Abstract
Background
The heart develops under reduced and varying oxygen concentrations, yet there is little understanding of oxygen metabolism in the normal and mal‐development of the heart. Here we used a novel reagent, the ODD‐Luc hypoxia reporter mouse (oxygen degradation domain, ODD) of Hif‐1α fused to Luciferase (Luc), to assay the activity of the oxygen sensor, prolyl hydroxylase, and oxygen reserve, in the developing heart. We tested the role of hypoxia‐dependent responses in heart development by targeted inactivation of Hif‐1α.
Methods and Results
ODD‐Luciferase activity was 14‐fold higher in mouse embryonic day 10.5 (E10.5) versus adult heart and liver tissue lysates. ODD‐Luc activity decreased in 2 stages, the first corresponding with the formation of a functional cardiovascular system for oxygen delivery at E15.5, and the second after birth consistent with complete oxygenation of the blood and tissues. Reduction of maternal inspired oxygen to 8% for 4 hours caused minimal induction of luciferase activity in the maternal tissues but robust induction in the embryonic tissues in proportion to the basal activity, indicating a lack of oxygen reserve, and corresponding induction of a hypoxia‐dependent gene program. Bioluminescent imaging of intact embryos demonstrated highest activity in the outflow portion of the E13.5 heart. Hif‐1α inactivation or prolonged hypoxia caused outflow and septation defects only when targeted to this specific developmental window.
Conclusions
Low oxygen concentrations and lack of oxygen reserve during a critical phase of heart organogenesis may provide a basis for vulnerability to the development of common septation and conotruncal heart defects.
Keywords: conotruncus, hypoxia, neural crest, persistent truncus arteriosus
Introduction
The prolyl hydroxylases (PHD; EGLN1‐3) and their substrates the hypoxia‐inducible transcription factor (HIF) function as evolutionarily conserved sensors and respondents to O2 deprivation, ie, tissue hypoxia.1–2 Embryonic development is a prototypical hypoxic response as all of the elements required for O2 delivery including the placenta, blood and blood vessels, must be generated de novo.3 That an intact hypoxia signaling system is required for mammalian development is demonstrated by the mid‐gestational lethality that occurs when Hif or Phd family members are inactivated in the mouse germ line.4 It has also been proposed that pathological O2 deprivation at these critical developmental stages, for example due to reductions in blood flow or inspired O2, may result in congenital defects in the cardiovascular and other systems.5 Quantifying tissue O2 concentration and O2 deprivation in normal and pathological developmental processes is problematic and usually relies on semi‐quantitative or qualitative chemical (nitroimidazoles) and biological (HIF‐1α) indicators. A further limitation of these indicators is that they do not directly report on the sufficiency of the O2 supply for the enzymatic reactions for which it is required.2 More recently, a mouse model was created to serve as a novel reagent to report on oxygen‐dependent PHD activity within tissues by incorporation of the Hif‐1α ODD into the Luciferase (Luc) coding sequence expressed from the Rosa26 locus.6 Luciferase activity can be accurately quantified over 6 logs of activity in tissue homogenates or in vivo through bioluminescent imaging.
Our interest has been in the role of O2 deficiency in the later stages of heart morphogenesis, in particular the septation of the heart and formation of the outlet structures that occurs at ≈E11.5 to 15.5 of mouse development.7 These processes are of particular interest given the relatively high prevalence and severity of congenital septation and outlet defects in humans.8 At E11.5 to 15.5, prior to the formation of a functional coronary vascular system, O2 is delivered to the cardiac tissues by passive diffusion from red blood cells within the chambers. We, and others,9–11 have proposed based on the semi‐quantitative indicators that the outlet myocardium and mesenchyme and ventricular septal myocardium are relatively hypoxic at these stages due to the tissue barrier to O2 diffusion. However, the role of O2 gradients in the normal and defective formation of the outlet structures has not been determined. In the current study we used ODD‐Luc mice as a novel quantitative indicator of O2 dependent PHD activity in normal mouse development and during maternal O2 deprivation, and conditional inactivation of Hif‐1α and maternal O2 deprivation to test the role of HIF and hypoxia signaling targeting a specific developmental window based on the measurements of ODD‐Luc activity.
Methods
Mice
Mice were handled in accordance with CWRU and University of Maryland‐Baltimore Institutional Animal Care and Use Committee guidelines. All mouse strains used in this study were obtained from Jackson Laboratory. ODD‐Luc6 (stock # 006206), in which the ODD of Hif‐1α is fused to firefly luciferase expressed from the Rosa26 locus and WT‐Luc12 with the same construct lacking ODD (stock # 005125). Both lines were maintained on an FVB background. Tamoxifen (TM) inducible β‐actinCre (CAGGCre‐ER™; stock # 004682),13 Rosa26 containing a LacZ reporter used to trace cell lineage (Stock # 003474) and Hif‐1αf/f, in which LoxP sites flank exon 2 of Hif‐1α (stock # 007561)14 were maintained on a C57BL/6J background. NCC specific Wnt1Cre15 mice were maintained on a Swiss Webster background. Mice were mated and genotyped by standard methods with primers listed at jax.org. In the mating between β‐actinCre+ males and Hif‐1αf/f females, 3 mg TM (MP Biomedical) in 100 μL of sunflower oil (Spectrum Chemical Corp) was injected i.p. to pregnant mice to induce the activity of Cre recombinase as previously described.13 To assess efficiency of Cre‐mediated recombination at the HIf‐1α locus, genomic DNA was isolated from embryonic heart and liver, PCR amplified with primers flanking the loxP sites (Table 1) and bands separated and quantified by gel electrophoresis and Syto60 staining (S11342, Invitrogen).
Table 1.
List of Primers
| Gene | Forward (5′ to 3′) | Reverse (5′ to 3′) |
|---|---|---|
| Mouse real‐time PCR | ||
| CCN1 | AAGCTTCCAGCCCAACTGTA | CAAACCCACTCTTCACAGCA |
| CCN2 | AGAGTGGAGCGCCTGTTCTA | GCAGCCAGAAAGCTCAAACT |
| BNIP3 | GGCGTCTGACAACTTCCACT | AACACCCAAGGACCATGCTA |
| IGFBP1 | TCGGAGATTTCCTCATCGTC | ATGTATGGGACGCAGCTTTC |
| GLUT1 | GGATCTCTCTGGAGCACAGG | TCCTCCTGGACTTCACTGCT |
| β‐actin | GACAGGATGCAG AAGGAG AT | GAAGCATTTGCGGTGGACGAT |
| β‐actinCre efficiency | ||
| GGATGAAAACATCTGCTTTGG | ACTGCCCCAACACAATACTTTT | |
| Chicken real‐time PCR | ||
| CCN1 | ATATGCGAAGTGAGGCCGTG | TTGGGGCGGTACTTCTTCAC |
| CCN2 | ATCTCGACCAGGGTCACCAA | GGGGTGCGAATGCACTTTTT |
| CCN3 | CCTGGGAGGCTTTGCTATGG | ACGGGTAGAAAAGCCCATTCC |
| BNIP3 | CCAGCGTCATGAAGAAAGGG | GGCGCCTCCCAATGTAAATC |
| IGFBP1 | AAATTGGCAAAGGCTCAGCAG | GCGGAATCTCCATCCAGTGAA |
| GLUT1 | AAGATGACAGCTCGCCTGATG | GCTCCTCATATCGGTACAGCC |
| β‐actin | GCCATGGATGATGATATTGCTGC | TGGCCCATACCAACCATCAC |
Hypoxic Stress
For acute exposure to hypoxia timed pregnant mice were placed in a hypoxia chamber (HypOxyc System, Kent Scientific). The O2 concentration was gradually decreased to 8% and maintained for 4 hours. As a control mice were placed in the chamber and exposed to room air. For chronic exposure to hypoxia mice were placed in a chamber under hypobaric hypoxia: 0.5 atmospheres (ATM), half of the normal O2 content at sea level (1 ATM). The pressure was slowly reduced through a servo‐controlled system and a vacuum line. Except where noted the mice were placed in the chamber at E10.5, returned to a normal environment at E13.5, and examined at E15.5. The mice had free access to food and water on a circadian light/dark cycle.
Measurement of Luciferase Activity
Pregnant mice were euthanized by CO2 inhalation followed by cervical dislocation and the uterine horns removed and embryos dissected in ice cold PBS. Tissues were homogenized in 1× Cell Culture Lysis Reagent (Promega) and clarified by centrifugation. Luciferase activity was measured in 20 μL of lysate in a 96‐well plate using the Luciferase Assay System (Promega) and a Flex Station 3 plate reader (Molecular Devices). Luminescence values were normalized to total protein and expressed as luminescence units (LU)/mg protein. Protein concentrations were measured with the Bio‐Rad Protein Assay Dye Reagent.
