Abstract
Hypoxia has been implicated as a crucial microenvironmental factor that induces cancer metastasis. We previously reported that hypoxia could promote gastric cancer (GC) metastasis, but the underlying mechanisms are not clear. Long noncoding RNAs (lncRNAs) have recently emerged as important regulators of carcinogenesis that act on multiple pathways. However, whether lncRNAs are involved in hypoxia-induced GC metastasis remains unknown. In this study, we investigated the differentially expressed lncRNAs resulting from hypoxia-induced GC and normoxia conditions using microarrays and validated our results through real-time quantitative polymerase chain reaction. We found an lncRNA, AK058003, that is upregulated by hypoxia. AK058003 is frequently upregulated in GC samples and promotes GC migration and invasion in vivo and in vitro. Furthermore, AK058003 can mediate the metastasis of hypoxia-induced GC cells. Next, we identified γ-synuclein (SNCG), which is a metastasis-related gene regulated by AK058003. In addition, we found that the expression of SNCG is positively correlated with that of AK058003 in the clinical GC samples used in our study. Furthermore, we found that the SNCG gene CpG island methylation was significantly increased in GC cells depleted of AK058003. Intriguingly, SNCG expression is also increased by hypoxia, and SNCG upregulation by AK058003 mediates hypoxia-induced GC cell metastasis. These results advance our understanding of the role of lncRNA-AK058003 as a regulator of hypoxia signaling, and this newly identified hypoxia/lncRNA-AK058003/SNCG pathway may help in the development of new therapeutics.
Introduction
Gastric cancer (GC) is one of the most common human cancers and the second leading cause of cancer-related mortality worldwide [1]. The major cause of death is metastasis, which greatly hinders treatment success [2]. Hypoxia is an important microenvironmental factor that induces such metastasis. In fact, hypoxic tumors are often aggressive and more likely to metastasize [3]. Therefore, investigating the underlying mechanisms of hypoxia-induced metastasis is critical. We have previously performed studies exploring the mechanism underlying GC pathogenesis under hypoxic stress, with a focus on protein-coding genes [4], [5], [6]. However, protein-coding genes account for less than 2% of the human genome, as most of the genome is transcribed into non-coding RNAs (ncRNAs) [7], [8]. Recently, long noncoding RNAs (lncRNAs) have been reported to be associated with hypoxia-induced cancer development [9] and to contribute to hypoxia-mediated HepatoCellular Carcinoma metastasis [10].
LncRNAs have been identified as a new class of functional ncRNAs and have kindled our interest. lncRNAs are defined as transcripts of longer than 200 nucleotides without evident protein-coding function [11]. Few studies have implicated lncRNAs in various cancers [12], [13], and several of the altered lncRNAs can result in the aberrant expression of nearby protein-coding genes, which may contribute to cancer development [14], [15], [16]. Studies have also demonstrated that lncRNAs play an important role in tumor development via various mechanisms, such as chromatin remodeling [17], DNA methylation [18], transcriptional regulation [19], and DNA damage repair [14], [20]. All related processes play a pivotal role in malignant transformation and cancer treatment. However, the function of most lncRNAs in cancer remains a mystery, and lncRNA profiles in hypoxic GC and hypoxia-responsive gene networks remain unknown.
Prototypical lncRNAs have been shown to be overexpressed in subsets of cancers, to interact with epigenetic complexes [17], to be recruited to DNA sequences as enhancers, and to regulate transcription [19], [21] and DNA methylation [18], [22]. To explore the role of lncRNAs in hypoxic GC, we identified a small number of lncRNAs and messenger RNAs (mRNAs) that are aberrantly expressed in GC under hypoxia compared with normoxia using microarrays. We also investigated the biological function of the hypoxia-upregulated AK058003 both in vivo and in vitro. The synuclein family consists of three distinct highly homologous genes, α-synuclein, β-synuclein, and γ-synuclein (SNCG), which have been well studied in connection with cancer [23]. Further analysis demonstrated that SNCG is overexpressed in several types of cancer [24]. In our study, the inhibition of lncRNA-AK058003 and SNCG was found to play an important role in hypoxia-induced metastasis and invasion, suggesting that manipulating new ncRNA functions can provide a therapeutic opportunity that is worth exploring.
Materials and Methods
Ethics Statement
For tissue specimens, signed informed consent was obtained from the patients who contributed dissected tissues. All GC cases were clinically and pathologically confirmed. The experimental procedures were approved by the Institutional Review Board of the Fourth Military Medical University and conformed to the Helsinki Declaration and local legislation. All animal experiments were performed in accordance with national guidelines for the care and use of laboratory animals and with the approval of the Institutional Committee for Animal Research.
