Abstract
Abnormal expression and function of chromatin regulators results in the altered chromatin structure seen in cancer. The chromatin regulator CTCF, its cofactor CHD8, and antagonistic paralogue BORIS have wide-ranging effects on gene regulation. Their concurrent expression and regulation was examined in benign, localized, and metastatic prostate cancer (PCa) arrays with extended follow-up using an automated quantitative imaging system, VECTRA. Epithelial staining was quantified and compared against a range of clinicopathologic variables. CHD8 expression was decreased in HGPIN, localized, and metastatic PCa compared to benign (P < .001). CHD8 promoter hypermethylation, assessed by Quantitative Pyrosequencing, occurred in over 45% of primary cancers in this population as well as the TGCA database. Treatment of cell lines with the demethylating agent 5-Aza-2′-deoxycytidine reinduced expression. An interesting dichotomy for CHD8 was observed within primary cancers, with higher nuclear protein expression associated with adverse clinical outcomes including extracapsular extension (P = .007), presence of metastases (P = .025) and worse PSA-recurrence free survival (P = .048). CHD8 outperformed Gleason score and predicted biochemical failure within intermediate grade prostate cancers. The BORIS/CTCF expression ratio increased in localized (P = .03) and metastatic PCa (P = .006) and was associated with higher Gleason score (P = .02), increased tumor volume (P = .02) and positive margins (P = .04). Per cell heterogeneity of expression revealed all protein expression to be more heterogeneous in cancerous tissue (both P < .001), especially high grade (P < .01). In the first detailed analysis in cancer, a marked loss of CHD8 expression and increased BORIS/CTCF ratio indicate frequent disruption of CTCF and its effector genes in PCa.
Abbreviations: 5-azadC, 5-Aza-2′deoxycytidine; BORIS, Brother of the regulator of imprinted sites/CTCFL; CTCF, CCCTC-binding factor; CHD8, Chromdomain helicase DNA-binding factor 8; HGPIN, High-grade prostatic intraepithelial neoplasia; Prostate cancer, PCa; PSA, Prostate specific antigen; SI, Simpson's Index; TSS, Transcription start site
Introduction
Prostate cancer (PCa) development is associated with epigenetic changes seen in both aging normal and cancerous tissues [1]. The factors that direct these changes remain elusive. Polycomb-group and other proteins play a role in regulating genes through their modification of chromatin structure. Recent data suggests a critical role of these proteins, notably EZH2, in the malignant prostate phenotype [2]. Three interrelated factors associated with the regulation of epigenetic marks include Chromodomain helicase DNA-binding protein 8 (CHD8), CCCTC-binding factor (CTCF), and Brother of the regulator of imprinted sites (BORIS). CHD8 and CTCF complex at CTCF binding sites and regulate gene expression through chromatin insulation, DNA methylation, and histone acetylation [3]. Conversely, BORIS antagonizes CTCF function by competing at CTCF binding sites [4], [5]. Given the critical role of these chromatin-regulating genes, the co-expression of these proteins in PCa development and progression was investigated utilizing a unique quantitative, per-cell expression analysis.
CTCF is an 11-zinc finger protein with multifaceted functions. In addition to acting as a classical transcription factor, its presence regulates chromatin structure and contributes to epigenetic homeostasis through the formation of “boundary elements” between hetero- and euchromatin [6]. With over 20,000 binding sites in the genome its regulatory action is complex and depends on the specific DNA sequence and interacting factors at CTCF binding sites [7]. CTCF loss of function epigenetically alters numerous cancer-associated genes. In various cancers lack of CTCF activity is associated with epigenetic repression of hTERT, pRb, p16INK4A, p14ARF, and p53 [7]. As a chromatin insulator, CTCF is known to have enhancer-blocking activity as demonstrated in the imprinted Igf2-H19 imprint control region [8]. Its function is opposed by its paralogue BORIS, also known as CTCFL, that has extensive homology to the CTCF DNA-binding motif [9]. While CTCF is known to protect and maintain DNA methylation marks, BORIS expression coincides with the loss of CpG methylation [4], [10], [11]. Their antagonistic function is seen at the MAGE A1 promoter, where CTCF acts as a transcriptional repressor and BORIS leads to gene activation [5]. BORIS may function as an oncogene and recent reports suggest its reactivation occurs in a variety of cancers, including the prostate [12].
The chromatin insulator function of CTCF is dependent on CHD8, an ATP-dependent chromatin remodeling enzyme [3], [13]. CHD8 co-localizes and interacts with CTCF at several gene insulator sites including the Igf2-H19 differentially methylated region (DMR), B-globin 5′HS5 insulator, and the c-myc and BRCA1 gene promoters. The presence of both factors is required for normal genetic and epigenetic regulation [3]. CHD8 is a target in gastric and colorectal cancers [14]. The CHD8-CTCF complex prevents the spread of transcriptionally inactive heterochromatin and a loss of CHD8 results in DNA hypermethylation and histone hypoacetylation near CTCF binding sites [3]. Functional studies of CHD8 have shown dichotomous roles with regard to cell cycle activity. The presence of CHD8 negatively regulates β-catenin signaling, suppresses p53-dependence [15], [16], and negatively regulates HOXA2 gene expression [17]. Conversely, CHD8 cooperates with androgen receptor to activate TMPRSS2 and is implicated in E2F-dependent gene transcription [18], [19]. The literature suggests a complex, and cryptic, role for CHD8 where losses and gains of function could have oncogenic-like gene regulation properties.
To analyze expression synchronously the VECTRA imaging system was employed, a quantitative tool which allows the automated selection and analysis of expression signals within cells, cellular subsets, and compartments. This study sought to investigate the compartmental coexpression of these proteins and their clinical significance in PCa. These analyses reveal significant decreases in CHD8, in part due to hypermethylation, and increases in BORIS-CTCF ratio in PCa development. However, a dichotomy of higher CHD8 expression is associated with adverse features, including increased risk of PSA recurrence. Using this powerful imaging tool, we find the CTCF regulatory pathway is frequently altered in PCa which may help to explain common changes in CTCF effector genes in cancer.
Methods
Tissue Microarray
The University of Wisconsin Institutional Review Board (IRB) provides ethical insight to clinical projects and reviews all human research protocols in accordance with federal regulations, state laws, and local and University policies. FFPE-patient tissues used in this study were from the archive of the Department of Pathology and Laboratory Medicine, University of Wisconsin-Madison. A tissue microarray was constructed consisting of 288 duplicate cores from prostate tissues of different disease groups: 48 benign prostate tissues (BPT) (from normal, non-adjacent tissue without evidence of disease), 50 high grade intraepithelial neoplasia (HGPIN) (tissue from HGPIN tissue blocks of cancer patients in this cohort), 84 localized PCa (pT2), 62 aggressive PCa (pT3), and 44 metastatic lesions (brain, lung, bone, omentum, testis, colon, bladder, and lymph nodes).
