Abstract
Patterns of linkage disequilibrium (LD) in the human genome are beginning to be characterized, with a paucity of haplotype diversity in “LD blocks,” interspersed by apparent “hot spots” of recombination. Previously, we cloned and physically characterized the low-density lipoprotein-receptor-related protein 5 (LRP5) gene. Here, we have extensively analysed both LRP5 and its flanking three genes, spanning 269 kb, for single nucleotide polymorphisms (SNPs), and we present a comprehensive SNP map comprising 95 polymorphisms. Analysis revealed high levels of recombination across LRP5, including a hot-spot region from intron 1 to intron 7 of LRP5, where there are 109 recombinants/Mb (4882 meioses), in contrast to flanking regions of 14.6 recombinants/Mb. This region of high recombination could be delineated into three to four hot spots, one within a 601-bp interval. For LRP5, three haplotype blocks were identified, flanked by the hot spots. Each LD block comprised over 80% common haplotypes, concurring with a previous study of 14 genes that showed that common haplotypes account for at least 80% of all haplotypes. The identification of hot spots in between these LD blocks provides additional evidence that LD blocks are separated by areas of higher recombination.
[Supplementary material: primers are available from our Web site: http://www-gene.cimr.cam.ac.uk/todd/human_data.shtml.]
Recent studies suggest that the genome is comprised of regions of strong intermarker linkage disequilibrium (LD), or haplotype blocks, interspersed by presumed recombination hot spots. Daly et al. (2001) showed that for a 500-kb region of Chromosome 5q31, 11 haplotype blocks spanned the interval from 3 to 100 kb in length. Tiret et al. (2002) studied the LD of 50 candidate genes for cardiovascular disease and observed gene-specific patterns of LD, concluding that all the sequence variation of a gene should be surveyed before attempting association studies with disease. Maniatis et al. (2002) observed blocks of LD on Chromosome 3q21. Patil et al. (2001) identified haplotype blocks on Chromosome 21 for which >80% of chromosomes were represented by a few common haplotypes. Gabriel et al. (2002) surveyed haplotype patterns in 51 autosomal regions, and observed LD blocks from <1 to 173 kb in European samples. Jeffreys et al. (2001) consolidated previous characterization of recombination patterns in the major histocompatibility complex (MHC) by analysis using sperm typing of a 216-kb subregion. They confirmed and detected hot spots of recombination in clusters: Within each cluster the hot-spot spacing was 1–7 kb, and between clusters it was 60–90 kb, with the rate of recombination varying between hot spots from 0.4–140 cM/Mb (Jeffreys et al. 2001). The LD patterns across the hot spots indicated that breaks in LD between LD blocks are due to chromosome regions of relatively high recombination.
Johnson et al. (2001) analyzed 14 genes, showing that the common haplotypes (each >5% frequency) accounted for 80% of the haplotypes that were observed in 400 chromosomes. Nickerson et al. (1998) screened a 9.7-kb region within the lipoprotein lipase (LPL) gene, encompassing exons 4–9, and concluded that this gene has a high haplotype diversity. However, Templeton et al. (2000a,b) showed that the haplotype diversity was not exceptional if a recombination hot spot in a 1.9-kb region close to exon 6 was taken into account.
Here we have determined the SNP profile of the low-density lipoprotein-receptor-related protein 5 (LRP5) gene (Hey et al. 1998), which maps to the putative insulin-dependent diabetes mellitus 4 (IDDM4) region on Chromosome 11q13 (Nakagawa et al. 1998). Studies of the LRP5 region in type 1 diabetes (T1D), using two microsatellite markers, D11S1917 and H0570polyA, indicated that the region may be associated with disease (Nakagawa et al. 1998). LRP5 comprises 23 exons spanning 160 kb of genomic sequence and encodes a 1615-amino-acid protein (Hey et al. 1998; Twells et al. 2001). LRP5, along with the homolog LRP6 (Brown et al. 1998), have recently been shown to mediate Wnt signaling (Pinson et al. 2000; Tamai et al. 2000; Wehrli et al. 2000). The LRP5 intracellular domain binds to axin, resulting in T-cell factor/lymphocyte enhancer factor (TCF/LEF) activation via β-catenin release (Mao et al. 2001; Nusse 2001). A mutation in exon 3 of the LRP5 gene has been shown to be responsible for high-bone-mass trait (Little et al. 2002), and numerous mutations identified in the autosomal recessive disorder osteoporosis–pseudoglioma syndrome (Gong et al. 2001). The LRP5 gene is therefore also a prime candidate for osteoporosis (Gong et al. 2001; Little et al. 2002). Previously, to investigate the possibility of association of the D11S987–D11S1917 region with T1D, we created a clone contig of 400 kb encompassing these two loci, sequenced the clones, and identified four genes: LRP5, C11orf23, C11orf24, and CGI-85 (Hey et al. 1998; Twells et al. 2001).
In the present study, we have identified 95 SNPs in the LRP5 gene and flanking regions spanning 269 kb. We chose 46 SNPs and typed them in 91 families (364 individuals) to assess the allele frequencies, LD between the markers, and haplotypes of this region. We also typed 32 microsatellites and 12 SNPs in 989 families to assess recombination across the entire IDDM4-linked region. This, in conjunction with the LD data, identified three to four hot spots of recombination: one in intron 1 of the LRP5 gene, one within intron 1 and/or intron 3–intron 5, and one within the region from intron 5 to intron 7 of LRP5, supporting the notion that homologous recombination events are unevenly distributed along chromosomes.
RESULTS
SNP Identification
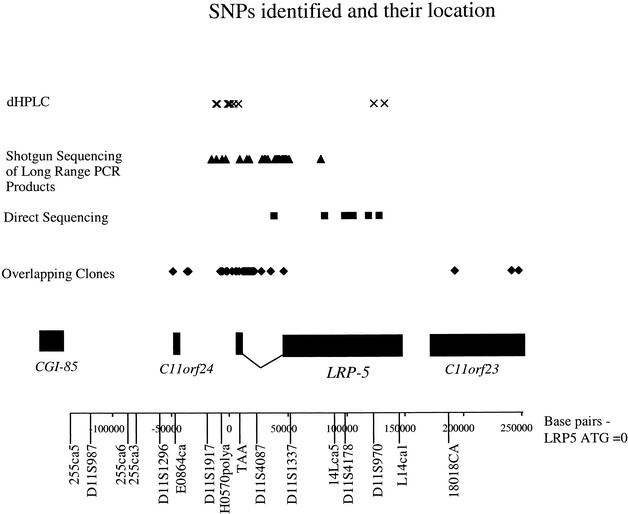
The identification of SNPs in the LRP5 region was achieved by examining sequence overlaps, direct sequencing, and dHPLC scanning. Sequence overlaps were compared as described in Methods, identifying a total of 38 SNPs (Table 1). The main focus of SNP identification was within the coding region and exon/intron boundaries of the LRP5 candidate gene, as well as putative regulatory elements based on mouse ortholog sequence we previously obtained (Twells et al. 2001). A total of 18 individuals were directly sequenced across the exons and exon/intron boundaries of LRP5, excluding exon 1, which had not been identified at this stage. In these, 11 new SNPs were identified, of which five are coding. Of these, two produce a predicted amino acid change: LRP5 g.95385G → A (Exon 9 V667M) and LRP5 30g.29264C → T (Exon 18 A1349V; Table 1). The 65-kb 5′ region of the LRP5 gene from D11S1917 to LRP5 exon 3 was shotgun-sequenced in two individuals, and an additional 31 SNPs were identified (Table 1).
