Abstract
To better understand the evolutionary forces that affect human genes, we sequenced 5055 expressed sequence tags from the chimpanzee and compared them to their human counterparts. In conjunction with intergenic chimpanzee DNA sequences and data on human single-nucleotide polymorphisms in the genes studied, this allows us to gauge the extent to which selection affects human genes at a genome-wide scale. The comparison to intergenic DNA sequences indicates that about 39% of silent sites in protein-coding regions are deleterious and subject to negative selection. Further, when the divergence between human and chimpanzee is compared with the extent of nucleotide polymorphisms among humans in the same sequences, there is significantly higher divergence in the 5′ untranslated regions (UTRs) but not in other parts of the transcript. This indicates that positive selection may have had a considerable influence on 5′UTRs. The dinucleotide CG (CpG) also exhibits a different substitution pattern within 5′UTRs as compared with other parts of the genome.
Comparison of the human genome to genomes from other species such as the mouse (Shabalina et al. 2001) and the pufferfish (Roest Crollius et al. 2000) has emerged as one of the major approaches toward understanding the evolutionary forces that shape the human genome (Rubin 2001). However, for several reasons, comparisons to species distantly related to humans cannot convey the entire picture. First, patterns of mutation, recombination, and selection are likely to have changed over the long time periods since these species shared common ancestors with humans. Second, DNA sequences that are under little or no selective constraints, such as intergenic, intronic, and untranslated regions, are so diverged in these organisms that they can hardly be aligned to human DNA sequences, if at all. Third, mutational hotspots such as the dinucleotide CpG or microsatellites experience such frequent changes that the mutational patterns that underlie the differences seen between distantly related species cannot always be accurately reconstructed. Thus, in addition to comparisons with highly diverged genomes, comparisons with genomes of closely related species are necessary in order to achieve a more complete understanding of the evolution of the human genome.
The African apes (chimpanzees, bonobos, and gorillas) are the closest living evolutionary relatives of humans. Of these, the chimpanzees, and their sibling species, the bonobos, differ from humans by an average of 1.2% in overall genomic DNA sequences (Chen and Li 2001; Ebersberger et al. 2002) and are estimated to have shared a common ancestor with humans 4.6–6.2 million yr ago (Chen and Li 2001). Gorillas differ from humans by an average of 1.6% in genomic DNA sequences and are estimated to have shared a common ancestor with humans, chimpanzees, and bonobos 6.2–8.4 million yr ago (Chen and Li 2001). Thus, on average, chimpanzees and bonobos are the species most closely related to humans. Of these, chimpanzees are both much more numerous and much better studied. Therefore, for the comparison of the human genome with a closely related genome, chimpanzees are the species of choice.
In order to obtain a large and relatively unbiased data set that provides insight into how genes have diverged between humans and chimpanzees, we sequenced 5055 chimpanzee expressed sequence tags (ESTs) and compared them with their human counterparts. The results show that unexpectedly high levels of purifying selection affect silent sites in protein-coding parts of human genes. The data furthermore indicate that 5′ untranslated parts of human genes have been the target of positive selection.
RESULTS
cDNA Sequencing
We constructed cDNA libraries from chimpanzee testis and brain and generated a total of 5055 ESTs of high quality. When clustered, they yielded 3139 unique chimpanzee cDNA contigs. We compared these sequences to the public databases and identified 2844 orthologous human DNA sequences. For 1845 of these sequence pairs, we could identify the coding sequence (CDS) and consequently also the untranslated regions (UTRs). From these sequence pairs, 1226 of the chimpanzee cDNA contigs contained CDS, 1071 3′UTRs, and 582 5′UTRs. This represents 2%–4% of all known human genes (Waterston et al. 2002) and can thus be expected to be a representative sample to compare the evolution of the different types of sites in human genes.
Untranslated Regions
For the 5′UTRs, the GC content is 53%, and 10.2% of the compared nucleotides occur in CpG contexts (Table 1). For the 3′UTRs, the GC content is 41.7% and 2.8% of the nucleotides are in a CpG context. The ratio of observed CpGs relative to the amount of expected CpGs, given the GC content of the DNA sequences, is 0.6 for 5′UTRs and 0.3 for 3′UTRs, and the genome average is about 0.2 (Lander et al. 2001). Thus, although CpGs are more frequent in both 5′UTRs and 3′UTRs than elsewhere in the genome, the number of CpG sites is drastically higher in the 5′UTRs than in the 3′UTRs.
Table 1.
