Abstract
We describe a method to select DNA encoding functional open reading frames (ORFs) from noncoding DNA within the context of a specific vector. Phage display has been used as an example, but any system requiring DNA encoding protein fragments, for example, the yeast two-hybrid system, could be used. By cloning DNA fragments upstream of a fusion gene, consisting of the β-lactamase gene flanked by lox recombination sites, which is, in turn, upstream of gene 3 from fd phage, only those clones containing DNA fragments encoding ORFs confer ampicillin resistance and survive. After selection, the β-lactamase gene can be removed by Cre recombinase, leaving a standard phage display vector with ORFs fused to gene 3. This vector has been tested on a plasmid containing tissue transglutaminase. All surviving clones analyzed by sequencing were found to contain ORFs, of which 83% were localized to known genes, and at least 80% produced immunologically detectable polypeptides. Use of a specific anti-tTG monoclonal antibody allowed the identification of clones containing the correct epitope. This approach could be applicable to the efficient selection of random ORFs representing the coding potential of whole organisms, and their subsequent downstream use in a number of different systems.
Only ∼1.5% of the human genome comprises functional ORFs encoded by genes (Lander et al. 2001; Venter et al. 2001). The remaining 98.5% comprises RNA genes, control elements, structural elements, repeat regions, and what has been termed junk DNA. One goal of the human genome project is the identification of all human genes, and consequently the polypeptides encoded by these genes. Attempts to carry this out in silico, using EST and whole-genome sequence information, analyzed with appropriate programs (e.g., Xu and Uberbacher 1997), are having some success; however, true functional analysis of the activities of the products encoded by these genes will always require access to the physical pieces of DNA containing these genes. This has been tackled for Caenorhabditis elegans by a systematic amplification of the open reading frames (ORFs) of all predicted genes (Reboul et al. 2001), with evidence for at least 17,300 genes in this organism, of which a high proportion have structures different to those predicted in silico. The cloning of these ORFs using a recombinatorial system (Hartley et al. 2000) allows easy transfer to different vectors, and a similar strategy has been proposed for the human genome (Brizuela et al. 2001). This provides the potential means to generate complete collections of gene products, if high-throughput methods to consistently produce proteins could be found. Such a complete collection could potentially represent all the polypeptides expressed by an organism, and the interrogation of such a collection could be carried out in a protein chip format (Zhu et al. 2001). However, this approach, apart from the considerable investment required, suffers from the problem that not all proteins can be easily expressed and purified. An alternative method would be to randomly fragment DNA enriched in coding sequences, and to rely on the variable expression of different polypeptides to provide overlapping fragmented representation of individual genes.
Such an approach could be particularly useful in phage display, a technology originally developed to select peptide epitopes recognized by antibodies (Parmley and Smith 1988, 1989; Cwirla et al. 1990; Balass et al. 1993; Smith and Scott 1993; Yayon et al. 1993), but subsequently expanded to include the display of antibodies (Marks et al. 1991; Griffiths and Duncan 1998; Hoogenboom et al. 1998) and many other proteins (for reviews, see Co et al. 1991; Rada et al. 1991; Saggio and Laufer 1993; Clackson and Wells 1994; Soumillion et al. 1994; Bradbury and Cattaneo 1995; Choo and Klug 1995; Perham et al. 1995; Burritt et al. 1996; Cortese et al. 1996; Iba and Kurosawa 1997; Lowman 1997). Although traditional phage display has been successfully applied to gene rich bacterial genomes (Jacobsson and Frykberg 1995, 1996, 1998; Jacobsson et al. 1997) and individual genes (Parmley and Smith 1989; Du Plessis et al. 1995; Petersen et al. 1995; Wang et al. 1995; Bluthner et al. 1996, 1999), to identify antibody epitopes or binding partners, it suffers from the problem that only one clone in 18, if starting with DNA encoding an ORF, will be correctly in frame (one clone in three will start correctly, one clone in three will end correctly, and one clone in two will have the correct orientation), although experiments with synthetic peptide libraries have indicated that stop codons do not necessarily prevent display (Carcamo et al. 1998). Although this high rate of nonfunctional inserts may be tolerable when starting with DNA from a single gene or even a small gene rich genome, in which complete functional representation can be obtained with relatively small libraries, it may become impractical if using more complex DNA sources.
In general, attempts to display random ORFs on filamentous phage, such as those encoded by cDNA fragments, have not been very successful, notwithstanding the development of vectors in which random fragments are displayed at the C terminus of a Jun peptide that interacts with Fos displayed at the N terminus of p3 (Crameri et al. 1994; Crameri and Blaser 1996), at the C terminus of p3 (Fuh and Sidhu 2000), p8 (Fuh et al. 2000), p6 (Jespers et al. 1995), or at the C terminus of an artificial protein that is able to replace p8 in filamentous phage (Weiss and Sidhu 2000), although successful display of a cDNA library was recently reported (Butteroni et al. 2000). Greater success appears to have been achieved with λ-based vectors for cDNA display (Santini et al. 1998; Beghetto et al. 2001). However, even though such C-terminal intracellular vectors increase the likelihood that ORFs will be displayed, they do not per se provide any selective pressure for ORFs.
This indicates the need for a selective step to filter DNA fragments encoding ORFs away from those that do not. Conceptually, the easiest way to do this would be to integrate an antibiotic selection step, in which DNA encoding an ORF permitted read-through into an antibiotic resistance gene, but DNA containing stop codons or frameshifts did not. Seehaus et al. (1992) have described a vector in which antibody genes were cloned upstream of a β-lactamase gene, with the rationale that only those antibody genes that were in frame would be capable of conferring ampicillin resistance by the creation of an antibody–lactamase fusion protein, whereas those that contained deletions or frameshifts would not. The extension of such a concept to the selection of ORFs generally, rather than just antibody genes, would have wide utility.
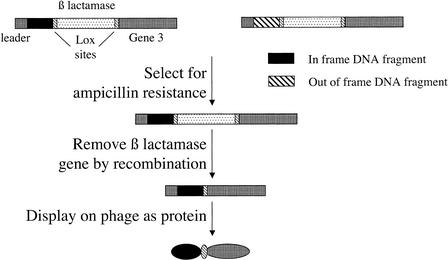
In this paper, we describe a vector that selects for ORFs directly within a vector with a subsequent functional context (phage display; see Fig. 1). This is carried out by cloning random fragments upstream of a β-lactamase gene flanked by two homologous lox sites in frame with gene 3. Only those phage carrying fragments in frame with the β-lactamase are able to confer ampicillin resistance. Once selection for ORFs has occurred, the lactamase gene can be removed by Cre-recombinase-induced recombination, allowing full display of selected fragments (see Fig. 1). We demonstrate the utility of this vector by showing that 100% of fragments randomly cloned into this vector contain ORFs, allowing us to identify epitopes from human tissue transglutaminase, an enzyme involved in protein cross-linking, recognized by a specific monoclonal antibody.
Figure 1.
The scheme for open reading frame selection. The scheme for selecting DNA fragments encoding ORFs. Random fragments are cloned upstream of a β-lactamase gene. Those fragments that are ORFs permit readthrough into the β-lactamase gene and confer ampicillin resistance. Those that are out of frame, or contain stop codons, do not survive. After selection on ampicillin, the β-lactamase gene can be removed by passage through bacteria expressing Cre recombinase. The selected ORF can then be displayed on phage.