Table 1.
SNPs Identified From the C11orf24–LRP5–C22orf23 Region, Showing the Method of Identification, Method of Genotyping, and Allele Frequency in 192 UK Individuals
| SNP name | Method of identificationa | dbSNP ss # | Typing methodb | Frequency |
| C11orf24 c.598G→A | ○ | 6905350 | Invader/RFLP(PstI) | 0.49 |
| EO864-6688 | ○ | 6905352 | RFLP/DraI | 0.49 |
| EO864-4419G→A | ○ | 6905353 | RFLP/NcoI | 0.29 |
| LRP5 g.-14447C→A |  |
6905427 | RFLP/DpnII | 0.009 |
| LRP5 g.-14279A→G | ♦ | 6905354 | Invader/cRFLP(NruI) | 0.34 |
| LRP5 g.-13901G→A |  |
6905428 | RFLP/HaeIII | 0.06 |
| LRP5 g.-13340-13366del-ins |  |
6905429 | Length polymorphism | 0.13 |
| LRP5 g.-11215T→A | ○ | 6905355 | NT | |
| LRP5 g.-11094G→A | ○ | 6905356 | NT | |
| LRP5 g.-10088G→A | ○ | 6905357 | Invader/RFLP(FauI) | 0.27 |
| LRP5 g.-9693G→A | ○ | 6905358 | Invader | 0.27 |
| LRP5 g.-9003T→C | ♦ | 6905359 | NT | |
| LRP5 g.-8870G→A | ♦ | 6905360 | NT | |
| LRP5 g.-5977C→T | ♦ | 6905361 | NT | |
| LRP5 g.-5880T→C | ♦ | 6905362 | NT | |
| LRP5 g.-5802G→C | ○ | 6905363 | Invader/RFLP(HincII) | 0.27 |
| LRP5 g.-5677C→T | ○ | 6905364 | Invader/RFLP(MspI) | 0.27 |
| LRP5 g.-5264G→A | ○ | 6905365 | Invader/RFLP(AflIII) | 0.27 |
| LRP5 g.-4317G→A |  |
6905430 | Invader | 0.20 |
| LRP5 g.-2945C→T |  |
6905431 | Invader | 0.20 |
| LRP5 g.-2713C→A |  |
6905432 | Invader | 0.10 |
| LRP5 g.-2517A→G |  |
6905433 | Invader | 0.06 |
| LRP5 g.-2366C→G |  |
6905434 | Invader | 0.08 |
| LRP5 g.-1792T→G |  |
6905435 | Invader | 0.10 |
| LRP5 g.-864A→G | ○ | 6905366 | Invader/RFLP(MspI) | 0.27 |
| LRP5 g.817C→T |  |
6905436 | Invader | 0.064 |
| LRP5 g.2221C→T | ○ | 6905367 | Invader/cRFLP(Hph) | 0.27 |
| LRP5 g.3103C→G | ○ | 6905368 | Invader/RFLP(pflMI) | 0.27 |
| LRP5 g.4780G→C | ○ | 6905369 | Invader/RFLP(EcoNI) | 0.34 |
| LRP5 g.5151C→T |  |
6905437 | Invader | 0.075 |
| LRP5 g.5257T→G | ○ | 6905370 | Invader/cRFLP(SnaBI) | 0.28 |
| LRP5 g.6088T→C | ♦ | 6905371 | NT | |
| LRP5 g.7374G→A | ○ | 6905372 | Invader/RFLP(AvaI) | 0.35 |
| LRP5 g.8003A→G | ○ | 6905373 | NT | |
| LRP5 g.8651T→G | ○ | 6905374 | NT | |
| LRP5 g.8924T→C | ○ | 6905375 | NT | |
| LRP5 g.10133insT | ○ | 6905376 | NT | |
| LRP5 g.10655A→T | ○ | 6905377 | NT | |
| LRP5 g.10674C→T | ○ | 6905378 | NT | |
| LRP5 g.11084T→C | ○ | 6905379 | NT | |
| LRP5 g.11263A→G | ○ | 6905380 | NT | |
| LRP5 g.11386A→G | ○ | 6905381 | NT | |
| LRP5 g.11886A→G | ♦ | 6905382 | NT | |
| LRP5 g.13279C→T | ○ | 6905383 | Invader/PvuII | 0.35 |
| LRP5 g.13963C→T | ♦ | 6905384 | Invader | 0.006 |
| LRP5 g.14250G→A | ○ | 6905385 | NT | |
| LRP5 g.14560C→T | ○ | 6905386 | NT | |
| LRP5 g.15531delG | ○ | 6905387 | NT | |
| LRP5 g.16868G→A | ○ | 6905388 | NT | |
| LRP5 g.17646G→T | ○ | 6905389 | Invader/BgIII | 0.42 |
| LRP5 g.23715C→A | ○ | 6905390 | NT | |
| LRP5 g.24964C→T | ♦ | 6905391 | Invader/RFLP(MnII) | 0.14 |
| LRP5 g.26659A→G | ♦ | 6905392 | NT | |
| LRP5 g.27081G→C | ♦ | 6905393 | NT | |
| LRP5 g.28047T→C | ♦ | 6905394 | NT | |
| LRP5 g.28149C→T | ♦ | 6905395 | Invader/RFLP(MboII) | 0.42 |
| LRP5 g.29652T→C | ♦ | 6905396 | NT | |
| LRP5 g.29925T→C | ♦ | 6905397 | NT | |
| LRP5 g.31502G→A | ○ | 6905398 | NT | |
| LRP5 g.31856G→A | ○ | 6905399 | Invader/RFLP(DpnI) | 0.094 |
| LRP5 g.34749G→A | □ | 6905400 | NT | |
| LRP5 g.35592T→C | ♦ | 6905401 | Invader | 0.44 |
| LRP5 g.37442A→C | ♦ | 6905402 | NT | |
| LRP5 g.37679G→A | ♦ | 6905403 | NT | |
| LRP5 g.38270G→A | ♦ | 6905404 | NT | |
| LRP5 g.38393C→T | ♦ | 6905405 | NT | |
| LRP5 g.38590C→T | ♦ | 6905406 | NT | |
| LRP5 g.38885C→A | ♦ | 6905407 | NT | |
| LRP5 g.39440A→G | ♦ | 6905408 | NT | |
| LRP5 g.39807T→G | ♦ | 6905409 | NT | |
| LRP5 g.40495C→G | ♦ | 6905410 | NT | |
| LRP5 g.42125G→T | ♦ | 6905411 | Invader | 0.44 |
| LRP5 g.43231G→A | ○ | 6905412 | NT | |
| LRP5 g.43404A→G | ♦ | 6905413 | NT | |
| LRP5 g.43718G→A | ♦ | 6905414 | NT | |
| LRP5 g.44602insC | ♦ | 6905415 | NT | |
| LRP5 g.45704G→A | ♦ | 6905416 | Invader | 0.44 |
| LRP5 g.47698G→A | ♦ | 6905417 | NT | |
| LRP5 g.75326G→A | □ | 6905418 | RFLP/HphI | 0.19 |
| LRP5 g.78477G→A | □ | 6905419 | NT | |
| LRP5 g.92423G→A |  |
6905438 | RFLP/Tsp451 | 0.017 |
| LRP5 g.95318G→A | □ | 6905420 | NT | |
| LRP5 g.95385G→A | □ | 6905421 | RFLP/HaeIII | 0.05 |
| LRP5.30g.5276C→TASN740 | □ | 6905422 | ARMS | 0.16 |
| LRP5 30g.5380C→T | □ | 6905425 | ARMS | 0.07 |
| LRP5 30g.6891C→T | □ | 6905426 | ARMS | 0.07 |
| LRP5 30g.19977T→G | □ | 6905423 | NT | |
| LRP5 30g.20149A→G | □ | 6905424 | NT | |
| LRP5 30g.24930C→T |  |
6905439 | cRFLP/NdeI | 0.31 |
| LRP5 30g.29264C→T | □ | 6905351 | RFLP/AciI | 0.16 |
| LRP5 30g.34160G→C |  |
6905440 | RFLP/RsaI | 0.08 |
| LRP5 30g.34295T→C |  |
6905441 | cRFLP/MspA11 | 0.006 |
| C11orf23 c.3297-3302del | ○ | 6905347 | Invader/length polymorphism | 0.25 |
| C11orf23 c.3488G→A | ○ | 6905348 | Invader/cRFLP(NlaIII) | 0.32 |
| C11orf23 c.3680G→A | ○ | 6905349 | Invader/ | 0.24 |
○ = Identified initially from cosmid/BAC overlaps.