Divergence Between Humans and Chimpanzees in Genic and Intergenic Regions
| % CpG | % GC | Loci | % Differences | Compared kb | ts/tve | ||
| mean | SE | ||||||
| 5′ UTRsa | 10.2 | 53 | 582 | ||||
| All | 1.12 | 0.040 | 68.7 | 1.80 | |||
| No CpG | 0.77 | 0.035 | 61.8 | 1.78 | |||
| CpG | 4.20 | 0.240 | 7.0 | 1.83 | |||
| CDSb | 5.3 | 51 | 1226 | ||||
| All | 0.45 | 0.010 | 426.7 | 3.67 | |||
| No CpG | 0.28 | 0.008 | 404.2 | 3.13 | |||
| CpG | 3.44 | 0.122 | 22.5 | 4.80 | |||
| nd sitesc | 4.8 | 47 | 1226 | ||||
| All | 0.22 | 0.009 | 272.8 | 2.75 | |||
| No CpG | 0.16 | 0.008 | 259.7 | 2.12 | |||
| CpG | 1.48 | 0.105 | 13.2 | 5.50 | |||
| 4d sitesd | 7.3 | 48 | 1226 | ||||
| All | 1.09 | 0.042 | 61.9 | 3.05 | |||
| No CpG | 0.54 | 0.030 | 57.3 | 2.36 | |||
| CpG | 8.16 | 0.407 | 4.5 | 3.87 | |||
| 3′UTRsa | 2.8 | 42 | 1071 | ||||
| All | 0.86 | 0.016 | 321.2 | 2.18 | |||
| No CpG | 0.63 | 0.014 | 312.0 | 1.81 | |||
| CpG | 8.85 | 0.297 | 9.1 | 3.68 | |||
| Intergenic | 3.3 | 47 | 5604 | ||||
| All | 1.28 | 0.010 | 1268.1 | 2.34 | |||
| No CpG | 0.89 | 0.008 | 1226.2 | 1.78 | |||
| CpG | 12.96 | 0.164 | 41.8 | 4.65 | |||
| Intronic | 3.5 | 48 | 2691 | ||||
| All | 1.20 | 0.014 | 597.7 | 2.42 | |||
| No CpG | 0.80 | 0.012 | 576.6 | 1.82 | |||
| CpG | 12.11 | 0.224 | 21.2 | 4.56 | |||
UTRs—untranslated regions.
CDS—coding sequence.
nd sites—non-degenerative sites.
4d sites—four-/dd degenerative sites.
ratio of transitions (ts) over transversions (tv).
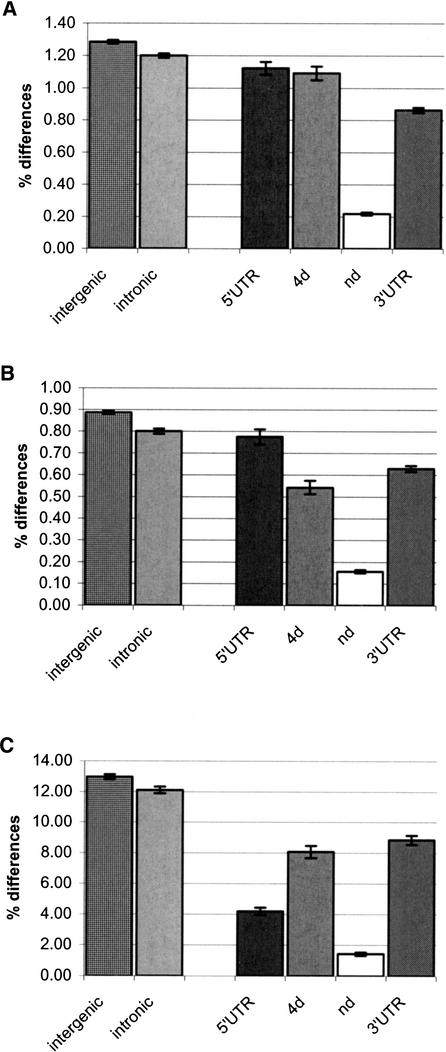
In the 5′UTRs, differences between human and chimpanzee cDNAs occur at 1.12% of the positions. CpG sites in the 5′UTR differ at 4.2% of positions, whereas non-CpG sites differ at 0.77% (Fig. 1). Thus, within 5′UTRs, differences occur approximately five times more often at CpG sites than at non-CpG sites. Transitional differences are 1.8 times more frequent than transversional differences in 5′UTRs. It is generally thought that the predominant type of mutation that affects methylated CpG sites is cytosine deamination, which results in transitional differences (Shen et al. 1994). However, we find no difference in the transition–transversion ratio between CpG and non-CpG positions in 5′UTRs.
Figure 1.
Divergence between humans and chimpanzees. Overall divergence (A) is given in percent for intergenic regions (Ebersberger et al. 2002), intronic sequences (Ebersberger et al. 2002), 5′UTRs, fourfold degenerate sites (4d), nondegenerate sites (nd), and 3′UTRs. (Below) Divergence is given for non-CpG sites (B) and CpG sites (C), respectively.
The 3′UTRs differ between humans and chimpanzees at 0.86% of positions. CpG sites and non-CpG sites differ at 8.85% and 0.63%, respectively (Fig. 1). Hence, in 3′UTR, CpG sites contain 14 times more differences than non-CpG sites. Transitional substitutions at non-CpG sites are 1.8 times more common than transversions, as in 5′UTR. However, in contrast to 5′UTRs, at CpG sites in 3′UTRs, transitions are 3.7 times more frequent than transversions. Thus, the transition bias is larger at CpG sites than at non-CpG sites.
In order to gauge to what extent 5′UTRs and 3′UTRs may evolve under functional constraints, we compared their divergence between humans and chimpanzees to the divergence of intergenic sequences (Ebersberger et al. 2002). The latter class of sequences exhibits slightly higher levels of divergence between humans and chimpanzees than intronic sequences and may evolve under little functional constraint (Fig. 1 and Table 1). The overall divergence at 5′UTRs does not differ significantly from intergenic sequences (t-test; P = 0.162, d.f. = 622). Similarly, the divergence of non-CpGs in 5′UTRs does not differ from the intergenic non-CpG divergence (t-test; P = 0.277, d.f. = 616). However, CpG sites in 5′UTR contain three times fewer differences than intergenic CpG sites (t-test; P < 10−6, d.f. = 891). Thus, although non-CpG sites in 5′UTRs seem to be under few constraints, CpG sites in 5′UTRs appear to be subject to substantial functional constraints. Alternatively, they may have a lower mutation rate than intergenic CpGs.