RESULTS
Design and Testing of pPAO2
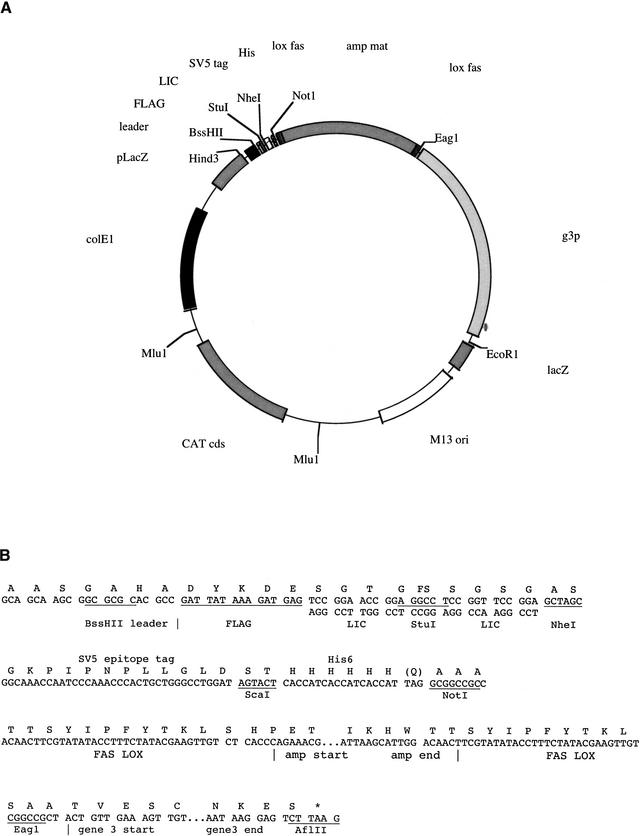
The essential features of pPAO2 are illustrated in Figure 2A. The vector was designed so that blunt-ended fragments could be cloned using either a blunt site (StuI) or the ligation-independent cloning (LIC; Aslanidis and de Jong 1990) strategy, in which no restriction enzymes are used, thus avoiding potential bias. Briefly, the vector contains an StuI site surrounded by two 12-nt-long palindromic sequences lacking dTMP. After cutting with StuI, the two blunt ends generated are degraded by the 3′ → 5′ exonuclease activity of T4 DNA polymerase in the presence of T4 DNA polymerase and dTTP. As a result of the sequence design, the exonuclease stops at nucleotide 13, which is dTMP, thus creating the short cohesive ends required for LIC adaptor mediated cloning. These cloning sites are upstream of a β-lactamase gene flanked by two lox recombination signals. In addition, the vector also carries chloramphenicol resistance in the backbone and contains tags for two commonly used monoclonals, FLAG (Hopp et al. 1988) and SV5 (Hanke et al. 1992), as well as a His6 tag for purification by immobilized metal affinity chromatography. The polylinker is out of frame with respect to the β-lactamase gene (indicated by FS in Fig. 2B), and can only be returned into frame if DNA containing an ORF with 3n + 2 nt is correctly cloned.
Figure 2.
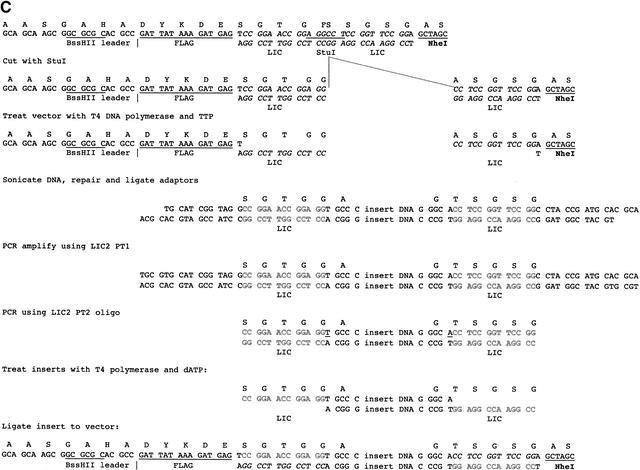
pPAO2, plasmid map, and polylinker sequence. (A) The plasmid map of pPAO2. (B) The polylinker sequence between the HindIII and EcoRI sites of pUC119. The amino acid sequence of the in-frame construct is given above the DNA sequence. FS indicates a frameshift, which can only be overcome if an in-frame DNA fragment is cloned via the LIC approach. (C) The LIC cloning procedure in detail.
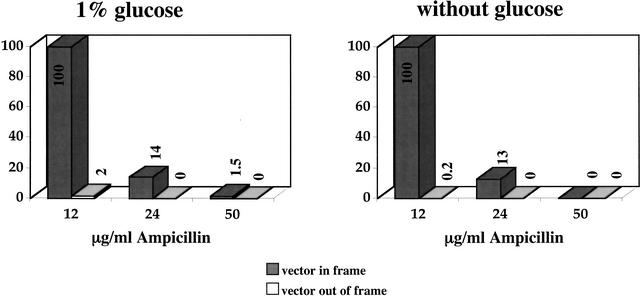
To examine the efficiency of the selection for ORFs, DNA encoding either a single-chain antibody fragment (D1.3), or an out-of-frame derivative, were cloned between the BssHII and NheI sites. As shown in Figure 3, by using an ampicillin concentration of 12 μg/mL in the absence of glucose, 100% of the in-frame clones survive, whereas only 0.2% of the out-of-frame clones survive. Increasing the concentration of ampicillin to 25 μg/mL reduced the percentage of in-frame clones surviving by 85%, and eliminated all out-of-frame clones, whereas the addition of glucose (which should inhibit transcription from the lac promoter) allowed more out-of-frame clones to survive at the lowest ampicillin concentration tested. On the basis of these results, 12 μg/mL ampicillin was used for all subsequent experiments.
Figure 3.
Only clones with open reading frames survive on ampicillin. The D1.3 scFv, or an out-of-frame derivative, were cloned into pPAO2, and bacteria were plated on different concentrations of chloramphenicol or ampicillin with or without 1% glucose. Chloramphenicol resistance is encoded by the backbone of the plasmid, whereas ampicillin resistance is generated by the presence of in-frame fusions (see Fig. 2). Clone survival on ampicillin is expressed as a percentage of the number of clones growing on chloramphenicol.
After selection on ampicillin, bacteria were harvested and infectious phagemids were prepared (Dente et al. 1983; Sambrook et al. 1989), this being a far more efficient method than transfection for DNA transfer between bacteria. The efficiency of the recombination-mediated removal of the β-lactamase gene was tested by infecting these phagemids into BS1365, an F‘ bacteria constitutively expressing Cre recombinase, and allowing recombination to occur overnight at 30°C. Phagemids were prepared from these bacteria, reinfected into DH5αF‘, and plated out onto chloramphenicol (24 μg/mL) or ampicillin (12 μg/mL) plates. The number of colonies growing on ampicillin plates was always <1% of those growing on chloramphenicol. To further confirm these results, 100 colonies were picked from the chloramphenicol plate and replated on either ampicillin (12 μg/mL) or chloramphenicol (24 μg/mL) plates. All colonies grew on the chloramphenicol plates, and none grew on the ampicillin plates, indicating that the β-lactamase gene had been efficiently removed by Cre recombinase. This was also confirmed by PCR, which showed the removal of the β-lactamase gene in the 20 clones tested (data not shown).
Although the ampicillin gene was removed by recombination, the lox recombination signal remains as a translated “linker” between the displayed protein and p3. Although this has not been a problem when present between VH and VL in scFvs (Sblattero and Bradbury 2000), three different D1.3 scFv phagemids were tested to see whether display efficiency was affected: pDAN5-D1.3 (a standard phage antibody vector; Sblattero and Bradbury 2000) and pPAO2-D1.3 before or after the removal of β-lactamase by recombination. The ability of the phage to bind to lysozyme (the antigen recognized by D1.3) was tested by ELISA. As can be seen in Table 1, the ELISA signals given by pDAN5-D1.3 and pPAO2-D1.3 after recombination were similar, indicating that the translated lox linker used did not inadvertently affect display. As expected, however, the presence of β-lactamase between D1.3 and p3 had a severe effect on display efficiency.
