Abstract
Current understanding of the epigenetic regulator roles in plant growth and development has largely derived from studies in the dicotyledonous model plant Arabidopsis thaliana. Rice (Oryza sativa) is one of the most important food crops in the world and has more recently becoming a monocotyledonous model plant in functional genomics research. During the past few years, an increasing number of studies have reported the impact of DNA methylation, non-coding RNAs and histone modifications on transcription regulation, flowering time control, and reproduction in rice. Here, we review these studies to provide an updated complete view about chromatin modifiers characterized in rice and in particular on their roles in epigenetic regulation of flowering time, reproduction, and seed development.
Keywords: chromatin, epigenetics, flowering time, histone modification, DNA methylation, non-coding RNA, reproduction, Oryza sativa
INTRODUCTION
Epigenetics is defined as nucleotide sequence-independent changes in the gene expression that are mitotically and/or meiotically heritable. The fundamental repeating unit of chromatin is nucleosome. The nucleosome contains 145–147 base pairs (bp) of DNA wrapped around an octamer of histone proteins, comprising two copies of each of the four core histones, H2A, H2B, H3, and H4 (McGinty and Tan, 2014). The linker histone H1 associates with DNA inbetween the two nucleosomes and participates in higher order chromatin structure formation and remodeling. The structure of chromatin can be subjected to panoply of epigenetic regulations including DNA methylation, histone covalent modifications, histone variants, and ATP-dependent chromatin remodeling. DNA methylation has been widely considered as a heritable epigenetic mark that regulates expression of genes in both plants and mammals (Law and Jacobsen, 2010; Furner and Matzke, 2011; Wu and Zhang, 2014). Histone modifications including methylation, acetylation, phosphorylation, ubiquitination, and sumoylation, play critical roles in regulating chromatin structure and gene expression, mainly by altering nucleosome stability and positioning that affect DNA accessibility for regulatory proteins or protein complexes involved in transcription, DNA replication and repair (Pikaard and Scheid, 2013; To and Kim, 2014; Van Lijsebettens and Grasser, 2014). ATP-dependent chromatin remodeling factors control relocation or dissociation of nucleosomes, and histone chaperones bind histones and play crucial roles in nucleosome assembly/disassembly in diverse chromatin metabolism and epigenetic regulation (Zhu et al., 2012; Gentry and Hennig, 2014).
Rice (Oryza sativa ) is a worldwide crop and represents a valuable model plant for monocots, to which many of our food crops belong. Compared to the extensively studied dicot model plant Arabidopsis thaliana, rice has only been more recently studied in epigenetic modifications (reviewed in Chen and Zhou, 2013). Genome-wide analyses of DNA methylations have revealed conservation as well as distinct differences between rice and Arabidopsis, and that a much higher level of DNA methylation is observed in association with more numerous transposable elements present in the rice genome (Yan et al., 2010; Zemach et al., 2010; Chodavarapu et al., 2012; Li et al., 2012). Genome-wide analyses by chromatin immunoprecipitation combined with high-throughput sequencing (ChIP-Seq) have shown that several types of histone modifications, e.g., histone H3 lysine 9 acetylation (H3K9ac) and H4K12ac, H3K4 di-/tri-methylation (H3K4me2/3), H3K27me3, and H3K36me3, are broadly distributed with distinct patterns within the rice genome (He et al., 2010; Malone et al., 2011; Du et al., 2013). In this review, we summarize and discuss regulators involved in different types of chromatin modifications and their roles in rice plant flowering time control and reproduction.
REGULATION OF DIFFERENT TYPES OF CHROMATIN MODIFICATIONS IN RICE
Different types of chromatin modifications are regulated by specific factors that are generally conserved in rice and other plant species (www.chromdb.org). So far, only some of the rice chromatin modifiers are functionally characterized by analysis of loss-of-function mutants and RNAi or overexpression transgenic plants (Table 1).
Table 1.
Chromatin modifiers functionally characterized in rice.
| Name | Gene locus | Molecularfunction | Biological role | Reference | |
|---|---|---|---|---|---|
| DNA methylation | OsMET1b/OsMET1-2 | LOC_Os07g08500 | DNA methyltransferase | Seed development | Hu et al. (2014), Yamauchi et al. (2014) |
| OsDRM2 | LOC_Os03g02010 | De novo DNA methyltransferase | Pleiotrpic effects on development | Moritoh et al. (2012), Pang et al. (2013) | |
| OsDDM1 | LOC_Os09g27060 | DNA methylation maintenance | Transposon repression, growth inhibition | Higo et al. (2012) | |
| DNA demethylation | OsROS1a | LOC_Os01g11900 | DNA demethylase | Plant reproduction | Zemach et al. (2010), Ono et al. (2012) |
| OsROS1c | LOC_Os05g37350 | DNA demethylase | Transposon activation | La et al. (2011) | |
| Histone methylation | SDG714 | LOC_Os01g70220 | H3K9 methyltransferase | Transposon repression, trichome development | Ding et al. (2007b) |
| SDG728 | LOC_Os05g41170 | H3K9 methyltransferase | Transposon repression, seed development | Qin et al. (2010) | |
| SDG725 | LOC_Os02g34850 | H3K36 methyltransferase | Hormone regulatory gene activation, flowering | Sui et al. (2012, 2013) | |
| SDG724 | LOC_Os09gl3740 | H3K36 methyltransferase | Flowering | Sun et al. (2012) | |
| SDG723/OsTrx1 | LOC_Os09g04890 | H3K4 methyltransferase | Flowering | Choi et al. (2014) | |
| Histone demethylation | JMJ706 | LOC_Os10g42690 | H3K9 demethylase | Floral organ development | Sun and Zhou (2008) |
| JMJ705 | LOC_Os01g67970 | H3K27 demethylase | Biotic stress response, plant reproduction | Li et al. (2013) | |
| JMJ703 | LOC_Os05g10770 | H3K4 demethylase | Stem elongation, transposon repression | Chen et al. (2013); Cui et al. (2013) | |
| JMJ701 | LOC_Os03g05680 | H3K4 demethylase | Flowering | Yokoo et al. (2014) | |
| Polycomb silencing | OsiEZ1/SDG718 | LOC_Os03g19480 | H3K27 methyltransferase | Flowering | Liu et al. (2014) |
| OsCLF/SDG711 | LOC_Os06gl6390 | H3K27 methyltransferase | Flowering | Liu et al. (2014) | |
| OsFIE1 | LOC_Os08g04290 | Drosophila ESC homolog | Pleiotrpic effects on development | Zhang et al. (2012b), Nallamilli et al. (2013), Folsom et al. (2014) | |
| OsFIE2 | LOC_Os08g04270 | Drosophila ESC homolog | Organ generation, reproduction | Luo et al. (2009), Li et al. (2014) | |
| OsEMF2b | LOC_Os09g13630 | Drosophila Su(z)12 homolog | Floral organ development | Yang et al. (2013), Conrad et al. (2014) | |
| Histone deacetylation | OsHDT1/HDT701 | LOC_Os05g51840 | H4 deacetylase | Biotic stress response, heterosis | Li et al. (2011a), Ding et al. (2012a) |
| OsSRT1 | LOC_Os04g20270 | H3K9 deacetylase | Cell death, transposon repression | Huang et al. (2007), Zhong et al. (2013) | |
| Others | CHD3/CHR729 | LOC_Os07g31450 | Chromodomain and PHD-domain protein | Pleiotrpic effects on development | Hu et al. (2012) |
| MEL1 | LOC_Os03g58600 | AGO-family protein | Meiosis progression | Nonomura et al. (2007), Komiya et al. (2014) | |
| SHO1 | LOC_Os04g43050 | Homolog of DICER-LIKE 4 | Pleiotrpic effects on development | Abe et al. (2010) | |
| SHL2 | LOC_Os01g34350 | RDR6 homolog | Floral organ development | Toriba et al. (2010) | |
| WAF1 | LOC_Os07g06970 | HEN1 homolog | Pleiotrpic effects on development | Abe et al. (2010) | |
| BRK1 | LOC_Os07g32480 | H2A phosphorylation | Meiosis progression | Wang et al. (2012) |
DNA METHYLATION
In plants, DNA methylation occurs at cytosine residues in symmetric, CG and CHG, as well as asymmetric, CHH, contexts (where H = A, T or C; Law and Jacobsen, 2010). In Arabidopsis, CG methylation is maintained by METHYLTRANSFERASE 1 (MET1; Saze et al., 2003), whereas CHG methylation is mediated by CHROMOMETHYLASE 3 (CMT3; Lindroth et al., 2001). The maintenance of CHH methylation is carried out by CMT2 and DOMAINS REARRANGED METHYLTRANSFERASE 2 (DRM2), an ortholog of mammalian Dnmt3 (Law and Jacobsen, 2010; Stroud et al., 2014). DRM2 is required for de novo cytosine methylation in both symmetric and asymmetric sequence contexts, which is guided to the target region by RNA-directed DNA methylation (RdDM) pathway (Cao and Jacobsen, 2002; Law and Jacobsen, 2010; Stroud et al., 2014). While Arabidopsis contains only one MET1 gene, rice has two MET1 genes, MET1a (also named OsMET1-1) and MET1b/OsMET1-2 (Teerawanichpan et al., 2004; Yamauchi et al., 2008). The transcripts of MET1b accumulate more abundantly than those of MET1a in all of the examined rice tissues, indicating that MET1b may play a more important role in maintaining DNA methylation (Yamauchi et al., 2008). Consistently, more recent studies demonstrate that MET1b is an essential gene and its loss causes genome-wide reduction of CG methylation in rice seedlings (Hu et al., 2014; Yamauchi et al., 2014). Rice contains also one DRM2 gene, OsDRM2, and the recombinant OsDRM2 protein expressed in Escherichia coli or Saccharomyces cerevisiae exhibits stochastic de novo DNA methyltransferase activity in vitro at CG, CHG, and CHH (Sharma et al., 2009; Pang et al., 2013). Interestingly, OsDRM2 was found to interact with the ATP-dependent RNA helicase, OseIF4A, in both in vitro and in vivo assays (Dangwal et al., 2013). The interaction specifically depends on the ubiquitin-associated domain of OsDRM2, pointing to a mechanism in which OsDRM2 is recruited to specific chromatin sites by eIF4A together with other cellular proteins for catalyzing DNA methylation (Dangwal et al., 2013). Similar to the Arabidopsis DECREASE IN DNA METHYLATION 1 (DDM1), which encodes a nucleosome remodeling ATPase, OsDDM1 is also necessary for maintenance of DNA methylation in transposons and repetitive sequences (Higo et al., 2012). The rice genome contains three putative CMT3 homologs (Sharma et al., 2009), yet their functions remain to be characterized.
