Abstract
Genotyping technologies need to be continually improved in terms of their flexibility, cost-efficiency, and throughput, to push forward genome variation analysis. To this end, we have leveraged the inherent simplicity of dynamic allele-specific hybridization (DASH) and coupled it to recent innovations of centrifugal arrays and iFRET. We have thereby created a new genotyping platform we term DASH-2, which we demonstrate and evaluate in this report. The system is highly flexible in many ways (any plate format, PCR multiplexing, serial and parallel array processing, spectral-multiplexing of hybridization probes), thus supporting a wide range of application scales and objectives. Precision is demonstrated to be in the range 99.8–100%, and assay costs are 0.05 USD or less per genotype assignment. DASH-2 thus provides a powerful new alternative for genotyping practice, which can be used without the need for expensive robotics support.
As the Human Genome Project moves forward, of major importance will be analysis of single nucleotide polymorphism (SNP) (Brookes 1999), the most abundant and simple form of DNA variation. Meaningful exploration of SNPs in most settings demands large-scale experimentation, and many groups are working towards adequately powered genotyping technologies to make this possible. Innovative designs involving enzymes for allele-specific cleavage, ligation, or polymerization, in conjunction with advanced implementations, have been brought forward in often fairly successful attempts to accurately score genotypes (Tsuchihashi and Dracopoli 2002). Highly effective systems have also been fashioned around the straightforward principle of DNA hybridization (Lay and Wittwer 1997; Wang et al. 1998; Howell et al. 1999). However, assay costs still remain high (typically at least tens of cents per assigned genotype), and throughputs limited (creating millions of genotypes remains daunting for most systems). Arguably, the highly sophisticated and/or multistep reaction chemistries of many of these procedures may be imposing fundamental limitations on how cheap, flexible, or prolific they can be made to be. Because of this, we chose to explore a genotyping strategy that emphasizes simplicity, namely Dynamic Allele-Specific Hybridization (DASH).
The core reaction principal of DASH is real-time (dynamic) tracking of allele-specific differences in the process of DNA denaturation. To achieve this, an oligonucleotide probe is first hybridized to the target DNA—a necessary component of essentially all genotyping methods. The target DNA comprises one strand of a PCR product immobilized onto a solid surface, and a single probe is used that is complementary to one of the target alleles. Probe hybridization is performed at low temperature so that it goes to completion regardless of which target allele(s) are present. After probe annealing, there follows a standard heating step wherein probe-target denaturation is followed dynamically. This reveals the precise melting temperature (Tm) at which the probe most rapidly falls off of the target. The observed Tms are reproducible, and they discriminate between probe-matched and probe-mismatched targets by as much as 4–15°C across all SNPs yet tested. Homozygous samples give single Tm outputs, while heterozygotes show two zones of rapid melting. Thus, from this simple reaction process, one can unambiguously score known alleles present in the assayed target DNAs. Previously, this assay concept was shown to be very precise (>99.9% accurate), and improved design principals were recently reported that overcome problematic secondary structures that may exist in some target molecules (Prince et al. 2001). Our experience shows that by instigating a basic pipeline around the published “first-generation DASH” procedure (Howell et al. 1999) (96-well single-plex sample processing, manual pipetting, intercalating dye fluorescence signals) it is trivial, with just a few staff to create several hundred thousand genotypes per year, across many hundred different SNPs, at a reagent cost of ∼50 cents per assigned genotype.
Looking to the future of genome variation analysis, there remains an unmet need for highly flexible single-user technologies that can produce millions of genotypes per year at a per assignment cost of a few cents or less. With this goal in mind, we have combined several recent technology innovations with the basic DASH concept, to create a new genotyping platform we refer to as “second generation DASH”, or DASH-2. The system is based upon “macro-arrays” on membranes (Jobs et al. 2002), and it uses “iFRET” (Howell et al. 2002) to produce strong fluorescence signals that may be spectrally multiplexed for increased throughput. PCRs are optionally multiplexed and small volume, keeping costs at an absolute minimum, and these reactions may be accessed from any plate format or density. DASH-2 is therefore extremely flexible in all respects, very cheap to execute, and suitable for ultrahigh-throughput application. The various components of DASH-2 have now been thoroughly tested, and the findings of those studies are reported here.
RESULTS
DASH-2 Concept
Figure 1 presents a schematic overview of the complete DASH-2 procedure. The architecture of DASH-2 was designed to (1) exploit the proven benefits of DASH (simplicity and robustness), (2) allow for maximal flexibility of experimental scale (number of samples assayed) and heterogeneity (number of SNPs tested), and (3) provide many options for multiplexing and small-volume processing (to keep costs low). By coupling DASH-2 to liquid handling and arraying robotics, ultrahigh-throughput genotyping may be achieved. When such robotics infrastructure is not available, the DASH-2 procedure nevertheless still supports medium-high-throughput studies that may be performed completely manually. As presented below, the individual components of the method were first evaluated separately. Once optimized, these steps were used in combination to execute various real-world trials of DASH-2 to meaningfully evaluate its functionality. This development work was undertaken exclusively upon real human SNPs (IDs from the Human Genome Variation Database: HGVbase (Fredman et al. 2002) representing the full spectrum of possible allelic base alternatives.