Table 1.
Display Efficiency With and Without β-Lactamase
| Construct | ELISA signals | |
| Lysozyme | BSA | |
| pDAN5 D1.3 | 0.934 ± 0.120 | 0.090 ± 0.021 |
| pPAO1 D1.3 before cre | 0.126 ± 0.013 | 0.086 ± 0.007 |
| pPAO1 D1.3 after cre | 0.878 ± 0.080 | 0.102 ± 0.038 |
ELISA signals generated for phage containing the D1.3 scFv cloned into a standard phage display vector (pDAN5), pPAO2 with β-lactamase still present (before cre), and after β-lactamase has been removed.
pPAO2 Can Select ORFs
To evaluate the efficiency of pPAO2 in filtering out ORFs from random DNA, a library of fragments from the tTG-encoding plasmid pET28b-tTG (Sblattero et al. 2000) was prepared. This plasmid can be considered to be a minigenome, containing four known genes accounting for ∼50% of the DNA, and containing an additional 62 ORFs >50 amino acids in length. The library was made by digesting the plasmid with DNAse to fragments of 100–300 bp, corresponding to 33–100 amino acids, repairing them with Pfu DNA polymerase, and ligating to LIC adaptors. After two amplification steps, the 12-nt single-stranded overhangs complementary to the pPAO2 cohesive ends were created using T4 DNA polymerase and dATP. The scheme for this cloning method is shown in Figure 2C. The library size range was similar to that of the majority of exons (Lander et al. 2001), which have been proposed to represent structural units (de Souza et al. 1996) within proteins, and should also exclude many nonfunctional ORFs. A small aliquot of the library was plated on chloramphenicol and ampicillin plates. The number of colonies obtained on the chloramphenicol plates was ∼80-fold greater than the number of colonies obtained on the ampicillin plates (see Table 2), indicating that strong selection had occurred. The library obtained after plating on ampicillin plates contained 7000 colonies. The β-lactamase gene from these was removed by infecting phagemids made from these colonies into BS1365 (which expresses Cre recombinase constitutively), and reinfecting phagemids produced by these bacteria into DH5αF‘.
Table 2.
Selection Efficiency of the Filter Vector
| Number of colonies | Ratio chloramphenicol/ampicillin | |
| Chloramphenicol (24 μg/mL) | Ampicillin (12 μg/mL) | |
| 32,000 | 400 | 80:1 |
The number of clones before (chloramphenicol) and after ampicillin preselection is given.
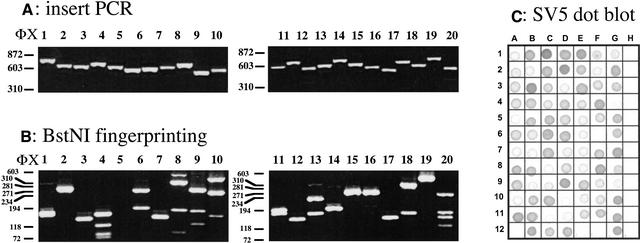
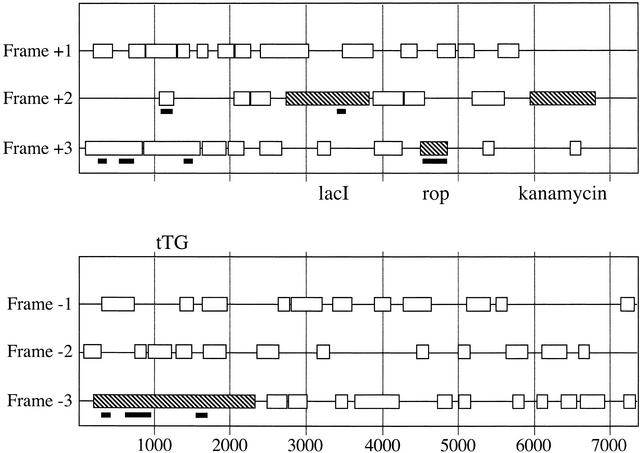
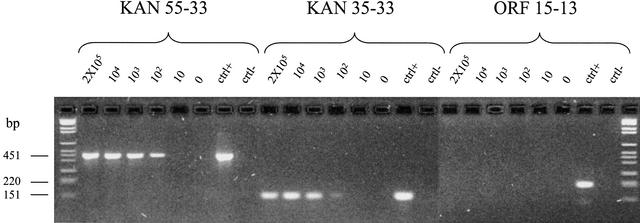
The characteristics of the resultant library were examined by amplifying several different inserts from randomly picked clones (see Fig. 4A) and treating them with BstNI to examine the diversity by fingerprinting (Fig. 4B). As can be seen, inserts are all of different sizes and show different digestion patterns, indicating that no single clone dominates. Then 96 colonies were plated out on a chloramphenicol IPTG plate and examined in a dot blot for expression of the SV5 tag (Fig. 4C). This is found between the cloned insert and p3 and will only be in frame if the cloned insert is in frame (see Fig. 2B). As can be seen in Figure 4C, at least 80 colonies showed SV5 binding (different signals are attributable to differences in leakage of the fusion protein out of the bacteria), indicating that a large proportion of the library now consists of ORFs. This was confirmed by sequencing 43 random colonies (Table 3), all of which were found to be in frame. The ORFs from which the clones were derived is indicated in the table, and represented graphically in Figure 5, in which all ORFs >50 amino acids (150 bp) are shown, and those that were found in the random sequencing are indicated by the thick black lines. Of the four functional ORFs in pET28b-tTG, three (lacI, tTG, and rop) were represented and comprised 84% of all the clones present. The kanamycin-resistance gene was not found among the 43 sequenced clones. To find out whether this ORF was found in the tTG ORF library, three nested primers amplifying fragments of 451 or 154 bp were designed. As shown in Figure 6, both the 451 and the 154 fragments were found when at least 100 templates were present in the PCR reaction, whereas a randomly picked ORF of no biological significance could not be detected in the whole library.
Figure 4.
Ampicillin selection generates randomly sized open reading frames. (A) 20 random clones taken from a plasmid-generated fragment library, after selection on ampicillin and passage through a Cre-expressing strain to remove β-lactamase, were amplified with primers spanning the cloning site. All fragments are different sizes, indicating that different random fragments have been cloned. (B) Clones from A were digested with BstNI. All clones show different fragment sizes. (C) The supernatants from 96 random clones from the same library were spotted onto nitrocellulose after induction of protein fragment expression with IPTG. As can be seen, >80% of these clones have signals indicating that an open reading frame has been selected.
Table 3.