DNA methylation can be removed passively through dilution during replication as well as actively through catalysis by demethylation enzymes (La et al., 2011; Ono et al., 2012). In Arabidopsis, active demethylation is catalyzed by REPRESSOR OF SILENCING 1 (ROS1; Gong et al., 2002; Agius et al., 2006), DEMETER (DME; Choi et al., 2002; Gehring et al., 2006), and DEMETER-LIKE 2 (DML2) and DML3 (Choi et al., 2002; Ortega-Galisteo et al., 2008). Phylogenetic analysis showed that the rice genome encodes six putative bi-functional DNA glycosylases that catalyze cytosine DNA demethylation: four ROS1 orthologs (ROS1a to ROS1d) and two DML3 orthologs (DML3a and DML3b), but no DME orthologs (Zemach et al., 2010). ROS1c has been shown to be involved in DNA demethylation and control of the retrotransposon Tos17 activity (La et al., 2011). Quantitative RT-PCR analysis revealed that ROS1a, ROS1d, and DML3a are expressed in different examined plant tissues, including anthers and pistils, whereas ROS1b and DML3b are scarcely expressed in these tissues (Ono et al., 2012). Future studies are necessary to investigate the role of these different genes in rice genome DNA methylation.
HISTONE METHYLATION
Histone methylation marks are established on lysine (K) and arginine (R) residues by distinct enzymes, namely histone lysine methyltransferases (HKMTs) and protein arginine methyltransferases (PRMTs), respectively (Liu et al., 2010; Yao and Shen, 2011). In general, H3K9, H3K27, and H4K20 methylations are associated with transcriptional repression, whereas methylation on H3K4 and H3K36 correlates with gene activation. Furthermore, each K residue can be mono-, di-, or tri-methylated, and different methylation status may have different functional implications (Yu et al., 2009).
All known plant HKMTs contain an evolutionarily conserved SET domain (reviewed in Berr et al., 2011). The rice genome encodes at least 37 SET domain proteins, grouped into distinct families (Ng et al., 2007; Huang et al., 2011; Thorstensen et al., 2011). To date, several members belonging to different families are characterized (Table 1). Analyses of SET DOMAIN GROUP 714 (SDG714) and its close homologs (e.g., SDG728) showed that these rice SDG proteins have either specific or redundant functions in regulating histone H3K9 methylation and retrotransposon repression (Ding et al., 2007a,b, 2010; Qin et al., 2010). Knockdown of SDG714 leads to decreased H3K9 methylation levels accompanied by a reduction of CG and CHG methylation, suggesting that H3K9 methylation and DNA methylation act closely together to stably repress the transposition of transposons to maintain genome stability (Ding et al., 2007b). Ectopic expression of SDG714 in Arabidopsis can cause a global elevation of H3K9me2 (Ding et al., 2010). Knockdown of SDG725 impairs deposition of H3K36me2/3 at several examined gene loci (Sui et al., 2012, 2013). SDG724 is also involved in H3K36me2/3 deposition (Sun et al., 2012). SDG723/OsTrx1 is a close homolog of the Arabidopsis H3K4-methyltransferase ATX1 and can methylate in vitro H3 within oligonucleosomes (Choi et al., 2014). The rice genome contains two genes encoding putative H3K27 methyltransferases, OsiEZ1/SDG718 (also named OsSET1) and OsCLF/SDG711, which likely work in protein complexes in Polycomb silencing pathway (see Section below).
Histone lysine methylation can be removed by histone demethylases, which consist of two classes: Lysine Specific Demethylase 1 (LSD1) and Jumonji C (jmjC) domain-containing proteins (Tsukada et al., 2006; Mosammaparast and Shi, 2010). LSD1, a flavin-dependent amine oxidase, has been the first histone demethylase reported (Shi et al., 2004) and Arabidopsis contains three LSD1 homologs, which are involved in flowering time regulation (Jiang et al., 2007; Liu et al., 2007; Shafiq et al., 2014). Three rice genes (Os02g0755200, Os04g0560300, and Os08g0143400) encode LSD1 homologs, but their functions remain uncharacterized. There are at least 20 jmjC domain-containing proteins in rice, and the first characterized JMJ706 specifically demethylates H3K9me2/me3 (Sun and Zhou, 2008). More recently, several other rice jmjC-encoding genes have been characterized. JMJ705 encodes a histone lysine demethylase that specifically removes H3K27me2/3, and the expression of JMJ705 is induced by stress signals and during pathogen infection (Li et al., 2013). For active histone marks, JMJ703 is involved in the removal of H3K4me1/me2/me3 (Chen et al., 2013; Cui et al., 2013), and JMJ701 in removal of H3K4me3 (Yokoo et al., 2014). So far, however, histone demethylase(s) involved in removal of H3K36 methylation is(are) unknown.
POLYCOMB SILENCING
Polycomb Group (PcG) proteins were first identified as master regulators and suppressors of homeotic genes in Drosophila melanogaster. Polycomb Repressive Complex 2 (PRC2) has four core components: ENHANCEROF ZESTE (E[z]), SUPPRESSOR OF ZESTE 12 (Su[z]12), EXTRA SEX COMBS (ESC), and the 55 kDa WD40-repeat protein N55 (Schuettengruber and Cavalli, 2009). PRC2 mediates H3K27me3 deposition via the catalytic subunit E[z], a SET-domain containing protein (Czermin et al., 2002). The four core subunits of the PRC2 complex are well conserved in animals as well as in plants (Chen and Rasmuson-Lestander, 2009; He et al., 2013). While in Drosophila all but one subunit is encoded by a single gene, most of the plant PRC2 core subunits are encoded by small gene families. In Arabidopsis, MEDEA (MEA)/FERTILIZATION INDEPENDENT SEED 1 (FIS1), CURLY LEAF (CLF), and SWINGER (SWN) are the three homologs of E[z]; FIS2, VERNALIZATION 2 (VRN2), and EMBRYONIC FLOWER 2 (EMF2) are the three homologs of Su[z]12; MULTICOPY SUPPRESSOR OF IRA1 (MSI1) to MSI5 are the five homologs of N55; and FERTILIZATION INDEPENDENT ENDOSPERM (FIE) is the only homolog of ESC. Remarkably, MEA/FIS1 and FIS2, which are important for endosperm and seed development in Arabidopsis, are absent from rice, and rice has two E[z] homologs: OsiEZ1/SDG718 and OsCLF/SDG711, two Su[z]12 homologs: OsEMF2a and OsEMF2b, but also two FIE homologs: OsFIE1 and OsFIE2 (Luo et al., 2009). Functional roles of some of these rice PcG proteins have been characterized (Table 1). The expression of OsiEZ1/SDG718 and OsCLF/SDG711 is induced by and represses flowering genes in long day and short day, respectively (Liu et al., 2014). While OsFIE2 is expressed broadly in all examined rice tissues, OsFIE1 is expressed specifically in the rice endosperm and its expression in vegetative tissues is likely to be silenced by promoter DNA methylation (Zhang et al., 2012b; Nallamilli et al., 2013). Furthermore, OsFIE1 is imprinted and only the maternal allele is expressed in endosperm (Luo et al., 2009). More recently, it was reported that OsFIE1 is responsive to temperature changes and its expression negatively correlates with the duration of the syncytial seed developmental stage during heat stress (Folsom et al., 2014). DNA methylation, H3K9me2 and/or H3K27me3 are likely involved in regulation of varied repressive status of OsFIE1 (Zhang et al., 2012b; Nallamilli et al., 2013; Folsom et al., 2014). Functional characterization of OsEMF2b revealed that PRC2 plays a major role in modulation of the expression of E-function MADS-box transcription factor genes required for floral organ specification and floral meristem determinacy (Luo et al., 2009; Yang et al., 2013; Conrad et al., 2014). Very importantly, OsFIE2 interacts with OsiEZ1/SDG718 and the OsFIE2-associated complex purified from transgenic rice suspension cells (containing OsEMF2b, OsCLF, OsiEZ1/SDG718) can methylate H3K27 in in vitro histone methyltransferase assay (Nallamilli et al., 2013).
HISTONE ACETYLATION
Histone lysine acetylation is generally associated with transcription activation and is dynamically regulated by the antagonistic activities between histone acetyltransferases (HATs) and histone deacetylases (HDACs; Chen and Tian, 2007). All four core histones can be acetylated and a nucleosome contains 26 putative acetylation sites (Lusser et al., 2001). Global analysis of lysine acetylation demonstrates the involvement of protein acetylation in diverse biological processes in rice (Nallamilli et al., 2014). The rice genome contains eight HATs and 19 HDACs (Hu et al., 2009; Liu et al., 2012). The eight HATs can be divided into four groups, namely the CREB-Binding Protein (CBP) group, the TAFII-associated factor (TAFII250) group, the GCN5-related N-terminal acetyltransferase (GNAT) group, and the MYST (named for the founding members MOZ, Ybf2/Sas3, Sas2, and Tip60) group (Liu et al., 2012). The 19 HDACs are grouped into three distinct families, namely the Reduced Potassium Deficiency 3 (RPD3) family, the Silent Information Regulator 2 (SIR2) family, and the type-II HDAC (HD2) family which is plant specific (Ma et al., 2013). Reversible and dynamic changes of H3 acetylation occurs at submergence-inducible genes, alcohol dehydrogenase 1 (ADH1) and pyruvate decarboxylase 1 (PDC1) in rice (Tsuji et al., 2006). Forward genetic analysis has identified a rice mutant, rice plasticity 1 (rpl1), which displays increased environment-dependent phenotypic variations and an elevation of overall H3K9 acetylation (Zhang et al., 2012a). Down-regulation of OsHDT1/HDT701, which encodes a histone H4 deacetylase, causes elevated levels of H4 acetylation and increased transcription of pattern recognition receptor (PRR) and defense-related genes (Ding et al., 2012a). Knockdown of OsSRT1, a member of SIR2-like HDAC family, results in an increase of H3K9 acetylation (H3K9ac), leading to DNA fragmentation and cell death, and the OsSRT1 protein binds to loci with relative low level of H3K9ac and regulates expression of many genes related to stress and metabolism as well as several families of transposable elements (Huang et al., 2007; Zhong et al., 2013).
READERS OF HISTONE MODIFICATIONS
Specific recognition of histone modifications by readers can recruit various components of the nuclear signaling network to chromatin, mediating fundamental processes such as gene transcription, DNA replication and recombination, DNA repair and chromatin remodeling (Musselman et al., 2012). Some readers are reported in Arabidopsis (reviewed in Berr et al., 2011), and more recent works have identified several novel chromodomain (CHD)- and/or plant homeodomain (PHD)-containing proteins as readers of H3K4me2/me3 and H3K36me3 (Bu et al., 2014; Lopez-Gonzalez et al., 2014; Molitor et al., 2014; Xu et al., 2014). Interestingly, the rice CHD3 protein can bind both the active mark H3K4me2 and the repressive mark H3K27me3 via its CHD and PHD domain, respectively (Hu et al., 2012). Knockdown of CHD3 caused reduction of H3K4me3 and H3K27me3 at many genes. It was thus suggested that the rice CHD3 may act as a bifunctional reader capable to recognize and modulate both H3K4 and H3K27 methylations (Hu et al., 2012).