Bioluminescence Imaging
Fifteen minutes after a single i.p. injection of d‐luciferin (150 mg/kg; Caliper Life Sciences) mice were anesthetized with 2.5% isofluorane and euthanized by cervical dislocation. The uterine horns were removed, individual embryos harvested and placed in supine position in a petri dish containing ice cold PBS. The skin between the forelimbs was gently removed to expose the heart and the embryos were placed in a light‐tight bioluminescence imaging chamber coupled to a high‐sensitivity charged coupled device camera (Xenogen IVIS‐200). The field of view was set to A and photons were collected with an exposure time of 30 seconds. Images were analyzed with Living Image‐3 software (Xenogen). The bioluminescence signal intensity is expressed as photons sec−1 (cm2)−1 steradian−1.
Quantitative RT‐PCR
Total RNA was isolated from E11.5 and E15.5 embryonic hearts and livers using RNAeasy mini kit (Qiagen). For E11.5 hearts 2 hearts were pooled for each sample. One microgram of total RNA was reverse transcribed (SSIII, Invitrogen) and analyzed by real‐time PCR using Fast SYBR Green PCR Master Mix (Applied Biosystems) and an Applied Biosystems StepOnePlus thermal cycler. Primers were designed with Primer3plus software (Table 1). Data was analyzed by the comparative Ct method, normalized to β‐actin and reported as fold‐change versus normoxia.
Morphologic and Histologic Analyses
Except where noted embryos were harvested at E15.5 for analysis. Pregnant mice were sacrificed by cervical dislocation and embryos dissected into ice‐cold PBS with 40 mmol/L KCl to relax the heart. In most embryos the chest was not opened so as to preserve the plane of sectioning. Embryos were fixed, sectioned in a coronal plane with respect to the embryo and stained with hematoxylin and eosin. In embryos in which the Rosa26‐LacZ reporter was used to fate map NCCs, tissues were reacted with X‐gal at 37°C overnight in whole mount or section using standard methods. Images were captured with a Spot RT digital camera and optimized with Adobe Photoshop software.
Hypoxia Exposure of Chick Embryos In Ovo and mRNA Analysis
Fertile White Leghorn (Gallus gallus) chicken eggs were obtained from Squire Valleevue Farm (Cleveland, OH). Eggs were incubated in a humidified room air incubator at 38°C until Hamburger‐Hamilton (HH) Stage 25 (ED 4). Eggs were then placed in a sealed chamber (Modular Incubator Chamber, Billups‐Rothenberg, Inc) with water and flushed with 7.5% O2/92.5% N2 for 10 minutes in order to exchange the inside air completely, and then the inlet and outlet tubes were clamped. Control eggs were incubated in the same chamber in room air. Embryos used for array studies were injected with DMOG (Sigma Inc) 10 mmol/L stock with 5 μL dripped onto the embryo after a piece of shell was removed. The eggs were covered with Parafilm prior to further incubation. After 6 hours of incubation in hypoxia the embryos were removed and RNA purified from the apical portions of the hearts using RNAeasy micro kit (Qiagen). Total RNA was reverse transcribed and hybridized against chicken microarrays (Affymetrix Chicken Gene 1.0 ST array). Gene expression in 3 control and 3 DMOG+hypoxia samples was measured for a total of 9 pairwise comparisons, with a threshold for fold‐change of 2‐fold. In additional experiments fold change of selected transcripts was validated by real‐time PCR as described above. In these experiments we observed no difference in embryos treated with DMOG+hypoxia (n=4) versus hypoxia alone (n=3) so these were combined into 1 group for statistical analysis.
Statistical Analysis
Data are expressed as mean±SEM. Differences between 2 groups were examined by Student unpaired t test. The Shapiro‐Wilk test was used to ensure that data was normally distributed. Multiple groups were compared by 1‐way ANOVA followed by Student t test with Bonferroni correction. Tests for interactions between developmental stage and hypoxic induction of ODD‐Luc were performed by 2‐way ANOVA using Holm‐Sidak method. Fisher's exact test was used to compare the prevalence of cardiac defects and survival frequencies. P<0.05 were considered statistically significant. SigmaPlot 12 software (Systat Software Inc) was used for these analyses.
Results
ODD‐Luc Activity During Normal Mouse Development and With Maternal O2 Deprivation
Luciferase activity was measured in ODD‐Luc mouse tissue homogenates throughout development as a reporter for O2‐dependent PHD activity. Activity normalized to total protein was highest in the E9.5 embryo (Figure 1A). Dissection of individual organs was not feasible at this stage due to their small size. In the embryonic heart, activity was highest at E10.5 and declined 3.5‐fold to E15.5 (P<0.001) and further decreased by 4.6‐fold between E17.5 and maturity (P<0.001; Figure 1B). The liver showed a similar pattern (Figure 1C); the activity was 2.2‐fold higher in E10.5 liver versus heart (P<0.001). The decline in activity in the liver occurred between E10.5 and 13.5. The developmental decline in luciferase activity was specific for the ODD of the modified Luciferase protein as there was no difference between E13.5 and adult heart (P=0.66) when wild type luciferase (WT‐Luc) was expressed from the Rosa26 locus (Figure 1D). Comparisons of WT‐Luc and ODD‐Luc activity suggest that ≈40% of the ODD‐Luc is degraded in E13.5 tissues versus 90% in adult tissues.
Figure 1.
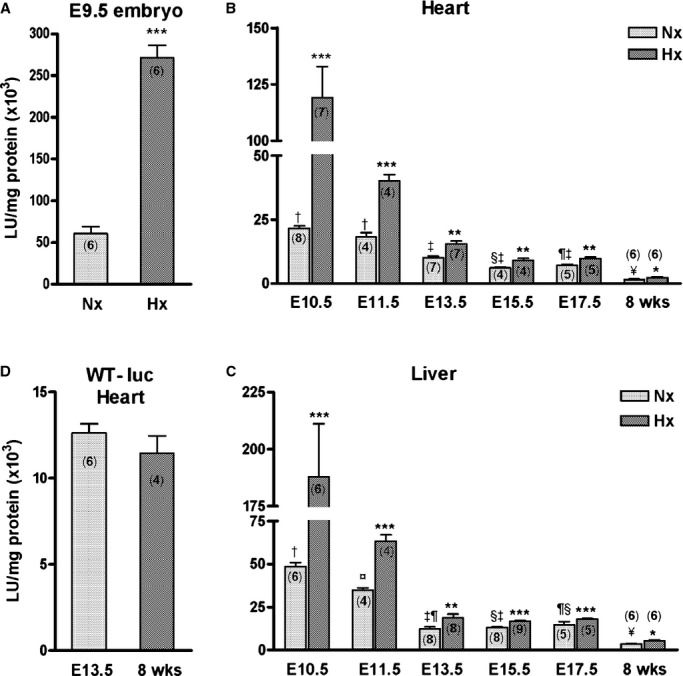
ODD‐Luc activity declines during normal development and is robustly induced by maternal O2 deprivation. Luciferase activity was measured in tissue lysates from (A) E9.5 whole embryo (B) heart and (C) liver from E10.5 to 17.5 and in the adult mouse (8 weeks). Pregnant ODD‐Luc mice from the same stages were exposed to hypoxia (8% O2 for 4 hours) and luciferase activity measured. These mice were homozygous for ODD‐Luc in the Rosa26 locus. D, Luciferase activity was measured in E13.5 and adult heart lysates from mice heterozygous for the wild‐type (WT) Luciferase in the Rosa26 locus. Luciferase activity was normalized to total protein and is expressed as mean±SEM LU/mg protein ×103. Number of samples in each group is indicated within the bar graph (n). Student t test was used for comparison of hypoxia (Hx) vs normoxia (Nx) samples. *P<0.05; **P<0.01; ***P<0.001. In the developmental series (B and C) basal luciferase activity (Nx) was analyzed by 1‐way ANOVA with Bonferroni correction for multiple comparisons; groups sharing the same symbol are not significantly different (P>0.05). For E13.5 vs E15.5 heart, P=0.06. Test for interaction between developmental stage and hypoxic induction of ODD‐Luc was performed using 2‐way ANOVA (P<0.001). LU indicates luminescence units; ODD‐Luc, oxygen degradation domain‐luciferase.