Figure 1.
DASH-2 schematic. DASH-2 examines products from optionally multiplexed, small-volume PCRs, performed in any density or format of plate (A). PCR products are transferred to membranes to create “macro-arrays” by specific capture of one strand on the membrane surface. In the absence of robotics, this is conveniently achieved by centrifugation (B). Using robotics (not illustrated), would allow staggered subarraying of many different plates onto a single membrane to achieve ultrahigh-throughput. Alkali rinsing (to remove PCR reagents and make targets single stranded) and saturation hybridization with allele-specific probe(s) entails immersion in buffer trays or enlisting special washing platforms, and more than one target per array feature may be interrogated in parallel by using differentially labeled (“spectrally multiplexed”) probes (C). Alternatively, probe cocktails could be employed to extract different assay (SNP) results from different features (not illustrated). Fluorescence signals are generated by means of iFRET (Howell et al. 2002). This entails using an allele-specific probe that is end-labeled with an acceptor-fluorophore, plus double-strand specific fluorescence dye to act as fluorescence donor (D). Dynamic tracking of probe-target denaturation is achieved by heating the membrane array in a controlled manner while monitoring fluorescence signal changes via a CCD camera (D). If using a spectrally multiplexed set of iFRET probes, the target-specific signals are separately visualized by imaging the array through appropriate optical filters on a rotating wheel (D). As Tms specific for different probe-target combinations are reached, the iFRET signals rapidly disappear. These transitions are plotted as the negative derivative of fluorescence versus temperature (E), and thereby the target DNA alleles are revealed as peaks at high (probe matched) and low (probe mismatched) temperatures. Heterozygous targets show two peaks of melting behavior. Serial processing of the membrane is also possible, and thus additional sets of data may be extracted from array features by rerunning the DASH-2 procedure from the alkali rinse onwards, using extra sets of iFRET probes (F). Abbreviations: T1–4, target PCR products for different SNPs; P1–4, probes specific for single alleles of targets T1–4; lines marked as λ1 and λ2, distinct iFRET emission wavelengths from different fluorophores on probes (red and blue respectively); green circles indicate SYBR Green I dye molecules that transfer energy (star shapes) to the probe fluorophores, as per iFRET chemistry, only when the probe is bound to the target.
PCR Considerations
The effectiveness of DASH-2 will depend upon how well PCRs can be set up quickly, in low volume, and multiplexed. We employ pipetting robotics to prepare PCR plates of dried-down DNA samples in standardized arrangements, in advance of running DASH assays. Small volume aliquots of complete PCR-mix are added to these at run time, and sample rehydration then occurs as PCR begins. Testing this system for 1–2-μL reactions in 1536-well plates and 2–4-μL reactions in 384-well plates showed it to perform just as well as when using DNA in solution. However, smaller volume amplifications suffer from excessive sample evaporation. Testing alternative plate designs and plastics, plus various sealing systems, suggested that much of the water loss was occurring through the vessel walls (data not shown). We empirically determined that polypropylene 384-well plates (ABgene), and 1536-well polypropylene plates (Greiner bio-one), closed with “Thermo-Seal” foils (VWR international), substantially limited this volume reduction, whether using dry-block or water-based thermocycling devices.
Regarding PCR multiplexing, DASH deliberately employs short PCR products of about 50–70 bp (as part of a secondary structure avoidance strategy; Prince et al. 2001) and these should be easier to multiplex than fragments of several hundred bp in length. To test routine multiplex PCR in practice, we tried 49 duplex PCRs, 12 triplex PCRs, 4 tetraplex PCRs, and 1 hexaplex PCR, assembled from among 74, 9, 8, and 6 SNPs, respectively, combining primer sets known to work at equivalent PCR annealing temperatures. Assay success or failure was determined by analyzing most of the resulting PCR products by first-generation DASH (Howell et al. 1999). Credible melting curves, suitable for genotype assignment, were seen for 78/97 (80%) of the duplexes, 31/33 (94%) of the triplexes, 13/16 (81%) of the tetraplexes, and 6/6 (100%) of the hexaplexes, indicating that routine PCR multiplexing on these levels is quite effective.