SMALL AND LONG NON-CODING RNAs
Non-coding small RNAs (sRNA) of 21–24 nucleotides (nt) in length as well as long non-coding RNAs (lncRNAs, >200 nt in length) are known to be involved in chromatin modifications and thus epigenetic inheritance (reviewed in Castel and Martienssen, 2013; Bond and Baulcombe, 2014). Genome-wide profiling have identified several hundreds of different sRNAs, and differences exist at their expression levels between different rice subspecies, reciprocal hybrids, different plant tissues, and under different growth conditions (Chen et al., 2010; He et al., 2010; Jeong et al., 2010; Zhang et al., 2014). Remarkably, the most abundant sRNAs identified in rice panicles are 24 nt in length and mainly correspond to transposon-associated or repeat-associated small interfering RNAs (siRNAs; Jeong et al., 2011). The most intriguing role of siRNAs is in repression of transposons and repeat elements in reproductive tissues and epigenomic reprogramming during gametogenesis (Gutierrez-Marcos and Dickinson, 2012; Castel and Martienssen, 2013; Bond and Baulcombe, 2014). ARGONAUTE (AGO) proteins play important roles in microRNA-mediated post-transcriptional gene silencing (PTGS) and siRNA-mediated RdDM (Vaucheret, 2008). A germ line specific AGO-encoding gene, MEIOSIS ARRESTED AT LEPTOTENE1 (MEL1), has been reported in rice, and the mel1 mutant shows chromosome abortion during early meiotic stages, leading to impaired male and female fertilities (Nonomura et al., 2007). More recently, forward genetic analysis has identified a lncRNA, which could be subsequently processed to small RNAs, as a key regulator of male fertility in rice (Ding et al., 2012b,c). Meanwhile, Zhou et al. (2012) reported that a spontaneous mutation of a small RNA could cause male sterility in rice. Nevertheless, the precise role of lncRNA and sRNA, particularly at rice chromatin structure levels, requires future investigations.
EPIGENETIC REGULATION OF RICE FLOWERING
Flowering represents the transition from vegetative to reproductive growth, a key developmental switch during the plant life cycle. Flowering time is precisely controlled by complex gene network that integrates environmental signals, such as day length (photoperiod), light intensity and quality, and ambient temperature, as well as endogenous cues involving plant hormones (Albani and Coupland, 2010; Shrestha et al., 2014). Photoperiod is one of the most predictable cues in nature, and according to photoperiod responsiveness plants can be categorized into three groups: long-day (LD) plants, short-day (SD) plants, and day-neutral plants. Arabidopsis is a facultative LD plant whose flowering is accelerated when grown under LD photoperiods. Furthermore, flowering of most Arabidopsis ecotypes is promoted by a prolonged exposure to the cold of winter (a process known as vernalization), which has an epigenetic basis of competence memory (Ream et al., 2012; Song et al., 2012). During recent years, many chromatin modifiers have been shown as involved in Arabidopsis flowering time regulation, with majority of them acting via the transcriptional regulation of FLOWERING LOCUS C (FLC), a key flowering repressor at which vernalization and autonomous pathways converge (Berr et al., 2011; He, 2012; Ietswaart et al., 2012). In contrast to Arabidopsis, rice is a facultative SD plant and does not require vernalization to induce flowering and does not contain a FLC homolog. The complex gene network of rice flowering pathways primarily consists of flowering activators, and remarkably several chromatin modifiers have been shown recently as involved in rice flowering time control (Figure 1).
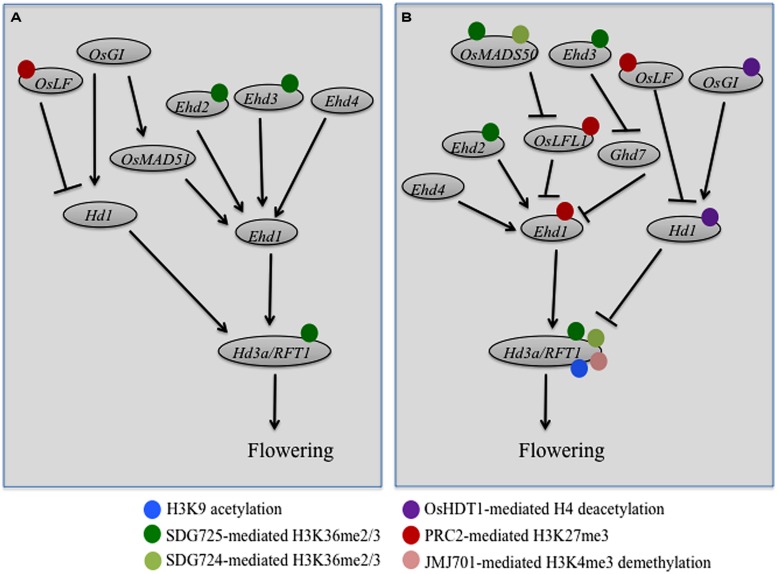
FIGURE 1.
Regulatory networks of genetic and epigenetic control of rice flowering under short-day (A) and long-day (B) photoperiod conditions. Rice flowering network is integrated by two florigen genes Hd3a and RFT1, which are regulated by at least two pathways: the Hd1-dependent and the Ehd1-dependent pathways. Expressions of Hd1 and Ehd1 are further regulated by more upstream genes as indicated by different names in the circles. Arrows indicate for transcriptional activation, whereas bars indicate for transcriptional repression. Different color spheres surrounding the flowering gene circles indicate for different regulations by the indicated histone modifications at the gene locus, currently described in literatures.
KEY TRANSCRIPTION FACTORS OF RICE FLOWERING PATHWAYS
Within the rice flowering pathways, the close paralogs Heading date 3a (Hd3a) and RICE FLOWERING LOCUS T1 (RFT1) are specifically upregulated upon the inductive SD photoperiods in leaf phloem tissue and encode small globular proteins named florigens, which move to the shoot apex to promote flowering (Tsuji et al., 2013; Sun et al., 2014). There are at least two pathways that control the Hd3a/RFT1 expression under either SD (Figure 1A) or LD (Figure 1B) photoperiods: the Early heading date 1 (Ehd1) and the Hd1 pathways (Tsuji et al., 2013; Sun et al., 2014). Ehd1 encodes a B-type transcription factor that plays a key role in activation of both Hd3a and RFT1 expression. The expression of Ehd1 is modulated by at least three different types of function factors (Sun et al., 2014). The first type comprises day length-independent activators, including Ehd2, also known as Rice Indeterminate1 (RID1) or Os Indeterminate1 (OsId1), and Ehd4, which encode two different zinc-finger transcription factors and act in both SD and LD conditions in Ehd1 induction (Figure 1). The second type comprises SD-preferential activators, including the PHD-finger factor Ehd3 and the MADS-box family transcription factor OsMADS51, which induce Ehd1 expression specifically in SD conditions (Figure 1A). And the third type comprises LD-preferential repressors, including Grain number, plant height, and heading date7 (Ghd7) that encodes a CCT-domain protein and LEC2-FUSCA3-Like 1 (OsLFL1) that encodes a B3-type transcription factor, both repress Ehd1 expression specifically in LD conditions (Figure 1B). Further upstream, the LD-preferential regulator OsMADS50 promotes flowering via repression of OsLFL1. Interestingly, Ehd3, which acts as an activator of Ehd1 to promote flowering in SD conditions (Figure 1A), displays a repressor function on Ghd7 and thus also promotes flowering in LD conditions (Figure 1B). The rice circadian clock related protein GIGANTEA (OsGI) activates the Ehd1 pathway partly via induction of OsMAD51 expression (Figure 1B). While the Ehd1 pathway is more unique to rice, the OsGI-Hd1-Hd3a pathway is very similar to the Arabidopsis GI-CO-FT pathway, composing of the respective orthologous proteins in the two plant species (Tsuji et al., 2013; Sun et al., 2014). An atypical helix-loop-helix (HLH) protein (OsLF) also is involved in the OsGI-Hd1-Hd3a pathway via Hd1 repression. Hd1 acts as an activator to promote rice flowering in SD conditions (Figure 1A) but as a suppressor of rice flowering in LD conditions (Figure 1B). Phytochrome signaling is crucial in conversion of Hd1 activity because mutation of Phytochrome B (PHYB) or phytochrome deficiency (e.g., in photoperiod sensitivity5 mutant) maintains Hd1 as an activator independent of day length. Under LD conditions, the red-light photoreceptor PHYB pathway may convert and maintain Hd1 as a repressor possible via post-translational modification and/or protein complex formation. Because of space limitation, the one who is interested in more details about rice flowering pathways can read the two excellent review articles here cited (Tsuji et al., 2013; Sun et al., 2014) and the original research papers referred therein.
ACTIVE CHROMATIN MARKS ARE INVOLVED IN RICE FLOWERING TIME REGULATION
Understanding how the rice flowering pathway genes are regulated in the chromatin context has great importance. Recent studies have found that histone acetylations, H3K4 and H3K36 methylations are involved in active transcription of several genes within the rice flowering pathways (Figure 1). It was reported that overexpression of the HD2-family HDAC gene OsHDT1 in hybrid rice leads to early flowering under LD conditions, probably through transcriptional repression of OsGI and Hd1 (Li et al., 2011a). Interestingly, the expression of OsHDT1 displays a circadian rhythm under SD conditions, peaked at the end of day, which coincides with rhythmic expression of OsGI and advances that of Hd1. Ectopic OsHDT1 expression in transgenic rice attenuates the overdominance rhythmic expression of OsGI and Hd1 in hybrid rice, which may explains the early flowering phenotype specifically observed in hybrid but not parental rice lines (Li et al., 2011a). Histone H4 acetylation levels were observed to positively correlate with the rhythmic expression of OsGI and Hd1, and OsHDT1 overexpression was shown to impair the acetylation increase at the peak time (Li et al., 2011a).
A positive DNA/histone methylation role in rice flowering promotion was first indicated by the study of the S-adenosyl-L-methionine synthetase gene mutants (Li et al., 2011b). S-Adenosyl-L-methionine is a universal methyl group donor for both DNA and protein methylations. Its deficiency caused late-flowering of rice plants and reduction of Ehd1, Hd3a, and RFT1 expression, which is associated with reduced levels of H3K4me3 and DNA CG/CHG-methylations at these flowering gene loci (Li et al., 2011b). More recently, it was reported that suppression of OsTrx1, an ortholog of the Arabidopsis H3K4-methyltransferase gene ATX1, delays rice flowering time under LD conditions (Choi et al., 2014). The OsTrx1 suppression did not affect the OsMADS50 and Hd1 pathways, but elevated Ghd7 expression and drastically reduced Ehd1, Hd3a and RFT1 expression, which is consistent with the plant late-flowering phenotype (Figure 1B). The PHD domain of OsTrx1 can bind to native histone H3 and the SET domain of OsTrx1 can methylate histone H3 from oligonucleosomes in vitro (Choi et al., 2014). Yet the role of OsTrx1 in histone methylation in vivo remains undemonstrated. Because the OsTrx1 and Ehd3 proteins bind each other, the authors propose that OsTrx1 may promote rice flowering via interaction with Ehd3 (Choi et al., 2014). Mutant characterization of Photoperiod sensitivity-14(Se14), which encodes the JmjC-domain protein JMJ701, revealed that H3K4me3 elevation at the RFT1 promoter region increases RFT1 expression, leading to rice plant early flowering under LD conditions (Yokoo et al., 2014). It is currently unknown whether or not OsTrx1 and JMJ701 could work as a couple in an antagonistic manner to control H3K4me3 levels at the RFT1 locus.