Analysis of Selected ORF Clones: Random Sequenced Clones
| Clone | Number | Origin | Start | End | ORF | Length |
| A2 | 1 | Escherichia coli | — | — | 161 | |
| G3 | 1 | pET | 2693 | 2870 | 2389–3024 | 178 |
| F9 | 1 | pET | 3531 | 3591 | LacI (2741–3856) | 61 |
| C5 | 2 | pET | 4528 | 4837 | Rop (4497–4856) | 310 |
| C3 | 1 | pET | 4594 | 4768 | rop | 175 |
| B6 | 1 | pET | 4603 | 4784 | rop | 187 |
| B3 | 2 | pET | 4618 | 4780 | rop | 163 |
| C9 | 2 | pET | 4627 | 4789 | rop | 163 |
| C11 | 1 | pET | 4630 | 4762 | rop | 133 |
| G1 | 1 | pET | 4630 | 4765 | rop | 136 |
| E12 | 1 | pET | 4630 | 4768 | rop | 139 |
| D3 | 1 | pET | 4630 | 4783 | rop | 154 |
| G11 | 2 | pET | 4630 | 4837 | rop | 208 |
| B4 | 2 | pET | 4630 | 4849 | rop | 220 |
| D1 | 1 | pET | 4636 | 4855 | rop | 220 |
| H1 | 1 | pET | 4636 | 4837 | rop | 202 |
| F3 | 1 | pET | 4639 | 4780 | rop | 142 |
| A1 | 1 | pET | 4651 | 4765 | rop | 115 |
| A3 | 1 | pET | 4551 | 4768 | rop | 118 |
| A7 | 5 | pET | 4752 | 4804 | rop | 154 |
| H8 | 1 | pET | 4654 | 4804 | rop | 151 |
| D12 | 1 | pET | 4669 | 4789 | rop | 121 |
| E6 | 1 | tTG | 1600 | 1483 | tTG (2330–198) | 118 |
| A5 | 1 | tTG | 928 | 745 | tTG | 184 |
| E10 | 1 | tTG | 874 | 733 | tTG | 142 |
| B3 | 2 | tTG | 724 | 612 | tTG | 113 |
| D4 | 1 | tTG | 385 | 265 | tTG | 121 |
| B2 | 1 | tTG- | 1089 | 1266 | 1067–1264 | 178 |
| D8 | 1 | tTG- | 1525 | 1600 | 855–1613 | 76 |
| F2 | 1 | tTG- | 1039 | 1222 | 877–1302 | 184 |
| B11 | 2 | tTG- | 568 | 730 | 7221–854 | 163 |
| D2 | 1 | tTG- | 262 | 379 | 7221–854 | 118 |
Detailed analysis of the 43 different sequenced clones, giving their lengths, the open reading frames from which they are derived, and the number of times they were isolated.
Figure 5.
Sequence analysis of ampicillin-selected open reading frame clones. The pET28b-tTG plasmid is represented linearly, with all ORFs >50 amino acids (150 bp) indicated. In addition, ORFs corresponding to kanamycin, tTG, lacI, and rop are hatched. Regions of the plasmid identified after sequencing 43 clones obtained after selection on ampicillin and removal of the β-lactamase after passage through a Cre-expressing strain are shown as black lines beneath the appropriate frame.
Figure 6.
Identification of kanamycin clones. Three kanamycin-specific primers were used in two separate PCR reactions (see Methods for details) with different numbers of estimated clones from the library indicated. Amplification is noted when 100 templates are present, indicating that the kanamycin sequence is present at an approximate level of 1/100. The 15–13 primers amplify an ORF of no biological significance that is not represented in the library. The bars on the left indicate the length of the amplification products.
Selecting and Screening Using the pPAO2 Library
To determine whether this approach to selecting ORFs provides libraries of ORFs suitable for subsequent selection or screening experiments, the library was selected on a commercial monoclonal antibody, CUB7402, known to recognize a linear epitope (860–768 in the plasmid) in human tissue transglutaminase. After a single round of selection, 17 positive clones were identified of which 15 were identical and overlapped the known epitope, whereas the remaining two clones, which had much weaker signals, corresponded to irrelevant sequences. As an alternative to selection, 96 randomly picked clones were tested by dot blot for binding to CUB7402, and two further clones were found to be positive, also corresponding to the known epitope (Table 4).
Table 4.
Analysis of Selected ORF Clones: Clones Selected or Positively Screened on mAb CUB (Epitope 860–768)
| Clone | Number | Origin | Start | End | ORF | Length |
| D9a | 15 | tTG | 982 | 739 | tTG | 244 |
| E4b | 1 | tTG | 991 | 766 | tTG | 226 |
| F6b | 1 | tTG | 883 | 700 | tTG | 184 |
| D12 | 1 | tTG | 412 | 262 | tTG | 151 |
| A4 | 1 | pET | 4651 | 4804 | rop | 154 |
Selected.
Positively screened.
Details of clones recognized by CUB, an anti-tTG monoclonal recognizing an epitope encoded by DNA 860–768 (reverse and complemented), are given.
DISCUSSION
Large-scale functional analysis of the gene products in individual genomes is a problem that requires access to DNA fragments encoding open reading frames cloned in appropriate vectors. Although this can be carried out in a directed fashion, using specific primers for each gene, and, indeed, projects are underway to do this (Brizuela et al. 2001; Reboul et al. 2001; see below), a random approach involving the creation of numerous random ORF fragments from each gene is an alternative strategy. Which strategy is better depends on the problem under study. Full-length gene products are required for full study of gene function, but the contributions of (sub)domains to enzymatic, binding or immunological function is usually better served by smaller fragments generated either randomly or by design, which are easier to express and purify. This is particularly true of the study of protein immunogenicity, which, with the use of synthetic or randomly encoded peptides, can be made independently of the original gene sequence, with the identification of antigenic epitopes by sequence comparison.
Phage peptide libraries have proved to be a very successful method to generate such random peptides, and have been extensively used to define linear antigenic epitopes recognized by both monoclonal (Parmley and Smith 1988, 1989; Cwirla et al. 1990; Balass et al. 1993; Smith and Scott 1993; Yayon et al. 1993) and polyclonal antibodies (Cortese et al. 1994, 1995, 1996). However, such selections often yield peptide sequences that cannot be found within the sequence of the antigen recognized by the antibody. Although such “mimotopes,” as they have been termed, can be correlated to discontinuous epitopes on the antigenic surface if the structure of the antigen is known (Luzzago et al. 1993), in most cases the structure is not known, and no useful information can be derived. Fragment libraries, as described above, have the advantage that if the fragments are big enough, and span the appropriate region, conformational epitopes can also be identified. In fact, when general synthetic peptide libraries have been compared with gene-specific libraries, epitopes were more easily and accurately identified using the gene fragment approach for a number of different antibodies (Fack et al. 1997; Matthews et al. 2002). However, this approach has two problems: First, the vast majority of clones are nonfunctional when making gene-specific display libraries because of the problem of correct frame; and second, new libraries need to be made for each experiment.
The use of the selection vector described here could overcome both of these problems. By ensuring that libraries contain only ORFs, functionality is enhanced: 100% of sequenced clones after ampicillin selection contained ORFs, and at least 80% of these were expressed in a dot blot. One concern with the approach described here is the potential for bias, and in fact, a large proportion of the randomly sequenced clones were derived from one ORF—rop. Whether this is because rop is particularly soluble, and so enhances lactamase expression, or whether there was earlier bias in the creation (e.g., from the known bias of DNAse [Herrera and Chaires 1994] or PCR), or manipulation (the two rounds of phagemid infection required to generate) of the library is unclear, and presently under investigation. The use of infection, rather than transfection, to propagate and select the library allows the sampling of far greater clone numbers because infection is far more efficient than transfection; however, it may itself lead to unintended bias. The fact that 100% of the sequenced clones contained ORFs indicates that the ampicillin concentration used for selection could perhaps be reduced, permitting the survival of a greater diversity of clones. However, it is significant, and interesting, that all the products of functional genes are represented in the small library we created, and that the vast majority of randomly sequenced clones corresponded to real gene products, as opposed to products of random nonbiological ORFs, indicating that in addition to selecting for ORFs, this method also has a bias for biologically significant ORFs. Why this is the case is not presently clear. It could be that portions of natural ORFs encode polypeptides that are more soluble than those derived from random ORFs, permitting the functional expression of antibiotic resistance. An alternative explanation relates to the overrepresentation of rop in the library. However, even if rop-encoding clones were completely eliminated from the analysis, 50% of clones were still derived from natural ORFs, as compared with the 19% expected by chance (∼3.8 kB of natural ORFs divided by ∼20 kB of all ORFs < 50 bp in the plasmid used).