Within 3′UTRs, overall divergence is lower than that at 5′UTRs (t-test; P = 2.6 × 10−6, d.f. = 818) and intergenic sequences (t-test; P < 10−6, d.f. = 1473). At non-CpG sites, the divergence is 0.63%, that is, significantly less than in the two other categories of sequences (t-tests: 3′UTR-intergenic: P < 10−6, d.f. = 1497; 3′UTR-5′UTR: P = 0.0031, d.f. = 771). Consequently, assuming that the mutation rates for non-CpG sites within the different categories of sequences are the same, about kg% of the substitutions within 3′UTR are lost as a result of purifying selection. Furthermore, CpG sites in 3′UTRs have accumulated fewer differences than intergenic CpGs (t-test; P < 10−6, d.f. = 1275), but the effect is not as drastic as for CpGs in 5′UTRs.
Because CpGs in 3′UTRs, 5′UTRs, introns, and intergenic regions are likely to be methylated to different extents, it cannot be assumed that they are subject to similar average mutation rates. For example, the fact that the transition/transversion ratio does not differ between non-CpG sites and CpG sites in the 5′UTRs, whereas it is higher at CpG sites in 3′UTRs (Table 1) could reflect differences in mutational pressures between these regions because of differential methylation. Thus, because the mutation rates are likely to differ for CpG sites in different regions of genes, it is not possible to tease apart possible mutation rate heterogeneity and selection acting on CpG sites in the different regions.
Coding Regions
In the coding regions, we contrast the nondegenerate (nd) sites, where all potential nucleotide changes result in amino acid replacements, with fourfold-degenerate (4d) sites, where no potential changes cause any amino acid replacement. From a total of 426.7 kb of coding region sequences analyzed, 272.8 kb are nd sites, whereas 61.9 kb are 4d sites. The GC content at nd sites is 47% and the CpG content 4.8%. At 4d sites, the GC content is similarly 48%, whereas the CpG content is 7.3%.
At nd sites, the overall divergence between humans and chimpanzees is 0.22%. The divergence at nd CpG sites is 1.5% and at non-CpG sites 0.16%. At 4d sites, the overall divergence between the species is 1.1%, roughly five times more than at nd sites. At CpG sites, the divergence is 8.2% and at non-CpG sites 0.54%. Thus, at 4d sites, CpG sites have accumulated 15.1 times more differences than non-CpG sites, whereas at nd sites, CpG sites differ 9.4-fold more than non-CpG sites.
If we use intergenic sequences as a neutral reference for the number of differences accumulated between humans and chimpanzees, we find that 4d sites have a slightly lower divergence than expected (t-test; P = 0.030, d.f. = 1324). In contrast, the divergence at nd sites is 0.22% and therefore, as expected, substantially lower than in intergenic regions, indicating that these sites evolve under extensive functional constraints.
However, when overall divergence is compared, the effects of mutation and selection are clearly compounded. For example, the fraction of CpG sites, for which divergence is substantially higher than at non-CpG sites, varies among the different categories of sites in coding regions (Table 1). Furthermore, as indicated earlier, the extent of methylation of CpG sites, and thus their mutation rate, may differ between different categories of sequences. In order to contrast the extent of functional constraints affecting different classes of sites, we therefore restrict our comparisons to non-CpG sites. We again find that nd non-CpG sites contain substantially fewer differences than the putatively neutral intergenic regions (t-test; P < 10−6, d.f. = 2370). The extent of the reduction indicates that 82% of the mutations have been eliminated as a result of purifying selection. Surprisingly, we find also that 4d non-CpG sites contain significantly fewer differences than intergenic sequence (F-test; P < 10−6, d.f. = 1353). The extent of this reduction indicates that 39% of mutations have been eliminated, presumably as a result of purifying selection (Fig. 1). Thus, not only sites that cause amino acid replacement in coding regions of human transcripts but also sites that have no direct effect on the primary structure of the encoded protein seem to be subject to substantial purifying selection.
Polymorphism vs. Divergence
In order to compare the level of human polymorphism to the level of human–chimpanzee divergence in a set of homologous gene sequences, the chimpanzee sequences were aligned to human mRNA-derived sequences. For these 1304 sequence pairs, we retrieved all entries on human single-nucleotide polymorphisms (SNPs). This left us with 136 orthologous 5′UTRs, 459 coding regions, and 228 3′UTRs for which both the chimpanzee–human divergence and the human nucleotide diversity could be estimated (Table 2, Fig. 2).
Table 2.
Estimates of Chimpanzee–Human Divergence and Human Diversity
| Differences (%) | Number of differences | Sequence length (kb) | Number of genes | Differences (%) non-CpG | Number of differences non-CpG | |
| Chimpanzee–human divergence | ||||||
| 5′UTRs | 1.18 ± 0.063 | 351 | 29.8 | 136 | 0.79 ± 0.055 | 203 |
| 4d sites | 1.06 ± 0.051 | 419 | 39.5 | 459 | 0.52 ± 0.038 | 191 |
| nd sites | 0.17 ± 0.010 | 287 | 175.4 | 549 | 0.11 ± 0.008 | 189 |
| 3′UTRs | 0.73 ± 0.034 | 465 | 63.6 | 228 | 0.53 ± 0.029 | 328 |
| Nucleotide diversity among humans (θW) | ||||||
| 5′UTRs | 0.040 ± 0.0142 | 43 | 0.029 ± 0.0127 | 28 | ||
| 4d sites | 0.074 ± 0.0166 | 113 | 0.055 ± 0.0148 | 82 | ||
| nd sites | 0.036 ± 0.0055 | 221 | 0.030 ± 0.0052 | 174 | ||
| 3′UTRs | 0.066 ± 0.0127 | 157 | 0.056 ± 0.0119 | 128 |
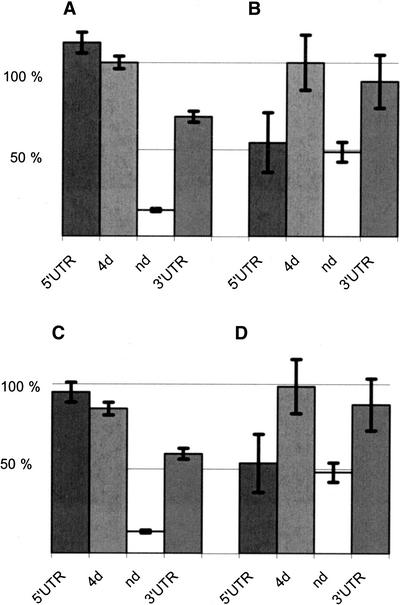
Figure 2.