H3K36me3 is generally considered as acting more downstream of H3K4me3 during transcription processes (Berr et al., 2011). The first H3K36-methyltransferase characterized in rice is SDG725, which has been shown to specifically methylate H3K36 from mononucleosomes in vitro and is required for H3K36me2/me3 deposition at chromatin regions of genes related to brassinosteroid biosynthesis or signaling pathways (Sui et al., 2012). Knockdown of SDG725 caused a rice plant late-flowering phenotype (Sui et al., 2012), and subsequent investigation revealed that SDG725 is necessary for H3K36me2/3 deposition at several flowering genes including Ehd3, Ehd2, OsMADS50, Hd3a, and RFT1 (Sui et al., 2013). Characterization of the late-flowering mutant named long vegetative phase 1 (lvp1) together with map-based cloning has uncovered SDG724 as an essential regulator of the OsMAD50-Ehd1-RFT1 pathway (Sun et al., 2012). The recombinant SDG724 protein can methylate H3 (with K site undetermined) from oligonucleosomes and the lvp1 mutant plants show global reduction of H3K36me2/me3 levels. Remarkably, ChIP analysis revealed specific reduction of H3K36me2/me3 at OsMADS50 and RFT1 but not at Ehd1 and Hd3a in the lvp1 mutant plants (Sun et al., 2012). Both the lvp1 (sdg724) mutant and the SDG725-knockdown mutant exhibit late-flowering phenotypes under either SD or LD conditions (Sun et al., 2012; Sui et al., 2013), pointing to a crucial role of H3K36me2/me3 in promoting rice plant flowering irrespective of photoperiods. It is noteworthy that in Arabidopsis the SDG8-mediated H3K36me2/me3 also plays a major role in flowering time control, but in that case in prevention of early flowering (Shafiq et al., 2014). Future studies are necessary to investigate mechanisms underlying the overlap and specific targets of SDG724 and SDG725 in the rice flowering time control.
REPRESSIVE CHROMATIN MARKS ARE INVOLVED IN RICE FLOWERING TIME REGULATION
The repressive mark H3K27me3 is known to play a key role in FLC repression in vernalization-induced Arabidopsis plant flowering (He, 2012; Ietswaart et al., 2012). Interestingly, recent studies have shown that H3K27me3 deposited by PRC2-like complexes also plays an important role in vernalization-independent rice flowering time control (Figure 1). Loss-of-function of the PRC2 gene OsEMF2b causes late-flowering, which is associated with an increase of OsLFL1 expression and a decrease of Ehd1 expression (Yang et al., 2013). The OsEMF2b protein physically interacts with OsVIL3 (named as OsVIL2 in Yang et al., 2013, but here corrected to the first nomenclature used in Zhao et al., 2010; also called LC2), a PHD-domain protein showing homologies to the Arabidopsis VIN3-group proteins including VERNALIZATION INSENSITIVE 3 (VIN3), VIN3-LIKE 1 (VIL1)/VRN5, and VIL2/VEL1. The Arabidopsis VIN3-group proteins are know to be associated and to work together with the PRC2 core complex (constituting the so-called PHD-PRC2 complexes) and the VIN3 expression is induced early during vernalization (reviewed in He, 2012; Ietswaart et al., 2012). Consistent with the idea that OsVIL3/LC2 works together with PRC2, knockdown of OsVIL3/LC2 results in rice late-flowering, increase of OsLFL1 and OsLF expression, and decrease of Ehd1 as well as Hd3a and RFT1 expression (Wang et al., 2013; Yang et al., 2013). The OsVIL3/LC2 protein binds at the OsLFL1 and OsLF chromatin regions and the H3K27me3 enrichments at OsLFL1 and OsLF are impaired in the osvil3/lc2 mutant (Wang et al., 2013; Yang et al., 2013). In addition to OsVIL3/LC2, OsVIL2 plays a similar but non-redundant role in rice flowering time control. Expression of both OsVIL3/LC2 and OsVIL2 is induced by SD conditions and the OsVIL3/LC2 and OsVIL2 proteins physically interact, thus leading to the proposition that the OsVIN3/LC2-OsVIL2 dimer may recruit PRC2 in H3K27me3 deposition and OsLF suppression in rice photoperiod flowering regulation (Wang et al., 2013). Very recently, OsiEZ1/SDG718 and OsCLF/SDG711 have been reported to display distinct roles in photoperiod regulation of flowering (Liu et al., 2014). While OsiEZ1/SDG718 is induced in SD conditions and represses OsLF to promote flowering (Figure 1A), OsCLF/SDG711 is induced in LD conditions and represses OsLF and Ehd1 to inhibit flowering (Figure 1B). The OsCLF/SDG711 protein has been shown to target OsLF and Ehd1 loci to mediate H3K27me3 deposition and gene repression (Liu et al., 2014).
EPIGENETIC REGULATION OF RICE REPRODUCTION AND SEED FORMATION
After flowering, plant sexual reproduction occurs in dedicated floral organs through sporogenesis, gametogenesis, embryo- and endosperm-genesis, resulting in seed formation. Studies in Arabidopsis have unraveled diverse epigenetic regulatory mechanisms as involved in different processes during floral organogenesis and plant sexual reproduction (Shen and Xu, 2009; Engelhorn et al., 2014; She and Baroux, 2014). Although more recent, studies in rice also have started to uncover multiple types of epigenetic modifiers involved in the regulation of plant reproduction (Figure 2).
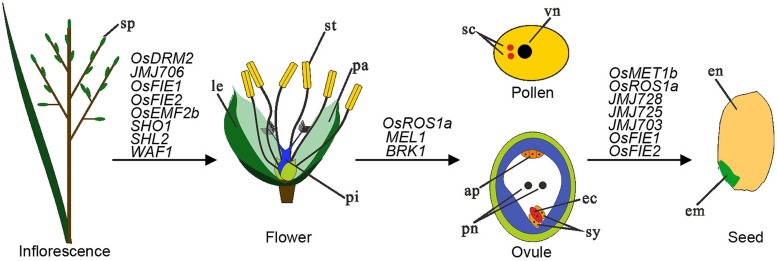
FIGURE 2.
Schematic representation of structures involved in rice reproduction together with chromatin modifier genes listed in regulation of three different steps. Inflorescence produces spikelets (sp) that generate numerous flowers. A mature flower contains different types of organs including lemma (le), palea (pa), stemen (st), and pistil (pi). The female gametophyte ovule is formed inside of ovary of the pistil and at maturation contains four different types of cells: antipodal (ap), polar nuclei (pn), synergid (sy), and egg cell (ec). The male gametophyte pollen is produced inside of anther of the stamen and at maturation contains two sperm cells (sc) and one vegetative cell (vn). Upon fertilization, one sperm cell fuses with egg cell to produce embryo (em) and the other sperm cell fuses with the two polar nuclei to produce endosperm (en), together forming a mature seed. Chromatin modifier genes playing important regulatory roles in floral organogenesis, gametophyte development, and fertilization/seed development are listed.
EPIGENETIC REGULATION IN RICE REPRODUCTION
Compared to those of Arabidopsis, the rice inflorescence and flower have greatly diverged structures that are regulated by a conserved genetic framework together with rice specific genetic mechanisms (Yoshida and Nagato, 2011). Several epialleles are found to affect rice plant reproduction. The metastable epigenetic silencing of DWARF1, which is associated with DNA methylation and H3K9me2 at the gene promoter region, causes dwarf tillers, compact panicles (inflorescences) and small round rice grains (Miura et al., 2009). The abnormal floral organ (afo) epimutation causes increased DNA methylation and suppression of the transcription factor gene OsMADS1, leading to pseudovivipary, a specific asexual reproductive strategy (Wang et al., 2010). The transcription factor gene SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 14 (SPL14), also known as IDEAL PLANT ARCHITECTURE 1 (IPA1) or WEALTHY FARMER’S PANICLE (WFP), promotes panicle branching and regulates a large number of genes, and differences in DNA methylation at the locus as well as the micro RNA 156 (OsmiR156) contribute to expression differences of SPL14/IPA1/WFP in different rice varieties (Jiao et al., 2010; Miura et al., 2010; Lu et al., 2013). Important roles of sRNA (both miRNAs and siRNAs) in rice floral organ development are also evidenced by mutants of several sRNA-pathway genes, including SHOOT ORGANIZATION 1 (SHO1) encoding a DICER-LIKE 4 homolog, SHOOTLESS 2 (SHL2) encoding a RDR6 homolog), and WAVY LEAF 1 (WAF1) encoding a HEN1 homolog (Abe et al., 2010; Toriba et al., 2010). lncRNAs are also reported as involved in plant reproductive process (Swiezewski et al., 2009; Heo and Sung, 2011). In rice, a point mutation that alter the secondary structure of the lncRNA called Long-Day-specific Male-fertility-Associated RNA (LDMAR) has been found to cause the photoperiod sensitive male sterility (Ding et al., 2012b).
Importance of DNA methylation in regulation of rice reproduction has been further supported by mutant studies. Targeted disruption of the DNA demethylase gene ROS1a leads to paternal allele transmission defect, presumably because of a male gametophytic defect prior to fertilization (Ono et al., 2012). Disruption of OsDRM2 led to pleiotropic developmental defects in both vegetative and reproductive stages including semi-dwarfed stature, reductions in tiller number, and complete sterility (Moritoh et al., 2012). Consistently, transcriptome analysis of isolated rice gametes by deep sequencing indicates that OsDRM2 is expressed in male cells but low in vegetative cells (Anderson et al., 2013).
