Abstract
Comparative sequence analyses on a collection of carefully chosen mammalian genomes could facilitate identification of functional elements within the human genome and allow quantification of evolutionary constraint at the single nucleotide level. High-resolution quantification would be informative for determining the distribution of important positions within functional elements and for evaluating the relative importance of nucleotide sites that carry single nucleotide polymorphisms (SNPs). Because the level of resolution in comparative sequence analyses is a direct function of sequence diversity, we propose that the information content of a candidate mammalian genome be defined as the sequence divergence it would add relative to already-sequenced genomes. We show that reliable estimates of genomic sequence divergence can be obtained from small genomic regions. On the basis of a multiple sequence alignment of ∼1.4 megabases each from eight mammals, we generate such estimates for five unsequenced mammals. Estimates of the neutral divergence in these data suggest that a small number of diverse mammalian genomes in addition to human, mouse, and rat would allow single nucleotide resolution in comparative sequence analyses.
[The multiple sequence alignment of the CFTR region and a spreadsheet with the calculations performed, will be available as supplementary information online at www.genome.org.]
Identification and characterization of noncoding functional elements in the human genome is currently an important goal for biomedical research (Pennacchio and Rubin 2001). Estimating functional constraint at the single nucleotide level is equally important, as it would permit evaluation of the importance of sites carrying single nucleotide polymorphisms (SNPs; Miller and Kumar 2001). Comparative sequence analysis, which rests on the principle that functionally important nucleotides are more highly conserved in evolution than nonfunctional ones, has been recognized as a powerful means to achieve these goals. Its basis is found in standard molecular evolutionary theory (Kimura 1983): Mutations that occur in functionally important regions are more likely to be deleterious and are more readily eliminated from the population by purifying selection than mutations in ‘junk DNA.’ Nonfunctional nucleotides therefore accumulate fixed mutations more quickly, and diverge more rapidly during the course of evolution, than do functionally constrained nucleotides (Sumiyama et al. 2001; Sidow 2002).
The effectiveness of comparative sequence analyses for identifying functional elements and for estimating constraint at the single nucleotide level is a function of the diversity of the aligned sequences. The underlying molecular evolutionary process that is leveraged in comparative sequence analyses suggests that the information content of a genome should be proportional to the amount of sequence divergence it contributes to the analysis. Thus, in this context, information content is equivalent to divergence, which can be measured as nucleotide substitution events. Currently, no quantitative framework exists for estimating information content from small amounts of genomic sequence data. We therefore set out to develop quantitative measures of sequence diversity and a framework for assessing the value of a candidate genome relative to already-sequenced genomes. We suggest that these estimates of genome information content are important criteria that should be considered in the selection of genomes to sequence.
The effectiveness of comparative sequence analyses is also a function of the chosen phylogenetic scope. We define the phylogenetic scope as the range of organisms being studied, denoted by their last common ancestor. Its effect on comparative sequence analyses is profound. Consider a study with a vertebrate scope in which fish, amphibian, avian, and mammalian sequences are compared. The long divergence times and high number of substitution events among these organisms permits identification of highly constrained elements with relatively few sequences (Göttgens et al. 2002). However, identification of more quickly evolving elements is hampered by a loss of reliably aligned positions. In addition, the chosen scope allows conclusions only about biology that is shared among the compared species, in that identifiable functional elements must have arisen before their last common ancestor. In a comparison using a vertebrate scope, elements that are specific to smaller clades (such as mammals), which are involved in specifying clade-specific characters (such as mammary glands), will remain unidentified. In contrast, an analysis of only primates, for example, allows alignment of the majority of genomic sequence and the potential for identifying primate-specific elements; low numbers of substitutions, however, require that a very large number of species be used in order to capture enough diversity to effectively identify functionally constrained regions. Given that the resources of the biomedical research community are limited, sensible criteria for the comparative value of candidate genomes for genome sequencing have to be established.
In this case study, we discuss a mammalian phylogenetic scope as an effective range in which to analyze functional elements relevant to human biology. We show that the divergence a mammalian genome would contribute to a comparative sequence analysis can be estimated from relatively small sets of genomic sequence data. We propose such estimates as a quantitative criterion for assessing the relative information content of candidate mammalian genomes. The information content of five genomes is evaluated relative to the genomes of human, mouse, and rat. We give estimates of how much additional divergence is necessary before the resolution of mammalian comparative analyses approaches that of individual nucleotides. On the basis of these estimates, we suggest that single nucleotide resolution within a mammalian scope is achievable with a small number of genomes if they are sufficiently diverse.