Staining and Image Analysis
Slide preparation and antigen retrieval were conducted as previously described [20]. Briefly, the slides were taken through routine deparaffinization and rehydration. Two triple stains (CTCF, CHD8, and E-cadherin; CTCF, BORIS, and E-cadherin) were performed from two TMA sections with antibodies against CTCF (sc-5916; Santa Cruz Biotech, Santa Cruz, CA), CHD8 (NB100-60418; Novus Biologicals, Littleton, CO), and BORIS (sc-98982 Santa Cruz Biotech, Santa Cruz, CA). E-cadherin antibodies (Cell Signaling Technology, Beverly, MA) were used to define the epithelial compartment for better tissue segmentation.
Stained slides were loaded onto the slide scanner. Slides were scanned as previously described [20]. Cores with < 5% epithelial component or loss of tissue were excluded from the analysis. Per-cell protein target signals were quantitated for individual cores using the VECTRA imaging system according to manufacturer's protocols (Caliper Life Sciences, Hopkinton, MA). The inForm 1.2 software was used to segment tissue subcellular compartments (nucleus vs. cytoplasm) and tissue compartments (epithelium vs. stroma).
Methylation Analysis of CHD8 in Human Prostate Tissues
We obtained 11 paired flash frozen samples of tumor and benign adjacent tissue from radical prostatectomy samples using an approved IRB protocol. DNA was generated and Quantitative Pyrosequencing was employed as we have previously described [21], to assess methylation across 2 CpG island regions previously suggested to be altered on methylation arrays [22]. These regions were: i) 6 CpGs spanning the transcription start site (TSS) of CHD8 [Chr14:21,907,003-21,906,863] and ii) 7 CpGs encompassing a CpG island 600 bp upstream of the transcription start site [Chr14:21,907,850-21,907,725]. Primers are available upon request.
Methylation Validation in The Cancer Genome Atlas Samples
To verify the patterns of methylation observed in cancer samples data was downloaded and analyzed form The Cancer Genome Atlas (TCGA) Project (https://tcga-data.nci.nih.gov/tcga/). The dataset contained methylation information for 49 solid prostate tissue normal (benign) and 336 prostate adenocarcinoma samples analyzed by the Illumina HumanMethylation450k Array. This dataset included information on the 6 CpGs analyzed at the CHD8 TSS, the 7 CpGs 600 bp upstream were not analyzed by the 450 k Array.
5-Aza-2′-Deoxycytidine Treatment and Methylation Analyses in Cell Lines
A panel of cells was screened for low expression of CHD8. Two prostate cancer cell lines (DU145 and LNCaP) and Hela were treated with increasing doses of 5-Aza-2′-deoxycytidine (0 to 100 μM) for 48 hours, RNA was isolated and Quantitative PCR was performed as previously described [21]. Briefly, CHD8 expression was analyzed by quantitative PCR using a CFX96 real-time PCR detection system (Bio-Rad) and SYBR Green PCR master mix (Applied Biosystems). Primers used as previously described [23]. Results were statistically compared using the t-test. DNA was generated, bisulfite treated, and Quantitative Pyrosequencing performed analyzing the same regions as the human prostate tissues above.
Statistical Analysis
Nuclear, cytoplasmic, and total expression of individual cores of various prostate tissues (benign, HGPIN, PCa, metastatic PCa) was statistically compared using Kruskal-Wallis test followed by Wilcoxon rank-sum tests. Pearson's r correlation analysis was used to quantify the relationship between CHD8/CTCF and BORIS/CTCF expressions. To compare protein expression in patient cancer samples with different clinicopathologic features (Gleason, pT stage, tumor volume, margins, SV involvement, extracapsular extension, and evidence of metastasis), Kruskal-Wallis test or Wilcoxon rank-sum test was used as appropriate, only primary tumor samples were used in this analysis. Duplicate PCa cores obtained from the same patient (73 cases) were averaged to provide a more precise estimation of protein expression in each biological replicate. Protein expression from 6 patients without clinical data was excluded from clinicopathologic analysis. Thus, 67 primary cancer patients in total were included in the clinicopathologic analysis after quality controls (Patient demographics, Table S1).
The spread and shape of per cell protein expressions for various tissues were characterized by calculating the coefficient of variation, skewness, and kurtosis. Further, these values were compared among varying pathological grades using the same non-parametric tests as described above. The Simpson's Diversity Index (Gini-Simpson transformation) [24] was used to analyze heterogeneity as outlined by Faratian et al. [25]. Briefly, per cell expression data of all samples was used to derive bins of equal percentages. Binning continuous expression values yield a score for each cell. Each cell from each core was then applied to the Gini-Simpson Index of diversity defined as:
Where n is the score of each binned cell and N is the total number of cells in each core. The SI scores for each core were then used to generate average diversity scores of Benign, HGPIN, localized PCa, and metastatic PCa.
Results
CHD8 Expression is Commonly Decreased in Localized and Metastatic PCa
The CTCF-CHD8 (BORIS) system was synchronously investigated given their associations (Figure 1A) using the VECTRA imaging system which allows the automated selection and analysis of cells, cellular subsets (epithelial vs. stromal) and subcellular compartments (nucleus vs. cytoplasm). Nuclear levels of CHD8 were 1.5 to 2.5 fold higher than cytoplasmic in all samples. Total, nuclear, and cytoplasmic expression of CHD8 was significantly decreased in HGPIN, metastatic lesions, and primary PCa compared to benign prostate tissue (Figure 1B) (all P < .001, HGPIN cytoplasmic not significant). To define reduced expression, a Receiver Operating Curve (ROC) was constructed using total CHD8 expression to identify the optimal cut point that maximized the sum of sensitivity and specificity for discriminating between benign versus cancer cores. ROC analysis demonstrated excellent discrimination between benign versus cancer (AUC 0.866, P < .0001) with a sensitivity and specificity of 84.3% and 76.3% respectively (Figure 1C). Using the optimal cut-off of 0.065 to define decreased core expression, 118/140 (84.3%) of localized PCa, 18/46 (39.1%) of HGPIN, and 29/39 (74.4%) of metastatic lesions demonstrated reduced expression of CHD8 in contrast to only 19/80 (23.8%) of all benign cores (Figure 1D). Therefore, decreased CHD8 expression is a common finding in PCa tissues.
