A. 2002. Mutational equilibrium model of genome size evolution. Theoret. Pop. Biol. 61: 531-544. [DOI] [PubMed] [Google Scholar]
- 23.Petrov D.A. and Hartl, D.L. 1997. Trash DNA is what gets thrown away: High rate of DNA loss in Drosophila. Gene 205: 279-289. [DOI] [PubMed] [Google Scholar]
- 24.___, 1998. High rate of DNA loss in the Drosophila melanogaster and Drosophila virilis species groups. Mol. Biol. Evol. 15: 293-302. [DOI] [PubMed] [Google Scholar]
- 25.Petrov D.A., Lozovskaya, E.R., and Hartl, D.L. 1996. High intrinsic rate of DNA loss in Drosophila. Nature 384: 346-349. [DOI] [PubMed] [Google Scholar]
- 26.Petrov D.A., Sangster, T.A., Johnston, J.S., Hartl, D.L., and Shaw, K.L. 2000. Evidence for DNA loss as a determinant of genome size. Science 287: 1060-1062. [DOI] [PubMed] [Google Scholar]
- 27.Pizon V., Cuny, G., and Bernardi, G. 1984. Nucleotide sequence organization in the very small genome of a tetraodontid fish, Arothron diadematus. Eur. J. Biochem. 140: 25-30. [DOI] [PubMed] [Google Scholar]
- 28.Poulter R., Butler, M., and Ormandy, J. 1999. A LINE element from the pufferfish (fugu) Fugu rubripes which shows similarity to the CR1 family of non-LTR retrotransposons. Gene 227: 169-179. [DOI] [PubMed] [Google Scholar]
- 29.Robertson H.M. 2000. The large srh family of chemoreceptor genes in Caenorhabditis nematodes reveals processes of genome evolution involving large duplications and deletions and intron gains and losses. Genome Res. 10: 192-203. [DOI] [PubMed] [Google Scholar]
- 30.Roest Crollius H., Jaillon, O., Dasilva, C., Ozouf-Costaz, C., Fizames, C., Fischer, C., Bouneau, L., Billault, A., Quetier, F., Saurin, W., et al. 2000. Characterization and repeat analysis of the compact genome of the freshwater pufferfish Tetraodon nigroviridis. Genome Res. 10: 939-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook J., Fritsch, E.F., and Maniatis, T., 1989. Molecular cloning: A laboratory manual Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 32.SantaLucia J. 1998. A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl. Acad. Sci. 95: 1460-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Swofford D.L., 2002. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods), Version 4, Sinaur Associates, Sunderland, MA.
- 34.Thompson J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. 1997. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tyler J.C. 1980. Osteology, phylogeny, and higher classification of the fishes of the order Plectognathi (Tetraodontiformes). NOAA Tech. Rep. NMFS Circ. 434: 1-422. [Google Scholar]
- 36.Tyler J.C. and Santini, F. 2002. Review and reconstructions of the tetraodontiform fishes from the Eocene of Monte Bolca, Italy, with comments on related Tertiary taxa. Studi e ricerche sui Giacimenti Terziari di Bolca. Mus. Civico di Storia Nat. di Verona 9: 117-119. [Google Scholar]
- 37.Verneau O., Renaud, F., and Catzeflis, F.M. 1991. DNA reassociation kinetics and genome complexity of a fish (Psetta maxima: Teleostei) and its gut parasite (Bothriocephalus gregarius: Cestoda). Comp. Biochem. Physiol. B 99: 883-886. [DOI] [PubMed] [Google Scholar]
- 38.Vinogradov A.E. 1998. Genome size and GC-percent in vertebrates as determined by flow cytometry: The triangular relationship. Cytometry 31: 100-109. [DOI] [PubMed] [Google Scholar]
- 39.Wright D.A., Ke, N., Smalle, J., Hauge, B.M., Goodman, H.M., and Voytas, D.F. 1996. Multiple non-LTR retrotransposons in the genome of Arabidopsis thaliana. Genetics 142: 569-578. [DOI] [PMC free article] [PubMed] [Google Scholar]