RESULTS
Multi-Species Sequence Alignment
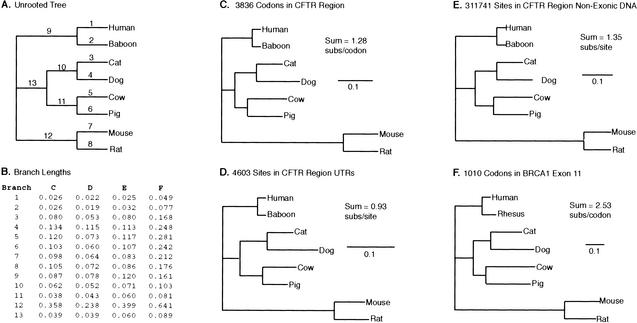
Most of our study was carried out on a comparative data set from nine mammals generated for the genomic segment harboring the Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) gene. This region includes 10 genes that encompass 129 exons as follows: TES, Caveolin 1, Caveolin 2, the cMET proto-oncogene, CAPZA2, ST7, WNT2, GASZ, CFTR, and CORTBP2. In human, the region extends for ∼1.8 Mb. A comprehensive characterization of the data set and of the comparative sequencing effort is to be reported elsewhere (J.W. Thomas, J.W. Touchman, R.W. Blakesley, G.G. Bouffard, S.M. Beckstrom-Sternberg, E.H. Margulies, M. Blanchette, A.C. Siepel, P.J. Thomas, J.C. McDowell, et al., in prep.). To quantitatively estimate divergence, we first generated a global alignment of the entire region for all species using MLAGAN, a new large-scale multiple sequence aligner (http://lagan.stanford.edu). As shown in a recent study that used this same region (Brudno et al. 2003) MLAGAN generates reliable multiple sequence alignments of very large stretches of orthologous genomic DNA. In the CFTR region, MLAGAN has >98% accuracy in aligning protein-coding exons. The alignment is comprised of 3,947,717 positions, representing a 2.1-fold expansion over the human sequence. The precise level of expansion is a function of the alignment method, and is partially explained by independent insertions and deletions in the lineages relating to the sequences. The chimp sequence was subsequently removed from the multiple sequence alignment due to the fact that it was incomplete at the time of analysis and is so closely related to the human that it adds very little divergence to our analysis. This left the following eight mammalian species as the focus of our study, that is, human, baboon, cat, dog, cow, pig, mouse, and rat. The evolutionary relationship of these species is shown as an unrooted tree in Figure 1A.
Figure 1.
The tree relating the organisms of this study, and the number of substitutions per site in each branch of the tree as estimated from our four data sets. (A) The unrooted topology relating eight mammals from which CFTR region sequence data are available. Numbers on branches are identifiers that correspond to the rows in B. (B) Likelihood estimates of the number of substitutions per site or per codon in each of the tree's branches. Note that substitutions per codon represent a cumulative measure of the number of nucleotide substitutions for each codon, consisting of three nucleotide sites. (C–F) The four data sets. Unrooted trees with branch lengths drawn proportional to the number of substitutions per site or per codon as listed in B. Data set and the number of sites or codons is shown at top. (Sum) Total length of the tree. Scale bar, 0.1 substitutions per site or per codon.
Estimates of Divergence in Mammalian Genomes
We first estimated the relative amount of divergence that a genome would contribute to the analysis of functional elements from different classes of sequence data. Specifically, we used data sets from three different classes of genomic elements as follows: protein-coding sequence, UTR, and non-exonic DNA. We tested the hypothesis that the amount of sequence divergence that has occurred along one particular branch of the organismal tree is correlated across these different data sets. For example, if the average rate of evolution of unconstrained DNA is twice the average rate of UTRs, then each branch in the tree estimated from unconstrained DNA should be twice as long as the corresponding branch in the tree estimated from the UTRs.
As the base data for testing this hypothesis, we generated subsets of the CFTR data comprised of all known protein-coding, UTR, or non-exonic sites. We also used an alignment of 1010 codons from BRCA1 from a recent study of mammalian phylogeny (Madsen et al. 2001). No BRCA1 data were available from baboon, so we used rhesus instead. Because baboon and rhesus share the same last common ancestor with human, we do not expect this replacement to substantially affect our results. In each subset of the data, we quantified the divergence among the species by maximum-likelihood estimation on the unrooted mammalian topology (Fig. 1; see Methods section). Sites in which at least one species had a gap were excluded from all analyses to minimize the proportion of sites in which homology is uncertain.