Luciferase activity was robustly induced in ODD‐Luc embryonic tissues with reduction of maternal inspired O2 concentration to 8% for 4 hours. The magnitude of induction was greatest in those tissues that had the highest basal activity (Figure 1A through 1C; P<0.001 for interaction between developmental stage and hypoxic induction of ODD‐Luc). Activity increased by 210‐, 86‐, and 139‐ ×103 luminescence units (LU)/mg protein in E9.5 embryo, E10.5 heart and E10.5 liver, respectively (P<0.001). In contrast, the activity increased in the adult heart and liver by only 0.6 and 1.8 ×103 LU/mg protein, respectively (P<0.05).
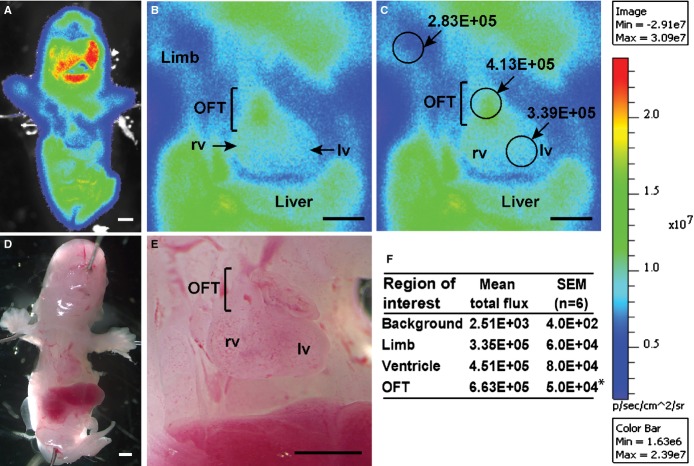
Bioluminescent imaging was used to determine the spatial distribution of ODD‐Luc activity in the E13.5 embryo. The substrate luciferin was administered to the pregnant mice and the embryos removed and imaged as described in Methods. Robust bioluminescent signal was observed in the ODD‐Luc embryos (Figure 2) while no activity was observed in wild type embryos or in ODD‐Luc mice in the absence of luciferin. The activity in the E13.5 heart was ≈200‐fold above background, similar to that measured in tissue homogenates. Within the heart a gradient of bioluminescent signal was observed with the outflow region of the heart exhibiting ≈50% greater signal as compared to the ventricles (Figures 2B, 2C, and 2F; P<0.001).
Figure 2.
Bioluminescent imaging of E13.5 ODD‐Luc embryo. Pregnant mice were injected i.p. with d‐luciferin (150 mg/kg) and embryos removed and imaged 15 minutes later in the supine position with a high sensitivity charged coupled device camera (Xenogen IVIS‐200) as described in Methods. A representative embryo from n=6 is shown with (A) low power and (B) high power images and (C) regions of interest with bioluminescent signal intensity shown. Scale bar shows range of signal from the field of view in photons sec−1 (cm2)−1 sr−1. D and E, are the corresponding bright field images. F, Mean±SEM of signals from n=6 embryos. Within the heart the bioluminescent signal is ≈200‐fold above background and highest within the OFT. *P<0.001 vs ventricle. All values are significantly above background (P<0.001; Student t test). Scale bars 500 μm. LV indicates left ventricle; ODD‐Luc, oxygen degradation domain‐luciferase; OFT, outflow tract; RV, right ventricle.
Conditional Inactivation of Hif‐1α in Specific Developmental Windows
Conditional inactivation of Hif‐1α, guided by ODD‐Luc as a reflection of PHD activity, was used to test the requirement for HIF‐1α in specific developmental windows. Hif‐1αf/f mice crossed with TM‐inducible β‐actinCre mice resulted in efficient recombination of the Hif‐1α floxed allele in E11.5 and E15.5 heart and liver of TM‐treated mice (Figure 3). Embryos from pregnant mice treated with TM at E10.5 were recovered at normal Mendelian ratios at E15.5 (Table 2). Four of 21 Hif‐1αf/f; β‐actinCre+ (conditional knock‐out (cKO) embryos examined at E15.5 (19%) had malposition of the aorta overriding a ventricular septal defect (VSD; Figure 4, Table 3). Another 3 of the 21 embryos (14%) had isolated VSD. All of the abnormal hearts had thinning of the ventricular myocardium. Twelve of the 21 embryos had open chest walls (thorachoschisis). This was observed in embryos with and without heart defects. The cKO embryos were smaller than their Cre− littermates. Littermates that were Cre− were structurally normal (Figure 4A). Embryos from mice injected with TM at E13.5 and examined at E17.5 did not display cardiac or extra‐cardiac structural defects (Table 3). Thus, abrogation of Hif‐1α after E10.5 causes cardiac outlet defects, while abrogation of HIf‐1α after E13.5 does not, establishing E10.5‐E13.5 as a critical window for HIF‐1α activity.
Figure 3.
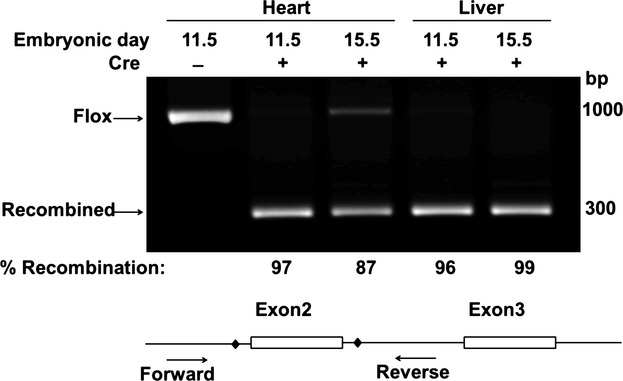
Highly efficient recombination at the Hif‐1α floxed locus with Tamoxifen (TM) induction of β‐actinCre. Cre mediated recombination results in nearly complete loss of the exon2 flanked by LoxP sites (diamond) in Introns 2 and 3 (compare Cre+ and Cre− lanes) in genomic DNA purified from E11.5 and E15.5 heart and liver. Shown is the average% recombination (n=2 each) calculated as the intensity of the recombined band divided by the combined intensity of the 2 bands and corrected for differences in band size. Hif indicates hypoxia‐inducible transcription factor.
Table 2.
Frequencies of Hif‐1α Conditional Knock‐Out Mice
| Wnt1Cre | β‐actinCre | |||||
|---|---|---|---|---|---|---|
| # Matings | # cKO | % cKO | # Matings | # cKO | % cKO | |
| E15.5 | 12 | 26 | 25 | 12 | 21 | 20 |
| PN 21 | 12 | 5 | 4.5 | ND | ND | ND |
Survival frequencies for Wnt1Cre at PN21 were significantly different from controls (P<0.0001; Fisher's exact test). E15.5 indicates embryonic day 15.5; ND, not done; PN 21, post‐natal day 21.
Figure 4.

Temporally targeted inactivation of Hif‐1α causes aorta overriding VSD. Control (Hif‐1αf/f; β‐actinCre−) and cKO (Hif‐1αf/f; β‐actinCre+) E15.5 littermate embryos from a mouse injected with 3 mg of TM at E10.5. Embryos are shown in whole mount (A and B) and in sections (C through J) from anterior to posterior with respect to the heart. The cKO embryo (B) is smaller than control (A). In the cKO the RVOT appears narrowed by mesenchyme (arrowhead) and infundibular muscle (D), and the aorta overrides a VSD (F and H). J, More posteriorly the atrio‐ventricular valves appear normal while the ventricular myocardium is thinned. Scale bars: 500 μm. Ao indicates aorta; cKO, conditional knock‐out; Hif, hypoxia‐inducible transcription factor; LV, left ventricle; LVOT, left ventricular outflow tract; pa, pulmonary artery; RV, right ventricle; RVOT, right ventricular outflow tract; TM, tamoxifen; VSD, ventricular septal defect.
Table 3.
Hif‐1α cKO and O2 Deprivation Cause Cardiac Defects
| Defect | Wnt1Cre | β‐actinCre Early | β‐actinCre Later | WT Hypoxia |
|---|---|---|---|---|
| Embryos examined | 26 | 21 | 16 | 15 |
| ASD* | — | — | 2 | |
| VSD* | — | 3 | 0 | 5 |
| PTA | 4 | — | 0 | — |
| DORV | 2 | — | 0 | 2 |
| Aorta over VSD | — | 4 | 0 | — |
The prevalence of cardiac defects in Wnt1Cre, β‐actinCre‐early and WT hypoxia groups were significantly different from controls (P<0.05). The prevalence of cardiac defects in β‐actinCre‐early vs. WT hypoxia were not significantly different (P=0.378). Data analyzed by Fisher's exact test. ASD indicates atrial septal defect; cKO, conditional knock‐out; DORV, double outlet right ventricle; Hif, hypoxia‐inducible transcription factor; PTA, persistent truncus arteriosus; VSD, ventricular septal defect; WT, wild type.