Figure 1.
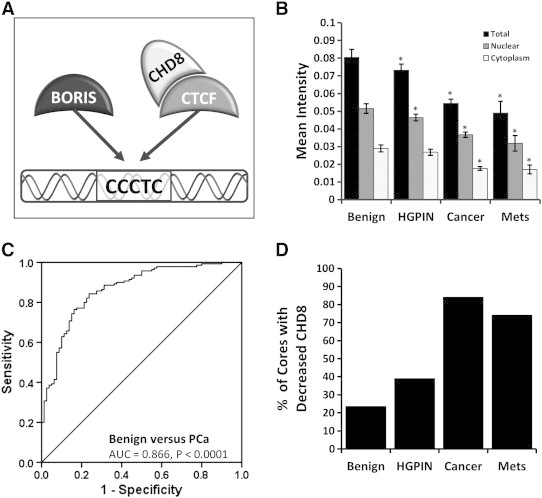
Quantitative analysis of CHD8 expression. Using the VECTRA automated image capture and analysis nuclear, cytoplasmic and total compartments were compared. Mean intensity of cores were compared for benign, HGPIN, cancer, and metastases with 95% confidence intervals. (A) Demonstrates the interactions of BORIS, CTCF, and CHD8; where BORIS and CTCF compete for similar binding sites, and CTCF and CHD8 complex at CTCF binding sites. (B) Significant decreases in CHD8 expression are seen in cancer and metastases cores for total, nuclear, and cytoplasmic expression (P < .001, cytoplasmic HGPIN not significant). (C) Total CHD8 expression demonstrated optimal discrimination between benign and cancer (AUC = 0.866, P < .0001) by ROC analysis, with a sensitivity and specificity of 84.3% and 76.3%. (D) Using the optimum cut-off obtained by ROC analysis, 84.3% of cancer and 74.4% of metastases cores demonstrated decreased CHD8.
Analysis of CHD8 Promoter-Associated CpG Hypermethylation in Human Prostate Tumor Samples
CHD8 contains a promoter CpG island making CHD8 promoter-associated hypermethylation one etiology for the decreased expression. One region spanning the CHD8 transcription start site (TSS) and another region 600 bp upstream were analyzed. At the region 600 bp upstream high levels of equivalent methylation was seen in all tumor and benign samples (data not shown). At the TSS CpG island, 5/11 (45%) of tumor samples demonstrated increased methylation across the first 4 CpG sites compared to benign (Figure 2A). To further assess this finding, we assessed methylation using 366 primary prostate tumors within the TGCA database. A significant increase in methylation across CpG 1 to 4 was also seen when tumor and normal tissues were compared (Wilcoxon Rank-sum all P < .001)(Figure 2B). In this validation dataset, 55%-67% of tumors showed increased methylation (compared to benign tissue mean) at CpGs 1 to 4 (data not shown).
Figure 2.
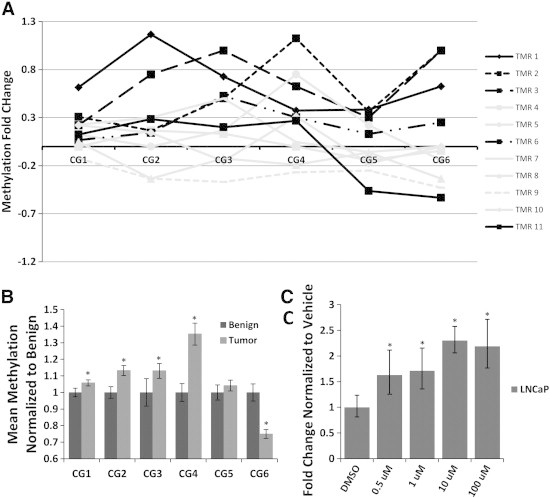
Methylation analysis of CHD8 in human PCa and 5-Aza-2′-deoxycytidine treatment of cancer cell lines. DNA methylation was analyzed by Quantitative Pyrosequencing across the CHD8 transcriptional start site. (A) Hypermethylation of CHD8 promoter-associated CpGs 1 to 4 was seen in 5/11 (45%) human tumor samples compared to matched benign. (B) Significant hypermethylation was seen at 4/6 of the promoter-associated CpGs in tumors compared to benign in TCGA PRAD samples (methylation 450 K array) (All P < .001). (C) 5-azadC treatment of LNCaP cells for 48 hours resulted in a 1.5- to 2.5-fold significant increase of CHD8 mRNA over treatment vehicle alone (all P < .05).
To assess the role of methylation in controlling CHD8 transcription, a panel of cell lines was analyzed for CHD8 expression. Cell lines with lower expression of CHD8 were treated with increasing doses of 5-aza-2′-deoxycytidine (5-azadC) for 48 hrs prior to harvesting. 5-azadC resulted in a dose-dependent increase in CHD8 expression compared to DMSO alone (all P < .05) for LNCaP (Figure 2C) and Hela (data not shown) cell lines. Similar treatment of DU145 did not result in increased expression of CHD8 mRNA (data not shown). Analysis of CHD8 CpG methylation across the TSS showed similar levels of methylation across all 3 cell types (Figure S1A). Additionally, all three cell types were highly methylated at a region 600 base pairs upstream of the TSS (Figure S1B). These data indicate that DNA hypermethylation is a common finding within the CHD8 promoter region and treatment with a demethylating agent results in increased CHD8 expression.
In Primary Tumor Samples, Increasing CHD8 Expression is Associated with Adverse Clinical Features and Outperforms Gleason Score in Predicting PSA Recurrence in Intermediate Grade Tumors
CHD8 expression was compared against a series of clinicopathologic variables including Gleason score, tumor stage, tumor volume, seminal vesicle involvement, positive margins, extracapsular extension and the presence of metastases (Table 1). Using CHD8 as a continuous variable, a surprising increase in total and nuclear expression was associated with extracapsular extension (P =.021 and P = .007 respectively) and metastases (P = .037 and P = .025 respectively) (Table 1). The analyses did not find any associations between Gleason score or tumor volume and CHD8 expression.
Table 1.