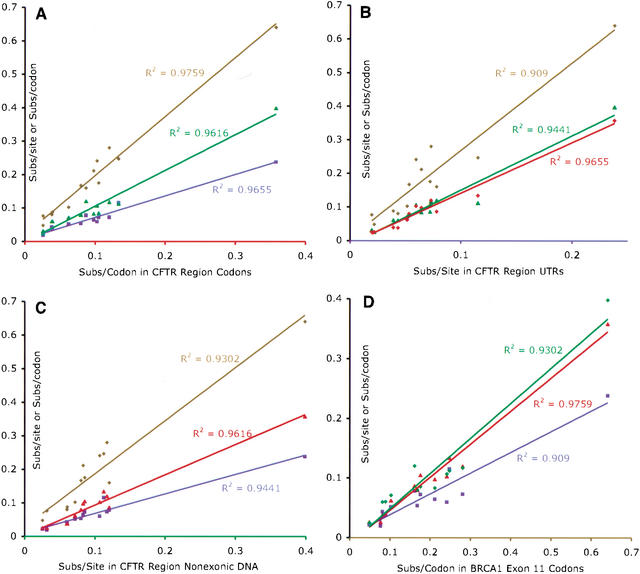
Figure 1B shows the lengths of individual branches, measured as the number of substitutions per site, or, in the case of protein-coding sequence, as the number of substitutions per codon. Figure 1, C–F represent the trees of the four data sets, with the branch lengths drawn to scale. Whereas the expected variation in constraint is reflected in the differences in the absolute branch lengths (compare scale bars), the relative distances among the mammals studied here appear stable (Fig. 1C–F). To quantify this stability, we assigned unique identifiers to each branch (Fig. 1A) and plotted the lengths of the same branches from different trees against each other. We then quantified the correlations between the data sets. The plots and the coefficients of determination (R-squared) between the branch lengths of each pair of trees are shown in Figure 2. All of the trees show a minimum coefficient of 90.1%, with the trees of BRCA1 codons and CFTR region codons corresponding at 97.6%. Thus, different regions of the genome and differently constrained sites have, on average, similar relative rates of evolution in the tree that relates the organisms.
Figure 2.
Correlations of the branch lengths in the tree of one data set with the branch lengths of the trees of the other data sets. Each point corresponds to the comparison of one branch's length in one data set vs. that branch's length in another data set. Data sets are color coded. (Red) CFTR region codons; (Blue) CFTR region UTRs; (Green) CFTR region non-exonic DNA; (Brown) BRCA1 codons. Trendlines and coefficients of determination were generated by regressing the branch lengths of one data set against the branch lengths of the other data sets. (A) CFTR region codons versus all others. (B) CFTR region UTRs vs. all others. (C) CFTR region non-exonic DNA vs. all others. (D) BRCA1 Exon 11 codons vs. all others.
Quantitative Assessment of Relative Genome Information Content
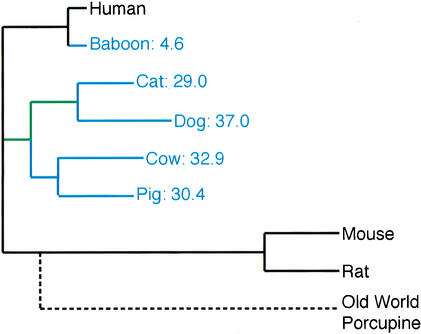
We now calculate the additional divergence each unsequenced mammal (dog, cat, pig, cow, baboon) would contribute if it were added to the tree of human, mouse, and rat. First, for each data set, we calculate the percentage of divergence that any given branch contributes to that data set by dividing the branch length by the sum of all branch lengths of that data set. For the CFTR codons, for example, branch 7 contributes 0.098 substitutions per codon to a total tree length of 1.276 substitutions per codon, which is equivalent to 7.68%. Then, for each branch, we average its normalized length from all four datasets. For example, branch 7 contributes 7.68% (CFTR codons), 6.93% (CFTR UTRs), 6.16% (CFTR non-exonic sites), and 8.39% (BRCA1 exon11), for an average of 7.29%. We then estimate the additional divergence each species would contribute by summing up all the lengths of the branches of its ancestral lineage up to the node connecting it with human, mouse, and rat. For dog, for example, we add the relative lengths of branches 4, 10, and 13, which equals 19.0%. Mouse, human, and rat (the sum of branches 1, 7, 8, 9, and 12; Fig. 1A), capture 51.4%. Thus, expressed as a fraction of what is captured by human, mouse, and rat, the dog would contribute an increase in genomic sequence diversity of 19.0%/51.4% = 37.0% (Fig. 3).
Figure 3.
Tree, drawn to scale, of the number of substitutions per site averaged over all data sets, with sequenced genomes in black and the other species of this study in blue. Estimates of the relative amount of additional substitutions per site that any one of the blue organisms would add are expressed as percentages of what is already present in the sequenced genomes of Human, Mouse, and Rat. Lineages shared by dog and cat since their last common ancestor with other mammals are in green. Old world porcupine is added for illustrating the potential of other mammalian genomes. The length of its branch (broken line) is based on BRCA1 exon 11.
Figure 3 presents these values for each of the mammals examined. The values quantify intuitions regarding relative information value and allow distinctions between species that are not intuitively obvious. For example, although it is clear without quantification that the baboon genome contains much less relative divergence than the other four mammals, we estimate that it provides sixfold less information than the cat genome, and nearly eightfold less information than the dog genome. Furthermore, the analyses suggest that the dog genome provides more divergence than the cat genome, and that the cow genome may provide a slight edge when contrasted with the value of the pig genome. However, the latter should not be overinterpreted, as small differences may disappear when more sequence data are analyzed.