Embryos with isolated ASD and/or VSD. β‐actinCre early: Dams were given Tamoxifen (3 mg) at E10.5 and embryos examined at E15.5. β‐actinCre later: Dams were given Tamoxifen (3 mg) at E13.5 and embryos examined at E17.5.
Inactivation of Hif‐1α in Neural Crest Cells (NCCs)
The previous experiments defined the role of Hif‐1α in a critical developmental window, but because the β‐actinCre is ubiquitously active, did not define its role in specific cell types that are required for development of the cardiac outlet. To address this, Wnt1Cre was used to inactivate Hif‐1α in NCCs that migrate into the cardiac outlet and are required for the formation of the aortico‐pulmonic septum,7 a region that this and a prior study10 suggested is particularly hypoxic. Embryos were examined at E15.5 to 16.5 after NCCs have migrated into the heart and septation of the heart and OFT are complete. Embryos were recovered at normal Mendelian ratios at E15.5 to 16.5 (Table 2). Six of 26 (23%) E15.5 to 16.5 Hif‐1αf/f; Wnt1Cre+ embryos had defects typical of NCC deficiency (Figure 5, Table 3), including exencephaly and cleft face (Figure 5B). Four of the embryos had persistent truncus arteriosus (PTA Type 1) reflecting failure of septation of a portion of the OFT (Figure 5D) and a single semi‐lunar valve separating the common arterial trunk from the ventricle (Figure 5F). More posteriorly abnormal persistence of the AV cushion mesenchyme is associated with ventricular and atrial septal defects, and ventricular myocardial thinning is also evident (Figures 5H and 5J). Two of the embryos had OFT defects classified as double outlet right ventricle (DORV), in which both aorta and pulmonary artery arise from the right ventricle. All of the abnormal cKO embryos and their hearts were smaller than their littermate controls.
Figure 5.
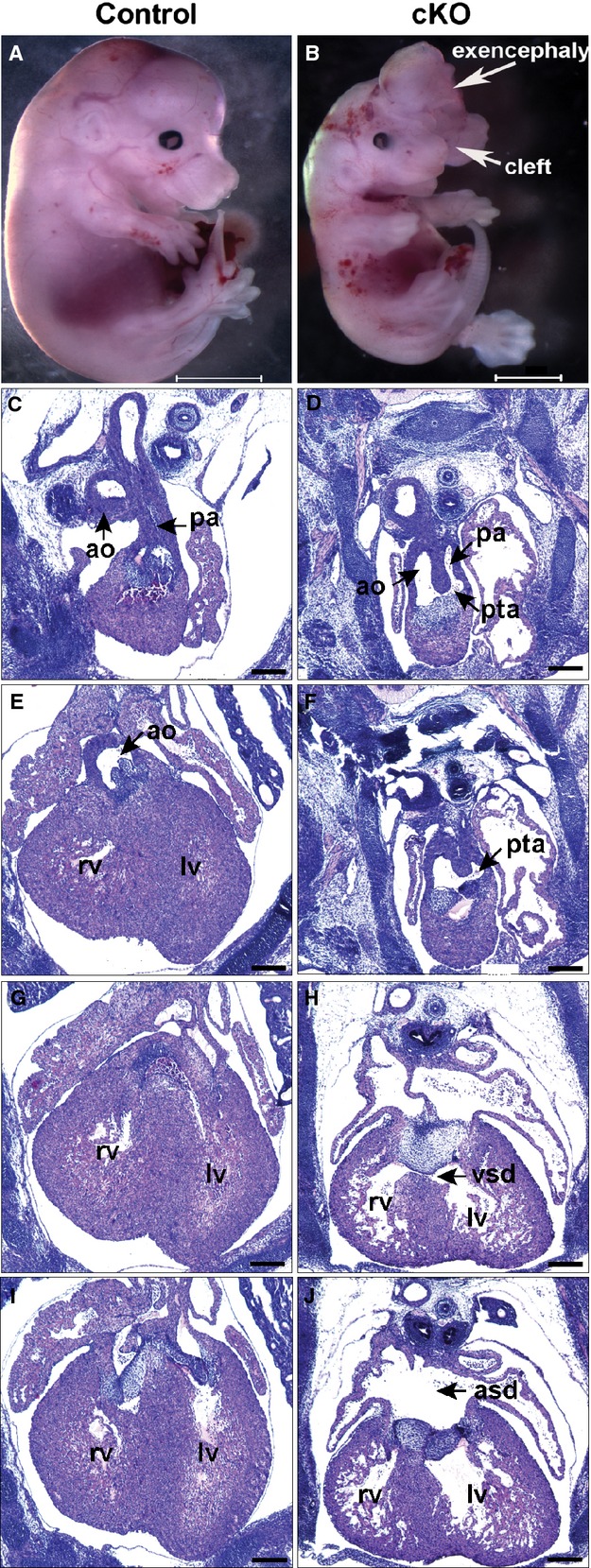
Persistent truncus arteriosus (PTA), exencephaly, and cleft face in E15.5 mouse embryo with Hif‐1α inactivation in neural crest cells (NCC). Control (Hif‐1αf/f; Wnt1Cre−) and cKO (Hif‐1αf/f; Wnt1Cre+) littermate embryos are shown in whole mount (A and B) and in sections (C through J) from anterior to posterior with respect to the heart. The cKO embryo (B) is smaller than control (A). The cKO exhibits PTA Type 1, in which the aorta (ao) and pulmonary artery (pa) share a common lumen originating above a single semilunar valve (D and F). H, More posteriorly is persistence of AV cushion mesenchyme with a VSD and (J) a large ASD. The ventricular myocardium is thinned. Scale bars: (A and B) 3000 μm (C through J) 500 μm. ASD indicates atrial septal defect; cKO, conditional knock‐out; Hif, hypoxia‐inducible transcription factor; LV, left ventricle; RV, right ventricle; VSD, ventricular septal defect.
Hif‐1αf/f mice were crossed with Rosa26‐LacZ mice to fate map NCCs migrating into the OFT using LacZ staining. In a Hif‐1αf/f; Wnt1Cre+ embryo with PTA, LacZ+ cells populate the great vessels and OFT, including the defective aortic‐pulmonic septum (Figures 6C and 6D; compare to control 6A and 6B). This suggests that HIF‐1α is not required within the NCCs for their migration into the cardiac OFT.
Figure 6.
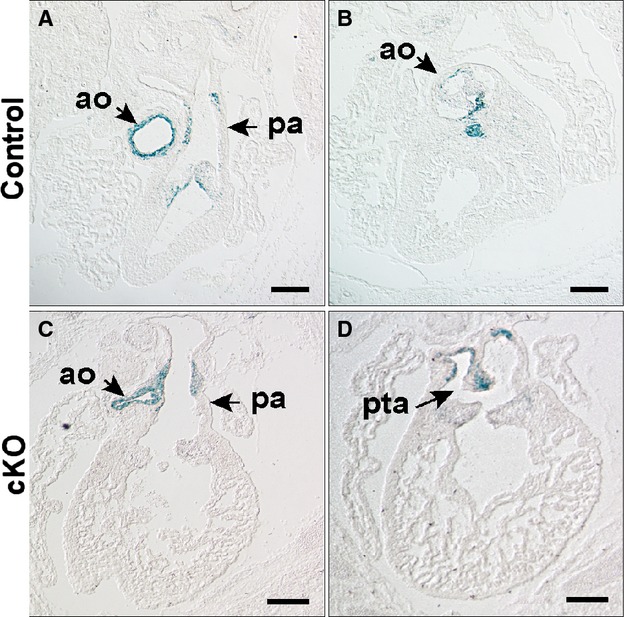
NCC fate map in an E15.5 embryo with Wnt1Cre mediated inactivation of Hif‐1α. NCC fate is mapped with histological detection of LacZ activity from the Rosa26 locus. Matched sections are from anterior to posterior. In control (A and B; Hif‐1αf/f; Wnt1Cre−) and cKO (C and D; Hif‐1αf/f; Wnt1Cre+) embryos, LacZ+ cells are present in the aorta, pulmonary artery and the semilunar valves. In the control embryo LacZ+ cells are evident in the rostral portion of the ventricular septum below the aortic valve (B). The outlet septum is absent in the cKO accounting for the outlet VSD. D, LacZ+ cells are observed in cKO in the abnormally positioned aortico‐pulmonic septum (arrow), which is incomplete resulting in PTA. Scale bars: 500 μm. Ao indicates aorta; cKO, conditional knock‐out; Hif, hypoxia‐inducible transcription factor; NCC, neural crest cells; pa, pulmonary artery; PTA, persistent truncus arteriosus; VSD, ventricular septal defect.