♦ = Identified initially from shotgun sequencing of 5-kb-long PCR products.
□ = Identified initially by directed PCR amplification and sequencing of the LRP5 gene.
 = Identified from dHPLC.
= Identified from dHPLC.
NT = not tested.
The above two sequencing efforts were achieved with sequencing 18 and 2 individuals, respectively. To increase the power to detect SNPs, we rescreened the LRP5 gene in an additional 24 individuals by dHPLC and sequenced the heteroduplex samples. The region analyzed was a rescreening of the 23,744 bp encompassing D11S1917, H0570POLYA, and the 5′ region and exon 1 of LRP5; thus it may contain regulatory elements of this gene (Fig. 1). In addition, all the exons of LRP5were screened (exon 1 within the 24.3-kb region), excluding exons 4, 14, 21, and 23, which failed to amplify. Ten new SNPs and an indel were identified from the dHPLC screening of the 23.7-kb region. Four new SNPs were identified in intronic regions of LRP5, close to exons 8 and 17 and two near exon 20. These are listed in Table 1. The total number of SNPs identified by all methods is 95.
Figure 1.
Map of the region and position of the SNPs identified.
The dHPLC in 24 individuals identified every SNP identified by the previous methods (18 SNPs) apart from LRP5 g.6088T → C, which had been identified from the shotgun sequencing of the long-range PCR products. This SNP was not pursued to confirm whether it was a true variant or a sequencing artifact. The SNP harvesting performed by dHPLC compared with direct sequencing was, therefore, very sensitive. The allele frequencies were ascertained in 182 UK parental chromosomes. The average allele frequency of the total number of SNPs identified by dHPLC in 24 individuals is 0.17. The average allele frequency in the 5′ region of the LRP5 gene (14 SNPs) is 0.17, in the introns (13 SNPs) 0.18, and in the three exonic SNPs 0.12. Overall, the SNPs identified by dHPLC average 1/911 nt in the 5′ region of LRP5. Of the coding region, the number of SNPs identified was an average of 1/804 nt. Three SNPs and a 3-bp deletion were identified from cDNA clone overlaps in the two flanking genes C11orf23 and C11orf24. These SNPs are all common, with an average allele frequency of 0.33. The C11orf24 SNP is coding, aa150Ala–Thr; one C11orf23 SNP is in the 5′-UTR, and the other two are in the 3′-UTR. The majority of SNPs are substitutions, with 6 indels.
For the LRP5 region, the nucleotide diversity, for the dHPLC-scanned samples, is θ = (2.7 × 10−4) ± (9 × 10−5). For the non coding regions (24,601 bp), θ = (2.6 × 10−4) ± (8.8 × 10−5). The coding region of LRP5 (4251 bp) has θ = (2.8 × 10−4) ± (1.4 × 10−4).
Linkage Disequilibrium
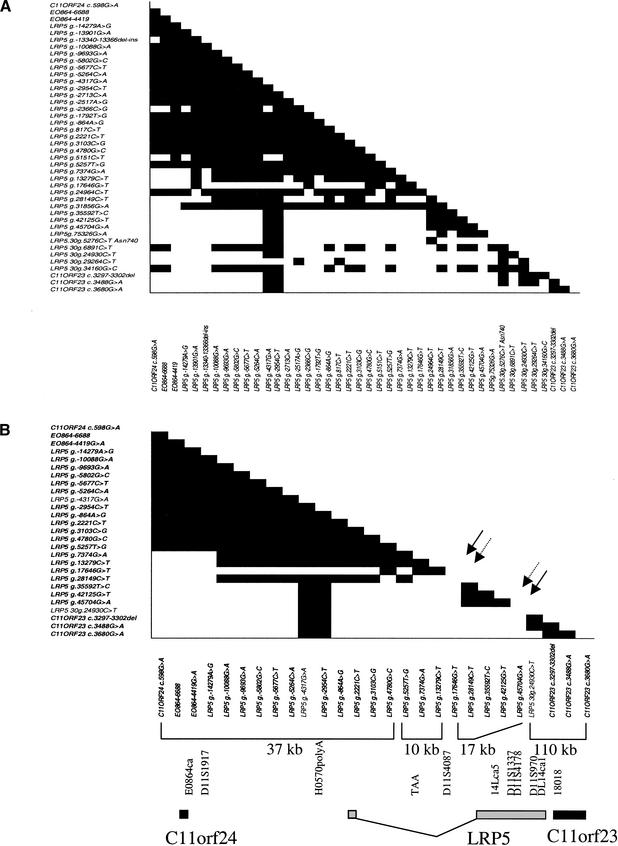
To analyze the LD in this region, 46 SNPs from the 95 identified were selected that span the region and typed in 91 UK multiplex families. The allele frequencies were ascertained as described in the previous section, and the SNPs with <0.03 frequency were omitted from further analysis. The remaining 42 SNPs were analyzed for LD using the measure ‖D‘‖ with the parental genotypes (364 chromosomes). Figure 2A shows the pairwise ‖D‘‖ values. We used the SNPs with minor allele frequency >0.20 to aid identification of “LD blocks” (Daly et al. 2001), as these are older SNPs, more likely to identify the block structure (Fig. 2B; Jeffreys et al. 2001). We used the criteria of an LD block having intermarker LD of ‖D‘‖ > 0.8 on average, which we have observed previously in studies of the CTLA4 gene and the INS region (H. Ueda, B. Barratt, J.A. Todd, unpubl.). Those studies also noted that the interblock LD is on average ‖D‘‖ < 0.3 with SNPs (H. Ueda, B. Barratt, J.A. Todd, unpubl.). A block of strong LD can be observed for the markers C11ORF24 c.598G → A–LRP5 g.5257T → G, a distance of 37 kb, called LD block 1. LD breaks down in the region LRP5 g.5257T → G–LRP5 g.28149C → T, in intron 1 of the LRP5 gene. A second block of very strong LD (‖D‘‖ > 0.9) is observed, LRP5 g.28149C → T–LRP5 g.45704G → A, spanning 17.6 kb of intron 1 to intron 3 of the LRP5 gene. The 3′ region of the LRP5 gene could not be characterized so well owing to the paucity of common SNPs identified within this larger genomic distance. LD appears to be lower from LRP5 g.45704G → A–LRP5 30g.24930C → T, from intron 3 of LRP5 to exon 17, with an average ‖D‘‖ of 0.72. A third LD block was identified from LRP5 30g.24930C → T to C11ORF23 c.3680G → A, encompassing from exon 17 of LRP5 to the 3′-UTR of the C11orf23 gene, ∼110 kb.