Array Creation
DASH-2 is based upon macro-arrays of PCR products (one strand, 5′-biotinylated) anchored onto streptavidin-coated membranes. For highest throughput application, robotic arraying devices may be used to transfer multiple plates of PCR products onto a single membrane, arranging the transferred DNA sets in a staggered manner, such as in 3 × 3, 4 × 4, or 5 × 5 high-density grids derived from 384-well formatted starting plates. Standard arraying operations such as these can be performed by any of a range of robotics platforms, and we have found the Microgrid II by BioRobotics (www.biorobotic.com) and the Qbot by Genetix (www.genetix.com) to perform equally well and highly robustly (data not shown). Alternatively, to create DASH-2 arrays from any starting plate without the use of robotics, one could a centrifugation approach (Jobs et al. 2002). That is, after PCR, a membrane is clamped above an opened PCR plate and the samples are centrifuged onto the array surface where they bind. To evaluate the practicality of this more novel approach to array creation, we attempted to transfer, by centrifugation, 0.5–5.0-μL volumes of a PCR product (5′-biotinylated on one strand), from portions of 384- and 1536-well microtiter plates onto a streptavidin-coated membrane. After alkali removal of the unbound strand, we assessed the DNA transfer by hybridization with a fluorescently labeled probe complementary to the bound strand (Fig. 2). For the 384-well plate, 5-μL volumes consistently transferred perfectly, creating evenly filled features. Smaller volumes yielded features that were progressively weaker in the center, with 0.5-μL volumes forming incomplete circular arcs. For the 1536-well plate, due to the smaller cross-sectional area of the wells, features became less than perfect only at sub-1-μL volumes. Fortunately, these feature-shape differences are unimportant for DASH-2 analysis, because the sum rather than the pattern of the measured pixel intensities within each feature area are interpreted as they change with time. Thus, up to the highest density and lowest volume PCR reactions typically used by researchers today, we found the centrifugal membrane array concept to be fully compatible with the needs of DASH-2, offering high-throughput potential without the use of robotics for array creation.
Figure 2.
The quality of membrane array features created by centrifugation is shown for different volumes of transfer solution, starting from 384-well plates (top image) and 1536-well plates (bottom image).
Executing DASH-2 on Arrays
Having established practicalities for PCR and array creation, we proceeded towards a full DASH-2 experiment. The signal generation mechanism was to be iFRET (Howell et al. 2002), which has previously shown to give particularly strong and unambiguous DASH signals. The iFRET chemistry entails utilizing a DNA-intercalating dye as a fluorescence resonance energy transfer (FRET) donor, plus a probe-bound fluorophore as a FRET acceptor. To be able to implement this on a membrane surface, issues of membrane auto-fluorescence first had to be addressed. Testing a range of membranes showed that autofluorescence was commonly high enough to be troublesome, and charged membranes had the additional complication that they interacted with the SYBR Green I dye used in iFRET, emitting strong background fluorescence (data not shown). By trial and error, we established that streptavidin coated inert polypropylene membranes could provide adequate target DNA binding and low autofluorescence in the DASH-2 procedure.
To conduct an initial DASH-2 experiment, 192 different genomic DNA samples (amplified for SNP000008200 [A/T] in single-plex PCRs) were prepared as eightfold replica arrangements of 2-μL volume in a 1536-well PCR plate. PCR products were centrifugally transferred from this plate to a binding membrane, and this was alkali rinsed and probed with a single iFRET probe for one of the target alleles. The membrane was then subjected to the heating phase of the DASH-2 procedure, and for each feature the probe–target melting curves were so determined (Fig. 3). Precisely equivalent denaturation profiles were seen across all eight replicas for each sample, showing uniformity of the DASH-2 system. Peak denaturation rates indicated unambiguous sample-specific melting temperatures (Tms), correlating to one of three expected genotype patterns previously seen when scoring this marker upon a first-generation DASH (Howell et al. 1999) system run with iFRET probes. Comparing the derived genotype calls for these 192 samples with known data derived by first-generation DASH, restriction digestion of PCR products (Jeffreys 1979), and Pyrosequencing (Nyren et al. 1993) studies indicated 100% correct genotype calls.
Figure 3.
DASH-2 melting curves are shown for all possible genotypes for marker SNP000008200[A/T]. Eight replicates of each genotype generated by the DASH-2 system (right) are compared to results produced by the first-generation DASH platform (left). The df/dT scale is arbitrary and platform dependent.
Multiplexing
Multiplexing is concerned with obtaining more results for little or no increased cost or effort, and there are several nonmutually exclusive ways this could be built into DASH-2. This can be viewed as “multiplexing of multiplexing”. PCR multiplexing is one obvious possibility, as discussed above. Other straightforward options would be parallel processing of plates when creating arrays, and conducting parallel buffer treatments of membrane arrays.
Aspects of the array structure and the membrane probing can also be designed to bring benefits of multiplexing. These would include (1) “spectral-multiplexing”; mixtures of iFRET probes with distinct emission spectra could be used in a single probing to extract more than one result per array feature carrying a multiplexed PCR target, (2) “intrafeature multiplexing”; complete sets of genotypes could be extracted from highly multiplexed PCR products by serial probing of membranes, and (3) “interfeature multiplexing”; multiple different target loci placed at different feature positions, could be extracted by hybridizing with a cocktail of many different probes. These various options were explored.