Human lncRNA Microarray
Various cell lines (three normoxia-induced GC cell lines and three corresponding hypoxia-induced GC cell lines) were used for human lncRNA microarray analysis. Briefly, total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA) and quantified using a NanoDrop ND-1000. The RNA integrity was assessed using standard denaturing agarose gel electrophoresis [25]. Approximately 5 μg total RNA from each sample was used to synthesize double-stranded cDNA, which was labeled and hybridiszd to the 12x135K Human lncRNA Expression Microarray (Arraystar, Rockville, MD). After hybridization and washing, array scanning was performed with an Axon GenePix 4000B microarray scanner. The acquired array images were analyzed with NimbleScan software (version 2.5). The expression data were normalized via quantile normalization and using the RMA algorithm included in the software. Differentially expressed lncRNAs and mRNAs were identified through fold-change filtering. Finally, hierarchical clustering was performed to show the differential lncRNA and mRNA expression pattern among samples.
Clinical Samples
Twenty pairs of human primary GC and matched noncancerous adjacent gastric tissues were collected at Xijing Hospital, Xi’an, China. Clinicopathologic features, including sex, age, tumor differentiation, tumor stage, and distant metastasis, are listed in Supplementary Table 2. All tissue samples were freshly frozen at − 80°C. Total RNA from the frozen tissues was isolated with TRIzol reagent according to the manufacturer’s instructions.
Lentivirus Infection and Construction of Stable Cell Lines with Downregulated lncRNA-AK058003
To observe the effects of AK058003 knockdown on invasion and metastasis in vivo and in vitro, four different small interfering RNAs (siRNAs) that targeted AK058003 RNA and a scrambled siRNA control were generously provided by GenePharma (Shanghai, China). The four siRNAs were transfected into SGC7901 and MKN45 cells using Lipofectamine 2000 (Invitrogen) following the manufacturer's protocol. Twenty-four hours after transfection, AK058003 expression levels were measured through reverse transcriptase polymerase chain reaction (RT-PCR), and we found that siRNA-AK1 with the sequence 5’-CCACCAGUUACCUGCAAUATT-3’ (sense) and 5’-UAUUGCAGGUAACUGGUGGTT-3’ (antisense) and siRNA-AK2 with the sequence 5’-GGAACAAAGAUGGUUUCUATT-3’ (sense) and 5’- UAGAAACCAUCUUUGUUCCTT-3’ (antisense) yielded the highest degree of AK058003 knockdown. Then, we designed and synthesized AK058003-targeting sequence and inserted this sequence into a Supersilencing Vector (GenePharma, Shanghai, China). An unrelated sequence lentiviral vector was used as a negative control. SGC7901 and MKN45 cells were then plated into six-well plates and allowed to adhere for 24 hours. Next, the lentivirus was transfected according to the manufacturer’s instructions. Stably transfected cells were selected with puromycin (Sigma-Aldrich, St. Louis, MO) and confirmed through fluorescence microscopy and RT-PCR.
Oligonucleotide Construction
siRNAs targeting SNCG were synthesized by GeneChem (Shanghai, China), and a scrambled siRNA control was used as a negative control (provided by GeneChem). The sequence for the SNCG siRNA was 5’-CCAAGGAGAATGTTGTACA-3’. The cells were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions.
Plasmid Construction
The full length of SNCG was amplified (forward primer: 5’-GATATGAATTCGCCACCATGGATGTCTTCAAGAAGGGCTTCTCC-3’; reverse primer: 5’-GTATCGGATCCCTAGTCTCCCCCACTCTGG-3’) and then cloned into the pEX-2 vector. After digestion with EcoRI and BamHI, the correctness of the construct was confirmed by sequencing. An empty vector was used as a negative control.
In Vitro Migration and Invasion Assays
For transwell migration assays, 5×104 cells in serum-free RPMI 1640 medium were added to the upper chamber of each insert (BD Biosciences, Franklin Lakes, NJ). For invasion assays, the chamber inserts were coated with 50 mg/l Matrigel (BD Biosciences, San Jose, CA). After 4 to 5 hours of incubation at 37°C, 1×105 cells in serum-free RPMI-1640 medium were added to the upper chamber. In both assays, medium supplemented with serum was used as a chemoattractant in the lower chamber. After incubation in a normoxia (37°C and 5% CO2) or hypoxia (37°C, 1% O2, 5% CO2, and 94% N2) chamber for 24 or 48 hours, the cells on the upper surface were removed, and the cells on the lower surface of the membrane were fixed in 100% methanol for 15 minutes, air dried, stained with 0.1% crystal violet, and counted under a microscope (Olympus Corp., Tokyo, Japan) to calculate relative numbers. Nine random fields were analyzed per insert. Each experiment was conducted in triplicate in three independent experiments.