It should be noted that internal branches of the tree are shared among species, as they constitute lineages of common ancestors. Consequently, the first genome to be sequenced from a clade will have a higher relative value than the second one. For example, the cat would contribute 29% additional divergence to the tree of human, mouse, and rat. However, if the dog were added first, then the cat would only contribute an additional 12% divergence due to its shared ancestry with the dog (branches 10 and 13 in Fig. 1A; green branches in Fig. 3).
As an indication that there is much mammalian diversity yet to be sampled, we show a branch connecting the old world porcupine to its most recent common ancestor with rodents (Fig. 3). It is drawn approximately to scale, on the basis of a study of mammalian phylogeny with BRCA1 exon 11 (Madsen et al. 2001). The length of the branch suggests that this species would contribute 70% additional divergence to the tree of human, mouse, and rat. It would therefore provide 15-fold more information than the baboon genome and as much as twofold more information than the dog genome. These are tentative estimates based only on BRCA1 codons from exon 11. For otherwise uncharacterized genomes, it would, of course, be important to obtain additional sequence data from several genes such as those in the CFTR region before embarking on a whole-genome sequencing effort. This will guard against the sampling of outlier genes with a relative rate of evolution that is not representative of the average in that lineage.
Estimates of Substitution Rates in Unconstrained Sites
Now that we are able to assess the relative amount of divergence a species would add to the tree of sequenced genomes, we turn to the question of how much total divergence would be needed to increase the resolution of comparative sequence analyses to the single nucleotide level. Any claim of the existence of evolutionary constraint in a nucleotide site is predicated upon the assumption that mutations could have been fixed if the site were not constrained. Therefore, power and resolution in quantification of constraint are a direct function of the neutral rate of evolution. Consider a site that appears conserved because it does not vary among several aligned sequences. If several neutral substitutions per site are expected, given the sampled sequences, then we infer this site to be constrained. However, if very few neutral substitutions per site are expected, a distinction between constrained and unconstrained sites cannot be made reliably.
We calculate the total amount of neutral evolution captured by our eight species by estimating the average number of fixed mutations that have occurred in presumably unconstrained positions. We used three methods for this purpose. Our first method, which is the traditional approach (Li 1997), is to estimate the amount of synonymous substitution in coding sequence. Using the estimates of the rate of evolution in the codons (Fig. 1) and the maximum likelihood ratios of synonymous to nonsynonymous rates of evolution, we calculated the amount of synonymous divergence in the codons of the CFTR region and in exon 11 of BRCA1. The amount of synonymous divergence is 1.31 substitutions per site in the codons of the CFTR region, and 2.09 substitutions per site in BRCA1 exon11 (Table 1). As there is a known, weak correlation of the rate of synonymous with nonsynonymous evolution (Li 1997), the larger synonymous divergence for exon 11 of BRCA1 is consistent with its large ratio of 0.63 of nonsynonymous to synonymous rates, compared with 0.17 for the codons of the CFTR region.
Table 1.
Estimates of Substitution Rates in Unconstrained Sites
| From synonymous substitutions in coding sequences | |
| 3,836 codons in CFTR Region | 1.31 subs/site |
| 1,010 codons in BRCA1 exon 11 | 2.09 subs/site |
| From extrapolation of pairwise distances in entire CFTR region | |
| 1,410,036 sites in Baboon/Human alignment | 1.50 subs/site |
| 776,169 sites in Cow/Pig alignment | 1.47 subs/site |
| 1,163,402 sites in Mouse/Rat alignment | 1.29 subs/site |
| 823,493 sites in Cat/Dog alignment | 1.32 subs/site |
| From multiple sequence alignment of CFTR region | |
| 311,741 non-exonic sites | 1.35 subs/site |
Our second method exploits the large fraction of unconstrained sites in an alignment between pairs of closely related mammals. We make the assumption that a majority of reliably aligned sites in non-exonic DNA is under minimal constraint. We have four pairs of closely related sequences as follows: human/baboon, cat/dog, cow/pig, and mouse/rat. Corresponding pairwise alignments of non-exonic DNA from the CFTR region yielded 1,410,036, 823,493, 1,163,402, and 776,169 ungapped positions, respectively. We estimate the maximum likelihood distance between each pair. Because we have shown that the relative amount of evolution captured by each branch is stable, we can extrapolate these estimates of pairwise divergence to estimate the neutral divergence over the entire tree. The human/baboon pair, for example, contributes an average of 4.4% to the total divergence in the tree of the eight mammals. The distance between the two species as estimated from the 1,410,036 ungapped non-exonic positions is 0.066 substitutions per site. Extrapolated over the entire tree, this yields 0.066 substitutions per site, divided by 0.044, which equals 1.50 substitutions per site. The distances, in substitutions per site, for the other three pairs are 0.216 (dog, cat), 0.255 (cow, pig), and 0.189 (mouse, rat). The corresponding estimates of the neutral amount of divergence are given in Table 1.