The use of this vector will allow the creation of far smaller libraries when targeting specific genes, and also allow the consideration of nontargeted libraries, which would encompass the coding potential of whole individual genomes. Such nonfocused libraries could use cDNA, genomic DNA, or best of all, collections of PCR-amplified ORFs corresponding to all known genes. The advantage of cDNA as a source is that ORFs are fully contiguous, and alternative splicing forms can be accessed. However, this is offset by the problem of overrepresentation of some messages, and under- or nonrepresentation of other messages, depending on the source of mRNA used to make the cDNA. Although these latter two problems may be overcome by using genomic DNA, this raises another problem: that of noncontiguous messages, which will eliminate epitopes spanning splice junctions. With the advent of facile cloning methods (Liu et al. 1998; Hartley et al. 2000), projects have been initiated to create libraries of all ORFs from different species (e.g., human and C. elegans; Brizuela et al. 2001; Reboul et al. 2001). Once completed, these will provide sources for random fragments that would overcome problems related to both cDNA and genomic DNA. Each ORF (and its alternative transcripts) would be present only once, and the cloning of random fragments from such collections into the selection vectors described here would provide a genome-wide resource of true protein fragments suitable for a number of different uses, including antibody specificity, enzyme activity, arrays (Holt et al. 2000a,b; MacBeath and Schreiber 2000), or protein interaction studies without the need to identify and clone individual well-expressing domains.
We have demonstrated the utility of this approach with filamentous phage display vectors and β-lactamase. However, a similar approach could be applied to expression systems, yeast two-hybrid vectors (Fields and Song 1989), protein complementation assays (Michnick 2001), or any other vector system requiring protein fragments. This is likely to complement the effort to analyze full-length ORFs, because not all ORFs are easily expressed within a particular context of interest, and this approach may also provide information on the roles of different domains within proteins, as has been recently shown for the F pilus (Harris et al. 1999). Although we have used ampicillin, the use of alternative antibiotic genes (e.g., chloramphenicol) would allow the selection of fragments stable in the intracelullar, as opposed to the periplasmic, environment. In fact, it is likely that different antibiotics will select different fragments, with combinations of antibiotic resistances providing the greatest coverage. Although we have used recombination to remove the selecting gene within the context of a functional vector, lox-based recombination, using appropriate pairs of sites (Siegel et al. 2001), could also be used to move selected ORFs from a selection vector to a recipient vector, in a fashion similar to that used for λ recombinase-based cloning systems (Hartley et al. 2000), but without the problems related to translation of the different-sized recombination signals.
Interestingly, the size of the library required to cover the coding potential of the human genome when starting from a collection of cloned ORFs would be relatively small (<108 clones assuming 100,000 human genes, with each gene represented by 1000 different clones) when compared with the sizes of phage antibody libraries that have been made by standard cloning methods (109–10; Vaughan et al. 1996; Sheets et al. 1998). The application of this technology to fragmented pathogenic organisms will permit the selection of antigenic epitopes recognized by patient sera, as has already been carried out using random peptide libraries (Cortese et al. 1994, 1995, 1996), with the possibility of identifying highly antigenic regions suitable as protein or DNA vaccines.
METHODS
Bacterial Strains
The bacterial strains used in this study were Dh5αF‘ (GIBCO BRL): F‘/endA1 hsd17 (rK− mK+) supE44 thi-1 recA1 gyrA (Nalr) relA1 Δ (lacZYA-argF)U169 deoR (F80dlacD[lacZ]M15); and BS1365: BS591 F‘ Kan (BS591: recA1 endA1 gyrA96 thi-1 D lacU169 supE44 hsdR17 [lambda1mm434 nin5 X1-cre] (Sauer and Henderson 1988).
Plasmid Construction
The phagemid pPAO2 (see Fig. 2) is a derivative of pDAN5 (Sblattero and Bradbury 2000) specifically modified to exploit the ligation-independent cloning method (Aslanidis and de Jong 1990). A new polylinker sequence was generated by PCR using primers LIC2 Vector For (5′-TTGCCGCTAGCTCCGG AACCGGAGGCCTCCGGTTCCGGACTCATCTTTATAA TCGGCATGCGCGCCGCTTGCTGC-3′) and M13 rev seq (5′-AGCGGATAACAATTTCACACA-3′) and pDAN5 as template. The PCR product was cloned into pDAN5 as a HindIII–NheI fragment.
Library Construction
For library construction, 15 μg of pET28-htTG (Sblattero et al. 2000) was digested into random fragments by adding 0.1 U of DNAse I in the presence of 50 mM Tris-HCl, 10 mM MnCl2 (pH 7.5) at 15°C for 1–2 min, and repaired by the addition of 5 U of Pfu DNA polymerase. Blunt-ended fragments of 100–300 bp were purified by electrophoresis in a 2% agarose gel and recovered from the gel using the Qiaquick Gel Extraction kit (QIAGEN). These DNA fragments were ligated to LIC linkers (oligo sense, 5′-TGCATCGGTAGGCCGGAACCGGAGGTG CCC-3′; oligo antisense, 5′-GGGCACCTCCGGTTCCGGC CTACCGATGCACGCA-3′) in a reaction mixture containing LIC Adaptors (20 μM), DNA fragments, 1× T4 DNA ligase buffer, and T4 DNA ligase at 15°C overnight. Unligated adaptors were removed using a 1-mL Sephacryl S-400 HR spin column (Pharmacia) as recommended by the supplier. The fragments with ligated adaptors were PCR-amplified using primers LIC2PT1 (5′-TGCGTGCATCGGTAG-3′) and LIC2PT2 (5′CCGGAACCGGAGG3′) before cloning. To create the single-stranded LIC tails in the plasmid, StuI-digested vector and adaptor-ligated inserts were treated with 2 U of T4 DNA polymerase in the presence of dTTP (0.5 mM) and dATP (0.5 mM), respectively. After incubation at 37°C for 20 min, the mixtures were heat-inactivated and purified using the Qiaquick PCR Purification kit (QIAGEN). For the large-scale ligation reaction, 15 μg of T4 DNA polymerase-treated vector was combined with 3 μg of T4 DNA polymerase-treated PCR products in a 100-μL volume. After incubation at room temperature for 1 h, the ligation mixture was extracted with an equal volume 50:50 phenol:chloroform followed by ethanol precipitation, and the resulting DNA pellet was resuspended in 20 μL of water. Each microliter of ligation reaction was then used to transform 50 μL of electrocompetent DH12S cells.
ELISA Analysis and Dot Blot
Phage ELISA (Marks et al. 1992) was used to identify lysozyme-binding D1.3 single-chain antibody fragments (scFvs) present in either pDAN5 or pPAO2 with or without β-lactamase.
The dot immunobinding assay was performed by spotting 1-μL aliquots from the supernatant of Escherichia coli colonies (see below) induced overnight with 1 mM IPTG onto a nitrocellulose (NC) sheet. The NC sheet was sequentially incubated with the SV5 tag mAb (Hanke et al. 1992) and a goat anti mouse Ig antiserum conjugated with alkaline phosphatase and revealed by the chromogenic substrate BCIP (5-bromo-4-chloro-3-indolyl-phosphate) and NBT (nitro blue tetrazolium).