Only 5 of 110 mice recovered at 3 weeks of age were of the genotype Hif‐1αf/f; Wnt1Cre+ (4.5% versus 25% expected; P<0.001; Table 2), indicating lethality between E16.5 and post‐natal day 21 (PN21) with incomplete penetrance. All 5 of the cKO mice were severely runted and euthanized. Structural heart defects were not detected in these mice and the cause of their poor health was not identified. Hif‐1αf/f; Wnt1Cre− mice had normal survival and no apparent defects.
Effect of Maternal O2 Deprivation on Gene Expression
To determine if the stage‐dependent induction of ODD‐Luc activity is reflective of the endogenous response to oxygen deprivation, pregnant mice were exposed to the same hypoxia protocol (8% O2 for 4 hours) and selected transcripts measured by qPCR. Transcripts were selected based on prior experiments in which Stage 25 (ED4) chicken eggs were incubated in 7.5% O2 for 6 hours with or without DMOG, an iron chelator that inhibits PHD activity. Hypoxia‐inducible gene expression in the embryonic chick hearts was measured by gene array and confirmed by real‐time PCR (Table 4). Ccn1+2 were most robustly induced by the hypoxic stress in the E11.5 mouse heart (Figure 7A). Ccn3 was expressed at a low level and not induced (data not shown). There was good correlation between induction of ODD‐Luc and Ccn1+2, Bnip3 and Glut1 in E11.5 versus E15.5 mouse heart. Igfbp1 was induced several‐fold in the embryonic mouse heart (Figure 7A) and liver (Figure 7B) with no relationship to the induction of ODD‐Luc activity. Ccn1+2 were modestly induced in the embryonic mouse liver (Figure 7B), but the induction of Ccn2 at E11.5 was not significant. These genes were not induced in the maternal tissues consistent with the minimal increase in ODD‐Luc activity.
Table 4.
Hypoxic Induction of Genes in Stage 25 Embryonic Chicken Heart
| Genes | Fold Increase | |
|---|---|---|
| Microarray* | qRT‐PCR* | |
| Glut1 | 2.7±0.7 | 2.0±0.2* |
| BNip3 | 2.7±0.6 | 2.1±0.2* |
| IGFBP1 | 4.5±1.0 | 5.9±0.9* |
| CCN1 | 5.3±1.1 | 3.7±0.4* |
| CCN2 | 3.6±0.4 | 4.6±0.3* |
| CCN3 | 7.5±2.3 | 8.0±0.8* |
Mean±SEM, *P<0.05 vs control.
Affymetrix 1.0 ST array (n=3 control, n=3 DMOG+hypoxia; n=9 for all pair‐wise comparisons).
qRT‐PCR indicates quantitative real‐time‐PCR (Hypoxia±DMOG, n=7 vs control n=6).
Figure 7.
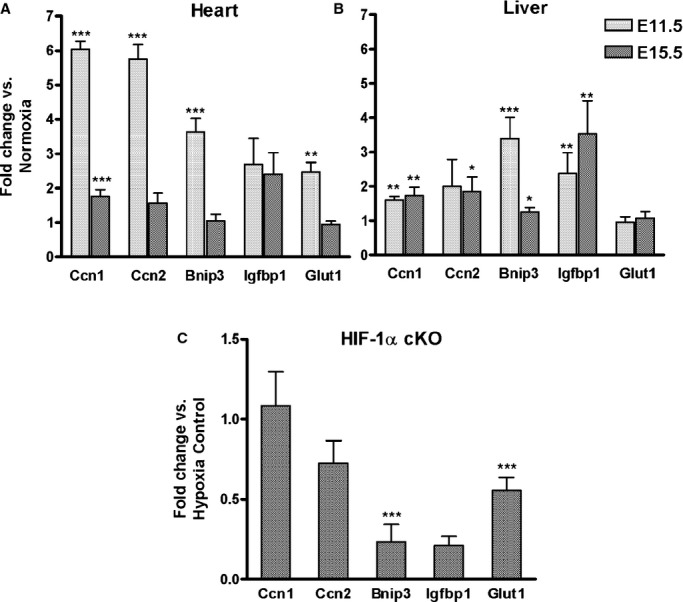
Effect of maternal O2 deprivation and Hif‐1α cKO on mRNA levels. A and B, Pregnant mice were exposed to hypoxia (8% O2 for 4 hours) or maintained in room air (21% O2, normoxia) at E11.5 or E15.5 and mRNA measured in (A) heart and (B) Liver by qPCR as described in Methods. Fold change vs normoxia was calculated using 2−ΔΔCt. (E11.5, n=5); E15.5, n=6). C, cKO of HIF‐1α: Pregnant mice were injected with 3 mg TM or vehicle at E10.5 and exposed to 8% O2 for 4 hours on E11.5. mRNAs were measured by qPCR in Hif‐1α cKO (Hif‐1αf/f; β‐actinCre+; n=4) E11.5 hearts and compared to values in a combined control group (Cre‐ with TM or vehicle; Cre+ with vehicle; n=12) using 2−ΔΔCt. β‐actin was used as the internal control and was unchanged by hypoxia. Mean±SEM. Data was analyzed by Student t test. *P<0.05; **P<0.01; ***P<0.001. cKO indicates conditional knock‐out; HIF, hypoxia‐inducible transcription factor; qPCR, quantitative polymerase chain reaction; TM, tamoxifen.
To determine if the hypoxic induction of these transcripts was HIF‐1‐dependent, transcript levels were measured in E11.5 hearts from embryos with cKO of Hif‐1α as described above (genotype: Hif‐1αf/f; β‐actinCre+, dam treated with TM). Cre+ embryos that were not exposed to TM were combined with Cre− embryos that were and were not exposed to TM to form the control group. All mice in these experiments were exposed to hypoxia (8% O2 for 4 hours) and the relative expression of mRNAs in cKO versus control embryos calculated and expressed as fold‐change. cKO of Hif‐1α markedly suppressed the hypoxic induction of Bnip3 and Glut1 but not Ccn1+2 in the E11.5 heart (Figure 7C). The hypoxic induction of Igfbp1 was highly variable in the E11.5 heart and its suppression by Hif‐1α cKO was not statistically significant.
Effect of Maternal O2 Deprivation on Cardiac Outlet Morphogenesis
To determine how O2 deprivation might affect heart development in this critical developmental window, pregnant mice were subjected to O2 deprivation (0.5 atmospheres [ATM], one‐half of normal) from E10.5 to 13.5 and heart morphology examined at E15.5 when the circulation has matured. Of 15 embryos examined the most common defect was isolated VSD associated with increased AV cushion mesenchyme (Table 3). Two of these embryos also had atrial septal defect (ASD). In 2 of the 15 embryos, the VSD was associated with mal‐position of the aorta partly originating from the RVOT and described as DORV (Figures 8B and 8D). VSD and ASD associated with increased AV cushion mesenchyme were present more posteriorly (Figures 8F and 8H). There was also thinning of the ventricular myocardium. In contrast the same regimen of O2 deprivation beginning at E13.5 resulted in no observable structural heart defects in embryos examined at E17.5 (n=21, Table 3), consistent with measures of ODD‐Luc activity and hypoxia‐inducible gene expression, establishing E10.5 to 13.5 as a critical developmental window for vulnerability of the embryo to O2 deprivation.
Figure 8.