Several modifiers of histone modifications are also critical for rice reproduction (Figure 2). Loss-of-function of the rice PRC2 gene OsEMF2b results in complete sterility, and severe floral organ defects and indeterminacy that resemble loss-of-function mutants in E-class floral organ specification genes (Conrad et al., 2014). The epimutation of OsFIE1 (Epi-df) that is caused by DNA hypomethylation, reduced H3K9me2 and increased H3K4me3 at the gene locus, leads to ectopic expression of OsFIE1, resulting in a dwarf stature, diverse floral defects, and alteration of H3K27me3 levels at hundreds of target genes (Zhang et al., 2012b). Mutation of the H3K27-demethylase gene JMJ705 also causes partial sterility (Li et al., 2013). The OsFIE2 RNAi lines display pleiotropic phenotypes including vegetative and reproductive organ formation, a decreased amount of pollen grains and a high proportion of male sterility (Li et al., 2014). These studies indicate that a balanced level of H3K27me3 is critical and that either its increase or decrease can cause rice reproduction defects. The other chromatin repressive mark H3K9me2/me3 is also important because mutation of the H3K9-demethylase gene JMJ706 impairs spikelet development, including defective floral morphology and altered organ number (Sun and Zhou, 2008). Pleiotropic defective phenotypes including panicle morphology, rachis branch and spikelet numbers have also been described for mutants of the H3K36-methyltransferase gene SDG725 and the H3K4-demethylase gene JMJ703 (Sui et al., 2012; Cui et al., 2013), indicating that chromatin active marks also play important function during rice reproduction.
While precise reproduction processes affected by many of the above mentioned modifiers remain to be elucidated, meiosis is found to be regulated by several epigenetic factors. The rice germline-specific AGO-family protein MEL1 binds preferentially 21 nt siRNAs derived mostly from intergenic regions (Komiya et al., 2014), and its loss-of-function impairs both sporophytic germ-cell development and meiosis (Nonomura et al., 2007). The mel1 mutant displays aberrant vacuolation of spore mother cells, and arrested chromosome condensation at early meiosis stages. H3K9me2 distribution as well as the localization of ZEP1, a component of transverse filaments of the rice synaptonemal complex, are affected in mel1, indicating for a role of MEL1 in chromatin structure organization and homologous chromosome synapsis in early meiosis (Nonomura et al., 2007; Komiya et al., 2014). Histone phosphorylation is also involved in rice meiosis process. The rice Bub1-Related Kinase 1 (BRK1) is required for H2A phosphorylation and the centromeric recruitment of SHUGOSHIN 1 (SGO1), which is likely essential for generating proper tension between the homologous kinetochores at metaphase I to facilitate the accurate segregation of homologous chromosomes at anaphase I (Wang et al., 2012).
EPIGENETIC REGULATIONS IN SEED DEVELOPMENT
Like other angiosperms, sexual double fertilization initiates rice seed development, giving rise to two fertilization products, the embryo and the endosperm. Epigenetic mechanisms are thought to have important contribution to plant hybrid vigor (heterosis), a phenomenon referring to the increased yield and biomass of hybrid offspring relative to the parents (Chen and Zhou, 2013; Groszmann et al., 2013). In line with this idea, divers epigenetic pathways are found as involved in seed development and seed quality control (Figure 2).
Genome-wide analyses in rice have revealed that sRNA expression, DNA methylation, and histone modifications (e.g., H3K9ac, H3K4me3, and H3K27me3) significantly differ between hybrids and their parents (He et al., 2010; Chodavarapu et al., 2012; Zhang et al., 2014). Remarkably, the amount of 24 nt siRNAs, with most of them likely involved in regulation of the starch and sucrose biosynthesis pathway, declines with the process of rice grain-filling and this decline is to a lower degree in inferior grains then superior grains (Peng et al., 2013). The siRNAs may act via or together with DNA methylation in heterochromatin silencing. In line with this idea, the maternal loss of ROS1a causes failure of early stage endosperm development, leading to incomplete embryogenesis producing irregular but viable embryos that failed to complete seed dormancy (Ono et al., 2012). While the met1a null mutant displays a normal phenotype, the met1b mutant exhibits abnormal seed phenotypes, which is associated with either viviparous germination or early embryonic lethality (Hu et al., 2014; Yamauchi et al., 2014). Levels of DNA methylation in met1b are broadly reduced at genome-wide scale and in particular at repetitive centromeric and transposon sequences as well as at the OsFIE1 gene locus in the embryos (Hu et al., 2014; Yamauchi et al., 2014).
OsFIE1 is an imprinted gene in rice endosperm but the osfie1 mutant does not display any autonomous endosperm proliferation without fertilization, differing from the Arabidopsis fie, mea and fis mutants that are generally recognized with an autonomous endosperm proliferation phenotype (Luo et al., 2009). Nevertheless, over-expression of OsFIE1 causes precocious cellularization and reduced seed size, and it has been proposed that that OsFIE1 has a role in regulating seed enlargement under heat stress (Folsom et al., 2014). In addition, OsFIE2 has a critical role in normal endosperm development and grain-filling. Down-regulation of OsFIE2 results in small seeds and partial loss of seed dormancy, likely because of down-regulation of genes encoding the starch synthesis rate limiting step enzymes and multiple storage proteins (Nallamilli et al., 2013). Future studies are necessary to precise similarities and differences of PRC2-mediated H3K27me3 repression mechanisms involved in seed development between Arabidopsis and rice.
Involvement of other histone methylation marks in seed development are also evidenced from mutant studies (Figure 2). Down-regulation of the H3K9-methyltransferase gene SDG728 reduces seed size and alters seed morphology (Qin et al., 2010). Loss-of-function of the H3K4-demethylase gene JMJ703 causes abnormal grain phenotypes, including reduced length, width, and thickness (Cui et al., 2013). Also, knockdown of the H3K36-methyltransferase gene SDG725 results in small seed size and reduced seed weight (Sui et al., 2012).
CONCLUSION REMARKS
The availability of full genome sequences and diverse improved powerful genomic and analytic tools have greatly advanced our knowledge about rice epigenetic modifiers and their biological roles. There are still a large number of modifiers uncharacterized, and molecular mechanisms of function of many chromatin modifiers remain to be investigated into details. It remains to be uncovered how the general histone modification and DNA methylation enzymes exert specific functions in plant growth and developmental processes and what effectors are involved. In particular, H3K27me3 is recognized as a crucial epigenetic mark associated with gene transcriptional repression, and the classical model proposes a sequential mode of action of the two Polycomb complexes: PRC2 is responsible H3K27me3 establishment, and PRC1 recognizes the H3K27me3 mark and further catalyzed downstream H2A monoubiquitination. While PRC1-like components and histone monoubiquitination have been recently studied in Arabidopsis (reviewed in Molitor and Shen, 2013; Feng and Shen, 2014), effectors acting together with H3K27me3 in rice remain unknown so far. Utilization of advanced technologies in proteomics, deep sequencing, and gene knockdown will facilitate future studies in functional characterization of interesting genes, investigation of protein complex composition and function, and gene networks controlling rice flowering and reproduction. The extensive agriculture breading has greatly enriched the rice germplasm resources with large collections of cultivated rice and their wild relatives. Comparative studies of different rice varieties and hybrids will likely impact on knowledge of genetics, epigenetics, and inheritance of agriculture traits as well as fundamental understanding of conservation and diversification of molecular mechanisms.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The work in authors’ laboratories were supported by National Basic Research Program of China (973 Program, grants no. 2012CB910500 and 2011CB944600), National Natural Science Foundation of China (grant no. 31300263), Science and Technology Commission of Shanghai Municipality (grant no. 13JC1401000), and the French Agence Nationale de la Recherche (ANR-12-BSV2-0013-02). The research was conducted within the context of the International Associated Laboratory Plant Epigenome Research, LIA PER.
REFERENCES
- Abe M., Yoshikawa T., Nosaka M., Sakakibara H., Sato Y., Nagato Y., et al. (2010). WAVY LEAF1, an ortholog of Arabidopsis HEN1, regulates shoot development by maintaining microRNA and trans-acting small interfering RNA accumulation in rice. Plant Physiol. 154 1335–1346 10.1104/pp.110.160234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agius F., Kapoor A., Zhu J. K. (2006). Role of the Arabidopsis DNA glycosylase/lyase ROS1 in active DNA demethylation. Proc. Natl. Acad. Sci. U.S.A. 103 11796–11801 10.1073/pnas.0603563103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albani M. C., Coupland G. (2010). Comparative analysis of flowering in annual and perennial plants. Curr. Top. Dev. Biol. 91 323–348 10.1016/S0070-2153(10)91011-9 [DOI] [PubMed] [Google Scholar]
- Anderson S. N., Johnson C. S., Jones D. S., Conrad L. J., Gou X., Russell S. D., et al. (2013). Transcriptomes of isolated Oryza sativa gametes characterized by deep sequencing: evidence for distinct sex-dependent chromatin and epigenetic states before fertilization. Plant J. 76 729–741 10.1111/tpj.12336 [DOI] [PubMed] [Google Scholar]
- Berr A., Shafiq S., Shen W. H. (2011). Histone modifications in transcriptional activation during plant development. Biochim. Biophys. Acta 1809 567–576 10.1016/j.bbagrm.2011.07.001 [DOI] [PubMed] [Google Scholar]
- Bond D. M., Baulcombe D. C. (2014). Small RNAs and heritable epigenetic variation in plants. Trends Cell Biol. 24 100–107 10.1016/j.tcb.2013.08.001 [DOI] [PubMed] [Google Scholar]