High-Content Screening Assay
Briefly, 5×103 cells were plated into each well of a 96-well plate and incubated at 37°C. After 24 hours, the culture medium was replaced with serum-free RPMI 1640 medium, and the cells were cultured for an additional 24 hours. The cells were then washed twice with ice-cold phosphate-buffered saline (PBS) and stained with Hoechst 33342 for 15 minutes in an incubator. The cells were subsequently washed twice with ice-cold PBS, and culture medium was added to each well. Cell motility was detected with a Cellomics ArrayScan VTI HCS (Thermo Scientific, Waltham, MA) according to the manufacturer’s instructions (five replicate wells per group).
Wound-Healing Assays
SGC7901-siAK or SGC7901-Scr and MKN45-siAK or MKN45-Scr cells were seeded in six-well plates and incubated until 90% confluence in serum-free medium before wounding. A 200-μl tip was used to make a vertical wound, and the cells were then washed three times with PBS to remove cell debris. Cell migration into the wounded area was monitored by microscopy at the designated times.
In Vivo Metastasis Assays
Nude mice were purchased from the Experimental Animal Center of the Fourth Military Medical University. For in vivo metastasis assays, 2×106 SGC7901 and MKN45 cells infected with a lentivirus containing AK058003 siRNA and a negative control were suspended in 0.2 ml PBS and injected into the tail vein of each mouse. After 6 weeks, the mice were sacrificed, and their tumor nodules were counted under a stereomicroscope (Olympus). The tumor tissues derived from various organs were then dissected and histologically examined. Each tumor cell line was injected into 10 mice.
Bisulfite Sequencing PCR Analyses
Genomic DNA was extracted from GC cells with the QIAamp DNA Mini Kit (Qiagen, Valencia, CA) and subjected to bisulfite modification using an EpiTect Bisulfite kit (Qiagen) according to the manufacturer’s protocol. We used Methyl Primer Express v1.0 to design primers on bisulfite-treated DNA.The primer is forward: 5'-GTTGTTTTGGGATAGGGGTT-3' and reverse: 5'-CCRCAAACAAAAAAATACAAA-3'. PCR was performed in a final volume of 25 ml containing ddH2O 19.5μl, 10 × PCR buffer 2.5μl, dNTP Mix 0.5μl, 0.5μl of each primer, 0.5μl rTaq, and 1μl DNA. PCR was carried out at 94°C for 5 minutes; 40 cycles at 94°C for 30 seconds, 58°C for 30 seconds, and 72°C for 30 seconds; and finally 72°C for 10 minutes. The PCR product was ligated into T Vector. After transformation, individual colonies were picked, and the insert was sequenced and analyzed by BiQ_Analyzer.
Statistical Analyses
The SPSS 12.0 program (SPSS Inc., Chicago, IL) was used for statistical analyses. The data are presented as the mean±standard error for at least three independent experiments. The differences between groups were analyzed using Student’s t test when comparing only two groups or one-way analysis of variance when comparing more than two groups. The chi-square test was used to analyze the relationship between SNCG expression and various clinicopathologic characteristics. AK058003 and SNCG expression levels in clinical GC tissues and corresponding adjacent nontumorous tissues were compared using the Wilcoxon signed-rank test. Correlations between AK058003 and SNCG expression in tissue specimens were explored using Pearson’s correlation. P < .05 was considered significant.
A detailed description of the materials and methods used in this study can be found in the Supporting Materials.
Results
lncRNA Expression Profile in Hypoxia-Induced GC Cells
To examine the overall impact of lncRNAs on hypoxic GC, we analyzed the expression profiles of lncRNAs and protein-coding RNAs in normoxia-induced and hypoxia-induced GC cells using microarray analysis. Scatter and volcano plots are shown in Figure S1. Hierarchical clustering showed the differential lncRNA and protein-coding RNA expression profiles between normoxia-induced and hypoxia-induced GC cells (Figure 1, A and B). We set a threshold of a fold change > 1.5, P < .05, and found that 84 lncRNAs were upregulated and 70 were downregulated in all hypoxia-induced GC cells compared with normoxia-induced GC cells (Figure 1, C and D; Supplementary Table 3, Supplementary Table 4). This finding indicated that the lncRNA expression profiles differed between the two groups.