Relative amounts of divergence and diversity in transcripts. The extent of differences between chimpanzee and human genes (A) within orthologous regions of 5′UTRs, fourfold degenerate sites (4d), nondegenerate sites (nd), and 3′UTRs and the extent of polymorphism among humans (θw) (B). Panels A and B are scaled such that changes at 4d sites have equal heights. Panel C shows the differences between the species normalized to the genome-wide divergence between humans and chimpanzees (Ebersberger et al. 2002), and panel D shows the extent of polymorphism normalized to the genome-wide nucleotide diversity (Sachidanandam et al. 2001).
If most differences observed both among humans and between humans and chimpanzees are selectively neutral, then the relative proportions of differences seen in the different parts of the genes should be the same for both within-species and between-species comparisons. However, the relative levels of diversity and divergence in 5′UTRs, in 3′UTRs, at nd sites, and at 4d sites clearly differ (Fig. 2).
If we compare the levels of diversity and divergence to those at 4d sites (which may be the least affected by selection within transcribed regions) we find that, within humans, differences at nd sites and 3′UTRs amount to 49% and 89%, respectively, of the differences at 4d sites. The corresponding fractions for the human–chimpanzee comparisons are 15% and 69%, respectively. The difference between the relative levels of diversity and divergence is not significant for 3′UTR (χ2 = 2.56, d.f. = 1, P = 0.11), but it is significant for nd sites (χ2 = 59.08, d.f. = 1, P < 0.001). This also holds true if CpG sites are excluded (χ2 = 20.75, d.f. = 1, P < 0.001; Table 2). Thus, for changes that affect amino acids in proteins, we observed more diversity within humans than expected, given the observed divergence between chimpanzee and human. This indicates that slightly deleterious amino acid mutations destined to be eliminated by purifying selection segregate in humans (Ohta 1976).
5′UTRs differ by 0.04% among humans and by 1.18% between humans and chimpanzees. Relative to the differences at 4d sites, this represents 48% and 111%, respectively. Thus, in stark contrast to nd sites, we observe more divergence than expected given the diversity at 5′UTRs (χ2 = 17.232, d.f. = 1, P < 0.001). The exclusion of CpG sites from the analysis increases the discrepancy to 53% and 152% of the differences at 4d sites (χ2 = 23.538, d.f. = 1, P < 0.001; Fig. 2).
Since our analyses indicate that 4d sites have been under negative selection, we also normalized the intraspecific diversity in the various parts of transcripts to a genome-wide estimate of diversity in humans of 0.075% (Sachidanandam et al. 2001). Similarly, we normalized the divergence to the genome-wide divergence between humans and chimpanzees of 1.24% (Ebersberger et al. 2002; Fig. 2C,D). Using this normalization (Fig. 2), we find that 4d site divergence amounts to 86% of the genome average, whereas 4d site diversity amounts to 111%, indicating that 4d sites are similar to nd sites and 3′UTRs in that slightly deleterious variants segregate in the human population. In contrast, the divergence in 5′UTRs represents 95% of the genome-wide divergence but only 53% of the diversity. Thus, also using genome-wide estimates of intergenic diversity and divergence, we find an excess of divergence at 5′UTRs.
DISCUSSION
CpGs Sites
A substantial proportion of the DNA sequence divergence between humans and chimpanzees is caused by changes at CpG sites (Ebersberger et al. 2002). For example, 28% of all transitional differences between humans and chimpanzees occur at CpG dinucleotides. The reason for this is that methylated CpG sites are hot spots for transitions (Shen et al. 1994). In addition, transversions are overrepresented at CpG dinucleotides (Nachman and Crowell 2000; Ebersberger et al. 2002).