Figure 2.
Plot of pairwise ‖D‘‖ values against distance (in base pairs) for 42 SNPs in 364 UK chromosomes. ‖D‘‖ values of 0.8 or more are shaded black. (A) Pairwise ‖D‘‖ values for all SNPs in the 300-kb region. (B) Pairwise ‖D‘‖ values for the 27 SNPs with allele frequencies >0.20. The arrows show the location of the recombination hot spots.
Recombination
In the UK, USA, and Sardinian data sets, the sex-averaged map from FCER1B to D11S916 is 23.7 cM, whereas the physical map, from ENSEMBL 1.1.3 is 18.1 Mb. Therefore, in this 18.1-Mb region overall there is on average 1.3 cM/Mb.
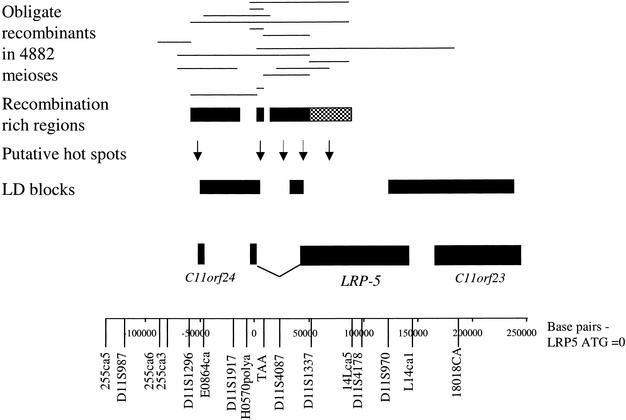
The obligate recombinants in the entire 18.1-Mb region were observed. In the 4882 meioses, this analysis detected 265 obligate recombinants, an average of 14.6 recombinants/Mb. In the 400-kb contig surrounding LRP5, the microsatellite and SNP markers spanned 300 kb. In this interval there were 15 obligate recombinants, an average of 50 recombinants/Mb (Fig. 3). Of these, 14 were localized between 255CA6 and 14LCA5, and one was localized between TAA and 18018CA. Thus, between 255CA5 and 14LCA5, there were ∼72.5 recombinants/Mb, and between 14LCA5 and 18018CA, there were up to 5 recombinants/Mb. Between 255CA5 and 14LCA5, nine recombinants could be localized precisely to within the interval LRP5g.3103C → G–14LCA5, with a further four recombinants overlapping this interval (Fig. 3). These five recombinants were localized between EO864–4419G → A–LRP5 g.17646G → T, D11S1296–14LCA5, TAA–18018CA, and D11S1296–TAA, respectively. Weighting these four for distance between the flanking markers (assuming an equal possible distribution by physical distance), plus the recombinants within the LRP5 g.3103C → G–14LCA5 interval, gives 9.1 recombinants between LRP5 g.3103C → G–14LCA5, a distance of 83.6 kb, resulting in 109 recombinants/Mb. Examination of this interval in more detail shows three subregions to which the recombinants can be mapped precisely: TAA–LRP5 g.7374G → A (601 bp), LRP5 g.17646G → T–D11S1337 (35.2 kb), and D11S1337–14LCA5 (33.8 kb). Again weighting recombinants for distance (assuming an even distribution of recombination) gives a value of 2178 recombinants/Mb for TAA–LRP5 g.7374G → A, 98 recombinants/Mb for LRP5 g.17646G → T–D11S1337, and 66 recombinants/Mb for D11S1337–14LCA5. Therefore, there appear to be several recombination hot spots within the region LRP5g.3103C → G–14LCA5, which is from intron 1 to between exons 7 and 8 of LRP5. This concurs with the LD data (Fig. 3), showing that there is a recombination hot spot in intron 1 of LRP5 that accounts for the decrease in LD between markers between LD blocks 1 and 2. Within the second hot-spot region (LRP5 g.17646G → T–D11S1337), there is an LD block, LRP5 g.28149C → T–LRP5 g.45704G → A; thus, it is possible the hot spot maps to either LRP5 g.17646G → T–LRP5 g.28149C → T or LRP5 g.45704G → A–D11S1337, or both. The third hot spot maps to the region between LD blocks 2 and 3 (D11S1337–14LCA5).
Figure 3.
Map of the LRP5 region showing the obligate recombinants observed in 4882 meioses and putative hot spots. (Solid arrow) Putative hot spot; (dashed arrow) either or both regions may contain a hot spot of recombination.
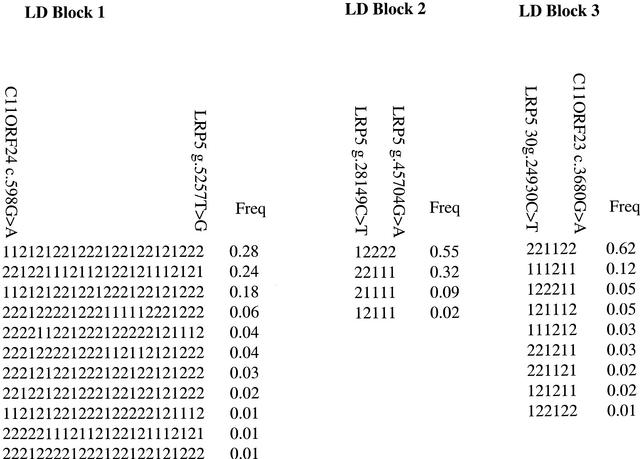
Haplotypes
Having identified three hot-spot regions flanking the LD blocks, we investigated the haplotypes in each LD block. The haplotypes were ascertained from 364 UK parental chromosomes using the program SNPHAP. The first LD block, C11ORF24 c.598G → A–LRP5 g.5257T → G, is a 37-kb region encompassing exon 4 of C11orf24 to intron 1 of LRP5. This has four haplotypes with frequency >3%, comprising 84% of the haplotypes (Fig. 4). The second LD block, LRP5 g.28149C → T–LRP5 g.45704G → A, spanning from intron 1 to intron 3 of LRP5, has 96% common haplotypes. The third LD block, from LRP5 30g.24930C → T–C11ORF23 c.3680G → A, encompassing from exon 17 of LRP5 to the 3′-UTR of the C11orf23 gene, ∼110 kb, has 84% of common haplotypes (Fig. 4). It is likely that this LD block extends from intron 7 of the LRP5 gene through to the 3′ region of C11orf23, based on the low recombination rate observed between 14Lca5 (in intron 7 of LRP5) and 18018CA (within C11orf23). However, the low number of SNPs with frequency >0.2, in the region precludes confirmation of the extent of this LD block.