Binding Capacity and Multiplexed Signal Quality
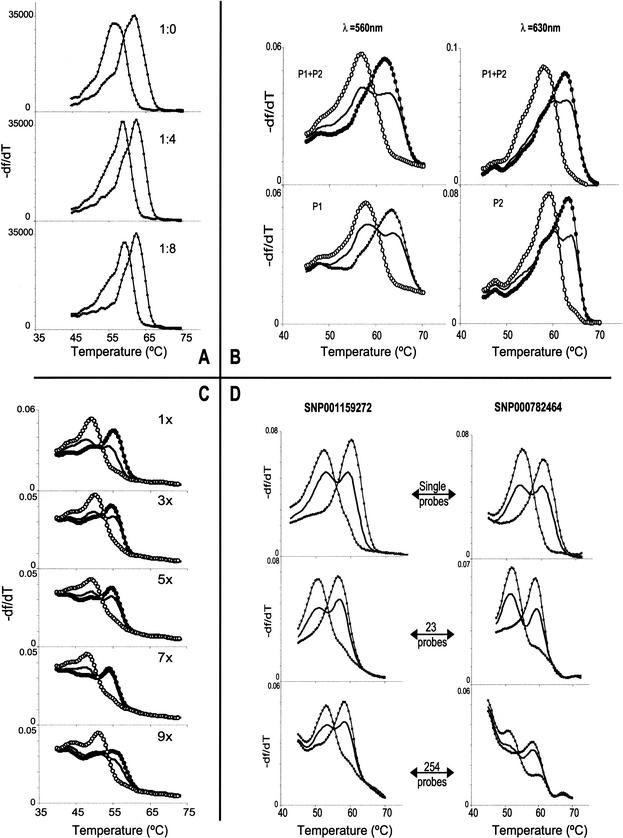
Should the membrane binding capacity or the signal quality in the presence of additional targets be nominal, then it would not be possible to derive reliable results from significantly PCR-multiplexed targets, either by using spectrally multiplexing probes or by serial probing. To explore these issues, various amounts of a 5′-biotinylated oligonucleotide carrying a 6-carboxy-X-rhodamine (ROX) label were centrifuged as 3-μL aliquots from a 1536-well plate onto a DASH-2 membrane. By simply measuring membrane-bound ROX fluorescence signals after rinsing the membrane, we found that for even up to 10 pmole (10–20-fold excess over a PCR product), the transferred DNA had not exceeded the membranes binding capacity. Next, to evaluate the practical relevance of this large binding capacity, we modeled a multiplexed target in a DASH-2 experiment by creating array features from 0.35 pmole (weak PCR equivalent) of synthetic oligonucleotides representing the two single-allele (homozygous) DNA targets (SNP000574314[A/C]). At the same time, a titration series of features equivalent to these two mock genotypes were made, also including in the transfer 0.35–2.9 pmole of another target oligonucleotide (SNP000574319[C/G]), so imposing competition (for binding and subsequent probe hybridization) equivalent to as much as an 8-plex PCR. Interrogating the membrane by DASH-2 for SNP000574314 yielded clear melting curves with no noticeable difference in absolute fluorescence intensities between any of the features (see Fig. 4A). These results indicate that the iFRET-based DASH-2 system provides sufficient target DNA binding capacity, absolute signal strength, and nonconfounding effects from coimmobilized targets, to support genotyping of at least ∼8-plex PCR products.
Figure 4.
Experiments into aspects of DASH-2 multiplexing are illustrated. Different genotypes are distinguished by line style, with equivalent genotypes per panel using the same line style. For each image the df/dT scale is in platform-dependent arbitrary units. (A) Robustness of signal strength and quality when multiplexing the PCR were demonstrated by modeling the presence of competing target molecules. The stated ratios indicated the amount of true target versus other DNAs coimmobilized within an array feature and assayed for the true target by DASH-2. (B) The viability of spectrally multiplexing iFRET probes is shown by these example genotypes, produced by DASH-2 analysis of a duplexed PCR. Targets were SNP000574304 and SNP000003618, and the matching probes were P1 (carrying Bodipy TMR: 560nm emission) and P2 (carrying ROX: 630nm emission), respectively. The probe combinations used for DASH-2 analysis are shown in each cell. The same DNA samples are assayed in the top and bottom cells. The left two cells were imaged through a 560-nm filter, while the right two cells were imaged through a 630-nm filter. (C) The potential for serial interrogation of membranes was established by comparing DASH-2 melting curves from replica arrayed samples after probing for the number of times indicated in the corner of each cell. (D) The extent to which probe cocktails may be used to examine different markers at different feature positions was explored by assaying arrayed single-plex PCRs for 24 SNPs (two examples shown, as left and right columns of cells); probe complexities are as indicated. Most markers suffered only a minimal loss of data quality regardless of the probe cocktail complexity (left column example), while three SNPs acquired extra early-melting fluorescence when interrogated by the 254-probe mixture (right column example).