Our third method is to use the alignment of non-exonic DNA positions for all eight mammals. This approach will be biased toward a slightly higher proportion of constrained sites, as many unconstrained sites are not reliably aligned. Accordingly, the number of ungapped positions drops to 311,741. The estimate of the total amount of evolution is 1.35 substitutions per site (Table 1). Thus, the three methods suggest that the neutral amount of evolution captured by these eight species ranges between 1.29 and 2.09 substitutions per site. The estimates from non-exonic DNA will systematically, though not grossly, underestimate the neutral rate due to the presence of sites that are under constraint. Exon 11 of BRCA1 may be slightly unusual because of its high rate of nonsynonymous evolution. It also has a relatively small number of sites. We therefore suggest that a reasonable estimate of the amount of neutral evolution is 1.5 substitutions per site for the eight mammals studied.
Single-Nucleotide Resolution and the Amount of Neutral Evolution
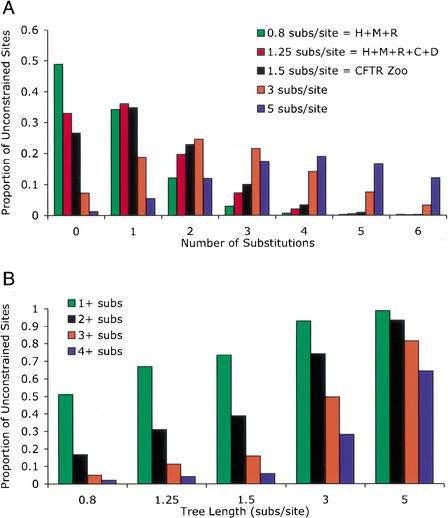
We now turn to the power of comparative analyses as a function of the total amount of neutral divergence captured by the species compared. We seek a confidence level for asserting that a site that appears fully conserved is conserved due to constraint, and not because it is a neutral site that has not been afforded the chance to change. To this end, we estimate the distribution of neutral substitution counts over the sites in a hypothetical alignment, using a Poisson model that accounts for CpG sites (see Methods). For a tree that captures 0.8 neutral substitutions per site, like that of human, mouse, and rat, we estimate that nearly 49% of unconstrained sites that are ungapped appear perfectly conserved (Fig. 4A). Clearly, it is impossible to state with confidence that an individual base is functionally significant merely upon the basis of the fact that it appears conserved in these three species.
Figure 4.
Estimates of the number of substitutions in unconstrained sites under a mixture of Poisson models (see Methods). (A) The proportion of unconstrained sites that have undergone 0, 1, 2, 3, 4, 5, or 6 substitutions when the average number of neutral substitutions over the tree relating the sequences is 0.8 (corresponding to human, mouse, and rat), 1.25 (corresponding to human, mouse, rat, cow, and dog), 1.5 (corresponding to all species of the CFTR region data set studied), and two larger arbitrary values, 3 and 5. The fraction of sites that has undergone 0 substitutions denotes those unconstrained sites that appear fully conserved. In the currently sequenced genomes (0.8 subs/site = H + M + R), this fraction is 49%. (B) the fraction of unconstrained sites that has undergone at least one, two, three, or four substitutions, as a function of the total length of the tree that relates the sequenced organisms.
If we expand this analysis to include more diverse trees, we see appreciable drops in this proportion with a concomitant increase in the proportion of sites that have experienced two, three, or more substitutions. We provide estimates of these expected proportions, both as a function of the number of substitution events expected (Fig. 4A), and as a function of the overall tree length (Fig. 4B). Inclusion of cow and dog, for example, reduces the expected proportion of unconstrained sites that appear perfectly conserved to 33%, and inclusion of all eight mammals to nearly 25%. Note that the presence of mutational hot or cold spots other than CpG sites will increase the expected proportions at the tails of the distribution. Mutational hot spots will tend to absorb more of the mutation events at the expense of cold spots. Thus, in the absence of knowing the identity of hot or cold spots, our general resolution will be lower than what the Poisson model predicts, with the severity of the decline in resolution proportional to the residual variability of mutation rates among sites.
DISCUSSION
We provide evidence that lineage-specific rates of evolution among mammals are highly correlated across different functional classes of sequence data and genomic location. Our estimates of the acceleration or deceleration of rates in rodents and primates, respectively, are broadly consistent with previous studies (Li 1997). In addition, our estimates of the rate of neutral evolution are consistent with previous estimates derived from coding sequences in fewer taxa. For example, the synonymous rate in 16,747 sites of primates, artiodactyls, and rodents is similar to that in the CFTR region codons (Ohta 1995). Furthermore, in a study of over 5000 human–mouse orthologs, the average synonymous rate extrapolates to 1.22 substitutions per site for our tree (Nembaware et al. 2002), which is similar to our estimate of 1.31 for the codons of the CFTR region. We conclude that relatively small samples of sequence data can be used to estimate the relative amount of additional sequence divergence that a candidate genome would provide.