Figure 4.
Haplotype blocks in the LRP5 region. Haplotypes are given with frequencies >0.01.
DISCUSSION
SNPs were characterized across the exons and 5′ region of the LRP5 gene in several stages. The comparison of clone overlaps has been used to identify millions of nontargeted SNPs from various regions (Dawson et al. 2001; Sachidanandam et al. 2001; Venter et al. 2001). In the LRP5 region, the clone overlaps only identified common SNPs, and one rare SNP, with an average minor allele frequency of 0.28. Eberle and Kruglyak (2000) showed that a greater number of rare SNPs is expected even if the population has followed a constant population model. The dHPLC of 24 individuals had >99.3% power to detect SNPs with an allele frequency of 0.1 (Zwick et al. 2000), and identified all SNPs except one that had been identified by other methods, in the region scanned by both. The approach we used, identifying SNPs in a cohort of individuals prior to genotyping the SNPs in a larger family-based study, will clearly miss rare variants, as observed by Nickerson et al. (2000) in a two-stage SNP detection and genotyping of the APOE gene, and by Glatt et al. (2001) in the sequencing of 450 samples for SLC6A4 and SLC18A2. It will be important to identify rare SNPs for any candidate gene, as has been demonstrated by the identification of rare variants at the Crohn disease locus, NOD2 (Hugot et al. 2001; Ogura et al. 2001).
In the LRP5 region, the overall nucleotide diversity was within the range of other genes studied (Cargill et al. 1999; Halushka et al. 1999; Thorstenson et al. 2001). The nucleotide diversity in the coding regions of LRP5 (θ = [2.8 × 10−4] ± [1.4 × 10−4]), was slightly lower than that observed for some other studies such as Cargill et al. (1999). Cargill et al. (1999) examined 106 genes, and the coding and noncoding diversity was similar ([5.0 × 10−4] ± [2.4 × 10−4], [5.2 × 10−4] ± [2.5 × 10−4], respectively). Similarly, Halushka et al. (1999), who scanned 75 genes for SNPs, reported an average nucleotide diversity of (8.0 × 10−4) ± (1.9 × 10−4) and (8.5 × 10−4) ± (2.0 × 10−4), for the coding and noncoding regions, respectively. However, in contrast, the ATM gene had a lower nucleotide diversity in its coding regions, (0.71 × 10−4) ± (0.61 × 10−4) than found in the above studies, or for the LRP5 gene, and the ATM coding nucleotide diversity was a 7.5-fold decrease compared with the noncoding regions (Thorstenson et al. 2001). This was also the case for the LPL gene, (21 × 10−4) ± (10 × 10−4), (5 × 10−4) ± (5 × 10−4), in the noncoding versus coding regions, respectively (Nickerson et al. 1998). Zwick et al. (2000) proposed that as the two large studies of Cargill and Halushka scanned proportionally fewer noncoding regions than the ATM and LPL genes, it is possible that had more noncoding regions been analyzed, the nucleotide diversity ratio would have increased. This was the case with 12 out of 15 of the autosomal genes scanned by Thorstenson et al. (2001). For LRP5, we screened 25,623 nt of noncoding DNA (including the regions flanking exons), which is larger than any of the above studies of individual genes, and yet the nucleotide diversity was similar to the coding regions of the LRP5 gene.
The magnitude of LD depends on a number of factors, including recombination, gene conversion, mutation, genetic drift, and selection, that give rise to the underlying haplotypes in the population studied. These empirical data show that the ‖D‘‖ LD is not directly ascertainable by distance, as expected. The three conserved blocks of LD were separated by potential hot spots of recombination, ascertained by recombination rate across the region. There was a 25-fold increase in recombination rate from intron 1 to intron 7 of LRP5, compared with the 3′ region of LRP5, and a 10-fold increase compared with the entire 18.1-Mb IDDM4 interval. This region of high recombination had three separate recombination-rich regions, ranging from 66 to 2178 recombinants/Mb. The first hot spot was within a 601-bp region of LRP5 intron 1. The second contained a 17-kb LD block from intron 1 to intron 3 of the LRP5 gene; thus, the hot spot either flanked this LD block in LRP5 intron 1 or LRP5 intron 3–intron 5 or comprised two hot spots, one in each region. The last hot spot was located within the region from intron 5 to intron 7 of LRP5. At the MHC, Jeffreys et al. (2001) noted that the recombination hot spots occur within discrete clusters, each individual hot spot 1–1.9 kb in width, with 60–90 kb of separation between clusters. At LRP5, the first hot spot in intron 1 was very finely mapped to a width of 601 bp, similar to the MHC hot spots observed by Jeffreys et al. (2001). The second hot spot region has a hot spot either in a 10.5-kb region, 20 kb distal to the first hot spot, proximal to LD block 2, and/or is located in a 7.1-kb region distal to LD block 2, 38.3 kb distal to hot spot 1. The last hot spot is within a 33.8-kb region, 45 kb from hot spot 1, and is bounded proximally by the same SNP that limits hot spot region 2 distally, so may be part of a cluster with hot spot region 2. The LD is higher for both these inter-LD block regions, an average of ‖D‘‖ = 0.65 and 0.72, respectively, compared with that observed at INS and CTLA4, which had inter LD-block ‖D‘‖ values of <0.3 (H. Ueda, B. Barratt, J.A. Todd, unpubl.).
LRP5 is similar to LPL in that both genes have hot spots of recombination within the gene (Templeton et al. 2000b). At LRP5, because of the multiple LD blocks, more than five haplotype tag SNPs (htSNPs; Johnson et al. 2001) will need to be tested to evaluate the association of LRP5 common haplotypes with T1D and other diseases (Nakagawa et al. 1998; Gong et al. 2001; Little et al. 2002). This is in contrast to the 14 genes previously studied by Johnson et al. (2001), in which the common haplotypes comprised 80% of all the haplotypes for each gene region, as each gene comprised one LD block. Consequently, our data indicate that a much larger number of genes will need to be characterized before a representative picture of gene-based haplotype diversity can be formed.
METHODS
Subjects
All the families were of white European origin with two parents and at least one diabetic child. When unaffected offspring were available, these were also included. The families comprise: 401 UK multiplex, 236 US multiplex, 80 Yorkshire UK simplex, 32 Southwest UK simplex, 50 Bristol UK simplex, 176 Sardinian simplex, and 14 Sardinian multiplex (Nakagawa et al. 1998). Informed consent was obtained from all subjects.
Microsatellite Identification and Genotyping
Microsatellite polymorphisms were identified by microsatellite rescue (Merriman et al. 1997) from a cosmid, PAC, and BAC clone contig spanning 400-kb interval around H0570POLYA (Twells et al. 2001). The microsatellites 255ca5, 255ca6, and 255ca3 were isolated from the clone RPCI-255M19; E0864CA from the cosmid E0864; 14LCA5 and 14LCA1from CITB-14L15; and 18018 from clone RPCI-18O18. TAA was identified from the shotgun sequence of cosmid clone B07185 (accession number AC024124), and is ∼23 kb distal to H0570POLYA. All the primers used in this study are available from our Web site: http://www-gene.cimr.cam.ac.uk/todd/human_data.shtml.