PCR Analysis of the Ampicillin-Resistant Transformants and DNA Sequencing
To characterize the transformants from different experiments, randomly picked recombinants were analyzed for inserts by PCR with primers M13 reverse, 5′-CAG GAA ACA GCT ACC-3′ (5′-end-specific), and AMP, 5′-TCGATGTAACCCACTCGTGC 3′ (3′-end-specific). Bacterial colonies were transferred into the PCR mixture by touching the colony using disposable pipette tips and pipetting up and down in the PCR mixture. Aliquots were analyzed by agarose gel electrophoresis. For BstNI fingerprinting, the PCR products were digested with BstNI and the different patterns of the resulting fragments were resolved on a 2% Metaphore agarose gel. DNA sequencing was carried out using Epicentre Sequiterm ExcelII Kits (Alsbyte) and analyzed using specific labeled M13 reverse and AMP-specific primers. Sequences were analyzed on a Li-Cor 4000L automatic sequencer.
To analyze the library for the presence of the kanamycin gene, three primers (kan 55, TGTATGGGAAGCCCGATG; kan 35, GCGATCGCGTATTTCGTC; and kan 33, GACTGAATCCG GTGAGAATG) were synthesised and used in a PCR with the library as template. Using kan 55 and kan 33 would be expected to give a band of 451 bp, whereas kan 35 and kan 33 would give a band of 154 bp, if the kan gene was present in the library. In addition, two other primers (orf13, CACCGG CATACTCTG; and orf15′, CATGCACCATTCCTT) corresponding to the largest ORF (2389–3024) were synthesized. These were expected to give a fragment of 220 bp (2596–2816).
β-lactamase Removal by Recombination
To induce the removal of the β-lactamase gene following selection on ampicillin plates, phagemids were prepared from pooled selected clones, as previously described (Sblattero and Bradbury 2000), and infected into BS1365 (bacteria constitutively expressing Cre) grown in 2xTY, 100 mg/mL kanamycin, 1% glucose at 37°C to OD550 = 0.5. Recombination was allowed to proceed by shaking at 30°C overnight. The following day, bacteria were diluted 1/20 in the same medium, grown to OD550 = 0.5 at 37°C, and infected with M13K07 helper phage at a multiplicity of infection of 20:1, and left without shaking at 37°C for 30 min before 2 h of further growth. Colonies were derived from these phagemids by infection into DH5αF‘. These represent preselected ORFs, displayed on phagemids, in which β-lactamase has been removed.
pPAO2 Library Selection
pPAO2 phagemids were selected on CUB7402, a commercial anti-tTG antibody (Neomarker). Rescue of phagemid particles was as described in Marks et al. (1991). Panning was performed by adding phages diluted in 2% nonfat milk in PBS (MPBS) to immunotubes (Nunc) coated with CUB7402 (10 μg/mL), washing 20 times with PBS, 0.1% Tween20 (PBST), and 20 times with PBS, followed by elution with 1 mL of E. coli cells at OD600 = 0.5 at 37°C for 30 min and overnight growth after addition of ampicillin, helper phage, and kanamycin. The panning procedure was repeated up to three times. After selection, 96 individual clones from each selection were screened for reactivity to the antibody used for selection by microtiter-plate ELISA.
Acknowledgments
This work was funded by a DOE grant, DE-FG02–98ER62647 awarded to A.R.M.B. We would like to thank Brian Sauer for the gift of BS1365.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
E-MAIL amb@lanl.gov; FAX (505) 667-2891.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.861503.
REFERENCES
- 1.Aslanidis C. and de Jong, P. 1990. Ligation-independent cloning of PCR products (LIC-PCR). Nucleic Acids Res. 18: 6069-6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balass M., Heldman, Y., Cabilly, S., Givol, D., Katchalski-Katzir, E., and Fuchs, S. 1993. Identification of a hexapeptide that mimics a conformation-dependent binding site of acetylcholine receptor by use of a phage-epitope library. Proc. Natl. Acad. Sci. 90: 10638-10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beghetto E., Pucci, A., Minenkova, O., Spadoni, A., Bruno, L., Buffolano, W., Soldati, D., Felici, F., and Gargano, N. 2001. Identification of a human immunodominant B-cell epitope within the GRA1 antigen of Toxoplasma gondii by phage display of cDNA libraries. Int. J. Parasitol. 31: 1659-1668. [DOI] [PubMed] [Google Scholar]
- 4.Bluthner M., Bautz, E.K., and Bautz, F.A. 1996. Mapping of epitopes recognized by PM/Scl autoantibodies with gene-fragment phage display libraries. J. Immunol. Methods 198: 187-198. [DOI] [PubMed] [Google Scholar]
- 5.Bluthner M., Schafer, C., Schneider, C., and Bautz, F.A. 1999. Identification of major linear epitopes on the sp100 nuclear PBC autoantigen by the gene-fragment phage-display technology. Autoimmunity 29: 33-42. [DOI] [PubMed] [Google Scholar]
- 6.Bradbury A. and Cattaneo, A. 1995. The use of phage display in neurobiology. Trends Neurosci. 18: 243-249. [PubMed] [Google Scholar]
- 7.Brizuela L., Braun, P., and LaBaer, J. 2001. FLEXGene repository: From sequenced genomes to gene repositories for high-throughput functional biology and proteomics. Mol. Biochem. Parasitol. 118: 155-165. [DOI] [PubMed] [Google Scholar]
- 8.Burritt J.B., Bond, C.W., Doss, K.W., and Jesaitis, A.J. 1996. Filamentous phage display of oligopeptide libraries. Anal. Biochem. 238: 1-13. [DOI] [PubMed] [Google Scholar]
- 9.Butteroni C., De Felici, M., Scholer, H.R., and Pesce, M. 2000. Phage display screening reveals an association between germline-specific transcription factor Oct-4 and multiple cellular proteins. J. Mol. Biol. 304: 529-540. [DOI] [PubMed] [Google Scholar]
- 10.Carcamo J., Ravera, M.W., Brissette, R., Dedova, O., Beasley, J.R., Alam-Moghe, A., Wan, C., Blume, A., and Mandecki, W. 1998. Unexpected frameshifts from gene to expressed protein in a phage-displayed peptide library. Proc. Natl. Acad. Sci. 95: 11146-11151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choo Y. and Klug, A. 1995. Designing DNA-binding proteins on the surface of filamentous phage. Curr. Opin. Biotechnol. 6: 431-436. [DOI] [PubMed] [Google Scholar]
- 12.Clackson T. and Wells, J.A. 1994. In vitro selection from protein and peptide libraries. TIBTECH 12: 173-184. [DOI] [PubMed] [Google Scholar]
- 13.Co M.S., Deschamps, M., Whitley, R.J., and Queen, C. 1991. Humanized antibodies for antiviral therapy. Proc. Natl. Acad. Sci. 88: 2869-2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cortese R., Felici, F., Galfre, G., Luzzago, A., Monaci, P., and Nicosia, A. 1994. Epitope discovery using peptide libraries displayed on phage. TIBTECH 12: 262-267. [DOI] [PubMed] [Google Scholar]