We also consider the resolution afforded by current comparative sequence analyses. Many examples of exciting insights generated by human–mouse pairwise alignments currently exist and are growing (Qiu et al. 2001; Mural et al. 2002). Such analyses are useful for discovering conserved stretches of nucleotides rather than individual positions, and succeed at identification of functional elements. Ideally, however, a set of sequenced mammalian genomes would allow precise estimates of constraint at the level of individual nucleotides. Current sequence data do not have enough resolution to distinguish important sites within a functional element from those that are not constrained but appear conserved. Being able to assert definitively that in any constrained element, positions x, y, and z are highly constrained, positions a, b, and c are under mild constraint, and all others are unconstrained, can provide precise hypotheses as to the distribution of functional base pairs within the element. This single-site level of resolution would also allow estimates of the potential impact of human SNPs in non-coding regions. Poisson modeling (Fig. 4) suggests that three substitutions per site, corresponding to a fourfold increase in sampling depth over current levels (∼0.8 substitutions per site captured by the human, mouse, and rat genomes) would begin to approach the level of single-nucleotide resolution. Ultimately, a tree capturing five neutral substitutions per site should be a goal, because virtually all neutral sites will have experienced at least one substitution, with the majority having undergone at least four substitutions.
Because this level of resolution is obtainable with mammalian genomes, we suggest that a mammalian scope will be an effective range in which to direct sequencing efforts. Mammals share many fundamental organismal features, including the biology of the reproductive systems and the structure of the central nervous system. The utility of mice, rats, and other mammals as experimental models of human biology and disease further attest to the relevance of a mammalian scope to human health and biology. A more narrow scope, such as primates, would provide much less resolution per sequenced genome, whereas broader scopes, such as vertebrates, lack enough similarity to identify many potentially important functional elements.
Our results and other studies of mammalian phylogeny suggest that there is much genome diversity yet to be sampled within the mammalian radiation (Madsen et al. 2001; Murphy et al. 2001a,b); we furthermore suggest that a reasonably small set of mammals may provide a sufficient amount of divergence to achieve single-base resolution. Animals such as tenrec, hedgehog, or fruit bat, which do not fit into the standard mold of experimental model organisms, might add considerable diversity to the current set of genome sequences. Sample datasets such as those of the CFTR region should be generated to estimate the potential value of candidate mammalian genomes.
METHODS
Construction of Data Sets
Sequence data of the CFTR region were generated at the NIH Intramural Sequencing Center. Sequences came from the following nine mammals: human, chimp, baboon, cat, dog, cow, pig, rat, and mouse (http://www.nisc.nih.gov/data/). At the time of analysis, the length of sequences ranged from 1,127,879 bases in pig to 1,877,426 bases in human. Virtually all of the sequence was finished, with the remainder being at a high-quality draft stage. The total alignment length, including only mammals, is 3,947,717 positions.
We divided the CFTR region into three subsets for the purposes of rate estimation as follows: protein coding, UTR, and non-exonic. To generate the protein-coding sequence subset, we parsed human mRNA sequence from each gene in the region, using UCSC gene annotations, into its exons, and translated each of these in frame. The exons were aligned by TBLASTN (WU-BLAST 2.0MP-WashU [09-Sep-2002] [W. Gish, unpubl.; http://blast.wustl.edu]) against the sequence data to generate precise start/stop coordinates of each exon in each genomic sequence. Exons without significant hits in all eight mammalian species examined were eliminated. These are a result of incomplete sequence data at either end of the locus (∼20 exons) or due to short coding sequences for which significant hits are difficult to distinguish (∼10 exons). All sequences were translated and inspected visually to ensure that the correct in-frame sequences had been extracted. Nearly six complete genes from the locus and half of a seventh gene are represented, comprising 3836 codons. We extracted 5′ and 3′ UTR sequence from the global alignment by using coordinates from the June 2002 freeze in the UCSC human genome browser (Kent et al. 2002). This generated a nucleotide alignment of 10,356 positions, yielding 4,603 sites that were ungapped in all sequences. (According to standard practice in phylogenetics, gapped sites were excluded to avoid potential local errors in the alignment.) A data set consisting of all non-exonic elements was similarly generated using the global alignment described below.