Association of CHD8 Expression With Patient Pathological Features.
| Variable | Number | Nucleus |
Cytoplasm |
Total |
|||
|---|---|---|---|---|---|---|---|
| Mean Intensity (SD) | p-value | Mean Intensity (SD) | p-value | Mean Intensity (SD) | p-value | ||
| Gleason⁎ | |||||||
| 3 + 3 or 3 + 4 | 38 | 3.80 (0.71) | 0.209 | 1.81 (0.47) | 0.589 | 5.61 (1.12) | 0.256 |
| 4 + 3/4 + 4/4 + 5 | 28 | 3.54 (0.77) | 1.75 (0. 54) | 5.28 (1.27) | |||
| Stage | |||||||
| T2 | 39 | 3.54 (0.64) | 0.054 | 1.68 (0.39) | 0.132 | 5.22 (0.99) | 0.064 |
| T3 | 11 | 3.73 (0.84) | 1.95 (0.68) | 5.68 (1.49) | |||
| T4 | 17 | 4.19 (0.78) | 1.98 (0.50) | 6.16 (1.20) | |||
| Tumor Volume | |||||||
| < 5 | 9 | 3.74 (0.42) | 0.891 | 1.73 (0.46) | 0.745 | 5.48 (0.84) | 0.987 |
| 5-20 | 31 | 3.65 (0.93) | 1.71 (0.52) | 5.36 (1.40) | |||
| > 20 | 24 | 3.68 (0.75) | 1.83 (0.53) | 5.51 (1.24) | |||
| SV involvement | |||||||
| Absent | 48 | 3.57 (0.71) | 0.10 | 1.68 (0.42) | 0.088 | 5.26 (1.09) | 0.089 |
| Present | 15 | 4.04 (0.96) | 2.06 (0.71) | 6.10 (1.60) | |||
| Margins Positive | |||||||
| No | 42 | 3.76 (0.62) | 0.119 | 1.84 (0.48) | 0.044 | 5.60 (1.05) | 0.085 |
| Yes | 24 | 3.51 (1.01) | 1.63 (0.56) | 5.14 (1.52) | |||
| Extracapsular | |||||||
| No | 42 | 3.47 (0.73) | 0.007 | 1.66 (0.46) | 0.0622 | 5.14 (1.15) | 0.021 |
| Yes | 23 | 4.04 (0.76) | 1.95 (0.57) | 5.99 (1.27) | |||
| Metastasis† | |||||||
| No | 50 | 3.59 (0.69) | 0.025 | 1.75 (0.49) | 0.111 | 5.34 (1.14) | 0.037 |
| Yes | 16 | 4.19 (0.78) | 1.98 (0.50) | 6.16 (1.20) | |||
One patient GS 3 + 5 not analyzed.
Metastasis refers to primary tumor samples with associated metastases.
Generating molecular predictors of treatment failure, especially within intermediate grade cancers is a pressing clinical need. An analysis of PSA recurrence after prostate removal, which indicates treatment failure, was performed. Patients with increased CHD8 expression exhibited an earlier PSA recurrence (P = .048) (Figure 3A). Gleason score provides weak separation of patients into risk categories for PSA recurrence (P = .163) (Figure 3B). Stratifying intermediate Gleason patients by CHD8 expression shows that patients with higher CHD8 expression had a greater risk of biochemical recurrence (P = .048) (Figure 3C). Therefore, although decreased in cancer compared to benign, increasing CHD8 levels in cancer cores are associated with adverse clinicopathologic variables and worse outcomes.
Figure 3.
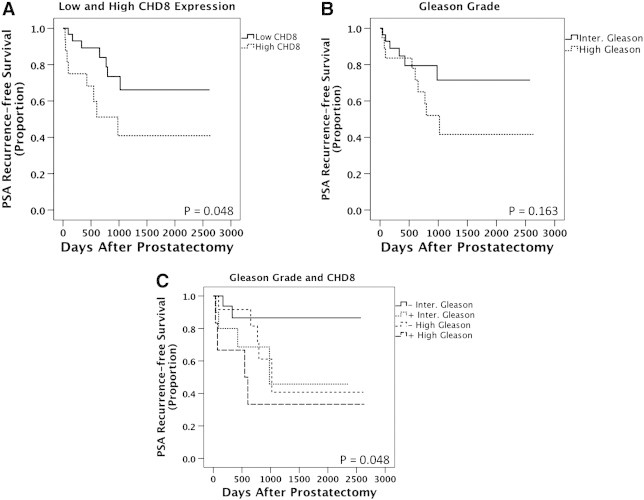
Kaplan Meier analyses comparing pathological Gleason Score versus CHD8. Biochemical recurrence after radical prostatectomy was examined in patient samples stained for CHD8. (A) Nuclear CHD8 significantly separates patients into risk categories for PSA recurrence using a Log rank analysis (P = .048). (B) Gleason score provides insignificant separation of patients for PSA recurrence using a Log rank analysis (P = .163). (C) Nuclear CHD8 is superior to pathological Gleason score when Gleason score was stratified into low versus high CHD8. The median value for nuclear CHD8 staining was 0.04025 in all benign and cancer samples. This value was subsequently used as a cut point to define high versus low nuclear CHD8 expression for the Log rank analysis.
BORIS/CTCF Expression Ratio is Increased in Cancer Samples and Correlates with Higher Gleason Score
CTCF is known to impact gene regulation through the management of chromatin organization and the maintenance of epigenetic marks [26]. CTCF expression levels measured by VECTRA were significantly decreased in metastatic PCa tumors (P < .001), but not localized PCa when compared to benign prostate tissues (P = .17) (Figure 4A). When CTCF expression within primary tumors was compared against clinicopathologic patient variables, decreased nuclear and total CTCF expression was associated with higher Gleason's Score (P = .01 and P = .001 respectively) and positive margins (P = .03 and P = .043 respectively) (Data not shown).
Figure 4.
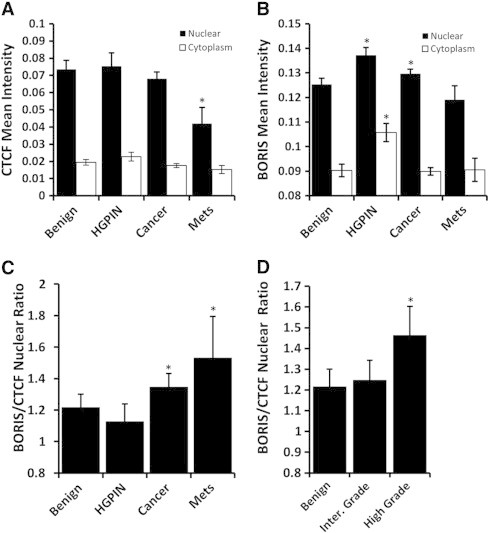
Quantitative analysis of CTCF and BORIS expression in human PCa. (A) Nuclear CTCF significantly decreases in metastases cores (P < .001) (B) BORIS nuclear expression significantly increased in cancer cores (P = .002); nuclear and cytoplasmic expression increased in HGPIN cores compared to benign (both P < .001). (C) The ratio of BORIS to CTCF expression was analyzed for differences among cores. Significant increases in the BORIS/CTCF ratio are seen among cancer and metastases cores compared to benign (P = .031 and P = .006 respectively). (D) Comparing the BORIS/CTCF ratio among cancer cores, high Gleason grade cancers have significantly higher BORIS/CTCF ratio than both benign and intermediate Gleason cores (P = .006 and P = .024 respectively).