Figure 1.
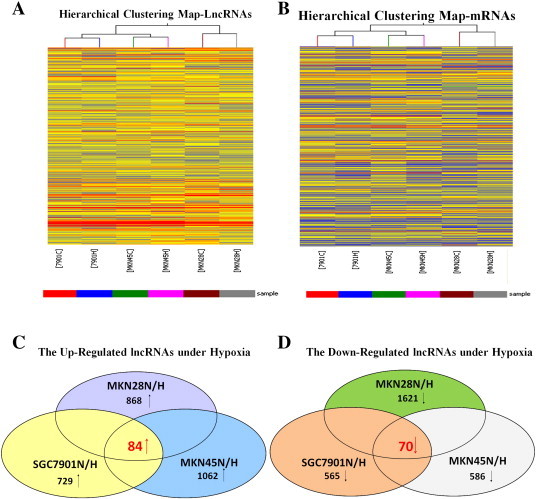
Differentially expressed lncRNAs (A) and mRNAs (B) were analyzed using hierarchical clustering. Hierarchical clustering analysis arranges samples into groups based on expression levels, which allows us to hypothesize the relationships between samples. The dendrogram shows the relationships between the lncRNA (A) and mRNA (B) expression patterns in the samples, that is, which samples are more similar in the expressing relationships. For every gene in each sample, the “Red” indicates high relative expression, and “Blue” indicates low relative expression. Actually, there is no inevitable relation between the lncRNA shown in A and mRNA presented in B expression patterns in the samples because they were analyzed independently. Schemas of the upregulated (C) and downregulated (D) lncRNAs, identified by microarray in the GC cells SGC7901, MKN45, and MKN28 under hypoxia. The overlapping areas represent the common lncRNAs in all three GC cell lines under hypoxia.
To validate the microarray findings, we randomly selected six lncRNAs from the differentially expressed lncRNAs with a fold change > 3 and analyzed their expression through real-time PCR with hypoxia-induced GC cells (after 24 hours in 1% O2 for the SGC7901, MKN45, and MKN28 GC cells) relative to normoxia-induced GC cells. The expression of most lncRNAs was consistent with the microarray results (Figure S2, Supplementary Table 1), indicating that a set of lncRNAs is frequently aberrantly expressed in hypoxia-induced GC cells.
Newly Identified AK058003 Frequently Upregulated in GC and Induced by Hypoxia in GC Cells
Among the differentially expressed lncRNAs among hypoxia-induced GC cells and normoxia-induced GC cells, we were particularly interested in lncRNA-AK058003 because its expression increased approximately 6.20±1.65-fold upon hypoxia treatment in all three cell lines. Thus, we studied the role of AK058003, which is an intronic antisense lncRNA.
Given that AK058003 is induced by hypoxia in GC cells, we next sought to determine whether AK058003 could be induced by hypoxia at different exposure times (after 4, 8, 16, 24, and 48 hours in 1% O2) in GC cells. We found that AK058003 was induced under hypoxia, with the most robust induction observed after 16 hours in 1% O2 for SGC7901 cells, 24 hours in 1% O2 for MKN45 cells, and 48 hours in 1% O2 for MKN28 cells (Figure 2, A–C). The results suggest that AK058003 can indeed be regulated by hypoxia in GC cells; however, no significant difference was observed in expression after 4 or 8 hours in 1% O2.
Figure 2.
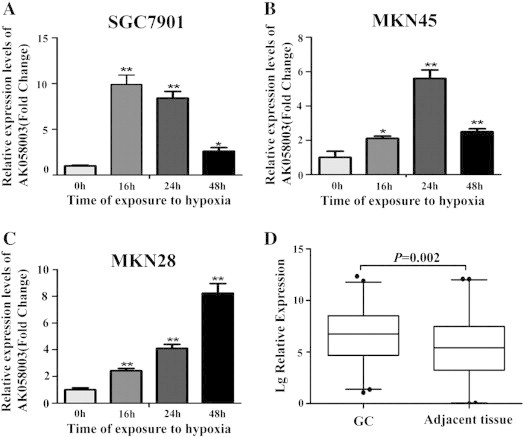
AK058003 is often upregulated in GC and is induced by hypoxia in GC cells. (A, B, C) AK058003 was induced under hypoxia, with the most robust induction observed after 16 hours in 1% O2 in (A) SGC7901 cells, 24 hours in 1% O2 in (B) MKN45 cells, and 48 hours in 1% O2 in (C) MKN28 cells. (D) RNA was extracted with TRIzol reagent from 95 pairs of human GC and adjacent tissues. AK058003 expression was assessed by real-time PCR. β-Actin was used as an internal control. The significant differences between samples were analyzed using the Wilcoxon signed-rank test (P = .002, n = 95).