Thus, one might assume that the amounts of CpG nucleotides in different classes of DNA sequences would largely determine the extent of their divergence between humans and chimpanzees. However, this is not the case. For example, although 3′UTRs contain about half as many CpG sites as CDSs, they have diverged almost twice as much. A reason for this may obviously be the different amounts of functional constraints that affect the sequences. This is reflected by the fact that also non-CpG sites in 3′UTRs have diverged about twice as much as in CDSs. However, additional factors must play a role because the ratio of divergence at CpG and non-CpG sites differs between different parts of the transcripts (Table 1). In particular, CpG sites in 5′UTRs appear to evolve in a different way from CpG sites in other parts of the transcripts. Although the ratios of the divergence at CpG and non-CpG sites are 12.3 and 14.0 in coding regions and 3′UTRs, respectively, and thus not too different from intronic and intergenic sequences where it is 15.1 and 14.6, respectively, the ratio is only 5.4 in the 5′UTRs (Table 1). Furthermore, although the transition/transversion ratio is substantially higher at CpG sites than at non-CpG sites in coding regions, 3′UTRs, introns, and intergenic regions, this is not the case in 5′UTRs. A possible explanation for this is that CpG islands, that is, regions of high CpG content located in upstream areas of about 70% (Davuluri et al. 2001) of genes, often extend into the first exon of genes (Pesole et al. 1997). Because CpGs in CpG islands may be conserved (Jones and Takai 2001) and not methylated in the germ line, they may have both a lower divergence and a lower transition/transversion ratio than CpG sites elsewhere. Indeed, 43% of the 5′UTRs compared exhibit CpG island features. CpGs within these 5′UTRs have a divergence of 3.2%, and 5′UTRs not carrying CpG island features have a divergence of 5.4% (χ2 = 20.6, P = 10−5,d.f. = 1). Thus, CpG islands are likely to be the cause of a large part, if not all, of the unusual patterns of evolution of CpGs in the 5′UTRs of human transcripts. However, further studies that also include nontranscribed upstream areas of genes are necessary to fully understand the patterns observed.
Constraints on 3′UTRs and Fourfold Degenerate Sites
Because differences in the extent of methylation in different regions of genes are likely to influence mutation rates at CpG sites, we exclude CpG sites in order to estimate the extent to which different regions of the transcripts are subject to functional constraints. Non-CpG sites in 3′UTRs exhibit 29% less divergence than non-CpG sites in intergenic regions, indicating that selective constraints act on 3′UTRs. Sequence motifs that influence mRNA stability and trafficking (Duret et al. 1993; Pesole et al. 1997) are putative targets of purifying selection in 3′UTRs.
At non-CpG 4d sites in protein-coding regions, we observe 39% lower divergence than in intergenic regions, in agreement with a study (Bustamante et al. 2002) that compared synonymous sites in functional human genes to pseudogenes. It is not clear why as much as 39% of 4d sites might be under purifying selection in humans. Because an overall codon usage bias is observed genome-wide in humans (Lander et al. 2001), the preference of certain isoacceptor tRNAs over others may play a role (Sharp and Li 1986). In humans, codon usage bias is positively correlated with expression breadth (Urrutia and Hurst 2001), which in turn covaries with expression levels (Duret and Mouchiroud 2000). If codon usage bias is the reason for the low divergence at 4d sites, its effect might be particularly strong for our data set, because genes highly expressed in brain or testis are likely to be overrepresented in our cDNA libraries. However, other factors such as conservation of mRNA secondary structures (Eyre-Walker and Bulmer 1993) or sequences involved in mRNA splicing (Cartegni et al. 2002) could also reduce divergence at 4d sites. Eventually, functional studies of a representative number of human transcripts carrying different synonymous codons will be necessary to elucidate the functional basis for the purifying selection that obviously affects a substantial proportion of synonymous sites in human genes.
Constraints on Amino Acid Substitutions
To estimate the extent to which selection affects protein-coding DNA sequences, we calculate the number of nucleotide substitutions that change amino acids per nucleotide sites that potentially change amino acids (Ka) and the number of substitutions that do not change amino acids per site that cannot change amino acids (Ks) and we compute the ratio between them (Ka/Ks; Kimura 1983; Eyre-Walker and Keightley 1999). Among the 1226 transcripts compared between humans and chimpanzees, the average Ka/Ks ratio is 0.22. In a previous study (Eyre-Walker and Keightley 1999) in which 46 genes retrieved from public databases were compared between chimpanzees and humans, the average Ka/Ks ratio was 0.63, thus indicating much fewer functional constraints than the present study. However, 10 of the 46 genes in the previous study encode components of the immune system that are known to evolve rapidly. Furthermore, a recent study (Fay et al. 2001) has estimated the proportion of human polymorphisms under strong constraint by excluding rare variants suspected to include slightly deleterious polymorphisms. For the remaining polymorphisms, expected to be able to reach fixation, a ratio of amino acid changes to silent changes (πa/πs) of 0.20 was found, almost identical to the Ka/Ks ratio found here for the human–chimpanzee comparison. Thus, we believe that the estimate from our data for genes selected at random from brain and testis transcripts represent a good overall estimate of the level of selective constraints under which human genes evolve.
Intriguingly, a study that compared 2820 rat and mouse coding regions revealed a Ka/Ks ratio of 0.19 (Makalowski and Boguski 1998), strikingly similar to the 0.22 observed between humans and chimpanzees. This is unexpected because the effective population size is generally thought to be larger in rodents than in humans, which should result in more efficient selection against deleterious variants (Ohta 1995). However, as the genes of our sample might be highly expressed, they might also be unusually conserved (Duret and Mouchiroud 2000). Further studies of the Ka/Ks ratios in various groups of mammals are necessary to arrive at an understanding of the relative contribution of different factors that influence the level of selection on the proteome.
Excess of Chimp–Human Divergence in 5′UTRs
When we normalize the extent of diversity among humans as well as divergence between humans and chimpanzees to 4d sites, we find that both nd sites and 3′UTRs exhibit more diversity than divergence. This may be due to slightly deleterious variants that segregate in the human population and are destined to become eliminated by selection. A rough estimate of the proportion of such slightly deleterious variants is 69% for amino acid polymorphisms and 23% for 3′UTRs (Fig. 2A,B).