Spectral Multiplexing
To explore the utility of spectrally multiplexed probes, duplex PCRs were performed for SNPs with high frequency alleles (SNP000574304[A/G] and SNP000003618[C/G]) upon 16 different genomic DNAs. Samples were transferred into three replica sections of a 1536-well plate, and a membrane array was created by centrifugation. The triplicate membrane portions were probed with either an equimolar mixture of two iFRET probes (one for each amplified marker), or separately with either one of the two probes. The probes carried different detection dyes, namely ROX and Bodipy TMR, which differ considerably in their emission spectra. A dynamic temperature ramp was applied, and the two marker-specific fluorescence signals were separately tracked by employing two band-pass optical filters. For Bodipy TMR fluorescence, the filter had a maximum at 560 nm with a 20-nm bandwith, and for the ROX filter the specifications were 630 and 30 nm. Figure 4B illustrates the data generated by this experiment. The observed genotype counts for the two markers were very different, 5:9:2 versus 1:8:7 (homozygotes matching the probe:heterozygotes:homozygotes mismatched to the probe), proving that each SNPs data was not corrupted by signals from the other assay. This, plus the near identity of the derived melting curves for the single-plex probings versus duplex-probings with spectral resolution, demonstrates that spectral duplexing of iFRET probes works well for DASH-2 analysis.
Intrafeature Multiplexing
Experiments were performed to establish how many times membranes could be reprobed to serially extract genotypes from individual features. Ten replica DASH-2 membranes containing single-plex PCR products for SNP000015168[A/T] were prepared. One membrane was processed to extract available genotypes, while nine were held back. One of the nine membranes plus the already processed membrane were then processed together through the DASH-2 procedure. By recycling in this way seven more times, each time adding one more virgin membrane to the processed set, a series of membranes were generated that had been serially processed between zero and nine times. Finally, all 10 membranes were processed one last time in parallel. Checks were made after the alkali rinse (i.e., before probing) to make sure that no residual probe (fluorescence) was carried through from earlier hybridizations, and the eventual DASH-2 melting curves were compared (Fig. 4C). It was apparent that data quality was relatively unchanged up to five repetitions. Between 5–10 runs the data quality decreased, although reliable genotypes could still be extracted from the very last run of the experiment. Thus, at least five serial hybridizations may be employed without loss of assay quality, and this could be used to extract multiple genotypes from individual array features carrying multiple target DNAs.
Interfeature Multiplexing
For diagnostic applications (testing many known mutations in an individual), or when examining different SNPs across different subportions of a PCR plate, it would be useful to be able to use a cocktail of iFRET probes such that each would automatically hybridize only to its cognate targets. To assess whether this might work in practice, a membrane was prepared from a 384-well PCR plate, carrying 16 sample DNAs amplified by single-plex PCRs for 24 different SNPs (SNP000007612[C/G], SNP000834421[C/T], SNP000782464[A/G], SNP001265242[G/T], SNP001265241[G/T], SNP000794409[A/G], SNP000783245[C/G], SNP000782459[C/G], SNP000782466[G/T], SNP001251211[A/C], SNP000080953[A/G], SNP001259858[A/T], SNP001257116[A/G], SNP000763322[A/G], SNP001257107[C/G], SNP000990182[A/G], SNP000108876[A/G], SNP000108526[A/G], SNP000108654[G/T], SNP000108052[A/G], SNP001159272[A/T], SNP000707079[A/G], SNP001164148[A/C], SNP000120603[A/G]). Cocktail probing was then attempted. First, the membrane was interrogated via DASH-2 with a probe mixture containing probes for 23 of the 24 assays (one probe omitted as a negative control). Second, the membrane was treated with alkali, and reprobed with a combination of the 23 SNP specific probes plus 231 unrelated probes (for other DASH-2 assays not present on the membrane). All probe mixtures contained 0.33 pmole/μL of each probe. To provide reference controls, the DNA samples were also scored for each of these individual SNPs by single-plex first-generation DASH using iFRET signal generation. The melting curves obtained for the 23 and 254 probe cocktails were very similar to results produced when assaying the SNPs individually (Fig. 4D), and the negative controls were negative. Specifically, for the 23-plex cocktail, the melting curves were essentially identical to the singly probed assays for all tested SNPs. For the 254-plex cocktail, the melting curves were highly similar to the single probings for 20 of the SNPs, while three assays had an additional fluorescence component that disappeared (denatured) at very low temperature. High-complexity probe cocktails can therefore occasionally generate some crosshybridization between noncognate probe-target pairs, but these duplexes denature early in the DASH-2 temperature gradient. Consequently, the additional signals do not excessively interfere with the melting profiles observed at the higher temperatures where true marker alleles are normally distinguished. Probe cocktails thus work well in DASH-2 analysis, and while the upper complexity limit is yet to be demonstrated, extrapolation from present results suggests that several thousand probes might be able to function together.