The above microsatellite markers were genotyped in the whole data set as well as: FCERIB, D11S1765, UT5017, D11S426, D11S480, D11S1883, D11S4205, D11S457, D11S1783, D11S913, D11S1258, PPP1CA, D11S4155, D11S987, D11S1296, D11S1917, H0570POLYA, D11S4087, D11S1337, D11S4178, D11S970, D11S971, D11S1314, and D11S916, using fluorescently labeled primers as described elsewhere (Reed et al. 1994).
SNP Identification
SNPs were identified in several stages. SNPs were identified from sequence overlaps. Sequence overlaps were compared between CITB-14L15 Contig 31 (accession no. AF283320; Twells et al. 2001) and the cosmids E0864, B07185, and F08180 (accession nos. AC024125, AC024124, and AC024126; Twells et al. 2001). These overlaps were 117,737 nt in total, encompassing Contig 31 2706–4180 nt, 4349–24,136 nt, and 24,413–120,664 nt. CITB-67M5 (accession no. AC024123; Twells et al. 2001) overlapped 79,701 nt with Contig 31, but have the same haplotype; therefore, no new SNPs were identified.
The second approach to SNP harvesting was direct sequencing of the exons and flanking exon/intron boundaries of the LRP5 gene. This was achieved by designing specific primers ranging from 500–800 bp flanking the regions of interest. LRP5 exon 1 was not screened, as it had not been identified at that stage. DNA samples from 18 individuals were analyzed. These consisted of 10 UK individuals and eight Sardinians. The UK samples were selected on the basis of their D11S1917–H0570POLYA haplotypes according to their TDT results in our previous study (Nakagawa et al. 1998). The UK samples are shown in Table 2. The Sardinian samples also comprise a selection of different haplotypes (Table 2).
Table 2.
Haplotype Content of the Individuals Screened for Polymorphisms, and Frequency of Haplotypes in the General Population Estimated as AFBAC Frequencies
| D11S1917 LRP5 g.5677C→T HO570POLYA | Number of UK chromosomes for direct sequencing | Number of Sardinian chromosomes for direct sequencing | UK chromosomes for dHPLC | Frequency of haplotypes for dHPLC | Parental frequency |
| 312 | 7 | 5 | 12 | 0.25 | 0.27 |
| 123 | 0 | 0 | 10 | 0.21 | 0.20 |
| 223 | 11 | 4 | 11 | 0.23 | 0.31 |
| 323 | 2 | 1 | 6 | 0.13 | 0.02 |
| 221 | 0 | 6 | 9 | 0.19 | 0.19 |
| Total | 20 | 16 | 48 | 1.01 | 0.98 |
Forward and reverse primers were tailed with sequences corresponding to the M13 Universal primer (5′-TGTAAA ACGACGGCCAGT-3′) and a modified reverse primer (5′-GCTATGACCATGATTACGCC-3′), respectively. The reaction volume was 50 μL with Perkin-Elmer 10× reaction buffer, 200 mM dNTPs, 0.5 μL Taq Gold (Perkin-Elmer Corp.), 50 ng of genomic DNA, and 20 pmole/mL of forward and reverse primers. The thermocycler conditions were 95°C for 12 min, then 35 cycles of 95°C for 30 sec, 57°C for 30 sec, and 68°C for 2 min, followed by 72°C for 6 min and 4°C stop. The PCR products were then purified for sequencing using QiaQuick strips or QiaQuick 96-well plates on the QIAGEN robot (QIAGEN). Direct BODIPY sequencing was performed according to Metzker et al. (1996).
The third method was a shotgun-sequencing strategy of two individuals who were homozygous for two haplotypes at D11S1917–H0570POLYA, 3–2 and 2–3. The genomic region between D11S1917 and LRP5 exon 6 (∼70 kb) was amplified with PCR products of ∼5 kb using the Expand Long Template PCR System (Boehringer Mannheim) with modifications (Barnes 1994). The PCR reaction was performed in a total reaction volume of 50 μL, containing 100 ng of genomic DNA template, 200 pmole/μL of forward and reverse primers, 500 μM dNTPs, 1× Buffer 3, and 0.5 μL of the enzyme mix. The thermocycling conditions were 35 cycles of 92°C for 30 sec, 60°C for 2 min, and 68°C for 10 min, followed by a 7-min extension at 72°C and 4°C finish using a Perkin Elmer 9600 DNA Thermal Cycler. PCR products were generated for each patient and pooled. The products were sheared by nebulization. Shotgun libraries were made and sequenced as described in Hey et al. (1998). The SNPs were identified visually with the aid of Consed (Gordon et al. 1998). Of the 19 long-range PCR primers designed, four failed to amplify -31-1, 31-6 (which includes LRP5 exon 1), 31-18 and 31-19 (Fig. 1). The total amount of PCR product was therefore 61.538 kb and ranged from CITB-14L15 Contig 31 8102–23,656 nt and 27,817–73,801 nt (LRP5 exon 3).
The fourth strategy was to scan 24 individuals by the dHPLC WAVE machine (Transgenomic Inc.). The 24 individuals were selected for their haplotypes at the D11S1917–LRP5 g.-5677C → T–H0570POLYA loci (Table 2). The region analyzed was a rescreening of the 25 kb encompassing D11S1917, H0570POLYA, and the 5′ region and exon 1 of LRP5. The region was repeat-masked and 49 primer sets were designed to the nonrepetitive regions, encompassing 24,269 bp (Contig 31 7524–31,793). Four repetitive regions were excluded, totaling 5398 nt, including an LTR of 3245 nt, which was 6 kb upstream of LRP5 exon 1 (−6190 to −9437). Therefore, a total region of 18,871 nt was screened. Primers were designed for PCR of ∼500 bp, and amplified in each of the 24 individuals according to Transgenomic Application Note 101 (Transgenomic Inc.). The PCR products were run through the WAVE machine according to Transgenomic Application Note 101. Samples with heteroduplex or an alternative homoduplex pattern were then directly sequenced using the PCR primers as described above.
SNP Genotyping
The Invader assay (Third Wave Technologies, Inc.) was carried out as described in Mein et al. (2000). RFLP and cRFLP analyses were also carried out as described in Mein et al. (2000). The ARMs assay was carried out as follows with a total reaction volume of 13 μL, 40 ng of DNA, 2 mM MgCl2, 50 ng of each primer, 0.2 U of Bioline Taq polymerase, 360 μM dNTPs, 20 ng of control primers (TGCCAAGTGGAGCACCCAA, GCATCTTGCTCTGTGCAGAT; M. Bunce and K. Welsh, pers. comm.). The thermocycling conditions are 96°C for 1 min; 5 cycles of 96°C for 35 sec, 70°C for 45 sec, 72°C for 35 sec; 21 cycles of 96°C for 25 sec, 65°C for 50 sec, 72°C for 40 sec; 4 cycles of 96°C for 35 sec, 55°C for 60 sec, and 72°C for 90 sec; 25°C for 5 min. The products are visualized after electrophoresis on a 1% agarose gel, 0.5× TBE.
Recombination, Haplotypes, and LD
The genetic multipoint map was calculated with ASPEX (D. Hinds and N. Risch; ftp://lahmed.stanford.edu/pub/aspex). The obligate recombinants were observed using the SHOWHAPLO program available from Frank Dudbridge (f.dudbridge@hgmp.mrc.ac.uk). Haplotypes were estimated with an EM-based algorithm, implemented in the program SNPHAP from the parents of 91 families available from http://www-gene.cimr.cam.ac.uk/clayton/software/.