In contrast, within 5′UTRs we observe more divergence than expected given the diversity. Because 4d sites are subject to purifying selection, reduced divergence at 4d sites could in principle account for this observation. However, when we restrict the analysis to non-CpG sites and add 39% to the divergence at 4d sites (our estimate of the extent of polymorphism removed by purifying selection), the excess of divergence in 5′UTRs remains (χ2 = 7.509, d.f. = 1, P = 0.006). Furthermore, when genome-wide estimates of diversity (Sachidanandam et al. 2001) and divergence (Chen and Li 2001; Ebersberger et al. 2002; Fujiyama et al. 2002) are used for normalization, the excess of divergence at 5′UTRs still remains. It should be noted, however, that the polymorphism data currently available are crude estimates of the human diversity. Thus, the estimates of the magnitude of the effects of selection have to be taken with caution.
The excess of divergence at 5′UTRs is an unexpected finding. It raises the possibility that 5′UTRs are subject to positive selection in humans and chimpanzees. This is interesting with regard to the recent finding that mRNA and protein levels have changed substantially in several tissues between humans and chimpanzees (Enard et al. 2002). Because transcriptional promoters often overlap with exons encoding 5′UTRs, and because sequences in 5′UTRs are often involved in the control of translation, it may be that DNA sequences that affect gene expression levels at the transcriptional and translational levels have been frequent targets of positive selection during human evolution. Further work that explores the functional properties of promoters and 5′UTRs in human and chimpanzees is necessary to test this suggestion.
METHODS
cDNA Libraries and Sequencing
We isolated total RNA from cerebral cortex of a female chimpanzee, and cerebral cortex and testis of a male chimpanzee using Trizol (Life Technologies). We purified the total RNA using RNAeasy columns (Qiagen) and isolated mRNA using Oligotex (Qiagen) according to the manufacturer's instructions. cDNA was synthesized according to the SMART Protocol (Clontech); ligated into the SfiI sites of the bacterial plasmid pUCHi, a derivative of pUC19 constructed in-house that contains SfiI sites, allowing directional cloning of cDNAs; and transformed into Epicurian Coli XL10-Gold (Stratagene). Plasmids were prepared using QIAprep (Qiagen) and sequenced from their 5′ ends using the M13 reverse primer with Big Dye Terminator Cycle sequencing (Perkin Elmer) and ABI 3700 sequencing machines. A total of 4011 cDNA sequences were obtained from the male chimp brain library, 992 from the testis library, and 52 from the female brain library.
Sequence Analyses
Nucleotides with a Phred quality score (Ewing et al. 1998) below 15 were masked. If three or more adjacent nucleotides were masked, the sequence was cut at that point and the longest of the resulting fragments used for further analysis. Vector sequences were removed and mitochondrial sequences were excluded. The resulting sequences were assembled into contigs using the TIGR Assembler (Sutton et al. 1995).
The average number of reads that cover each base in the contigs was 1.49. An empirical upper limit of the sequencing error rate can be gauged from the most frequent transcript, which occurred 30 times among the cDNA clones (average clone length 488 bp). None of the clones carried a mismatch. Thus, as a rough estimate the DNA sequences determined contain less than 1 mismatch per 10,000 bp.
Repeats were masked using RepeatMasker (see http://repeatmasker.genome.washington.edu/) and the resultant cDNA sequences were used to search dbEST, the GenBank sections for primate and high-throughput sequencing (htg), unigene cluster consensus sequences (Coward et al. 2002), the human genome sequence assembly (July 2001), and Refseq (http://www.ncbi.nlm.nih.gov/) using the BLAST program (Altschul et al. 1990) on the HUSAR platform (http://genome.dkfz-heidelberg.de). The best BLAST result from each database with an e-value smaller or equal to 0.01 was realigned to the unmasked chimpanzee sequence using Bestfit (Wisconsin GCG-package). These alignments were generated by scoring matches with 1, mismatches with −2, the opening of a gap with −4, and the extension of a gap with −1, whereas gap extensions were penalized only for the first 50 bp. From these alignments, the one with the highest alignment score (≥30) was used for further analysis. This sieving process left us with 2890 alignments; from those, all alignments with 95% or less identity were checked manually, leaving us with 2844 alignments.
The database entries for 1103 of these genes contained a gene annotation that was used to extract the coding region. If the database entry did not contain a gene annotation and was not an EST entry, Genscan (Burge and Karlin 1997) was applied to the human genome sequence starting 50 kb before and ending 50 kb after the alignment to the chimpanzee cDNA contig. Identified coding regions were extracted and collected in a local database, which was again searched with the chimpanzee cDNA contig using BLAST. We thus identified 742 additional CDSs. The average divergence within 4d sites, 3′UTRs, 5′UTRs, and nd sites was similar for the genes for which the CDS was identified using Genscan and the previously annotated ones (data not shown).
If the cDNA contig was longer than the coding region, everything before the start codon of the CDS was assigned as 5′UTRs (582 5′UTRs), and everything behind the stop codon as 3′UTRs (1071 3′UTRs). These alignments were used to count substitutional differences in the 3′UTRs, 5′UTRs, and coding regions between human and chimpanzee. In order to estimate the average percentage of nonsynonymous and synonymous differences, we considered sites that were either nondegenerate or fourfold degenerate in both species.
A nucleotide was counted as being within a CpG dinucleotide if the chimpanzee and/or the human sequence contained a CG. A 5′UTR sequence was considered to be within a CpG island if the ratio of observed-to-expected CpG dinucleotides (given the base composition) was higher than 0.6 and the GC content was above 50% over the entire 5′UTR alignment (Gardiner-Garden and Frommer 1987).