Trial Applications
To tie together all the above developments, and evaluate them for cost and precision in a real application, we used DASH-2 to analyze 1494 samples. Among these, 44 DNA samples were duplicated on different plates as an internal control for genotype calling accuracy, and 42 water controls were also included. The samples were scored for six SNPs (SNP000574304[A/G], SNP000003618[C/G], SNP000007612[C/G], SNP000015168[A/T], SNP000063124[C/T], and SNP000003288[C/T]). The experiment employed three-plex PCRs of 2-μL volume, performed in 384-well plates. Two sets of four derived membranes (each set carrying the different three-plex PCR products) were serially probed three times with single iFRET probes for their component SNPs. This compact study of only eight 384-well plates was thus structured to yield 9216 different genotypes (less duplicates and controls).
Results from this experiment were uniform across all plates and assays, and negative controls were all negative. A total of 45 samples repeatedly failed to provide any, or anything but weak, melting curve signals, and hence, this was assumed to be due to low concentration or degraded DNA stocks that were difficult to PCR amplify. Other than these, 25 singleton failures were observed, giving a sample dropout rate of <0.3%. Genotyping precision was estimated in three ways. First, Hardy Weinberg Equilibrium was assessed for each marker, and no significant deviations were observed. Second, we compared a number of derived genotypes (329 DNAs typed for two SNPs) to separately determined and triply-confirmed assignments obtained by other methods (first-generation DASH, restriction digestion of PCR products; Jeffreys 1979; and Pyrosequencing; Nyren et al. 1993). Among these 658 genotypes, >99.8% identical calls were made by the DASH-2 system (difference due to a single genotype discrepancy). Third, we examined the results obtained for the 528 internal replicates built into the experiment (264 duplicates, comprising 44 samples scored for six SNPs). For these we found that the calls agreed in 100% of the cases. Thus, the accuracy of the DASH-2 system is in the range 99.8–100%, matching the precision established previously for the first-generation DASH system (Prince et al. 2001).
DISCUSSION
Basic DASH chemistry, executed under standard reaction conditions with optimal assay design rules implemented by “DFold” software (D. Fredman, M. Jobs, and A.J. Brookes, in prep.), has previously been shown to be highly robust and able to genotype well in excess of 95% of all real SNP sequences with a precision of over 99.9%. By building new innovations into this DASH chemistry, and transferring the procedure to an array implementation, we have now created the powerful and highly flexible “DASH-2” system.
DASH-2 is based upon the use of macro-arrays of target DNAs. This offers a range of advantages, not least (1) target DNA processing and probing is reduced to direct immersion of membranes in suitable buffer solutions, (2) multiple membranes may be parallel-processed during their creation and interrogation, (3) membranes containing multiplexed PCR products may be serially interrogated many times to extract all available genotypes, (4) monitoring of the dynamic assay step (probe-target denaturation) entails simple real-time CCD imaging of the full array, and (5) array features of any scale or density can be processed equivalently. Creating these arrays is made simple by the use of low-cost centrifugation procedures. Alternatively, if one wished to employ robotic arraying devices, throughput could be increased further by condensing PCR products from multiple plates onto individual membranes in a staggered arrangement, thereby significantly reducing downstream membrane processing effort.
For a signal generation system, DASH-2 takes advantage of the proven benefits of iFRET. Previous work demonstrated that iFRET eliminates background signals due to target DNA secondary structures. We have now shown that iFRET is effective as a way to achieve spectral multiplexing in DASH-2 analysis. Our data specifically demonstrate spectral duplexing, but given available dyes and the generality of iFRET, it is likely that up to fourfold spectral multiplexing may be possible, particularly if applying mathematical deconvolution of signals.
Fundamentally, the power of DASH-2 emerges from its many and varied uses of multiplexing—a principle that is endorsed by other notable methods such as the MASDA assay (Shuber et al. 1997). DASH-2 multiplexing options include, (1) PCR multiplexing, (2) parallel processing of array preparation and buffer treatments, (3) use of spectrally multiplexed sets of iFRET probes to extract several marker results from each array feature, (4) use of probe cocktails to derive results from features carrying different target loci, and (5) repeated probing to serially extract data from features derived from highly multiplexed PCRs. This “multiplexing of multiplexing” underpins the methods flexibility, enables high-throughput application, and helps to keep genotyping costs very low. The fact that the method only requires a single end-labeled fluorescent iFRET probe also ensures cost efficiency. By way of example, in the final experiment we report, the plastic-ware plus enzyme plus reagent cost for the full set of 9216 genotypes was only 460 USD. That equates to only 0.05 USD per genotype. Minimal further multiplexing, PCR volume reduction, and the use of 1536-well plates (or roboticized subgridding) would easily bring this cost down several fold to ∼0.01–0.02 USD per genotype—at least an order of magnitude cheaper than most other fully reported systems.