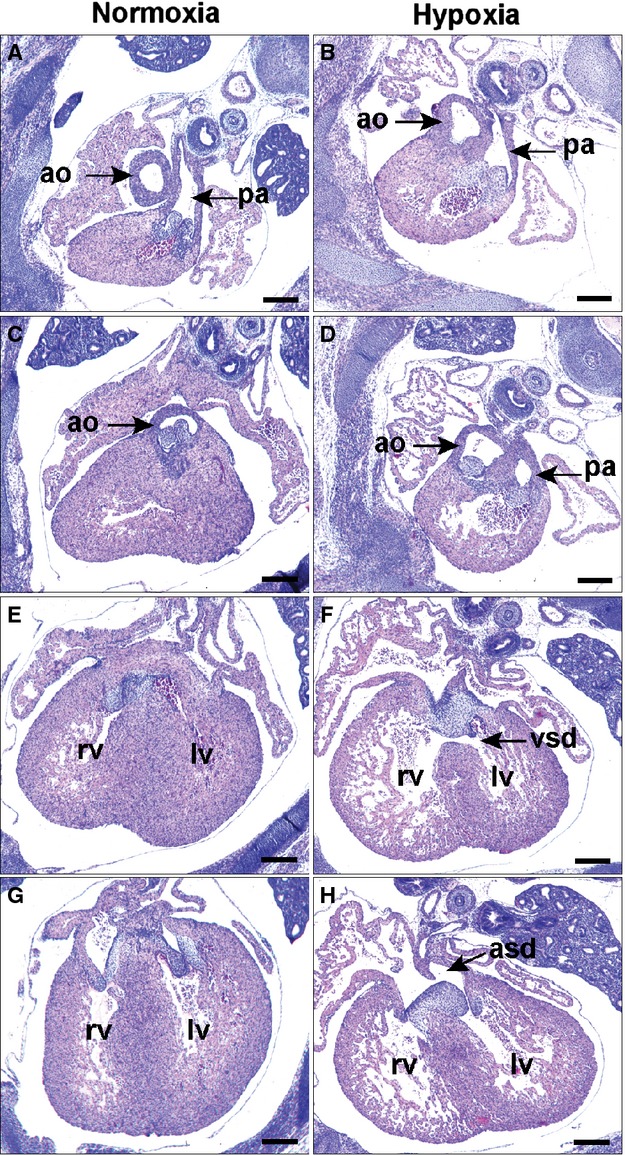
Effect of maternal O2 deprivation on cardiac outlet morphogenesis. Pregnant mice were housed at 0.5 ATM (half of the normal O2 content) from E10.5 to 13.5 and returned to normal conditions until E15.5. Sections shown are from E15.5 normoxic (A, C, E, and G) and hypoxic (B, D, F, and H) embryos from anterior to posterior with respect to the heart. B and D, The aorta of the hypoxic embryo is mal‐positioned originating partly from the RVOT in DORV morphology. More posteriorly is (F) VSD and (H) ASD with persistence of the AV cushion mesenchyme. Scale bars: 500 μm. Ao indicates aorta; ASD, atrial septal defect; ATM, atmospheres; DORV, double outlet right ventricle; LV, left ventricle; pa, pulmonary artery; RV, right ventricle; VSD, ventricular septal defect.
Discussion
The mammalian embryo must generate de novo all of the elements required for O2 delivery to the tissues, a necessary prerequisite to its continued growth and development.3–4 Here we have used a novel model, the ODD‐Luc mouse, as a quantitative reporter of the O2 supply to the developing embryo and contrasted that with the mature mouse. The 40‐fold decline in ODD‐Luc activity from E9.5 to maturity likely reflects improved O2 delivery to the tissues and occurs in 2 phases. The decline from E9.5 to 15.5 is likely due to maturation of the placenta for O2 exchange, red blood cells for O2 transport and organ blood vessels for convective replacing diffusional O2 delivery to the tissues.4 The decline from E17.5 to maturity likely reflects the complete oxygenation of the arterial blood that occurs after birth with entry of air into the lungs and closure of the fetal arterio‐venous shunts. That these differences in basal ODD‐Luc activity reflect O2 deprivation is supported by the striking differences of the tissues to maternal O2 deprivation (8% O2 for 4 hours). ODD‐Luc was induced in proportion to resting levels, with 15‐ to 100‐fold greater induction in the embryonic versus adult tissues, demonstrating the lack of O2 reserve in the developing embryo. In contrast the mature cardiovascular system is able to buffer against reductions in O2 due, for example, to the higher basal concentration of O2 in the blood and cooperative binding of O2 to hemoglobin. The developmental and hypoxia‐induced changes in ODD‐Luc activity reported here are similar to the 7‐fold induction of ODD‐Luc activity originally reported for this construct under hypoxic stress in vitro6 though the time scales are different. ODD‐Luc turnover is a function of PHD activity,6 which is a function of the concentration of the enzyme and co‐factors (O2)16 when the substrate (ODD‐luciferase) is in excess. It is thus possible that developmental differences in the expression and activity of components of the PHD‐dependent degradation pathway account for some of the differences in ODD‐Luc activity. The relationship between O2 concentration and ODD‐Luc activity could be further tested in vitro; however, modeling O2 delivery to embryos in vitro is problematic.17
Inactivation of Hif‐1α by administration of TM at E10.5 resulted in septation and conotruncal heart defects modeling human defects, while inactivation of Hif‐1α after E13.5 did not. This supports the ODD‐Luc measurements and suggests a critical role for oxygen sensing and HIF‐1α at this stage of morphogenesis. This, to our knowledge, is the first demonstration that directly interfering with the master regulator of hypoxic transcriptional responses can cause these common heart defects in a mouse model. Prior studies of germ line knockout of the HIF transcriptional repressor Cited2 demonstrated similar types of defects18–19 sensitive to Hif‐1α gene dosage.11 However, Cited2 transcriptional inhibition is not specific to HIF and perturbations of other signaling pathways may cause the heart defects.20–21 That the defects occur in the outlet and septal tissues is consistent with the current and prior studies9–11 showing that these tissues are relatively hypoxic compared to the ventricular myocardium at these stages of development. This is likely due to the thick mesenchymal and myocardial tissue in the OFT and septum forming a greater than several hundred micron barrier to the diffusional delivery of O2, well above the estimated ≈100 μm limit for diffusion of oxygen through tissues.22 In contrast, the ventricular chambers at these stages are highly trabeculated facilitating O2 diffusion, while at later stages the coronary arterial system develops for the convective transport of oxygen throughout the cardiac tissues. However, given that both inlet and outlet cardiac defects are observed in these experiments, it is also possible that hypoxia signaling through HIF plays a role earlier in development, eg, in the second heart field, a contributor to both regions of the heart (reviewed in reference23). The role proposed here for hypoxia and HIF‐1 in the remodeling of the OFT mesenchyme and myocardium has a parallel to that of skeletal development, in which HIF‐1 is required for the survival of cells in the centrally located avascular and hypoxic mesenchymal chondrocytes24 (reviewed in reference 25). It is thus possible that the thoracic skeletal defects observed in the current study are due to the ablation of Hif‐1α within the developing bones. Because β‐actinCre will inactivate Hif‐1α in tissues throughout the embryo, it is also possible that the cardiac and skeletal defects are secondary to effects on other HIF‐dependent tissues. In particular the placenta is thought to require a hypoxic stimulus and HIF‐1 for its proper development,26–29 and perturbations in placental development could impair O2 delivery particularly affecting those cardiac and other tissues with the least O2 reserve. Another possibility is an effect on the liver, where Erythropoeitin transcription (and thus red blood cell production) is proposed to be under the combinatorial control of HIF‐1, GATA and RARs early in development30 when the liver is most hypoxic (references,30–31 and this study).
That HIF‐1α is required in cells that directly contribute to the heart and great vessels was demonstrated by inactivation of Hif‐1α in NCCs using Wnt1Cre. This caused conotruncal defects the most common being PTA, along with exencephaly and cleft face, prototypical of neural crest cell deficiency (reviewed in7–8). The Type 1 PTA observed in the current study results from failure in NCC‐dependent septation of the proximal portion of the truncus, resulting in a single semi‐lunar valve above which is a common arterial trunk, while more distally the artery is septated into aortic and pulmonic channels. This is the most common type of PTA in humans. Two of the embryos had DORV, thought to represent a more limited failure of NCC dependent fusion of the more proximal portion of the OFT cushion mesenchyme, ie, below the level of the forming semi‐lunar valves.32 In the Wnt1Cre cross, fate mapping with Rosa26‐LacZ showed that NCCs had migrated into the OFT in an embryo with PTA. This suggests that HIF‐1α is not required within NCCs for their migration. A prior study of germline Hif‐1α inactivated mice had inferred that it is required for the migration of the NCCs into the OFT33; if so the current study would suggest that this is a non‐cell autonomous function, for example signaling by hypoxic tissues to recruit NCCs into the OFT. A study of a different line of germline Hif‐1α inactivated mice observed increased cell death (cell type undefined) in the OFT mesenchyme,34 consistent with our prior study using adenoviral mediated forced expression of HIF‐1α in the chicken OFT indicating a pro‐survival function.35 However, the precise mechanisms of action of HIF‐1 in this context requires defining its gene targets (discussed below).