- Bu Z., Yu Y., Li Z., Liu Y., Jiang W., Huang Y., et al. (2014). Regulation of Arabidopsis flowering by the histone mark readers MRG1/2 via interaction with CONSTANS to modulate FT expression. PLoS Genet. 10:e1004617 10.1371/journal.pgen.1004617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X., Jacobsen S. E. (2002). Role of the Arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr. Biol. 12 1138–1144 10.1016/S0960-9822(02)00925-9 [DOI] [PubMed] [Google Scholar]
- Castel S. E., Martienssen R. A. (2013). RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat. Rev. Genet. 14 100–112 10.1038/nrg3355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., He G., He H., Chen W., Zhu X., Liang M., et al. (2010). Expression analysis of miRNAs and highly-expressed small RNAs in two rice subspecies and their reciprocal hybrids. J. Integr. Plant Biol. 52 971–980 10.1111/j.1744-7909.2010.00985.x [DOI] [PubMed] [Google Scholar]
- Chen Q., Chen X., Wang Q., Zhang F., Lou Z., Zhang Q., et al. (2013). Structural basis of a histone H3 lysine 4 demethylase required for stem elongation in rice. PLoS Genet. 9:e1003239 10.1371/journal.pgen.1003239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Rasmuson-Lestander A. (2009). Regulation of the Drosophila engrailed gene by Polycomb repressor complex 2. Mech. Dev. 126 443–448 10.1016/j.mod.2009.01.004 [DOI] [PubMed] [Google Scholar]
- Chen X., Zhou D. X. (2013). Rice epigenomics and epigenetics: challenges and opportunities. Curr. Opin. Plant Biol. 16 164–169 10.1016/j.pbi.2013.03.004 [DOI] [PubMed] [Google Scholar]
- Chen Z. J., Tian L. (2007). Roles of dynamic and reversible histone acetylation in plant development and polyploidy. Biochim. Biophys. Acta 1769 295–307 10.1016/j.bbaexp.2007.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodavarapu R. K., Feng S., Ding B., Simon S. A., Lopez D., Jia Y., et al. (2012). Transcriptome and methylome interactions in rice hybrids. Proc. Natl. Acad. Sci. U.S.A. 109 12040–12045 10.1073/pnas.1209297109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S. C., Lee S., Kim S. R., Lee Y. S., Liu C., Cao X., et al. (2014). Trithorax group protein Oryza sativa TRITHORAX1 controls flowering time in rice via interaction with EARLY HEADING DATE 3. Plant Physiol. 164 1326–1337 10.1104/pp.113.228049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y., Gehring M., Johnson L., Hannon M., Harada J. J., Goldberg R. B., et al. (2002). DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in Arabidopsis. Cell 110 33–42 10.1016/S0092-8674(02)00807-3 [DOI] [PubMed] [Google Scholar]
- Conrad L. J., Khanday I., Johnson C., Guiderdoni E., An G., Vijayraghavan U., et al. (2014). The Polycomb group gene EMF2B is essential for maintenance of floral meristem determinacy in rice. Plant J. 80 883–894 10.1111/tpj.12688 [DOI] [PubMed] [Google Scholar]
- Cui X., Jin P., Gu L., Lu Z., Xue Y., Wei L., et al. (2013). Control of transposon activity by a histone H3K4 demethylase in rice. Proc. Natl. Acad. Sci. U.S.A. 110 1953–1958 10.1073/pnas.1217020110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czermin B., Melfi R., Mccabe D., Seitz V., Imhof A., Pirrotta V. (2002). Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal polycomb sites. Cell 111 185–196 10.1016/S0092-8674(02)00975-3 [DOI] [PubMed] [Google Scholar]
- Dangwal M., Malik G., Kapoor S., Kapoor M. (2013). De novo methyltransferase, OsDRM2, interacts with the ATP-dependent RNA helicase, OseIF4A, in rice. J. Mol. Biol. 425 2853–2866 10.1016/j.jmb.2013.05.021 [DOI] [PubMed] [Google Scholar]
- Ding B., Bellizzi Mdel R., Ning Y., Meyers B. C., Wang G. L. (2012a). HDT701, a histone H4 deacetylase, negatively regulates plant innate immunity by modulating histone H4 acetylation of defense-related genes in rice. Plant Cell 24 3783–3794 10.1105/tpc.112.101972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J., Lu Q., Ouyang Y., Mao H., Zhang P., Yao J., et al. (2012b). A long noncoding RNA regulates photoperiod-sensitive male sterility, an essential component of hybrid rice. Proc. Natl. Acad. Sci. U.S.A. 109 2654–2659 10.1073/pnas.1121374109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J., Shen J., Mao H., Xie W., Li X., Zhang Q. (2012c). RNA-directed DNA methylation is involved in regulating photoperiod-sensitive male sterility in rice. Mol. Plant 5 1210–1216 10.1093/mp/sss095 [DOI] [PubMed] [Google Scholar]
- Ding B., Zhu Y., Bu Z. Y., Shen W. H., Yu Y., Dong A. W. (2010). SDG714 regulates specific gene expression and consequently affects plant growth via H3K9 dimethylation. J. Integr. Plant Biol. 52 420–430 10.1111/j.1744-7909.2010.00927.x [DOI] [PubMed] [Google Scholar]
- Ding B., Zhu Y., Gao J., Yu Y., Cao K., Shen W.-H., et al. (2007a). Molecular characterization of three rice SET-domain proteins. Plant Sci. 172 1072–1078 10.1016/j.plantsci.2007.03.009 [DOI] [Google Scholar]
- Ding Y., Wang X., Su L., Zhai J., Cao S., Zhang D., et al. (2007b). SDG714, a histone H3K9 methyltransferase, is involved in Tos17 DNA methylation and transposition in rice. Plant Cell 19 9–22 10.1105/tpc.106.048124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z., Li H., Wei Q., Zhao X., Wang C., Zhu Q., et al. (2013). Genome-wide analysis of histone modifications: H3K4me2 H3K4me3 H3K9ac, and H3K27ac in Oryza sativa L. Japonica. Mol. Plant 6 1463–1472 10.1093/mp/sst018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelhorn J., Blanvillain R., Carles C. C. (2014). Gene activation and cell fate control in plants: a chromatin perspective. Cell. Mol. Life Sci. 71 3119–3137 10.1007/s00018-014-1609-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J., Shen W. H. (2014). Dynamic regulation and function of histone monoubiquitination in plants. Front. Plant Sci. 5:83 10.3389/fpls.2014.00083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folsom J. J., Begcy K., Hao X., Wang D., Walia H. (2014). Rice FERTILIZATION-INDEPENDENT ENDOSPERM 1 regulates seed size under heat stress by controlling early endosperm development. Plant Physiol. 165 238–248 10.1104/pp.113.232413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furner I. J., Matzke M. (2011). Methylation and demethylation of the Arabidopsis genome. Curr. Opin. Plant Biol. 14 137–141 10.1016/j.pbi.2010.11.004 [DOI] [PubMed] [Google Scholar]
- Gehring M., Huh J. H., Hsieh T. F., Penterman J., Choi Y., Harada J. J., et al. (2006). DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell 124 495–506 10.1016/j.cell.2005.12.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentry M., Hennig L. (2014). Remodelling chromatin to shape development of plants. Exp. Cell Res. 321 40–46 10.1016/j.yexcr.2013.11.010 [DOI] [PubMed] [Google Scholar]
- Gong Z., Morales-Ruiz T., Ariza R. R., Roldán-Arjona T., David L., Zhu J.-K. (2002). ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell 111 803–814 10.1016/S0092-8674(02)01133-9 [DOI] [PubMed] [Google Scholar]
- Groszmann M., Greaves I. K., Fujimoto R., Peacock W. J., Dennis E. S. (2013). The role of epigenetics in hybrid vigour. Trends Genet. 29 684–690 10.1016/j.tig.2013.07.004 [DOI] [PubMed] [Google Scholar]
- Gutierrez-Marcos J. F., Dickinson H. G. (2012). Epigenetic reprogramming in plant reproductive lineages. Plant Cell Physiol. 53 817–823 10.1093/pcp/pcs052 [DOI] [PubMed] [Google Scholar]
- He C., Huang H., Xu L. (2013). Mechanisms guiding Polycomb activities during gene silencing in Arabidopsis thaliana. Front. Plant Sci. 4:454 10.3389/fpls.2013.00454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He G., Zhu X., Elling A. A., Chen L., Wang X., Guo L., et al. (2010). Global epigenetic and transcriptional trends among two rice subspecies and their reciprocal hybrids. Plant Cell 22 17–33 10.1105/tpc.109.072041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y. (2012). Chromatin regulation of flowering. Trends Plant Sci. 17 556–562 10.1016/j.tplants.2012.05.001 [DOI] [PubMed] [Google Scholar]
- Heo J. B., Sung S. (2011). Vernalization-mediated epigenetic silencing by a long Intronic noncoding RNA. Science 331 76–79 10.1126/science.1197349 [DOI] [PubMed] [Google Scholar]
- Higo H., Tahir M., Takashima K., Miura A., Watanabe K., Tagiri A., et al. (2012). DDM1 (Decrease in DNA Methylation) genes in rice (Oryza sativa). Mol. Genet. Genomics 287 785–792 10.1007/s00438-012-0717-5 [DOI] [PubMed] [Google Scholar]
- Hu L., Li N., Xu C., Zhong S., Lin X., Yang J., et al. (2014). Mutation of a major CG methylase in rice causes genome-wide hypomethylation, dysregulated genome expression, and seedling lethality. Proc. Natl. Acad. Sci. U.S.A. 111 10642–10647 10.1073/pnas.1410761111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Liu D., Zhong X., Zhang C., Zhang Q., Zhou D.-X. (2012). CHD3 protein recognizes and regulates methylated histone H3 lysines 4 and 27 over a subset of targets in the rice genome. Proc. Natl. Acad. Sci. U.S.A. 109 5773–5778 10.1073/pnas.1203148109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Qin F., Huang L., Sun Q., Li C., Zhao Y., et al. (2009). Rice histone deacetylase genes display specific expression patterns and developmental functions. Biochem. Biophys. Res. Commun. 388 266–271 10.1016/j.bbrc.2009.07.162 [DOI] [PubMed] [Google Scholar]
- Huang L., Sun Q., Qin F., Li C., Zhao Y., Zhou D. X. (2007). Down-regulation of a SILENT INFORMATION REGULATOR2-related histone deacetylase gene, OsSRT1, induces DNA fragmentation and cell death in rice. Plant Physiol. 144 1508–1519 10.1104/pp.107.099473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Liu C., Shen W.-H., Ruan Y. (2011). Phylogenetic analysis and classification of the Brassica rapa SET-domain protein family. BMC Plant Biol. 11:175 10.1186/1471-2229-11-175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ietswaart R., Wu Z., Dean C. (2012). Flowering time control: another window to the connection between antisense RNA and chromatin. Trends Genet. 28 445–453 10.1016/j.tig.2012.06.002 [DOI] [PubMed] [Google Scholar]
- Jeong D. H., German M. A., Rymarquis L. A., Thatcher S. R., Green P. J. (2010). Abiotic stress-associated miRNAs: detection and functional analysis. Methods Mol. Biol. 592 203–230 10.1007/978-1-60327-005-2_14 [DOI] [PubMed] [Google Scholar]