To help the research community avail itself of DASH genotyping, suggested assay designs (PCR primer and hybridization probe sequences) are being assembled, via “DFold” software (D. Fredman, M. Jobs, and A.J. Brookes, in prep.), for all known human SNPs represented in the public HGVbase human polymorphism database (Fredman et al. 2002). These designs will be presented as part of the records in that database. An experimentally validated list of proven cSNPs (variants that alter amino acid sequences) plus DASH assay reagents is also being constructed, built upon all available entries in that database. In terms of practical assistance, novel SNP assay design help and quality-controlled validated kits of DASH reagents may be obtained from a dedicated DASH-support company (DynaMetrix, www.dynametrix-ltd.com). These components may be used with first-generation DASH (Howell et al. 1999) protocols for low-medium-throughput studies (several thousand genotypes per week per assay device) or with the multiplexed DASH-2 protocols presented here for high-throughput applications (one million or more genotypes per week per assay device).
In conclusion, DASH-2 provides a powerful new alternative for genotyping practice. It is flexible and cheap enough for all to use, and it does not require expensive robotics or assay devices to be purchased. These benefits stem directly from the elemental simplicity of the underlying DASH reaction concept, which perhaps also has potential for expression array analyses, (re)sequencing, and DNA fingerprinting. We additionally envisage further implementation improvements towards DASH-3 (e.g., nano-scale and microbead versions) that could bring even faster and cheaper genotyping possibilities in the relatively near future.
METHODS
PCR
Polymerase Chain Reacion (PCR) primers for DASH-2 assays were designed using the DFold software (DynaMetrix Ltd, UK: D. Fredman, M. Jobs, and A.J. Brookes, in prep.). All PCR reaction mixes were scaled from the following basic protocol. A 1-μL reaction contained 0.15 pmole 5′-biotinylated primer, 0.75 pmole of nonlabeled primer, 0.03 units AmpliTaq Gold® DNA polymerase (PE Corp.), 1× AmpliTaq Gold® Buffer, 3 mM MgCl2, 5% Dimethylsulphoxide, and 0.2 mM of each dNTP. Genomic DNA ranged from 1 ng down to 0.25 ng per reaction and never less, thereby negating any risk of stochastic preferential amplification of one allele due to low target copy number. Thermal-cycling consisted of an initial 10-min activation step of 94°C for 10 min, followed by 40 cycles of 94°C for 15 sec and an assay-specific annealing temperature for 30 sec. PCRs were performed in 1–5-μL volumes in 384-well polypropylene plates (ABgene) or 1–2-μL volumes in the 1536-well polypropylene plates (Greiner bio-one). The 384-well plates were heat-sealed using the Easy-Peel heat sealing foil (ABgene) and the 1536-well plates were sealed with Thermo-seal heat sealing foil (VWR international). Thermal cycling was performed on a 384 MultiBlock System (Thermo-Hybaid) or in a water bath-based thermal cycling unit developed in-house. For multiplex PCR, primer combinations were done such as to maintain the same reagent concentrations as for regular PCR, and thermal cycling was performed as described above.
Transfer and Binding to Membrane
Sample transfer from a microtiter plate to a membrane was accomplished via centrifugation as previously described (Jobs et al. 2002). In brief, a streptavidin-coated polypropylene membrane (DynaMetrix Ltd, UK) was prewet in HEN buffer (0.1 M HEPES, 10 mM EDTA, 50 mM NaCl, pH 7.5) and placed on top of the open wells of a microtiter plate (384 wells or 1536 wells). A compression pad (HB-TD-SFOAM, Hybaid) was then placed on top of the membrane, and the arrangement was pressed together in a clamping device. The device was then placed in a microtiter plate centrifuge (B4i Jouan, Inc) and centrifuged at 1500 rpm (rotor s20) for 30 sec. The device was left at room temperature for 30 min (to allow the biotinylated PCR products to bind to the strepatividin-coated membrane). Finally, the clamping device was inverted and briefly centrifuged to return the remaining fluid back into the microtiter plate wells.
Probe Hybridization
Membranes were submerged in a 0.1 M NaOH bath for 2 min to remove nonbiotinylated PCR product strands. They were then rinsed once in HEN for neutralization, and placed separately on 8 × 12-cm glass plates (slightly larger than the membrane). A 1.5-mL HEN solution containing 0.5 pmole/μL of the appropriate oligonucleotide probe was added onto each membrane, and a second glass plate was placed on top to form a hybridization chamber. The individually sandwiched membranes were heated to 85°C on a flat PCR block (PCR express, Thermo Hybaid) and air cooled to room temperature. A final rinsing in HEN was then applied to remove excess probe.