The parental genotypes of 91 multiplex families were used to calculate Lewontin’s D‘ (Lewontin 1995) using PWLD (http://www-gene.cimr.cam.ac.uk/clayton/software/) with the stata package (http://www.stata.com). ‖D‘‖ was calculated as ‖D/Dmax‖ and ranges from 0 to 1. SNPs with a frequency <0.03 (within the 95% CI for a frequency of 5%) were not included for further analysis, as in this strategy of testing SNPs for disease association, there will not be enough power with the number of families (or case/controls) for the likely genetic effects (OR < 3; Johnson et al. 2001). The LD of the region was then examined using ‖D‘‖ with the markers of allele frequency >0.2. The region was divided into apparent blocks of LD according to Figure 2B, as there was discontinuous LD across the gene region. The haplotypes from each block were generated with SNPs >0.03 frequency, using the program SNPHAP (v0.2-; http://www-gene.cimr.cam.ac.uk/clayton/software/).
WEB SITE REFERENCES
ftp://lahmed.stanford.edu/pub/aspex; the ASPEX program (D. Hinds and N. Risch).
http://www-gene.cimr.cam.ac.uk/clayton/software/; SNPHAP and PWLD (D. Clayton).
http://www-gene.cimr.cam.ac.uk/todd/human_data.shtml; primers used in this work.
http://www.stata.com; the stata package.
Acknowledgments
We thank the Wellcome Trust, the Juvenile Diabetes Research Foundation, and Diabetes UK for their support. This work was also supported by a grant from Merck Research Laboratories. R.C.J.T. held a Diabetes UK R.D. Lawrence Fellowship. We thank the anonymous reviewers for their helpful comments.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
E-MAIL rebecca.twells@cimr.cam.ac.uk; FAX (44) 1223 762 102.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.563703.
REFERENCES
- 1.Barnes W. 1994. PCR amplification of up to 35-kb DNA with high fidelity and high yield from λ bacteriophage templates. Proc. Natl. Acad. Sci. 91: 2216-2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown S., Twells, R., Hey, P.J., Cox, R.D., Levy, E.R., Soderman, A.R., Metzker, M.L., Caskey, C.T., Todd, J.A., and Hess, J.F. 1998. Isolation and characterization of LRP6, a novel member of the low density lipoprotein receptor gene family. Biochem. Biophys. Res. Commun. 248: 879-888. [DOI] [PubMed] [Google Scholar]
- 3.Cargill M., Altschuler, D., Ireland, J., Sklar, P., Ardlie, K., Patil, N., Shaw, N., Lane, C.R., Lim, E.P., Kalyanaraman, N., et al. 1999. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat. Genet. 22: 231-238. [DOI] [PubMed] [Google Scholar]
- 4.Daly M.J., Rioux, J.D., Schaffner, S.F., Hudson, T.J., and Lander, E.S. 2001. High-resolution haplotype structure in the human genome. Nat. Genet. 29: 229-232. [DOI] [PubMed] [Google Scholar]
- 5.Dawson E., Chen, Y., Hunt, S., Smink, L.J., Hunt, A., Rice, K., Livingston, S., Bumpstead, S., Bruskiewich, R., Sham, P., et al. 2001. A SNP resource for human Chromosome 22: Extracting dense clusters of SNPs from the genomic sequence. Genome Res. 11: 170-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eberle M.A. and Kruglyak, L. 2000. An analysis of strategies for discovery of single-nucleotide polymorphisms. Genet. Epidemiol. 19: S29-S35. [DOI] [PubMed] [Google Scholar]
- 7.Gabriel S.B., Schaffner, S.F., Nguyen, H., Moore, J.M., Roy, J., Blumenstiel, B., Higgins, J., DeFelice, M., Lochner, A., Faggart, M., et al. 2002. The structure of haplotype blocks in the human genome. Science 296: 2225-2229. [DOI] [PubMed] [Google Scholar]
- 8.Glatt C.E., DeYoung, J.A., Delgado, S., Service, S.K., Giacomini, K.M., Edwards, R.H., Risch, N., and Freimer, N.B. 2001. Screening a large reference sample to identify very low frequency sequence variants: Comparisons between two genes. Nat. Genet. 27: 435-438. [DOI] [PubMed] [Google Scholar]
- 9.Gong Y., Slee, R.B., Fukai, N., Rawadi, G., Roman-Roman, S., Reginato, A.M., Wang, H., Cundy, T., Glorieux, F.H., Lev, D., et al. 2001. LDL receptor-related protein 5 (LRP5) affects bone accrual and development. Cell 107: 513-523. [DOI] [PubMed] [Google Scholar]
- 10.Gordon D., Abajian, C., and Green, P. 1998. Consed: A graphical tool for sequence finishing. Genome Res. 8: 195-202. [DOI] [PubMed] [Google Scholar]
- 11.Halushka M.K., Fan, J., Bentley, K., Hsie, L., Shen, N., Weder, A., Cooper, R., Lipshutz, R., and Chakravarti, A. 1999. Patterns of single-nucleotide polymorphisms in candidate genes for blood-pressure homeostasis. Nat. Genet. 22: 239-247. [DOI] [PubMed] [Google Scholar]
- 12.Hey P.J., Twells, R.C., Phillips, M.S., Nakagawa, Y., Brown, S.D., Kawaguchi, Y., Cox, R., Xie, G., Dugan, V., Hammond, H., et al. 1998. Cloning of a novel member of the low-density lipoprotein receptor family. Gene 216: 103-111. [DOI] [PubMed] [Google Scholar]
- 13.Hugot J.P., Chamaillard, M., Zouali, H., Lesage, S., Cezard, J.P., Belaiche, J., Almer, S., Tysk, C., O’Morain, C.A., Gassull, M., et al. 2001. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature 411: 599-603. [DOI] [PubMed] [Google Scholar]
- 14.Jeffreys A.J., Kauppi, L., and Neumann, R. 2001. Intensely punctate meiotic recombination in the class II region of the major histocompatibility complex. Nat. Genet. 29: 217-222. [DOI] [PubMed] [Google Scholar]
- 15.Johnson G.C., Esposito, L., Barratt, B.J., Smith, A.N., Heward, J., Di Genova, G., Ueda, H., Cordell, H.J., Eaves, I.A., Dudbridge, F., et al. 2001. Haplotype tagging for the identification of common disease genes. Nat. Genet. 29: 233-237. [DOI] [PubMed] [Google Scholar]
- 16.Lewontin R.C. 1995. The detection of linkage disequilibrium in molecular sequence data. Genetics 140: 377-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Little R., Carulli, J., Del Mastro, R., Dupuis, J., Osborne, M., Folz, C., Manning, S.P., Swain, P.M., Zhao, S.C., Eustace, B., et al. 2002. A mutation in the LDL receptor-related protein 5 gene results in the autosomal dominant high-bone-mass trait. Am. J. Hum. Genet. 70: 11-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maniatis N., Collins, A., Xu, C.-F., McCarthy, L.C., Hewett, D.R., Tapper, W., Ennis, S., Ke, X., and Morton, N.E. 2002. The first linkage disequilibrium (LD) maps: Delineation of hot and cold blocks by diplotype analysis. Proc. Natl. Acad. Sci. 99: 2228-2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mao J., Wang, J., Liu, B., Pan, W., Farr, G.H., Flynn, C., Yuan, H., Takada, S., Kimelman, D., Li, L., et al. 2001. Low-density lipoprotein receptor-related protein-5 binds to axin and regulates the canonical Wnt signaling pathway. Mol. Cell 7: 801-809. [DOI] [PubMed] [Google Scholar]