- Jeong D.-H., Park S., Zhai J., Gurazada S. G. R., De Paoli E., Meyers B. C., et al. (2011). Massive analysis of rice small RNAs: mechanistic implications of regulated microRNAs and variants for differential target RNA cleavage. Plant Cell 23 4185–4207 10.1105/tpc.111.089045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D., Yang W., He Y., Amasino R. M. (2007). Arabidopsis relatives of the human lysine-specific Demethylase1 repress the expression of FWA and FLOWERING LOCUS C and thus promote the floral transition. Plant Cell 19 2975–2987 10.1105/tpc.107.052373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., Wang Y., Xue D., Wang J., Yan M., Liu G., et al. (2010). Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 42 541–544 10.1038/ng.591 [DOI] [PubMed] [Google Scholar]
- Komiya R., Ohyanagi H., Niihama M., Watanabe T., Nakano M., Kurata N., et al. (2014). Rice germline-specific Argonaute MEL1 protein binds to phasiRNAs generated from more than 700 lincRNAs. Plant J. 78 385–397 10.1111/tpj.12483 [DOI] [PubMed] [Google Scholar]
- La H., Ding B., Mishra G. P., Zhou B., Yang H., Bellizzi M. R., et al. (2011). A 5-methylcytosine DNA glycosylase/lyase demethylates the retrotransposon Tos17 and promotes its transposition in rice. Proc. Natl. Acad. Sci. U.S.A. 108 15498–15503 10.1073/pnas.1112704108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law J. A., Jacobsen S. E. (2010). Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11 204–220 10.1038/nrg2719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Huang L., Xu C., Zhao Y., Zhou D. X. (2011a). Altered levels of histone deacetylase OsHDT1 affect differential gene expression patterns in hybrid rice. PLoS ONE 6:e21789 10.1371/journal.pone.0021789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Han Y., Tao F., Chong K. (2011b). Knockdown of SAMS genes encoding S-adenosyl-L-methionine synthetases causes methylation alterations of DNAs and histones and leads to late flowering in rice. J. Plant Physiol. 168 1837–1843 10.1016/j.jplph.2011.05.020 [DOI] [PubMed] [Google Scholar]
- Li S., Zhou B., Peng X., Kuang Q., Huang X., Yao J., et al. (2014). OsFIE2 plays an essential role in the regulation of rice vegetative and reproductive development. New Phytol. 201 66–79 10.1111/nph.12472 [DOI] [PubMed] [Google Scholar]
- Li T., Chen X., Zhong X., Zhao Y., Liu X., Zhou S., et al. (2013). Jumonji C domain protein JMJ705-mediated removal of histone H3 lysine 27 trimethylation is involved in defense-related gene activation in rice. Plant Cell 25 4725–4736 10.1105/tpc.113.118802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Zhu J., Hu F., Ge S., Ye M., Xiang H., et al. (2012). Single-base resolution maps of cultivated and wild rice methylomes and regulatory roles of DNA methylation in plant gene expression. BMC Genomics 13:300 10.1186/1471-2164-13-300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindroth A. M., Cao X., Jackson J. P., Zilberman D., Mccallum C. M., Henikoff S., et al. (2001). Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science 292 2077–2080 10.1126/science.1059745 [DOI] [PubMed] [Google Scholar]
- Liu C., Lu F., Cui X., Cao X. (2010). Histone methylation in higher plants. Annu. Rev. Plant Biol. 61 395–420 10.1146/annurev.arplant.043008.091939 [DOI] [PubMed] [Google Scholar]
- Liu F., Quesada V., Crevillen P., Baurle I., Swiezewski S., Dean C. (2007). The Arabidopsis RNA-binding protein FCA requires a LYSINE-SPECIFIC DEMETHYLASE 1 homolog to downregulate FLC. Mol. Cell 28 398–407 10.1016/j.molcel.2007.10.018 [DOI] [PubMed] [Google Scholar]
- Liu X., Luo M., Zhang W., Zhao J., Zhang J., Wu K., et al. (2012). Histone acetyltransferases in rice (Oryza sativa L.): phylogenetic analysis, subcellular localization and expression. BMC Plant Biol. 12:145 10.1186/1471-2229-12-145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Zhou C., Zhao Y., Zhou S., Wang W., Zhou D.-X. (2014). The rice enhancer of zeste [E(z)] genes SDG711 and SDG718 are respectively involved in long day and short day signaling to mediate the accurate photoperiod control of flowering time. Front. Plant Sci. 5:591 10.3389/fpls.2014.00591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Gonzalez L., Mouriz A., Narro-Diego L., Bustos R., Martinez-Zapater J. M., Jarillo J. A., et al. (2014). Chromatin-dependent repression of the Arabidopsis floral integrator genes involves plant specific PHD-containing proteins. Plant Cell 26 3922–3938 10.1105/tpc.114.130781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z., Yu H., Xiong G., Wang J., Jiao Y., Liu G., et al. (2013). Genome-wide binding analysis of the transcription activator IDEAL PLANT ARCHITECTURE 1 reveals a complex network regulating rice plant architecture. Plant Cell 25 3743–3759 10.1105/tpc.113.113639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M., Platten D., Chaudhury A., Peacock W. J., Dennis E. S. (2009). Expression, imprinting, and evolution of rice homologs of the Polycomb group genes. Mol. Plant 2 711–723 10.1093/mp/ssp036 [DOI] [PubMed] [Google Scholar]
- Lusser A., Kolle D., Loidl P. (2001). Histone acetylation: lessons from the plant kingdom. Trends Plant Sci. 6 59–65 10.1016/S1360-1385(00)01839-2 [DOI] [PubMed] [Google Scholar]
- Ma X., Lv S., Zhang C., Yang C. (2013). Histone deacetylases and their functions in plants. Plant Cell Rep. 32 465–478 10.1007/s00299-013-1393-6 [DOI] [PubMed] [Google Scholar]
- Malone B. M., Tan F., Bridges S. M., Peng Z. (2011). Comparison of four ChIP-Seq analytical algorithms using rice endosperm H3K27 trimethylation profiling data. PLoS ONE 6:e25260 10.1371/journal.pone.0025260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinty R., Tan S. (2014). “Histone, nucleosome, and chromatin structure,” in Fundamentals of Chromatin eds Workman J. L., Abmayr S. M. (New York: Springer; ), 1–28. [Google Scholar]
- Miura K., Agetsuma M., Kitano H., Yoshimura A., Matsuoka M., Jacobsen S. E., et al. (2009). A metastable DWARF1 epigenetic mutant affecting plant stature in rice. Proc. Natl. Acad. Sci. U.S.A. 106 11218–11223 10.1073/pnas.0901942106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K., Ikeda M., Matsubara A., Song X. J., Ito M., Asano K., et al. (2010). OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet. 42 545–549 10.1038/ng.592 [DOI] [PubMed] [Google Scholar]
- Molitor A. M., Bu Z., Yu Y., Shen W. H. (2014). Arabidopsis AL PHD-PRC1 complexes promote seed germination through H3K4me3-to-H3K27me3 chromatin state switch in repression of seed developmental genes. PLoS Genet. 10:e1004091 10.1371/journal.pgen.1004091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molitor A., Shen W.-H. (2013). The Polycomb complex PRC1: composition and function in plants. J. Genet. Genomics 40 231–238 10.1016/j.jgg.2012.12.005 [DOI] [PubMed] [Google Scholar]
- Moritoh S., Eun C. H., Ono A., Asao H., Okano Y., Yamaguchi K., et al. (2012). Targeted disruption of an orthologue of DOMAINS REARRANGED METHYLASE 2, OsDRM2, impairs the growth of rice plants by abnormal DNA methylation. Plant J. 71 85–98 10.1111/j.1365-313X.2012.04974.x [DOI] [PubMed] [Google Scholar]
- Mosammaparast N., Shi Y. (2010). Reversal of histone methylation: biochemical and molecular mechanisms of histone demethylases. Annu. Rev. Biochem. 79 155–179 10.1146/annurev.biochem.78.070907.103946 [DOI] [PubMed] [Google Scholar]
- Musselman C. A., Lalonde M. E., Cote J., Kutateladze T. G. (2012). Perceiving the epigenetic landscape through histone readers. Nat. Struct. Mol. Biol. 19 1218–1227 10.1038/nsmb.2436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nallamilli B. R., Edelmann M. J., Zhong X., Tan F., Mujahid H., Zhang J., et al. (2014). Global analysis of lysine acetylation suggests the involvement of protein acetylation in diverse biological processes in rice (Oryza sativa). PLoS ONE 9:e89283 10.1371/journal.pone.0089283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nallamilli B. R. R., Zhang J., Mujahid H., Malone B. M., Bridges S. M., Peng Z. (2013). Polycomb group gene OsFIE2 regulates rice (Oryza sativa) seed development and grain filling via a mechanism distinct from Arabidopsis. PLoS Genet. 9:e1003322 10.1371/journal.pgen.1003322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng D. W., Wang T., Chandrasekharan M. B., Aramayo R., Kertbundit S., Hall T. C. (2007). Plant SET domain-containing proteins: structure, function and regulation. Biochim. Biophys. Acta 1769 316–329 10.1016/j.bbaexp.2007.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonomura K.-I., Morohoshi A., Nakano M., Eiguchi M., Miyao A., Hirochika H., et al. (2007). A germ cell-specific gene of the ARGONAUTE family is essential for the progression of premeiotic mitosis and meiosis during sporogenesis in rice. Plant Cell 19 2583–2594 10.1105/tpc.107.053199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono A., Yamaguchi K., Fukada-Tanaka S., Terada R., Mitsui T., Iida S. (2012). A null mutation of ROS1a for DNA demethylation in rice is not transmittable to progeny. Plant J. 71 564–574 10.1111/j.1365-313X.2012.05009.x [DOI] [PubMed] [Google Scholar]
- Ortega-Galisteo A. P., Morales-Ruiz T., Ariza R. R., Roldan-Arjona T. (2008). Arabidopsis DEMETER-LIKE proteins DML2 and DML3 are required for appropriate distribution of DNA methylation marks. Plant Mol. Biol. 67 671–681 10.1007/s11103-008-9346-0 [DOI] [PubMed] [Google Scholar]
- Pang J., Dong M., Li N., Zhao Y., Liu B. (2013). Functional characterization of a rice de novo DNA methyltransferase, OsDRM2, expressed in Escherichia coli and yeast. Biochem. Biophys. Res. Commun. 432 157–162 10.1016/j.bbrc.2013.01.067 [DOI] [PubMed] [Google Scholar]
- Peng T., Du Y., Zhang J., Li J., Liu Y., Zhao Y., et al. (2013). Genome-wide analysis of 24-nt siRNAs dynamic variations during rice superior and inferior grain filling. PLoS ONE 8:e61029 10.1371/journal.pone.0061029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikaard C. S., Scheid O. M. (2013). Epigenetic regulation in plants. Cold Spring Harb. Perspect. Biol. 6:a019315 10.1101/cshperspect.a019315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin F. J., Sun Q. W., Huang L. M., Chen X. S., Zhou D. X. (2010). Rice SUVH histone methyltransferase genes display specific functions in chromatin modification and retrotransposon repression. Mol. Plant 3 773–782 10.1093/mp/ssq030 [DOI] [PubMed] [Google Scholar]
- Ream T. S., Woods D. P., Amasino R. M. (2012). The molecular basis of vernalization in different plant groups. Cold Spring Harb. Symp. Quant. Biol. 77 105–115 10.1101/sqb.2013.77.014449 [DOI] [PubMed] [Google Scholar]
- Saze H., Mittelsten Scheid O., Paszkowski J. (2003). Maintenance of CpG methylation is essential for epigenetic inheritance during plant gametogenesis. Nat. Genet. 34 65–69 10.1038/ng1138 [DOI] [PubMed] [Google Scholar]