Human–Chimpanzee Divergence
Because humans and chimpanzees are so closely related, multiple substitutions at the same site are highly unlikely. Therefore, we measured divergence by dividing the number of substitutions by the number of base pairs compared for a given sequence (Nei and Kumar 2000). The significance of differences in the average divergence of different sequence categories was assessed with a t-test assuming unequal variances.
Human Diversity Data
Of the 1845 orthologous human sequences for which we could identify the CDS, 1304 were mRNA-derived sequences. For simplicity, we used only these mRNA-derived sequences, which were repeat masked and compared with dbSNP (http://www.ncbi.nlm.nih.gov/SNP/) using BLAST. For identification of SNPs, alignments were required to be over 95% identical and at least 50 bp long. SNPs derived from EST data mining were excluded as they may contain false-positive SNPs that would cause an inflated diversity in 3′UTRs because they are derived mainly from 3′ ends of transcripts.
We tried to account for the fact that different average numbers of chromosomes were sampled for SNP discovery in 5′UTRs, 3′UTRs, and coding regions by two different approaches. First, we accepted only SNPs for which 20 or fewer chromosomes were sampled because, for some regions, SNPs tended to be discovered in very small samples. Second, we used only SNPs detected by reduced representation shotgun sequencing (Altshuler et al. 2000) and in BAC overlaps (Sachidanandam et al. 2001). Both approaches resulted in approximately equal numbers of chromosomes being sampled for 5′UTRs, 3′UTRs, and coding regions and showed a similar relation of diversity among these regions for nondegenerate and fourfold-degenerate sites (data not shown) as the entire data in Figure 2. Similar levels of diversity in these regions have also been seen by others (Cargill et al. 1999; Halushka et al. 1999; Sunyaev et al. 2000).
WEB SITE REFERENCES
http://genome.dkfz-heidelberg.de; Husar Analysis Package.
http://repeatmasker.genome.washington.edu; RepeatMasker server home page.
http://www.ncbi.nlm.nih.gov/NCBI Web site.
http://www.ncbi.nlm.nih.gov/SNP; dbSNP.
Acknowledgments
We thank C.D. Bustamante, J.C. Fay, M. Kohn, E.S. Lander, M. Przeworski, S.E. Ptak, and C.I. Wu for discussion and critical comments and T. Kitano for analysis support.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
E-MAIL paabo@eva.mpg.de; FAX 49-341-9952-555.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.944903.
REFERENCES
- 1.Altschul S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403-410. [DOI] [PubMed] [Google Scholar]
- 2.Altshuler D., Pollara, V.J., Cowles, C.R., Van Etten, W.J., Baldwin, J., Linton, L., and Lander, E.S. 2000. An SNP map of the human genome generated by reduced representation shotgun sequencing. Nature 407: 513-516. [DOI] [PubMed] [Google Scholar]
- 3.Burge C. and Karlin, S. 1997. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268: 78-94. [DOI] [PubMed] [Google Scholar]
- 4.Bustamante C.D., Nielsen, R., and Hartl, D.L. 2002. A maximum likelihood method for analyzing pseudogene evolution: Implications for silent site evolution in humans and rodents. Mol. Biol. Evol. 19: 110-117. [DOI] [PubMed] [Google Scholar]
- 5.Cargill M., Altshuler, D., Ireland, J., Sklar, P., Ardlie, K., Patil, N., Shaw, N., Lane, C.R., Lim, E.P., Kalyanaraman, N., et al. 1999. Characterization of single-nucleotide polymorphisms in coding regions of human genes [published erratum appears in Nat. Genet. 1999 Nov.23(3):373]. Nat. Genet. 22: 231-238. [DOI] [PubMed] [Google Scholar]
- 6.Cartegni L., Chew, S.L., and Krainer, A.R. 2002. Listening to silence and understanding nonsense: Exonic mutations that affect splicing. Nat. Rev. Genet. 3: 285-298. [DOI] [PubMed] [Google Scholar]
- 7.Chen F.C. and Li, W.H. 2001. Genomic divergences between humans and other hominoids and the effective population size of the common ancestor of humans and chimpanzees. Am. J. Hum. Genet. 68: 444-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coward E., Haas, S.A., and Vingron, M. 2002. SpliceNest: Visualizing gene structure and alternative splicing based on EST clusters. Trends Genet. 18: 53-55. [Google Scholar]
- 9.Davuluri R.V., Grosse, I., and Zhang, M.Q. 2001. Computational identification of promoters and first exons in the human genome. Nat. Genet. 29: 412-417. [DOI] [PubMed] [Google Scholar]
- 10.Duret L. and Mouchiroud, D. 2000. Determinants of substitution rates in mammalian genes: Expression pattern affects selection intensity but not mutation rate. Mol. Biol. Evol. 17: 68-74. [DOI] [PubMed] [Google Scholar]
- 11.Duret L., Dorkeld, F., and Gautier, C. 1993. Strong conservation of non-coding sequences during vertebrates evolution: Potential involvement in post-transcriptional regulation of gene expression. Nucleic Acids Res. 21: 2315-2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ebersberger I., Metzler, D., Schwarz, C., and Paabo, S. 2002. Genome-wide comparison of DNA sequences between humans and chimpanzees. Am. J. Hum. Genet. 70: 1490-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Enard W., Khaitovich, P., Klose, J., Zollner, S., Heissig, F., Giavalisco, P., Nieselt-Struwe, K., Muchmore, E., Varki, A., Ravid, R., et al. 2002. Intra- and interspecific variation in primate gene expression patterns. Science 296: 340-343. [DOI] [PubMed] [Google Scholar]