The CTCF paralogue, BORIS, shares a common zinc-finger DNA-binding domain with divergent N- and C- termini. BORIS and CTCF have competing effects on gene expression and epigenetic regulation [5], [27]. In contrast to CTCF, BORIS expression was increased in HGPIN and primary cancer cores compared to benign (P < .001 and P = .002 respectively) (Figure 4B). No clinicopathologic correlates were noted.
The competing role for BORIS/CTCF suggests the ratio of their expression may contain functional significance and greater predictive power than the expression of these proteins individually as seen in ovarian cancer [28]. Analyzing all prostate tissues, a weak correlation was seen between BORIS and CTCF (Pearson's r = .224, P < .001). BORIS/CTCF levels increased significantly in cancer and metastases compared to benign (P = .03 and P = .006 respectively) (Figure 4C). High Gleason cancers have a significantly greater BORIS/CTCF ratio than both benign tissues and intermediate Gleason cancers (P = .006 and P = .024 respectively) (Figure 4D). The ratio of nuclear and total expression was calculated and found to be associated with Gleason's grade, positive surgical margins, and tumor volume (nuclear P = .024, P = .027, P = .020, respectively) (Table 2, for full table see supplementary data, Table S2).
Table 2.
Association of BORIS/CTCF Expression Ratios with Patient Pathological Features.
| Variable | Number | Nucleus |
Cytoplasm |
Total |
|||
|---|---|---|---|---|---|---|---|
| Mean Intensity (SD) | p-value | Mean Intensity (SD) | p-value | Mean Intensity (SD) | p-value | ||
| Gleason⁎ | |||||||
| 3 + 3 or 3 + 4 | 38 | 1.22 (0.41) | 0.024⁎ | 2.96 (1.21) | 0.019⁎ | 1.59 (0.54) | 0.021⁎ |
| 4 + 3/4 + 4/4 + 5 | 28 | 1.53 (0.64) | 3.88 (2.06) | 2.03 (0.93) | |||
| Tumor volume | |||||||
| < 5 | 9 | 0.96 (0.38) | 0.020⁎ | 2.28 (0.89) | 0.029⁎ | 1.25 (0.49) | 0.022⁎ |
| 5-20 | 31 | 1.38 (0.67) | 3.35 (2.13) | 1.82 (0.97) | |||
| > 20 | 24 | 1.36 (0.42) | 3.54 (1.38) | 1.81 (0.58) | |||
| Margins positive | |||||||
| No | 42 | 1.23 (0.42) | 0.027⁎ | 3.12 (1.31) | 0.204 | 1.63 (0.56) | 0.041⁎ |
| Yes | 24 | 1.56 (0.69) | 3.78 (2.24) | 2.06 (1.00) | |||
One patient GS 3 + 5 not analyzed.
Exclusion of CHD8 and/or BORIS Alterations is Seen in a Majority of Cancers
Our analysis permits a comparison of the expression of each of the chromatin regulators in the CTCF/CHD8/BORIS pathway within specific tumors. We speculated that inactivation of one gene in this pathway might exclude alterations in the other related genes in the CTCF complex. To first test this, the cBioPortal for Cancer Genomics (www.cbioportal.org/public-portal/) was queried for alterations in these 3 genes and the CTCF/CHD8/BORIS pathway was genetically altered in 15% of PCa (Table S3). Similar low rates of concurrent alteration were seen in other cancers. This included amplification of BORIS, homozygous deletion of CTCF, and mutation of CHD8. We extended this analysis to our data by segregating gene expression into quartiles in the entire dataset using all cores (benign, HGPIN, cancer, and metastases). Expression alterations in CTCF were rarely seen and thus were not analyzed. Segregating all 73 patient tumor samples from our data into expression quartiles we found 26/73 (36%) exhibit decreased and 8/73 (11%) exhibit increased CHD8. Analyzing BORIS expression, 19/73 (26%) exhibit increases and 7/73 (10%) show decreased expression. Expressional alterations occurred in 51/73 (70%) of the tumors in total (Figure 5A). Comparatively, alterations in both genes were rarely seen (9/73; 12%%). Odds ratio analysis indicated that these alterations showed a tendency toward mutual exclusivity (OR, 0.46).
Figure 5.
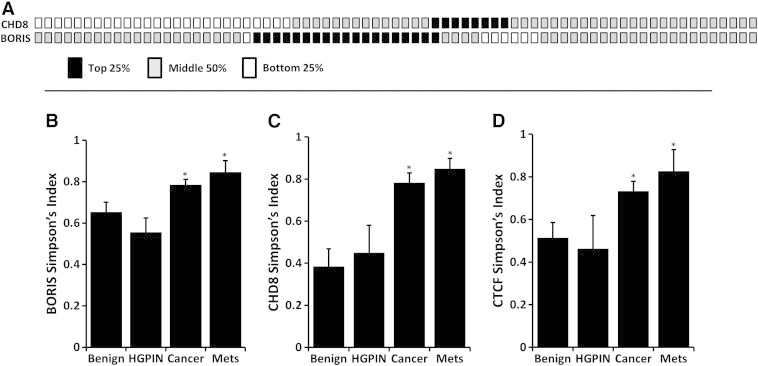
CHD8 and BORIS expression alterations are exclusive and exhibit significantly greater heterogeneity in malignant cores. (A) Using expression of all cores, the expression data for CHD8 and BORIS was divided into quartiles. Each column box-pair represents a primary tumor sample (total 73). When cancer cores are separated into quartiles of protein expression, CHD8 is frequently decreased (26/73; 36%) while BORIS is frequently increased (19/73; 26%) in primary cancer. Rare cancers (12%) exhibited both alterations in concert indicating that these pathway alterations infrequently occur in concert. An analysis of heterogeneity of cores was performed using the Simpson's Diversity Index (SI). BORIS (B), CHD8 (C), and CTCF (D) all had significantly higher heterogeneity of protein expression in cancer and metastases cores compared to benign (all P < .005) as measured by Simpson's Diversity Index.