DASH-2 Analysis
Membranes, carrying the bound probe-target duplexes, were soaked for 30 min in 0.5× HE buffer (0.1 M HEPES, 10 mM EDTA, pH 8.0) containing a 1:10,000 dilution of supplied stock SYBR Green I dye (Molecular Probes). They were then individually sandwiched between two glass plates and placed into a DASH-2 genotyping device (DynaMetrix Ltd). The device consisted of a dark box, a heating platform, a light source, and a CCD camera with appropriate optical filters. Fluorescence values were collected as the membrane assembly was heated from 35–85°C (with a heating rate of 2–3°C/min) by imaging every 0.5°C.
Genotype Calling
Melting curves were generated for each array feature by purpose built software (DynaMetrix Ltd). Denaturation events were most readily visualized by analysis of a plot of the negative derivative of the fluorescence versus temperature profile. Presence of a single high-temperature peak indicated the sample was homozygous for the allele complementary to the allele present in the oligonucleotide probe. A single low-temperature peak indicated homozygosity for the alternative allele. A curve with peaks at both temperatures indicated that the sample was heterozygous.
WEB SITE REFERENCES
www.biorobotic.com; suppliers of lab-robotic equipment.
www.genetix.com; suppliers of lab-robotic equipment.
www.dynametrix-ltd.com; supplies and supports SNP genotyping by Dynamic Allele-Specific Hybridization—DASH.
Acknowledgments
We gratefully acknowledge the Swedish Research Council & Carl Tryggers Foundation for Scientific Research for funding that helped with aspects of our research undertakings, and we thank the Karolinska Institute Center for Genomics and Bioinformatics and Pharmacia Corporation for provision of enabling infrastructure. General technical assistance by various members of our laboratory is appreciated, and in particular, we thank David Fredman, Daniel Pederson, Johan Klingberg, and Mark Reimers for innovative software contributions.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
E-MAIL Anthony.Brookes@cgb.ki.se; FAX 46 8 324826.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.801103.
REFERENCES
- 1.Brookes A.J. 1999. The essence of SNPs. Gene 234: 177-186. [DOI] [PubMed] [Google Scholar]
- 2.Fredman D., Siegfried, M., Yuan, Y.P., Bork, P., Lehvaslaiho, H., and Brookes, A.J. 2002. HGVbase: A human sequence variation database emphasizing data quality and a broad spectrum of data sources. Nucleic Acids Res. 30: 387-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Howell W.M., Jobs, M., and Brookes, A.J. 2002. iFRET: An improved fluorescence system for DNA-melting analysis. Genome Res. 12: 1401-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Howell W.M., Jobs, M., Gyllensten, U., and Brookes, A.J. 1999. Dynamic allele-specific hybridization. A new method for scoring single nucleotide polymorphisms. Nat. Biotechnol. 17: 87-88. [DOI] [PubMed] [Google Scholar]
- 5.Jeffreys A.J. 1979. DNA sequence variants in the G γ-, A γ-, δ- and β-globin genes of man. Cell 18: 1-10. [DOI] [PubMed] [Google Scholar]
- 6.Jobs M., Howell, W.M., and Brookes, A.J. 2002. Creating arrays by centrifugation. Biotechniques 32: 1322-1324., 1326, 1329. [DOI] [PubMed] [Google Scholar]
- 7.Lay M.J. and Wittwer, C.T. 1997. Real-time fluorescence genotyping of factor V Leiden during rapid-cycle PCR. Clin. Chem. 43: 2262-2267. [PubMed] [Google Scholar]
- 8.Nyren P., Pettersson, B., and Uhlen, M. 1993. Solid phase DNA minisequencing by an enzymatic luminometric inorganic pyrophosphate detection assay. Anal. Biochem. 208: 171-175. [DOI] [PubMed] [Google Scholar]
- 9.Prince J.A., Feuk, L., Howell, W.M., Jobs, M., Emahazion, T., Blennow, K., and Brookes, A.J. 2001. Robust and accurate single nucleotide polymorphism genotyping by dynamic allele-specific hybridization (DASH): Design criteria and assay validation. Genome Res. 11: 152-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shuber A.P., Michalowsky, L.A., Nass, G.S., Skoletsky, J., Hire, L.M., Kotsopoulos, S.K., Phipps, M.F., Barberio, D.M., and Klinger, K.W. 1997. High-throughput parallel analysis of hundreds of patient samples for more than 100 mutations in multiple disease genes. Hum. Mol. Genet. 6: 337-347. [DOI] [PubMed] [Google Scholar]
- 11.Tsuchihashi Z. and Dracopoli, N.C. 2002. Progress in high-throughput SNP genotyping methods. Pharmacogenomics J. 2: 103-110. [DOI] [PubMed] [Google Scholar]
- 12.Wang D.G., Fan, J.B., Siao, C.J., Berno, A., Young, P., Sapolsky, R., Ghandour, G., Perkins, N., Winchester, E., Spencer, J., et al. 1998. Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science 280: 1077-1082. [DOI] [PubMed] [Google Scholar]