The 2 different approaches to HIF‐1α loss‐of‐function and the one gain‐of‐function (hypoxic stress) each produced a somewhat different spectrum of conotruncal defects classified as PTA, malposition of the aorta over‐riding a VSD (a cardinal feature of Tetralogy of Fallot), and DORV, while they all had septation defects. While the conotruncal defects are separated into specific subtypes, they represent a continuum of defects involving the muscular infundibulum (OFT myocardial remnant) added from the second heart field and NCC‐dependent septation (reviewed in reference36). Of note, the NCC septation defect PTA type 1 was not observed in the Hif‐1α cKO mice. This could relate to the timing of Hif‐1 inactivation in these experiments after E10.5. TM‐inducible inactivation of Hif‐1α prior to E10.5 caused embryonic lethality prior to septation of the OFT, similar to germline or early myocardial inactivation of Hif‐1α,26,33–34,37 precluding use of this approach. The interplay between the different cell types in the OFT, as for example NCC signaling to the second heart field and ventricular myocardium, and dynamic tissue remodeling, creates great complexity in the generation of these structures. Defining the molecular basis of hypoxia signaling through HIF‐1 in the formation and mal‐formation of these structures will require definition of its gene targets in each context.
In the current study septation and contronucal heart defects were partially penetrant with incidences of 20% to 50%, consistent with models suggesting that combinations of genetic susceptibility and environmental stressors38 cause human CHD at an incidence of ≈0.8%. The partial penetrance of the heart defects in the current study is likely due to compensation from other loci. It could also in part be due to Cre inefficiency, though this seems unlikely as: (1) both Cre drivers13,15 and the flox allele14 have been extensively validated (and Figure 3) and (2) the full penetrance of the post‐natal lethality with Wnt1Cre, the basis of which was not defined. NCCs migrate throughout the body and contribute to the development of the thymus, thyroid and parathyroid glands, nervous tissue, teeth, head, and neck structures.39 Effects on any of these organs could negatively impact the growth and survival of the mice.
In this study we identified CCNs as robustly induced by hypoxia in the E11.5 mouse heart and Stage 25 chicken heart. These genes have been shown to be induced by hypoxia in other cell types in vitro.40 The degree of induction mirrored ODD‐Luc activity validating each as an indicator of hypoxic stress. Interestingly the hypoxic induction of CCN1+2 was not affected by HIF‐1 cKO, while hypoxic induction of other classic HIF‐dependent genes such as BNip3 and Glut1 were markedly suppressed. This is consistent with a prior in vitro study showing that hypoxic induction of CCN1+2 is HIF‐independent.41 It is also of interest to note that there were differences between the mouse embryonic heart and liver in hypoxic gene induction at stages when they experienced similar hypoxic stress in vivo as indicated by ODD‐Luc activity. While the mechanistic basis for these differences remains to be determined, it is consistent with the premise that the induction of gene transcription by hypoxic stress depends on cell type‐specific basal expression42 (reviewed in reference 43). In absolute terms, CCNs were more highly expressed in the heart than in the liver (data not shown). The functional role of these genes in each organ's response to normal developmental and stress‐induced hypoxia will require targeted loss‐of‐function studies. Ccn1 is an intriguing HIF target as it is expressed in the embryonic heart in regions that are identified as hypoxic44 and the present study shows that these regions, OFT and ventricular septum, are defective when Hif‐1α is inactivated. Mice null for CCn1 have Atrio‐ventricular septal defects (AVSD),44 while the specific function of CCNs in heart development remains to be defined.45
A role for O2 deprivation as a contributing factor to human congenital heart defects (CHD) has been proposed based on their increased prevalence in humans living at high altitudes and with other conditions that may reduce O2 supply to the embryo such as placental insufficiency, drugs causing transient uterine artery constriction and anemia.5 A longstanding hypothesis to explain CHD is the “threshold and multi‐factorial” model.36,46 O2 deprivation or reactive oxygen species may condition the embryo to increase sensitivity to chemical teratogens, environmental stressors and genetic defects. The current study using the ODD‐Luc mouse as a novel reporter of embryonic tissue oxygenation demonstrates the stage‐dependent susceptibility of the embryo to reductions in maternal inspired O2, and should be useful for quantifying the presumed O2‐depriving effects of other animal models of teratogenesis. Loss of HIF activity or maternal O2 deprivation in the critical window of organogenesis may cause common septation and conotruncal heart defects, providing a compelling rationale for examining the relationship between O2 delivery and tissue oxygenation in human CHD.
Sources of Funding
This work was supported by NIH grant R01 HL65314 to Fisher and R01‐CA154624 to Martin.
Disclosures
None.
Acknowledgments
We thank Qiu Doughman, Jessica Jenkins, and Alex Lloyd for technical assistance, Dr Michiko Watanabe for scientific advice and Dr Thomas Dick for use of the hypobaric chamber.
References
- 1.Kaelin WG, Jr, Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell. 2008; 30:393-402. [DOI] [PubMed] [Google Scholar]
- 2.Prabhakar NR, Semenza GL. Adaptive and maladaptive cardiorespiratory responses to continuous and intermittent hypoxia mediated by hypoxia‐inducible factors 1 and 2. Physiol Rev. 2012; 92:967-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fisher SA, Burggren WW. Role of hypoxia in the evolution and development of the cardiovascular system. Antioxid Redox Signal. 2007; 9:1339-1352. [DOI] [PubMed] [Google Scholar]
- 4.Dunwoodie SL. The role of hypoxia in development of the mammalian embryo. Dev Cell. 2009; 17:755-773. [DOI] [PubMed] [Google Scholar]
- 5.Webster WS, Abela D. The effect of hypoxia in development. Birth Defects Res C. 2007; 81:215-228. [DOI] [PubMed] [Google Scholar]
- 6.Safran M, Kim WY, O'Connell F, Flippin L, Gunzler V, Horner JW, DePinho RA, Kaelin WG., Jr Mouse model for noninvasive imaging of HIF prolyl hydroxylase activity: assessment of an oral agent that stimulates erythropoietin production. Proc Natl Acad Sci USA. 2006; 103:105-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hutson MR, Kirby ML. Model systems for the study of heart development and disease. Cardiac neural crest and conotruncal malformations. Semin Cell Dev Biol. 2007; 18:101-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gruber PJ, Epstein JA. Development gone awry: congenital heart disease. Circ Res. 2004; 94:273-283. [DOI] [PubMed] [Google Scholar]
- 9.Sugishita Y, Watanabe M, Fisher SA. Role of myocardial hypoxia in the remodeling of the embryonic avian cardiac outflow tract. Dev Biol. 2004; 267:294-308. [DOI] [PubMed] [Google Scholar]
- 10.Barbosky L, Lawrence DK, Karunamuni G, Wikenheiser JC, Doughman YQ, Visconti RP, Burch JB, Watanabe M. Apoptosis in the developing mouse heart. Dev Dyn. 2006; 235:2592-2602. [DOI] [PubMed] [Google Scholar]
- 11.Xu B, Doughman Y, Turakhia M, Jiang W, Landsettle CE, Agani FH, Semenza GL, Watanabe M, Yang YC. Partial rescue of defects in Cited2‐deficient embryos by HIF‐1alpha heterozygosity. Dev Biol. 2007; 301:130-140. [DOI] [PubMed] [Google Scholar]
- 12.Safran M, Kim WY, Kung AL, Horner JW, DePinho RA, Kaelin WG., Jr Mouse reporter strain for noninvasive bioluminescent imaging of cells that have undergone Cre‐mediated recombination. Mol Imaging. 2003; 2:297-302. [DOI] [PubMed] [Google Scholar]