CTCF, BORIS, and CHD8 Expression is Significantly More Heterogeneous in Higher Grade Cancers
The majority of human tumors display startling heterogeneity in many morphological and physiological features, but the ability to objectively quantitate this aspect has been lacking. VECTRA was utilized to assess per cell expression within benign and tumor cores and heterogeneity scores for each core was calculated using the Simpson's Diversity Index (SI), a formula generally used in the population sciences [24]. The SI uses population size to normalize diversity analyses to a uniform scoring system for comparison. SI analyses revealed BORIS, CHD8, and CTCF all demonstrate increased heterogeneity of protein expression in primary cancer and metastases compared to benign (all P < .001) (Figure 5, B–D respectively). Within primary PCa cores, increased heterogeneity was also seen in higher grade cancer (CHD8, P = .001, CTCF P < .001, and BORIS, P = .001). Similar increased heterogeneity was seen when the coefficient of variation was calculated for these proteins (data not shown).
Discussion
CTCF is a transcriptional regulator and member of the BORIS and CTCF gene family. It has well known insulator activity where binding to a transcriptional insulator element serves to block enhancer-promoter interactions [29] and can also act as a classical transcription factor [30]. In addition, CTCF is uniquely involved in epigenetic regulation including many cell cycle and cancer specific genes [7], [10]. The function of CTCF is modulated at a number of levels, including at the protein level by cofactors and competitive inhibitors. This study analyzed the synchronous expression of CTCF, its cofactor CHD8, and the antagonistic CTCF paralogue BORIS using VECTRA imaging technology. Increased heterogeneity in higher grade cancers was found and the coexpression of these chromatin regulators was largely independent of each other. A striking decrease in CHD8 expression is a major finding in the majority of primary PCas and metastatic deposits. An interesting contrast in CHD8 expression was also discovered with increased levels being associated with more adverse clinical variables and PSA recurrence-free survival. BORIS/CTCF levels also correlated with worse pathologic variables. This unique approach suggests a significant role for alterations in these chromatin factors in PCa.
There has been increased interest in the CHD family [13], [14] given the finding of inactivation of other family members in disease including the recent findings of somatic mutation of CHD5 [31] and deletion of CHD1 [32]. CHD8 exhibits a functional dichotomy operating in both growth inhibitory and promoting roles including chromatin remodeling, WNT signaling, CTCF insulator activity, p53-mediated apoptosis, androgen receptor mediated gene activity, and regulation of cell cycle genes [33]. In the current study, the expression of CHD8 significantly decreased in HGPIN, cancer, and metastases compared to benign tissue. Using a cut-off determined by ROC analysis maximizing sensitivity and specificity, 84% of localized and 74% of metastatic PCa tumors exhibited decreased CHD8 expression. One potential significant implication of decreased CHD8 may be a loss of the CTCF-CHD8 complex, which serves to stabilize regulation of CTCF effector genes. Ishihara et al. found losses of CHD8 altered DNA methylation and histone acetylation around CTCF binding sites, adjacent to heterochromatin, in HeLa and hepatoma cell lines [3].
An interesting dichotomy of expression was discovered using imaging analysis. In tumor tissues, increased CHD8 expression was associated with the presence of metastases and extracapsular extension (Table 1). Extending this analysis to examine risk of PSA recurrence after radical prostatectomy, we found that CHD8 expression served to discriminate between indolent intermediate grade cancers and those at higher risk of recurrence. Markers that predict a worse outcome for intermediate grade cancers are an area of intense clinical interest. The dichotomy in expression suggests a loss of function is important for cancer development, but progression is facilitated by higher expression. Alternatively, higher levels seen with cancer progression may indicate a nonfunctional or altered protein.
Loss of CHD8 may alter CTCF function, as well as aberrant expression of the CTCF paralogue and cancer-testis antigen BORIS. We found BORIS expression to be significantly increased in cancer compared to benign prostate tissues. Recently published findings in prostate tumor tissues and cell lines support this observation [12]. The identical 11-zinc finger DNA-binding domains of CTCF and BORIS may result in sibling rivalry for binding sites but their divergent amino- and carboxy-terminal domains result in antagonistic gene regulation functions [11]. Exploiting this competition, a recent study in ovarian cancer found BORIS/CTCF ratio of expression levels correlate with advanced stage and DNA hypomethylation levels [28]. An analysis of the ratio of BORIS/CTCF expression in prostate cancer demonstrated a significant increase in cancer that correlated with higher Gleason's grade, positive surgical margins, and increased tumor volume. Aberrant BORIS expression may work alternatively or in concert with CHD8 decreases to disrupt CTCF action within the genome.
Our analysis permits a comparison of expression of each of the chromatin regulators in the CTCF/CHD8/BORIS pathway within specific tumors. By investigating the highest or lowest quartiles of expression we determined that CHD8 down-regulation and BORIS up-regulation occurred commonly in 56% of tumors analyzed. Furthermore, decreased CHD8 expression rarely occurred with BORIS amplification (Figure 5A). The mechanisms underlying CTCF deregulation in disease are complex and include loss of heterozygosity, mutation, post-translational modification, and methylation at CTCF target sites. Using the cBioPortal, a similar tendency for multiple tumor types (breast, ovarian, uterine, colon) that demonstrate BORIS amplification to not contain alterations in CTCF or CHD8 was seen, suggesting if one component of the pathway is altered, others are not required (Table S2).
We performed an analysis of CHD8 TSS hypermethylation and find this occurs in roughly 45% of tumors examined. TCGA cohorts analyzed revealed that four of the six CpGs analyzed are significantly hypermethylated in tumor samples. This confirms data seen in a recent high-throughput methylation array analysis of prostate tumors [22]. Treatment of cancer cell lines with a methyltransferase inhibitor (5-azadC) resulted in increased expression of CHD8 mRNA in LNCaP indicating DNA methylation plays a role in CHD8 regulation (Figure 2C). Other mutational events or epigenetic marks, such as histone modifications, may also play a role in reducing CHD8 expression. Therefore, epigenetic silencing of CHD8 may be one mechanism for alterations of expression.
Tumor development can be regarded as a process of Darwinian evolution. Selection forces required for the emergence of malignancies and increase genetic and epigenetic instability generate clonal populations. The VECTRA analysis platform is a powerful objective tool for analyzing patterns of protein expression in tissues. To capitalize on this extensive quantitation we used per cell data to analyze the heterogeneity of protein expression in tissue cores. The coefficient of variation and the Simpson's Diversity Index, a measure originally developed to measure ecological diversity, were applied to capture this aspect. We find PCa cores contain increased heterogeneity for all proteins. In addition, cancers with higher Gleason score are significantly more heterogeneous. This suggests that heterogeneity of expression translates into phenotypic diversity; this may be a potential biomarker in the future.