Alignments
ClustalW (Thompson et al. 1994) was used to align the concatenated protein sequences, and the alignment was curated manually to eliminate any regions of uncertain homology. To obtain the nucleotide alignment corresponding to this amino acid alignment, the genomic sequence of each exon included in the final alignment was aligned according to the amino acid alignment. To globally align the CFTR region sequences, we used MLAGAN, a large-scale multiple sequence alignment program (Brudno et al. 2003), with default parameters. Repeats and low-complexity elements were masked using RepeatMasker (A.F.A. Smit and P. Green, unpubl.; http://ftp.genome.washington.edu/RM/RepeatMasker.html). Masked sequence is used by MLAGAN in an initial step to prevent potential alignment artifacts due to repetitive elements, but for the final alignment steps, unmasked sequence is used. To create a dataset of non-exonic sequence, all portions of the global alignment corresponding to any human exon, as annotated by UCSC, were excised from the alignment, with the remaining portions concatenated. This generated a global non-exonic alignment of 3,913,385 positions, in which 311,741 positions contained no gap in any sequence. We generated pairwise alignments by extracting each pair of lineage-specific sequences from the non-exonic global alignment. These pairwise alignments yielded 823,493, 776,169, 1,410,036, and 1,163,402 ungapped positions for the carnivores, hoofed mammals, primates, and rodents, respectively.
Branch Length/Rate Estimations and Tree Correlations
We used the PAML software package, v3.13, (Yang 1997) (http://abacus.gene.ucl.ac.uk/software/paml.html), to estimate branch lengths, assuming the unrooted species topology presented in Figure 1A. The codeml procedure was used to estimate synonymous and nonsynonymous rates of substitution in both the protein-coding alignment generated from the CFTR data and the BRCA1 alignment. Model conditions include a single dN/dS ratio for all lineages, estimating codon frequencies from nucleotide frequencies at each of the codon positions, and no molecular clock. Branch lengths for the UTR, global non-exonic, and pairwise non-exonic alignments were estimated by baseml using the HKY85 model (Hasegawa et al. 1985), assuming no molecular clock and no rate variation among sites. Full parameter files used are available upon request. Note that both of these procedures eliminate gapped positions (i.e., alignment positions that have a gap character in any of the sequences) in branch length estimation. To estimate the similarity of the trees generated from each of the data sets, we first assigned unique identifiers to each branch, as demonstrated in Figure 1A. We then performed a simple linear regression of branch lengths versus branch lengths for each pair of trees; the R-squared values for each of these regressions is presented in Figure 2.
Estimation of Neutral Substitution Rates
The ratios of synonymous to nonsynonymous substitution rates as provided by PAML were used to calculate synonymous substitution rates per codon. To convert these rates to synonymous substitutions per site, they were adjusted by the fraction of synonymous sites per codon as estimated by PAML for the BRCA1 and CFTR codon alignments (24.7% and 27.9% synonymous sites, respectively). This was done to translate the codon-based estimates to site-based estimates for comparison with other neutral sites that do not evolve in a codon context.
The total tree length estimated from the non-exonic sequence is provided as substitutions per site.
Finally, we extrapolated the rate estimates from each pairwise alignment to represent the neutral rate over the entire tree. To do so, we simply divide each pairwise distance estimate by the percentage of the total tree length represented by that pair for our average complete tree (Fig. 3).
Poisson Modeling
To estimate the distribution of substitution counts across sites in an alignment with a known amount of neutral divergence, a Poisson model that takes into account CpG mutation rates was used. The substitution process is modeled as a weighted average of two Poisson distributions, one for each population of sites: CpG (assumed to be present at 20% of their expected frequency; Sved and Bird 1990) and non-CpG. The substitution rates (Poisson parameters) in these populations are calculated such that CpG sites mutate at 10 times the rate of non-CpG sites, subject to the restriction that the overall mutation rate of the two combined populations remains identical to the estimate for the entire alignment. Weights were assigned as the proportion of each population of sites within the alignment. Implementation of these models is available in the Supplementary material.
WEB SITE REFERENCES
http://abacus.gene.ucl.ac.uk/software/paml.html; PAML.
http://blast.wustl.edu; WU-BLAST.
http://ftp.genome.washington.edu/RM/RepeatMasker.html; RepeatMasker.
http://genome.ucsc.edu/; UCSC Genome Bioinformatics Home.
http://lagan.stanford.edu; LAGAN Alignment Toolkit Web site.
http://www.nisc.nih.gov/data/; NISC Comparative Sequencing Data Freezes.
Acknowledgments
G.C. is a Howard Hughes Medical Institute Predoctoral Fellow. M.B. is supported by an NSF graduate fellowship. E.D.G. and the NISC Comparative Sequencing Program were supported by funds from the National Human Genome Research Institute. The following individuals were key contributors within the NISC Comparative Sequencing Program: Jim Thomas (BAC isolation and mapping); Jeff Touchman and Bob Blakesley (BAC sequencing); Gerry Bouffard, Steve Beckstrom-Sternberg, Pam Thomas, Jenny McDowell, and Baishali Maskeri (computational analyses). We thank Chuong Do and Michael Kim for help with developing our alignment tools. We also thank Eric Stone for helpful discussion regarding the Poisson modeling, Midori Hosobuchi and Christopher D. Brown for critical reading of the manuscript, and two anonymous reviewers for insightful comments on a previous draft.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
E-MAIL arend@stanford.edu; FAX (650) 725-4905.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1064503.