- 20.Mein C.A., Barratt, B., Dunn, M.G., Siegmund, T., Smith, A.N., Esposito, L., Nutland, S., Stevens, H.E., Wilson, A.J., Phillips, M.S., et al. 2000. Evaluation of single nucleotide polymorphism typing with invader on PCR amplicons and its automation. Genome Res. 10: 330-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merriman T., Twells, R., Merriman, M., Eaves, I., Cox, R., Cucca, F., McKinney, P., Shield, J., Baum, D., Bosi, E., et al. 1997. Evidence by allelic association-dependent methods for a type 1 diabetes polygene (IDDM6) on Chromosome 18q21. Hum. Mol. Genet. 6: 1003-1010. [DOI] [PubMed] [Google Scholar]
- 22.Metzker M.L., Lu, J., and Gibbs, R.A. 1996. Electrophoretically uniform fluorescent dyes for automated DNA sequencing. Science 271: 1420-1422. [DOI] [PubMed] [Google Scholar]
- 23.Nakagawa Y., Kawaguchi, Y., Twells, R.C., Muxworthy, C., Hunter, K.M., Wilson, A., Merriman, M.E., Cox, R.D., Merriman, T., Cucca, F., et al. 1998. Fine mapping of the diabetes-susceptibility locus, IDDM4, on Chromosome 11q13. Am. J. Hum. Genet. 63: 547-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nickerson D.A., Taylor, S., Weiss, K.M., Clark, A.G., Hutchinson, R.G., Stengard, J., Salomaa, V., Vartiainen, E., Boerwinkle, E., and Sing, C.F. 1998. DNA sequence diversity in a 97-kb region of the human lipoprotein lipase gene. Nat. Genet. 19: 233-240. [DOI] [PubMed] [Google Scholar]
- 25.Nickerson D.A., Taylor, S., Fullerton, S.M., Weiss, K.M., Clark, A.G., Stengard, J.H., Salomaa, V., Boerwinkle, E., and Sing, C.F. 2000. Sequence diversity and large-scale typing of SNPs in the human apolipoprotein E gene. Genome Res. 10: 1532-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nusse R. 2001. Making head or tail of Dickkopf. Nature 411: 255-256. [DOI] [PubMed] [Google Scholar]
- 27.Ogura Y., Bonen, D.K., Inohara, N., Nicolae, D.L., Chen, F.F., Ramos, R., Britton, H., Moran, T., Karaliuskas, R., Duerr, R.H., et al. 2001. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature 411: 603-606. [DOI] [PubMed] [Google Scholar]
- 28.Patil N., Berno, A., Hinds, D., Barrett, W.A., Doshi, J.M., Hacker, C.R., Kautzer, C.R., Lee, D.H., Marjoribanks, C., McDonough, D.P., et al. 2001. Blocks of limited haplotype diversity revealed by high-resolution scanning of human Chromosome 21. Science 294: 1719-1723. [DOI] [PubMed] [Google Scholar]
- 29.Pinson K.I., Brennan, J., Monkley, S., Avery, B.J., and Skarnes, W.C. 2000. An LDL-receptor-related protein mediates Wnt signalling in mice. Nature 407: 535-538. [DOI] [PubMed] [Google Scholar]
- 30.Reed P.W., Davies, J., Copeman, J.B., Bennett, S.T., Palmer, S.M., Pritchard, L.E., Gough, S.C., Kawaguchi, Y., Cordell, H.J., Balfour, K.M., et al. 1994. Chromosome-specific microsatellite sets for fluorescence-based, semi-automated genome mapping. Nat. Genet. 7: 390-395. [DOI] [PubMed] [Google Scholar]
- 31.Sachidanandam R., Weissman, D., Schmidt, S.C., Kakol, J.M., Stein, L.D., Marth, G., Sherry, S., Mullikin, J.C., Mortimore, B.J., Willey, D.L., et al. 2001. A map of human genome sequence variation containing 142 million single nucleotide polymorphisms. Nature 409: 928-933. [DOI] [PubMed] [Google Scholar]
- 32.Tamai K., Semenov, M., Kato, Y., Spokony, R., Liu, C., Katsuyama, Y., Hess, F., Saint-Jeannet, J.P., and He, X. 2000. LDL-receptor-related proteins in Wnt signal transduction. Nature 407: 530-535. [DOI] [PubMed] [Google Scholar]
- 33.Templeton A.R., Weiss, K., Nickerson, D.A., Boerwinkle, E., and Sing, C.F. 2000a. Cladistic structure within the human lipoprotein lipase gene and its implications for phenotypic association studies. Genetics 156: 1259-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Templeton A.R., Clark, A.G., Weiss, K.M., Nickerson, D.A., Boerwinkle, E., and Sing, C.F. 2000b. Recombinational and mutational hotspots within the human lipoprotein lipase gene. Am. J. Hum. Genet. 66: 69-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thorstenson Y.R., Shen, P., Tusher, V.G., Wayne, T.L., Davis, R.W., Chu, G., and Oefner, P.J. 2001. Global analysis of ATM polymorphism reveals significant functional constraint. Am. J. Hum. Genet. 69: 396-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tiret L., Poirier, O., Nicaud, V., Barbaux, S., Herrmann, S.M., Perret, C., Raoux, S., Francomme, C., Lebard, G., Tregouet, D., et al. 2002. Heterogeneity of linkage disequilibrium in human genes has implications for association studies of common diseases. Hum. Mol. Genet. 11: 419-429. [DOI] [PubMed] [Google Scholar]
- 37.Twells R.C., Metzker, M., Brown, S.D., Cox, R., Garey, C., Hammond, H., Hey, P.J., Levy, E., Nakagawa, Y., Philips, M.S., et al. 2001. The sequence and gene characterization of a 400-kb candidate region for IDDM4 on Chromosome 11q13. Genomics 72: 231-242. [DOI] [PubMed] [Google Scholar]
- 38.Venter J.C., Adams, M., Myers, E.W., Li, P.W., Mural, R.J., Sutton, G.G., Smith, H.O., Yandell, M., Evans, C.A., Holt, R.A., et al. 2001. The sequence of the human genome. Science 291: 1304-1351. [DOI] [PubMed] [Google Scholar]
- 39.Wehrli M., Dougan, S., Caldwell, K., O'Keefe, L., Schwartz, S., Vaizel-Ohayon, D., Schejter, E., Tomlinson, A., and DiNardo, S. 2000. arrow encodes an LDL-receptor-related protein essential for wingless signalling. Nature 407: 527-530. [DOI] [PubMed] [Google Scholar]
- 40.Zwick M., Cutler, D.J., and Chakravarti, A. 2000. Patterns of genetic variation in Mendelian and complex traits. Annu. Rev. Genom. Hum. Genet. 1: 387-407. [DOI] [PubMed] [Google Scholar]