In conclusion, we demonstrate for the first time that CHD8 expression is frequently altered in PCa and its expression more accurately predicts PSA-recurrence with Gleason score over Gleason score alone. Interestingly, CHD8 is decreased in cancer, however higher expression is seen with more adverse clinical features. This dichotomy may be explained, in part, by the diverse functions of CHD8. The use of imaging technology allows a number of novel observations to be performed. The higher heterogeneity of protein expression seen in cancer and metastases may convey plasticity in CHD8 expression. Early decreases in CHD8 may conceivably be advantageous to tumor development, while tumor progression involves a subsequent shift to higher CHD8 within the tumor. Changes in CTCF/BORIS may accompany CHD8 expressional alterations, further contributing to neoplastic development. Frequent alterations of the CHD8/CTCF/BORIS pathway suggest a crucial function in early PCa development. Biologically, this pathway affects chromatin and epigenetic regulation [3], [10], key factors in early neoplasia. These findings warrant future study of the functional consequences of CHD8, CTCF, and BORIS expression alterations in the prostate.
Acknowledgments
The authors have declared that no competing interests exist.
This work was supported by the National Institutes of Health 5R01CA097131 [DJ], and ND was supported by the NCI Cancer Biology Training Grant T32 CA009135.
The authors thank the University of Wisconsin Translational Research Initiatives in Pathology laboratory, in part supported by the UW Department of Pathology and Laboratory Medicine and UWCCC grant P30 CA014520, for use of its facilities and services.
The results shown here are in part based upon data generated by the TCGA Research Network: http://cancergenome.nih.gov/
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.neo.2014.10.003.
Appendix A. Supplementary data
Methylation of Cancer Cell Lines at CHD8 Regulatory Regions.
Despite no significant changes in CHD8 expression in DU145 with 5-azadC treatment (A), methylation levels were similar to LNCaP and HELA. B) Methylation analyses by quantitative pyrosequencing at a CpG region spanning from the promoter into exon 1 exhibited moderate methylation. C) Methylation analyses at a CpG region 600 BPs upstream of the TSS revealed heavy methylation across all three cell lines with DU145 and LNCaP exhibiting slightly greater methylation.
Table S1. Clinicopathological Characteristics of Cancer Samples
Table S2. Association of BORIS/CTCF Expression Ratios With Patient Pathological Features.
Table S3. Rates of genetic alteration in CTCF/BORIS/CHD8 genes in various cancers.
References
- 1.Damaschke N.A., Yang B., Bhusari S., Svaren J.P., Jarrard D.F. Epigenetic susceptibility factors for prostate cancer with aging Prostate. 2013;73:1721–1730. doi: 10.1002/pros.22716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Varambally S., Dhanasekaran S.M., Zhou M., Barrette T.R., Kumar-Sinha C., Sanda M.G., Ghosh D., Pienta K.J., Sewalt R.G., Otte A.P. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 3.Ishihara K., Oshimura M., Nakao M. CTCF-dependent chromatin insulator is linked to epigenetic remodeling. Mol Cell. 2006;23:733–742. doi: 10.1016/j.molcel.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 4.Loukinov D.I., Pugacheva E., Vatolin S., Pack S.D., Moon H., Chernukhin I., Mannan P., Larsson E., Kanduri C., Vostrov A.A. BORIS, a novel male germ-line-specific protein associated with epigenetic reprogramming events, shares the same 11-zinc-finger domain with CTCF, the insulator protein involved in reading imprinting marks in the soma. Proc Natl Acad Sci U S A. 2002;99:6806–6811. doi: 10.1073/pnas.092123699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vatolin S., Abdullaev Z., Pack S.D., Flanagan P.T., Custer M., Loukinov D.I., Pugacheva E., Hong J.A., Morse H., Schrump D.S. Conditional expression of the CTCF-paralogous transcriptional factor BORIS in normal cells results in demethylation and derepression of MAGE-A1 and reactivation of other cancer-testis genes. Cancer Res. 2005;65:7751–7762. doi: 10.1158/0008-5472.CAN-05-0858. [DOI] [PubMed] [Google Scholar]
- 6.Merkenschlager M., Odom D.T. CTCF and cohesin: linking gene regulatory elements with their targets. Cell. 2013;152:1285–1297. doi: 10.1016/j.cell.2013.02.029. [DOI] [PubMed] [Google Scholar]
- 7.Fiorentino F.P., Giordano A. The tumor suppressor role of CTCF. J Cell Physiol. 2012;227:479–492. doi: 10.1002/jcp.22780. [DOI] [PubMed] [Google Scholar]
- 8.Kurukuti S., Tiwari V.K., Tavoosidana G., Pugacheva E., Murrell A., Zhao Z., Lobanenkov V., Reik W., Ohlsson R. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc Natl Acad Sci U S A. 2006;103:10684–10689. doi: 10.1073/pnas.0600326103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hore T.A., Deakin J.E., Marshall Graves J.A. The evolution of epigenetic regulators CTCF and BORIS/CTCFL in amniotes. PLoS Genet. 2008;4:e1000169. doi: 10.1371/journal.pgen.1000169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klenova E.M., Morse H.C., Ohlsson R., Lobanenkov V.V. The novel BORIS + CTCF gene family is uniquely involved in the epigenetics of normal biology and cancer. Semin Cancer Biol. 2002;12:399–414. doi: 10.1016/s1044-579x(02)00060-3. [DOI] [PubMed] [Google Scholar]
- 11.Pugacheva EM, Suzuki T, Pack SD. The structural complexity of the human BORIS gene in gametogenesis and cancer. PLoS One. 2010;5:e13872. doi: 10.1371/journal.pone.0013872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheema Z., Hari-Gupta Y., Kita G.X., Farrar D., Seddon I., Corr J., Klenova E. Expression of the cancer-testis antigen BORIS correlates with prostate cancer. Prostate. 2014;74:164–176. doi: 10.1002/pros.22738. [DOI] [PubMed] [Google Scholar]
- 13.Marfella C.G., Imbalzano A.N. The Chd family of chromatin remodelers. Mutat Res. 2007;618:30–40. doi: 10.1016/j.mrfmmm.2006.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim M.S., Chung N.G., Kang M.R., Yoo N.J., Lee S.H. Genetic and expressional alterations of CHD genes in gastric and colorectal cancers. Histopathology. 2011;58:660–668. doi: 10.1111/j.1365-2559.2011.03819.x. [DOI] [PubMed] [Google Scholar]