- 14.Ewing B., Hillier, L., Wendl, M.C., and Green, P. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8: 175-185. [DOI] [PubMed] [Google Scholar]
- 15.Eyre-Walker A. and Bulmer, M. 1993. Reduced synonymous substitution rate at the start of enterobacterial genes. Nucleic Acids Res. 21: 4599-4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eyre-Walker A. and Keightley, P.D. 1999. High genomic deleterious mutation rates in hominids. Nature 397: 344-347. [DOI] [PubMed] [Google Scholar]
- 17.Fay J.C., Wyckoff, G.J., and Wu, C.I. 2001. Positive and negative selection on the human genome. Genetics 158: 1227-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujiyama A., Watanabe, H., Toyoda, A., Taylor, T.D., Itoh, T., Tsai, S.F., Park, H.S., Yaspo, M.L., Lehrach, H., Chen, Z., et al. 2002. Construction and analysis of a human–chimpanzee comparative clone map. Science 295: 131-134. [DOI] [PubMed] [Google Scholar]
- 19.Gardiner-Garden M. and Frommer, M. 1987. CpG islands in vertebrate genomes. J. Mol. Biol. 196: 261-282. [DOI] [PubMed] [Google Scholar]
- 20.Halushka M.K., Fan, J.B., Bentley, K., Hsie, L., Shen, N., Weder, A., Cooper, R., Lipshutz, R., and Chakravarti, A. 1999. Patterns of single-nucleotide polymorphisms in candidate genes for blood-pressure homeostasis. Nat. Genet. 22: 239-247. [DOI] [PubMed] [Google Scholar]
- 21.Jones P.A. and Takai, D. 2001. The role of DNA methylation in mammalian epigenetics. Science 293: 1068-1070. [DOI] [PubMed] [Google Scholar]
- 22.Kimura M., 1983. The neutral theory of molecular evolution. Cambridge University Press, Cambridge, UK.
- 23.Lander E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860-921. [DOI] [PubMed] [Google Scholar]
- 24.Makalowski W. and Boguski, M.S. 1998. Evolutionary parameters of the transcribed mammalian genome: An analysis of 2,820 orthologous rodent and human sequences. Proc. Natl. Acad. Sci. 95: 9407-9412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nachman M.W. and Crowell, S.L. 2000. Estimate of the mutation rate per nucleotide in humans. Genetics 156: 297-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nei M. and Kumar, S., 2000. Molecular evolution and phylogenetics. Oxford University Press, New York, NY.
- 27.Ohta T. 1976. Role of very slightly deleterious mutations in molecular evolution and polymorphism. Theor. Popul. Biol. 10: 254-275. [DOI] [PubMed] [Google Scholar]
- 28.___, 1995. Synonymous and nonsynonymous substitutions in mammalian genes and the nearly neutral theory. J. Mol. Evol. 40: 56-63. [DOI] [PubMed] [Google Scholar]
- 29.Pesole G., Liuni, S., Grillo, G., and Saccone, C. 1997. Structural and compositional features of untranslated regions of eukaryotic mRNAs. Gene 205: 95-102. [DOI] [PubMed] [Google Scholar]
- 30.Roest Crollius H., Jaillon, O., Bernot, A., Dasilva, C., Bouneau, L., Fischer, C., Fizames, C., Wincker, P., Brottier, P., Quetier, F., et al. 2000. Estimate of human gene number provided by genome-wide analysis using Tetraodon nigroviridis DNA sequence. Nat. Genet. 25: 235-238. [DOI] [PubMed] [Google Scholar]
- 31.Rubin G.M. 2001. The draft sequences. Comparing species. Nature 409: 820-821. [DOI] [PubMed] [Google Scholar]
- 32.Sachidanandam R., Weissman, D., Schmidt, S.C., Kakol, J.M., Stein, L.D., Marth, G., Sherry, S., Mullikin, J.C., Mortimore, B.J., Willey, D.L., et al. 2001. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature 409: 928-933. [DOI] [PubMed] [Google Scholar]
- 33.Shabalina S.A., Ogurtsov, A.Y., Kondrashov, V.A., and Kondrashov, A.S. 2001. Selective constraint in intergenic regions of human and mouse genomes. Trends Genet. 17: 373-376. [DOI] [PubMed] [Google Scholar]
- 34.Sharp P.M. and Li, W.H. 1986. Codon usage in regulatory genes in Escherichia coli does not reflect selection for ‘rare’ codons. Nucleic Acids Res. 14: 7737-7749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen J.C., Rideout, W.M., III, and Jones, P.A. 1994. The rate of hydrolytic deamination of 5-methylcytosine in double-stranded DNA. Nucleic Acids Res. 22: 972-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sunyaev S.R., Lathe, W.C., Ramensky, V.E., and Bork, P.E. 2000. SNP frequencies in human genes an excess of rare alleles and differing modes of selection. Trends Genet. 16: 335-337. [DOI] [PubMed] [Google Scholar]
- 37.Sutton G.G., White, O., Adams, M.D., and Kerlavage, A.R. 1995. TIGR Assembler: A new tool for assembling large shotgun sequencing projects. Genome Sci. Technol. 1: 9-19. [Google Scholar]
- 38.Urrutia A.O. and Hurst, L.D. 2001. Codon usage bias covaries with expression breadth and the rate of synonymous evolution in humans, but this is not evidence for selection. Genetics 159: 1191-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Waterston R.H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J.F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520-562. [DOI] [PubMed] [Google Scholar]