- 15.Cortese R., Monaci, P., Nicosia, A., Luzzago, A., Felici, F., Galfre, G., Pessi, A., Tramontano, A., and Sollazzo, M. 1995. Identification of biologically active peptides using random libraries displayed on phage. Curr. Opin. Biotechnol. 6: 73-80. [DOI] [PubMed] [Google Scholar]
- 16.Cortese R., Monaci, P., Luzzago, A., Santini, C., Bartoli, F., Cortese, I., Fortugno, P., Galfre, G., Nicosia, A., and Felici, F. 1996. Selection of biologically active peptides by phage display of random peptide libraries. Curr. Opin. Biotechnol. 7: 616-621. [DOI] [PubMed] [Google Scholar]
- 17.Crameri R. and Blaser, K. 1996. Cloning Aspergillus fumigatus allergens by the pJuFo filamentous phage display system. Int. Arch. Allergy Immunol. 110: 41-45. [DOI] [PubMed] [Google Scholar]
- 18.Crameri R., Jaussi, R., Menz, G., and Blaser, K. 1994. Display of expression products of cDNA libraries on phage surfaces. A versatile screening system for selective isolation of genes by specific gene-product/ligand interaction. Eur. J. Biochem. 226: 53-58. [DOI] [PubMed] [Google Scholar]
- 19.Cwirla S.E., Peters, E.A., Barrett, R.W., and Dower, W.J. 1990. Peptides on phage: A vast library of peptides for identifying ligands. Proc. Natl. Acad. Sci. 87: 6378-6382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dente L., Cesareni, G., and Cortese, R. 1983. pEMBL: A new family of single stranded plasmids. Nucleic Acids Res. 11: 1645-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Souza S.J., Long, M., Schoenbach, L., Roy, S.W., and Gilbert, W. 1996. Intron positions correlate with module boundaries in ancient proteins. Proc. Natl. Acad. Sci. 93: 14632-14636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Du Plessis D.H., Romito, M., and Jordaan, F. 1995. Identification of an antigenic peptide specific for bluetongue virus using phage display expression of NS1 sequences. Immunotechnology 1: 221-230. [DOI] [PubMed] [Google Scholar]
- 23.Fack F., Hügle-Dörr, B., Song, D., Queitsch, I., Petersen, G., and Bautz, E.K. 1997. Epitope mapping by phage display: Random versus gene-fragment libraries. J. Immunol. Methods 206: 43-52. [DOI] [PubMed] [Google Scholar]
- 24.Fields S. and Song, O. 1989. A novel genetic system to detect protein–protein interactions. Nature 340: 245-246. [DOI] [PubMed] [Google Scholar]
- 25.Fuh G. and Sidhu, S.S. 2000. Efficient phage display of polypeptides fused to the carboxy-terminus of the M13 gene-3 minor coat protein. FEBS Lett. 480: 231-234. [DOI] [PubMed] [Google Scholar]
- 26.Fuh G., Pisabarro, M.T., Li, Y., Quan, C., Lasky, L.A., and Sidhu, S.S. 2000. Analysis of PDZ domain-ligand interactions using carboxyl-terminal phage display. J. Biol. Chem. 275: 21486-21491. [DOI] [PubMed] [Google Scholar]
- 27.Griffiths A.D. and Duncan, A.R. 1998. Strategies for selection of antibodies by phage display. Curr. Opin. Biotechnol. 9: 102-108. [DOI] [PubMed] [Google Scholar]
- 28.Hanke T., Szawlowski, P., and Randall, R.E. 1992. Construction of solid matrix-antibody–antigen complexes containing simian immunodeficiency virus p27 using tag-specific monoclonal antibody and tag-linked antigen. J. Gen. Virol. 73: 653-660. [DOI] [PubMed] [Google Scholar]
- 29.Harris R.L., Sholl, K.A., Conrad, M.N., Dresser, M.E., and Silverman, P.M. 1999. Interaction between the F plasmid TraA (F-pilin) and TraQ proteins. Mol. Microbiol. 34: 780-791. [DOI] [PubMed] [Google Scholar]
- 30.Hartley J.L., Temple, G.F., and Brasch, M.A. 2000. DNA cloning using in vitro site-specific recombination. Genome Res. 10: 1788-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herrera J.E. and Chaires, J.B. 1994. Characterization of preferred deoxyribonuclease I cleavage sites. J. Mol. Biol. 236: 405-411. [DOI] [PubMed] [Google Scholar]
- 32.Holt L.J., Bussow, K., Walter, G., and Tomlinson, I.M. 2000a. By-passing selection: Direct screening for antibody–antigen interactions using protein arrays. Nucleic Acids Res. 28: E72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holt L.J., Enever, C., de Wildt, R.M., and Tomlinson, I.M. 2000b. The use of recombinant antibodies in proteomics. Curr. Opin. Biotechnol. 11: 445-449. [DOI] [PubMed] [Google Scholar]
- 34.Hoogenboom H.R., de Bruine, A.P., Hufton, S.E., Hoet, R.M., Arends, J.W., and Roovers, R.C. 1998. Antibody phage display technology and its applications. Immunotechnology 4: 1-20. [DOI] [PubMed] [Google Scholar]
- 35.Hopp T., Prickett, K., Price, V., Libby, R., March, C., Cerretti, D., Urdal, D., and Conlon, P. 1988. A short polypeptide marker sequence useful for recombinant protein identification and purification. BioTech. 6: 1204-1210. [Google Scholar]
- 36.Iba Y. and Kurosawa, Y. 1997. Comparison of strategies for the construction of libraries of artificial antibodies. Immunol. Cell Biol. 75: 217-221. [DOI] [PubMed] [Google Scholar]
- 37.Jacobsson K. and Frykberg, L. 1995. Cloning of ligand-binding domains of bacterial receptors by phage display. Biotechniques 18: 878-885. [PubMed] [Google Scholar]
- 38.___, 1996. Phage display shot-gun cloning of ligand-binding domains of prokaryotic receptors approaches 100% correct clones. Biotechniques 20: 1070-1081. [DOI] [PubMed] [Google Scholar]
- 39.___, 1998. Gene VIII-based, phage-display vectors for selection against complex mixtures of ligands. Biotechniques 24: 294-301. [DOI] [PubMed] [Google Scholar]
- 40.Jacobsson K., Jonsson, H., Lindmark, H., Guss, B., Lindberg, M., and Frykberg, L. 1997. Shot-gun phage display mapping of two streptococcal cell-surface proteins. Microbiol. Res. 152: 121-128. [DOI] [PubMed] [Google Scholar]
- 41.Jespers L.S., Messens, J.H., De Keyser, A., Eeckhout, D., Van Den Brande, I., Gansemans, Y.G., Lauwereys, M.J., Vlasuk, G.P., and Stanssens, P.E. 1995. Surface expression and ligand-based selection of cDNAs fused to filamentous phage gene VI. Biotechnology (NY) 13: 378-382. [DOI] [PubMed] [Google Scholar]
- 42.Lander E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860-921. [DOI] [PubMed] [Google Scholar]
- 43.Liu Q., Li, M.Z., Leibham, D., Cortez, D., and Elledge, S.J. 1998. The univector plasmid-fusion system, a method for rapid construction of recombinant DNA without restriction enzymes. Curr. Biol. 8: 1300-1309. [DOI] [PubMed] [Google Scholar]
- 44.Lowman H.B. 1997. Bacteriophage display and discovery of peptide leads for drug development. Annu. Rev. Biophys. Biomol. Struct. 26: 401-424. [DOI] [PubMed] [Google Scholar]
- 45.Luzzago A., Felici, F., Tramontano, A., Pessi, A., and Cortese, R. 1993. Mimicking of discontinuous epitopes by phage-displayed peptides, I. Epitope mapping of human H ferritin using a phage library of constrained peptides. Gene 128: 51-56. [DOI] [PubMed] [Google Scholar]