- 13.Hayashi S, McMahon AP. Efficient recombination in diverse tissues by a tamoxifen‐inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Dev Biol. 2002; 244:305-318. [DOI] [PubMed] [Google Scholar]
- 14.Ryan HE, Poloni M, McNulty W, Elson D, Gassmann M, Arbeit JM, Johnson RS. Hypoxia‐inducible factor‐1alpha is a positive factor in solid tumor growth. Cancer Res. 2000; 60:4010-4015. [PubMed] [Google Scholar]
- 15.Jiang X, Rowitch DH, Soriano P, McMahon AP, Sucov HM. Fate of the mammalian cardiac neural crest. Development. 2000; 127:1607-1616. [DOI] [PubMed] [Google Scholar]
- 16.Pan Y, Mansfield KD, Bertozzi CC, Rudenko V, Chan DA, Giaccia AJ, Simon MC. Multiple factors affecting cellular redox status and energy metabolism modulate hypoxia‐inducible factor prolyl hydroxylase activity in vivo and in vitro. Mol Cell Biol. 2007; 27:912-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.New DA, Coppola PT. Effects of different oxygen concentrations on the development of rat embryos in culture. J Reprod Fertil. 1970; 21:109-118. [DOI] [PubMed] [Google Scholar]
- 18.Bamforth SD, Braganca J, Eloranta JJ, Murdoch JN, Marques FI, Kranc KR, Farza H, Henderson DJ, Hurst HC, Bhattacharya S. Cardiac malformations, adrenal agenesis, neural crest defects and exencephaly in mice lacking Cited2, a new Tfap2 co‐activator. Nat Genet. 2001; 29:469-474. [DOI] [PubMed] [Google Scholar]
- 19.Barbera JP, Rodriguez TA, Greene ND, Weninger WJ, Simeone A, Copp AJ, Beddington RS, Dunwoodie S. Folic acid prevents exencephaly in Cited2 deficient mice. Hum Mol Genet. 2002; 11:283-293. [DOI] [PubMed] [Google Scholar]
- 20.Bamforth SD, Braganca J, Farthing CR, Schneider JE, Broadbent C, Michell AC, Clarke K, Neubauer S, Norris D, Brown NA, Anderson RH, Bhattacharya S. Cited2 controls left‐right patterning and heart development through a Nodal‐Pitx2c pathway. Nat Genet. 2004; 36:1189-1196. [DOI] [PubMed] [Google Scholar]
- 21.Lopes Floro K, Artap ST, Preis JI, Fatkin D, Chapman G, Furtado MB, Harvey RP, Hamada H, Sparrow DB, Dunwoodie SL. Loss of Cited2 causes congenital heart disease by perturbing left‐right patterning of the body axis. Hum Mol Genet. 2011; 20:1097-1110. [DOI] [PubMed] [Google Scholar]
- 22.Popel AS, Goldman D, Vadapalli A. Modeling of oxygen diffusion from the blood vessels to intracellular organelles. Adv Exp Med Biol. 2003; 530:485-495. [DOI] [PubMed] [Google Scholar]
- 23.Kelly RG. The second heart field. Curr Top Dev Biol. 2012; 100:33-65. [DOI] [PubMed] [Google Scholar]
- 24.Schipani E, Ryan HE, Didrickson S, Kobayashi T, Knight M, Johnson RS. Hypoxia in cartilage: HIF‐1alpha is essential for chondrocyte growth arrest and survival. Genes Dev. 2001; 15:2865-2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maes C, Carmeliet G, Schipani E. Hypoxia‐driven pathways in bone development, regeneration and disease. Nat Rev Rheumatol. 2012; 8:358-366. [DOI] [PubMed] [Google Scholar]
- 26.Ryan HE, Lo J, Johnson RS. HIF‐1alpha is required for solid tumor formation and embryonic vascularization. EMBO J. 1998; 17:3005-3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adelman DM, Gertsenstein M, Nagy A, Simon MC, Maltepe E. Placental cell fates are regulated in vivo by HIF‐mediated hypoxia responses. Genes Dev. 2000; 14:3191-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Withington SL, Scott AN, Saunders DN, Lopes Floro K, Preis JI, Michalicek J, Maclean K, Sparrow DB, Barbera JP, Dunwoodie SL. Loss of Cited2 affects trophoblast formation and vascularization of the mouse placenta. Dev Biol. 2006; 294:67-82. [DOI] [PubMed] [Google Scholar]
- 29.Takeda K, Ho VC, Takeda H, Duan LJ, Nagy A, Fong GH. Placental but not heart defects are associated with elevated hypoxia‐inducible factor alpha levels in mice lacking prolyl hydroxylase domain protein 2. Mol Cell Biol. 2006; 26:8336-8346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Makita T, Duncan SA, Sucov HM. Retinoic acid, hypoxia, and GATA factors cooperatively control the onset of fetal liver erythropoietin expression and erythropoietic differentiation. Dev Biol. 2005; 280:59-72. [DOI] [PubMed] [Google Scholar]
- 31.Sugishita Y, Leifer DW, Agani F, Watanabe M, Fisher SA. Hypoxia‐responsive signaling regulates the apoptosis‐dependent remodeling of the embryonic avian cardiac outflow tract. Dev Biol. 2004; 273:285-296. [DOI] [PubMed] [Google Scholar]
- 32.Bradshaw L, Chaudhry B, Hildreth V, Webb S, Henderson DJ. Dual role for neural crest cells during outflow tract septation in the neural crest‐deficient mutant Splotch(2H). J Anat. 2009; 214:245-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Compernolle V, Brusselmans K, Franco D, Moorman A, Dewerchin M, Collen D, Carmeliet P. Cardia bifida, defective heart development and abnormal neural crest migration in embryos lacking hypoxia‐inducible factor‐1alpha. Cardiovasc Res. 2003; 60:569-579. [DOI] [PubMed] [Google Scholar]
- 34.Iyer NV, Kotch LE, Agani F, Leung SW, Laughner E, Wenger RH, Gassmann M, Gearhart JD, Lawler AM, Yu AY, Semenza GL. Cellular and developmental control of O2 homeostasis by hypoxia‐inducible factor 1 alpha. Genes Dev. 1998; 12:149-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu H, Fisher SA. Hypoxia‐inducible transcription factor‐1alpha triggers an autocrine survival pathway during embryonic cardiac outflow tract remodeling. Circ Res. 2008; 102:1331-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Friedman JM. The principles of teratology: are they still true? Birth Defects Res A Clin Mol Teratol. 2010; 88:766-768. [DOI] [PubMed] [Google Scholar]
- 37.Krishnan J, Ahuja P, Bodenmann S, Knapik D, Perriard E, Krek W, Perriard JC. Essential role of developmentally activated hypoxia‐inducible factor 1alpha for cardiac morphogenesis and function. Circ Res. 2008; 103:1139-1146. [DOI] [PubMed] [Google Scholar]
- 38.Lage K, Greenway SC, Rosenfeld JA, Wakimoto H, Gorham JM, Segre AV, Roberts AE, Smoot LB, Pu WT, Pereira AC, Mesquita SM, Tommerup N, Brunak S, Ballif BC, Shaffer LG, Donahoe PK, Daly MJ, Seidman JG, Seidman CE, Larsen LA. Genetic and environmental risk factors in congenital heart disease functionally converge in protein networks driving heart development. Proc Natl Acad Sci USA. 2012; 109:14035-14040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kulesa PM, Bailey CM, Kasemeier‐Kulesa JC, McLennan R. Cranial neural crest migration: new rules for an old road. Dev Biol. 2010; 344:543-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wolf N, Yang W, Dunk CE, Gashaw I, Lye SJ, Ring T, Schmidt M, Winterhager E, Gellhaus A. Regulation of the matricellular proteins CYR61 (CCN1) and NOV (CCN3) by hypoxia‐inducible factor‐1alpha and transforming‐growth factor‐beta3 in the human trophoblast. Endocrinology. 2010; 151:2835-2845. [DOI] [PubMed] [Google Scholar]
- 41.Meyuhas R, Pikarsky E, Tavor E, Klar A, Abramovitch R, Hochman J, Lago TG, Honigman A. A key role for cyclic AMP‐responsive element binding protein in hypoxia‐mediated activation of the angiogenesis factor CCN1 (CYR61) in tumor cells. Mol Cancer Res. 2008; 6:1397-1409. [DOI] [PubMed] [Google Scholar]
- 42.Xia X, Kung AL. Preferential binding of HIF‐1 to transcriptionally active loci determines cell‐type specific response to hypoxia. Genome Biol. 2009; 10:R113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lendahl U, Lee KL, Yang H, Poellinger L. Generating specificity and diversity in the transcriptional response to hypoxia. Nat Rev Genet. 2009; 10:821-832. [DOI] [PubMed] [Google Scholar]
- 44.Mo FE, Lau LF. The matricellular protein CCN1 is essential for cardiac development. Circ Res. 2006; 99:961-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Katsube K, Sakamoto K, Tamamura Y, Yamaguchi A. Role of CCN, a vertebrate specific gene family, in development. Dev Growth Differ. 2009; 51:55-67. [DOI] [PubMed] [Google Scholar]
- 46.Wright S. The results of crosses between inbred strains of guinea pigs, differing in number of digits. Genetics. 1934; 19:537-551. [DOI] [PMC free article] [PubMed] [Google Scholar]