- 15.Nishiyama M., Oshikawa K., Tsukada Y., Nakagawa T., Iemura S., Natsume T., Fan Y., Kikuchi A., Skoultchi A.I., Nakayama K.I. CHD8 suppresses p53-mediated apoptosis through histone H1 recruitment during early embryogenesis. Nat Cell Biol. 2009;11:172–182. doi: 10.1038/ncb1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishiyama M., Skoultchi A.I., Nakayama K.I. Histone H1 recruitment by CHD8 is essential for suppression of the Wnt-β-catenin signaling pathway. Mol Cell Biol. 2012;32:501–512. doi: 10.1128/MCB.06409-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yates J.A., Menon T., Thompson B.A., Bochar D.A. Regulation of HOXA2 gene expression by the ATP-dependent chromatin remodeling enzyme CHD8. FEBS Lett. 2010;584:689–693. doi: 10.1016/j.febslet.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 18.Menon T., Yates J.A., Bochar D.A. Regulation of androgen-responsive transcription by the chromatin remodeling factor CHD8. Mol Endocrinol. 2010;24:1165–1174. doi: 10.1210/me.2009-0421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Subtil-Rodríguez A., Vázquez-Chávez E., Ceballos-Chávez M., Rodríguez-Paredes M., Martín-Subero J.I., Esteller M., Reyes J.C. The chromatin remodeller CHD8 is required for E2F-dependent transcription activation of S-phase genes. Nucleic Acids Res. 2014;42:2185–2196. doi: 10.1093/nar/gkt1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang W., Hennrick K., Drew S. A colorful future of quantitative pathology: validation of Vectra technology using chromogenic multiplexed immunohistochemistry and prostate tissue microarrays. Hum Pathol. 2013;44:29–38. doi: 10.1016/j.humpath.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 21.Desotelle J., Truong M., Ewald J., Weeratunga P., Yang B., Huang W., Jarrard D. CpG island hypermethylation frequently silences FILIP1L isoform 2 expression in prostate cancer. J Urol. 2013;189:329–335. doi: 10.1016/j.juro.2012.08.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kobayashi Y., Absher D.M., Gulzar Z.G., Young S.R., McKenney J.K., Peehl D.M., Brooks J.D., Myers R.M., Sherlock G. DNA methylation profiling reveals novel biomarkers and important roles for DNA methyltransferases in prostate cancer. Genome Res. 2011;21:1017–1027. doi: 10.1101/gr.119487.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodríguez-Paredes M., Ceballos-Chávez M., Esteller M., García-Domínguez M., Reyes J.C. The chromatin remodeling factor CHD8 interacts with elongating RNA polymerase II and controls expression of the cyclin E2 gene. Nucleic Acids Res. 2009;37:2449–2460. doi: 10.1093/nar/gkp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peet R.K. The Measurement of Species Diversity. Annual Review of Ecology and Systematics. 1974;5:285–307. [Google Scholar]
- 25.Faratian D., Christiansen J., Gustavson M., Jones C., Scott C., Um I., Harrison D.J. Heterogeneity mapping of protein expression in tumors using quantitative immunofluorescence. J Vis Exp. 2011:e3334. doi: 10.3791/3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Phillips J.E., Corces V.G. CTCF: master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hong J.A., Kang Y., Abdullaev Z., Flanagan P.T., Pack S.D., Fischette M.R., Adnani M.T., Loukinov D.I., Vatolin S., Risinger J.I. Reciprocal binding of CTCF and BORIS to the NY-ESO-1 promoter coincides with derepression of this cancer-testis gene in lung cancer cells. Cancer Res. 2005;65:7763–7774. doi: 10.1158/0008-5472.CAN-05-0823. [DOI] [PubMed] [Google Scholar]
- 28.Woloszynska-Read A., Zhang W., Yu J., Link P.A., Mhawech-Fauceglia P., Collamat G., Akers S.N., Ostler K.R., Godley L.A., Odunsi K. Coordinated cancer germline antigen promoter and global DNA hypomethylation in ovarian cancer: association with the BORIS/CTCF expression ratio and advanced stage. Clin Cancer Res. 2011;17:2170–2180. doi: 10.1158/1078-0432.CCR-10-2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bell A.C., West A.G., Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–396. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- 30.Klenova E.M., Nicolas R.H., Paterson H.F., Carne A.F., Heath C.M., Goodwin G.H., Neiman P.E., Lobanenkov V.V. CTCF, a conserved nuclear factor required for optimal transcriptional activity of the chicken c-myc gene, is an 11-Zn-finger protein differentially expressed in multiple forms. Mol Cell Biol. 1993;13:7612–7624. doi: 10.1128/mcb.13.12.7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robbins C.M., Tembe W.A., Baker A., Sinari S., Moses T.Y., Beckstrom-Sternberg S., Beckstrom-Sternberg J., Barrett M., Long J., Chinnaiyan A. Copy number and targeted mutational analysis reveals novel somatic events in metastatic prostate tumors. Genome Res. 2011;21:47–55. doi: 10.1101/gr.107961.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang S., Gulzar Z.G., Salari K., Lapointe J., Brooks J.D., Pollack J.R. Recurrent deletion of CHD1 in prostate cancer with relevance to cell invasiveness. Oncogene. 2012;31:4164–4170. doi: 10.1038/onc.2011.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sakamoto I., Kishida S., Fukui A., Kishida M., Yamamoto H., Hino S., Michiue T., Takada S., Asashima M., Kikuchi A. A novel beta-catenin-binding protein inhibits beta-catenin-dependent Tcf activation and axis formation. J Biol Chem. 2000;275:32871–32878. doi: 10.1074/jbc.M004089200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Methylation of Cancer Cell Lines at CHD8 Regulatory Regions.
Despite no significant changes in CHD8 expression in DU145 with 5-azadC treatment (A), methylation levels were similar to LNCaP and HELA. B) Methylation analyses by quantitative pyrosequencing at a CpG region spanning from the promoter into exon 1 exhibited moderate methylation. C) Methylation analyses at a CpG region 600 BPs upstream of the TSS revealed heavy methylation across all three cell lines with DU145 and LNCaP exhibiting slightly greater methylation.
Table S1. Clinicopathological Characteristics of Cancer Samples
Table S2. Association of BORIS/CTCF Expression Ratios With Patient Pathological Features.
Table S3. Rates of genetic alteration in CTCF/BORIS/CHD8 genes in various cancers.