REFERENCES
- 1.Brudno M., Do, C.B., Cooper, G.M., Kim, M.F., Davydov, E., Sidow, A., Batzoglou, S., and NISC Comparative Sequencing Program 2003. LAGAN and Multi-LAGAN: Efficient tools for large-scale multiple alignment of genomic DNA. Genome Res. 13: 721-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Göttgens B., Barton, L.M., Chapman, M.A., Sinclair, A.M., Knudsen, B., Grafham, D., Gilbert, J.G., Rogers, J., Bentley, D.R., and Green, A.R. 2002. Transcriptional regulation of the stem cell leukemia gene (SCL)—comparative analysis of five vertebrate SCL loci. Genome Res. 12: 749-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hasegawa M., Kishino, H., and Yano, T. 1985. Dating of the human–ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 22: 160-174. [DOI] [PubMed] [Google Scholar]
- 4.Kent W.J., Sugnet, C.W., Furey, T.S., Roskin, K.M., Pringle, T.H., Zahler, A.M., and Haussler, D. 2002. The human genome browser at UCSC. Genome Res. 12: 996-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kimura M., 1983. The neutral theory of molecular evolution. Cambridge University Press, Cambridge, UK.
- 6.Li W.-H., 1997. Molecular evolution. Sinauer Associates, Sunderland, MA.
- 7.Madsen O., Scally, M., Douady, C.J., Kao, D.J., DeBry, R.W., Adkins, R., Amrine, H.M., Stanhope, M.J., de Jong, W.W., and Springer, M.S. 2001. Parallel adaptive radiations in two major clades of placental mammals. Nature 409: 610-614. [DOI] [PubMed] [Google Scholar]
- 8.Miller M.P. and Kumar, S. 2001. Understanding human disease mutations through the use of interspecific genetic variation. Hum. Mol. Genet. 10: 2319-2328. [DOI] [PubMed] [Google Scholar]
- 9.Mural R.J., Adams, M.D., Myers, E.W., Smith, H.O., Miklos, G.L., Wides, R., Halpern, A., Li, P.W., Sutton, G.G., Nadeau, J., et al. 2002. A comparison of whole-genome shotgun-derived mouse chromosome 16 and the human genome. Science 296: 1661-1671. [DOI] [PubMed] [Google Scholar]
- 10.Murphy W.J., Eizirik, E., Johnson, W.E., Zhang, Y.P., Ryder, O.A., and O'Brien, S.J. 2001a. Molecular phylogenetics and the origins of placental mammals. Nature 409: 614-618. [DOI] [PubMed] [Google Scholar]
- 11.Murphy W.J., Eizirik, E., O'Brien, S.J., Madsen, O., Scally, M., Douady, C.J., Teeling, E., Ryder, O.A., Stanhope, M.J., de Jong, W.W., et al. 2001b. Resolution of the early placental mammal radiation using Bayesian phylogenetics. Science 294: 2348-2351. [DOI] [PubMed] [Google Scholar]
- 12.Nembaware V., Crum, K., Kelso, J., and Seoighe, C. 2002. Impact of the presence of paralogs on sequence divergence in a set of mouse–human orthologs. Genome Res. 12: 1370-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohta T. 1995. Synonymous and nonsynonymous substitutions in mammalian genes and the nearly neutral theory. J. Mol. Evol. 40: 56-63. [DOI] [PubMed] [Google Scholar]
- 14.Pennacchio L.A. and Rubin, E.M. 2001. Genomic strategies to identify mammalian regulatory sequences. Nat. Rev. Genet. 2: 100-109. [DOI] [PubMed] [Google Scholar]
- 15.Qiu Y., Cavelier, L., Chiu, S., Yang, X., Rubin, E., and Cheng, J.F. 2001. Human and mouse ABCA1 comparative sequencing and transgenesis studies revealing novel regulatory sequences. Genomics 73: 66-76. [DOI] [PubMed] [Google Scholar]
- 16.Sidow A. 2002. Sequence first. Ask questions later. Cell 111: 13. [DOI] [PubMed] [Google Scholar]
- 17.Sumiyama K., Kim, C.B., and Ruddle, F.H. 2001. An efficient cis-element discovery method using multiple sequence comparisons based on evolutionary relationships. Genomics 71: 260-262. [DOI] [PubMed] [Google Scholar]
- 18.Sved J. and Bird, A. 1990. The expected equilibrium of the CpG dinucleotide in vertebrate genomes under a mutation model. Proc. Natl. Acad. Sci. 87: 4692-4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thompson J.D., Higgins, D.G., and Gibson, T.J. 1994. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang Z. 1997. PAML: A program package for phylogenetic analysis by maximum likelihood. CABIOS 13: 555-556. [DOI] [PubMed] [Google Scholar]