- Schuettengruber B., Cavalli G. (2009). Recruitment of Polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development 136 3531–3542 10.1242/dev.033902 [DOI] [PubMed] [Google Scholar]
- Shafiq S., Berr A., Shen W. H. (2014). Combinatorial functions of diverse histone methylations in Arabidopsis thaliana flowering time regulation. New Phytol. 201 312–322 10.1111/nph.12493 [DOI] [PubMed] [Google Scholar]
- Sharma R., Mohan Singh R. K., Malik G., Deveshwar P., Tyagi A. K., Kapoor S., et al. (2009). Rice cytosine DNA methyltransferases – gene expression profiling during reproductive development and abiotic stress. FEBS J. 276 6301–6311 10.1111/j.1742-4658.2009.07338.x [DOI] [PubMed] [Google Scholar]
- She W., Baroux C. (2014). Chromatin dynamics during plant sexual reproduction. Front. Plant Sci. 5:354 10.3389/fpls.2014.00354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W. H., Xu L. (2009). Chromatin remodeling in stem cell maintenance in Arabidopsis thaliana. Mol. Plant 2 600–609 10.1093/mp/ssp022 [DOI] [PubMed] [Google Scholar]
- Shi Y., Lan F., Matson C., Mulligan P., Whetstine J. R., Cole P. A., et al. (2004). Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119 941–953 10.1016/j.cell.2004.12.012 [DOI] [PubMed] [Google Scholar]
- Shrestha R., Gomez-Ariza J., Brambilla V., Fornara F. (2014). Molecular control of seasonal flowering in rice, arabidopsis and temperate cereals. Ann. Bot. 114 1445–1458 10.1093/aob/mcu032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Angel A., Howard M., Dean C. (2012). Vernalization – a cold-induced epigenetic switch. J. Cell Sci. 125 3723–3731 10.1242/jcs.084764 [DOI] [PubMed] [Google Scholar]
- Stroud H., Do T., Du J., Zhong X., Feng S., Johnson L., et al. (2014). Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat. Struct. Mol. Biol. 21 64–72 10.1038/nsmb.2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui P., Jin J., Ye S., Mu C., Gao J., Feng H., et al. (2012). H3K36 methylation is critical for brassinosteroid-regulated plant growth and development in rice. Plant J. 70 340–347 10.1111/j.1365-313X.2011.04873.x [DOI] [PubMed] [Google Scholar]
- Sui P., Shi J., Gao X., Shen W. H., Dong A. (2013). H3K36 methylation is involved in promoting rice flowering. Mol. Plant 6 975–977 10.1093/mp/sss152 [DOI] [PubMed] [Google Scholar]
- Sun C., Chen D., Fang J., Wang P., Deng X., Chu C. (2014). Understanding the genetic and epigenetic architecture in complex network of rice flowering pathways. Protein Cell 5 889–898 10.1007/s13238-014-0068-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C., Fang J., Zhao T., Xu B., Zhang F., Liu L., et al. (2012). The histone methyltransferase SDG724 mediates H3K36me2/3 deposition at MADS50 and RFT1 and promotes flowering in rice. Plant Cell 24 3235–3247 10.1105/tpc.112.101436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q., Zhou D. X. (2008). Rice jmjC domain-containing gene JMJ706 encodes H3K9 demethylase required for floral organ development. Proc. Natl. Acad. Sci. U.S.A. 105 13679–13684 10.1073/pnas.0805901105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiezewski S., Liu F., Magusin A., Dean C. (2009). Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 462 799–802 10.1038/nature08618 [DOI] [PubMed] [Google Scholar]
- Teerawanichpan P., Chandrasekharan M. B., Jiang Y., Narangajavana J., Hall T. C. (2004). Characterization of two rice DNA methyltransferase genes and RNAi-mediated reactivation of a silenced transgene in rice callus. Planta 218 337–349 10.1007/s00425-003-1112-6 [DOI] [PubMed] [Google Scholar]
- Thorstensen T., Grini P. E., Aalen R. B. (2011). SET domain proteins in plant development. Biochim. Biophys. Acta 1809 407–420 10.1016/j.bbagrm.2011.05.008 [DOI] [PubMed] [Google Scholar]
- To T. K., Kim J. M. (2014). Epigenetic regulation of gene responsiveness in Arabidopsis. Front. Plant Sci. 4:548 10.3389/fpls.2013.00548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toriba T., Suzaki T., Yamaguchi T., Ohmori Y., Tsukaya H., Hirano H. Y. (2010). Distinct regulation of adaxial-abaxial polarity in anther patterning in rice. Plant Cell 22 1452–1462 10.1105/tpc.110.075291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji H., Saika H., Tsutsumi N., Hirai A., Nakazono M. (2006). Dynamic and reversible changes in histone H3-Lys4 methylation and H3 acetylation occurring at submergence-inducible genes in rice. Plant Cell Physiol. 47 995–1003 10.1093/pcp/pcj072 [DOI] [PubMed] [Google Scholar]
- Tsuji H., Taoka K.-I., Shimamoto K. (2013). Florigen in rice: complex gene network for florigen transcription, florigen activation complex, and multiple functions. Curr. Opin. Plant Biol. 16 228–235 10.1016/j.pbi.2013.01.005 [DOI] [PubMed] [Google Scholar]
- Tsukada Y., Fang J., Erdjument-Bromage H., Warren M. E., Borchers C. H., Tempst P., et al. (2006). Histone demethylation by a family of JmjC domain-containing proteins. Nature 439 811–816 10.1038/nature04433 [DOI] [PubMed] [Google Scholar]
- Van Lijsebettens M., Grasser K. D. (2014). Transcript elongation factors: shaping transcriptomes after transcript initiation. Trends Plant Sci. 19 717–726 10.1016/j.tplants.2014.07.002 [DOI] [PubMed] [Google Scholar]
- Vaucheret H. (2008). Plant ARGONAUTES. Trends Plant Sci. 13 350–358 10.1016/j.tplants.2008.04.007 [DOI] [PubMed] [Google Scholar]
- Wang J., Hu J., Qian Q., Xue H.-W. (2013). LC2 and OsVIL2 promote rice flowering by photoperoid-induced epigenetic silencing of OsLF. Mol. Plant 6 514–527 10.1093/mp/sss096 [DOI] [PubMed] [Google Scholar]
- Wang K., Tang D., Hong L., Xu W., Huang J., Li M., et al. (2010). DEP and AFO regulate reproductive habit in rice. PLoS Genet. 6:e1000818 10.1371/journal.pgen.1000818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Tang D., Luo Q., Jin Y., Shen Y., Wang K., et al. (2012). BRK1, a Bub1-related kinase, is essential for generating proper tension between homologous kinetochores at metaphase I of rice meiosis. Plant Cell 24 4961–4973 10.1105/tpc.112.105874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Zhang Y. (2014). Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell 156 45–68 10.1016/j.cell.2013.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Gan E. S., Zhou J., Wee W. Y., Zhang X., Ito T. (2014). Arabidopsis MRG domain proteins bridge two histone modifications to elevate expression of flowering genes. Nucleic Acids Res. 42 10960–10974 10.1093/nar/gku781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi T., Johzuka-Hisatomi Y., Terada R., Nakamura I., Iida S. (2014). The MET1b gene encoding a maintenance DNA methyltransferase is indispensable for normal development in rice. Plant Mol. Biol. 85 219–232 10.1007/s11103-014-0178-9 [DOI] [PubMed] [Google Scholar]
- Yamauchi T., Moritoh S., Johzuka-Hisatomi Y., Ono A., Terada R., Nakamura I., et al. (2008). Alternative splicing of the rice OsMET1 genes encoding maintenance DNA methyltransferase. J. Plant Physiol. 165 1774–1782 10.1016/j.jplph.2007.12.003 [DOI] [PubMed] [Google Scholar]
- Yan H., Kikuchi S., Neumann P., Zhang W., Wu Y., Chen F., et al. (2010). Genome-wide mapping of cytosine methylation revealed dynamic DNA methylation patterns associated with genes and centromeres in rice. Plant J. 63 353–365 10.1111/j.1365-313X.2010.04246.x [DOI] [PubMed] [Google Scholar]
- Yang J., Lee S., Hang R., Kim S. R., Lee Y. S., Cao X., et al. (2013). OsVIL2 functions with PRC2 to induce flowering by repressing OsLFL1 in rice. Plant J. 73 566–578 10.1111/tpj.12057 [DOI] [PubMed] [Google Scholar]
- Yao X., Shen W. (2011). Crucial function of histone lysine methylation in plant reproduction. Chin. Sci. Bull. 56 3493–3499 10.1007/s11434-011-4814-3 [DOI] [Google Scholar]
- Yokoo T., Saito H., Yoshitake Y., Xu Q., Asami T., Tsukiyama T., et al. (2014). Se14, encoding a JmjC domain-containing protein, plays key roles in long-day suppression of rice flowering through the memethylation of H3K4me3 of RFT1. PLoS ONE 9:e96064 10.1371/journal.pone.0096064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H., Nagato Y. (2011). Flower development in rice. J. Exp. Bot. 62 4719–4730 10.1093/jxb/err272 [DOI] [PubMed] [Google Scholar]
- Yu Y., Bu Z., Shen W.-H., Dong A. (2009). An update on histone lysine methylation in plants. Progr. Nat. Sci. 19 407–413 10.1016/j.pnsc.2008.07.015 [DOI] [Google Scholar]
- Zemach A., Kim M. Y., Silva P., Rodrigues J. A., Dotson B., Brooks M. D., et al. (2010). Local DNA hypomethylation activates genes in rice endosperm. Proc. Natl. Acad. Sci. U.S.A. 107 18729–18734 10.1073/pnas.1009695107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C. C., Yuan W. Y., Zhang Q. F. (2012a). RPL1, a gene involved in epigenetic processes regulates phenotypic plasticity in rice. Mol. Plant 5 482–493 10.1093/mp/ssr091 [DOI] [PubMed] [Google Scholar]
- Zhang L., Cheng Z., Qin R., Qiu Y., Wang J. L., Cui X., et al. (2012b). Identification and characterization of an epi-allele of FIE1 reveals a regulatory linkage between two epigenetic marks in rice. Plant Cell 24 4407–4421 10.1105/tpc.112.102269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Peng Y., Wei X., Dai Y., Yuan D., Lu Y., et al. (2014). Small RNAs as important regulators for the hybrid vigour of super-hybrid rice. J. Exp. Bot. 65 5989–6002 10.1093/jxb/eru337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S. Q., Hu J., Guo L. B., Qian Q., Xue H. W. (2010). Rice LEAF INCLINATION 2, a VIN3-like protein, regulates leaf angle through modulating cell division of the collar. Cell Res. 20 935–947 10.1038/cr.2010.109 [DOI] [PubMed] [Google Scholar]
- Zhong X., Zhang H., Zhao Y., Sun Q., Hu Y., Peng H., et al. (2013). The rice NAD(+)-dependent histone deacetylase OsSRT1 targets preferentially to stress- and metabolism-related genes and transposable elements. PLoS ONE 8:e66807 10.1371/journal.pone.0066807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., Liu Q., Li J., Jiang D., Zhou L., Wu P., et al. (2012). Photoperiod- and thermo-sensitive genic male sterility in rice are caused by a point mutation in a novel noncoding RNA that produces a small RNA. Cell Res. 22 649–660 10.1038/cr.2012.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Dong A., Shen W. H. (2012). Histone variants and chromatin assembly in plant abiotic stress responses. Biochim. Biophys. Acta 1819 343–348 10.1016/j.bbagrm.2011.07.012 [DOI] [PubMed] [Google Scholar]