- 46.MacBeath G. and Schreiber, S.L. 2000. Printing proteins as microarrays for high-throughput function determination. Science 289: 1760-1763. [DOI] [PubMed] [Google Scholar]
- 47.Marks J.D., Hoogenboom, H.R., Bonnert, T.P., McCafferty, J., Griffiths, A.D., and Winter, G. 1991. By-passing immunization. Human antibodies from V-gene libraries displayed on phage. J. Mol. Biol. 222: 581-597. [DOI] [PubMed] [Google Scholar]
- 48.Marks J.D., Griffiths, A.D., Malmqvist, M., Clackson, T., Bye, J.M., and Winter, G. 1992. By-passing immunization: Building high affinity human antibodies by chain shuffling. BioTechnology 10: 779-783. [DOI] [PubMed] [Google Scholar]
- 49.Matthews L.J., Davis, R., and Smith, G.P. 2002. Immunogenically fit subunit vaccine components via epitope discovery from natural peptide libraries. J. Immunol. 169: 837-846. [DOI] [PubMed] [Google Scholar]
- 50.Michnick S.W. 2001. Exploring protein interactions by interaction-induced folding of proteins from complementary peptide fragments. Curr. Opin. Struct. Biol. 11: 472-477. [DOI] [PubMed] [Google Scholar]
- 51.Parmley S.F. and Smith, G.P. 1988. Antibody-selectable filamentous fd phage vectors: Affinity purification of target genes. Gene 73: 305-318. [DOI] [PubMed] [Google Scholar]
- 52.___, 1989. Filamentous fusion phage cloning vectors for the study of epitopes and design of vaccines. Adv. Exp. Med. Biol. 251: 215-218. [DOI] [PubMed] [Google Scholar]
- 53.Perham R.N., Terry, T.D., Willis, A.E., Greenwood, J., di Marzo Veronese, F., and Appella, E. 1995. Engineering a peptide epitope display system on filamentous bacteriophage. FEMS Microbiol. Rev. 17: 25-31. [DOI] [PubMed] [Google Scholar]
- 54.Petersen G., Song, D., Hugle-Dorr, B., Oldenburg, I., and Bautz, E.K. 1995. Mapping of linear epitopes recognized by monoclonal antibodies with gene-fragment phage display libraries. Mol. Gen. Genet. 249: 425-431. [DOI] [PubMed] [Google Scholar]
- 55.Rada C., Gupta, S.K., Gherardi, E., and Milstein, C. 1991. Mutation and selection during the secondary response to 2-phenyloxazolone. Proc. Natl. Acad. Sci. 88: 5508-5512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reboul J., Vaglio, P., Tzellas, N., Thierry-Mieg, N., Moore, T., Jackson, C., Shin-i, T., Kohara, Y., Thierry-Mieg, D., Thierry-Mieg, J., et al. 2001. Open-reading-frame sequence tags (OSTs) support the existence of at least 17,300 genes in C. elegans. Nat. Genet. 27: 332-336. [DOI] [PubMed] [Google Scholar]
- 57.Saggio I. and Laufer, R. 1993. Biotin binders selected from a random peptide library expressed on phage. Biochem. J. 293: 613-616.. Erratum 295: 903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sambrook J., Fritsch, E.F., and Maniatis, T., 1989. Molecular cloning: A laboratory manual Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 59.Santini C., Brennan, D., Mennuni, C., Hoess, R.H., Nicosia, A., Cortese, R., and Luzzago, A. 1998. Efficient display of an HCV cDNA expression library as C-terminal fusion to the capsid protein D of bacteriophage λ. J. Mol. Biol. 282: 125-135. [DOI] [PubMed] [Google Scholar]
- 60.Sauer B. and Henderson, N. 1988. The cyclization of linear DNA in Escherichia coli by site-specific recombination. Gene 70: 331-341. [DOI] [PubMed] [Google Scholar]
- 61.Sblattero D. and Bradbury, A. 2000. Exploiting recombination in single bacteria to make large phage antibody libraries. Nat. Biotechnol. 18: 75-80. [DOI] [PubMed] [Google Scholar]
- 62.Sblattero D., Berti, I., Trevisiol, C., Marzari, R., Tommasini, A., Bradbury, A., Fasano, A., Ventura, A., and Not, T. 2000. Human recombinant tissue transglutaminase ELISA: An innovative diagnostic assay for celiac disease. Am. J. Gastroenterol. 95: 1253-1257. [DOI] [PubMed] [Google Scholar]
- 63.Seehaus T., Breitling, F., Dubel, S., Klewinghaus, I., and Little, M. 1992. A vector for the removal of deletion mutants from antibody libraries. Gene 114: 235-237. [DOI] [PubMed] [Google Scholar]
- 64.Sheets M.D., Amersdorfer, P., Finnern, R., Sargent, P., Lindqvist, E., Schier, R., Hemmingsen, G., Wong, C., Gerhart, J.C., and Marks, J.D. 1998. Efficient construction of a large nonimmune phage antibody library; the production of panels of high affinity human single-chain antibodies to protein antigens. Proc. Natl. Acad. Sci. 95: 6157-6162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Siegel R.W., Jain, R., and Bradbury, A. 2001. Using an in vivo phagemid system to identify non-compatible loxP sequences. FEBS Lett. 505: 467-473. [DOI] [PubMed] [Google Scholar]
- 66.Smith G.P. and Scott, J.K. 1993. Libraries of peptides and proteins displayed on filamentous phage. Methods Enzymol. 217: 228-257. [DOI] [PubMed] [Google Scholar]
- 67.Soumillion P., Jespers, L., Bouchet, M., Marchand-Brynaert, J., Sartiaux, P., and Fastrez, J. 1994. Phage display of enzymes and in vitro selection for catalytic activity. Appl. Biochem. Biotechnol. 47: 175-190. [DOI] [PubMed] [Google Scholar]
- 68.Vaughan T.J., Williams, A.J., Pritchard, K., Osbourn, J.K., Pope, A.R., Earnshaw, J.C., McCafferty, J., Hodits, R.A., Wilton, J., and Johnson, K.S. 1996. Human antibodies with sub-nanomolar affinities isolated from a large non-immunised phage display library. Nat. Biotechnol. 14: 309-314. [DOI] [PubMed] [Google Scholar]
- 69.Venter J.C., Adams, M.D., Myers, E.W., Li, P.W., Mural, R.J., Sutton, G.G., Smith, H.O., Yandell, M., Evans, C.A., Holt, R.A., et al. 2001. The sequence of the human genome. Science 291: 1304-1351. [DOI] [PubMed] [Google Scholar]
- 70.Wang L.F., Du Plessis, D.H., White, J.R., Hyatt, A.D., and Eaton, B.T. 1995. Use of a gene-targeted phage display random epitope library to map an antigenic determinant on the bluetongue virus outer capsid protein VP5. J. Immunol. Methods 178: 1-12. [DOI] [PubMed] [Google Scholar]
- 71.Weiss G.A. and Sidhu, S.S. 2000. Design and evolution of artificial M13 coat proteins. J. Mol. Biol. 300: 213-219. [DOI] [PubMed] [Google Scholar]
- 72.Xu Y. and Uberbacher, E.C. 1997. Automated gene identification in large-scale genomic sequences. J. Comput. Biol. 4: 325-338. [DOI] [PubMed] [Google Scholar]
- 73.Yayon A., Aviezer, D., Safran, M., Gross, J.L., Heldman, Y., Cabilly, S., Givol, D., and Kathcalski-Katzir, E. 1993. Isolation of peptides that inhibit binding of basic fibroblast growth factor to its receptor from a random phage-epitope library. Proc. Natl. Acad. Sci. 90: 10643-10647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhu H., Bilgin, M., Bangham, R., Hall, D., Casamayor, A., Bertone, P., Lan, N., Jansen, R., Bidlingmaier, S., Houfek, T., et al. 2001. Global analysis of protein activities using proteome chips. Science 293: 2101-2105. [DOI] [PubMed] [Google Scholar]