Next, we assessed AK058003 expression in 95 pairs of human primary GC tissues and adjacent gastric tissues using quantitative RT-PCR to determine AK058003 expression in GC tissues. AK058003 expression was remarkably upregulated in GC tissues compared with noncancerous gastric tissues (Figure 2D), indicating that AK058003 upregulation is common in GC.
Effect of AK058003 on GC Cell Migration and Invasion and Hypoxia-Induced Migration and Invasion
The frequent AK058003 upregulation in hypoxic GC cells implies that AK058003 may play a role in hypoxia-induced GC. To test this hypothesis, the effects of reduced AK058003 expression on cell proliferation, migration, and invasion were investigated in two GC cell lines. Four different siRNA molecules were tested for their knockdown efficiencies, and the two most efficient of these molecules (siRNA-AK1 and siRNA-AK2) were selected for subsequent studies (Figure 3, A and B). We first established SGC7901 and MKN45 cell lines that stably repressed AK058003 expression by using an AK058003 siRNA-lentivirus (si-AK) vector, as verified by fluorescence microscopy (Figure 3C) and RT-PCR (Figure 3D). However, 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide and colony-forming assays showed that AK058003 downregulation did not significantly affect cell proliferation (Figure S3).
Figure 3.
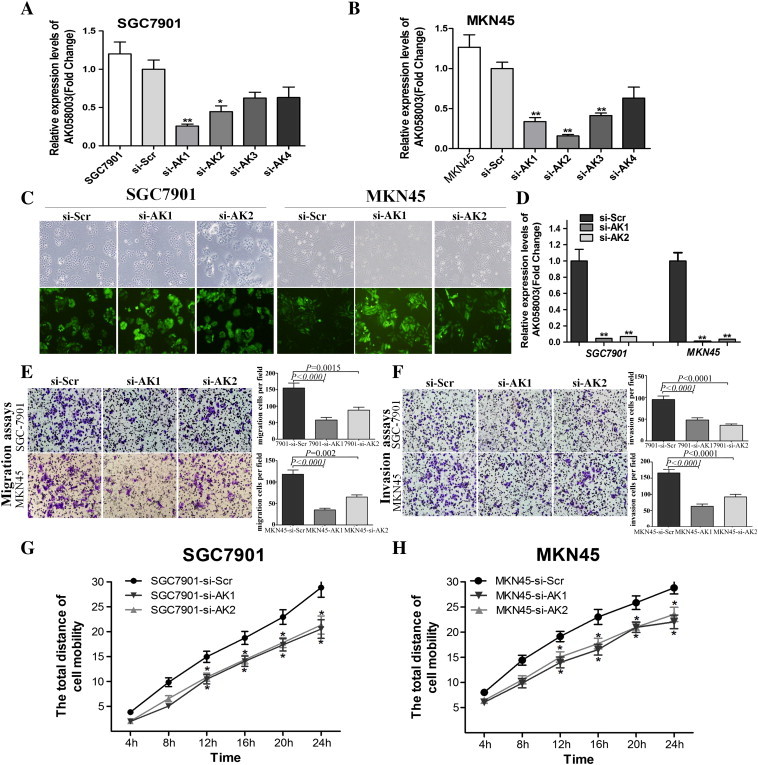
AK058003 promotes GC cell migration and invasion. (A, B) AK058003 expression following knockdown by four different siRNA (si-AK1, si-AK2, si-AK3, and si-AK4) in the GC cell lines SGC7901 (A) and MKN45 (B). (C) Observation of the infection efficiency of AK058003 (si-AK1 and si-AK2) and scrambled siRNA lentivirus (si-Scr) by fluorescence microscopy. (D) After transfection, the cells were collected, and AK058003 expression was assessed by RT- PCR. (E, F) Transwell migration (E) and invasion (F) assays of SGC7901 and MKN45 cells were performed after transfection with AK058003 si-AK1 and si-AK2 or a scrambled siRNA control (si-Scr). (G, H) Cell mobility was examined with a Cellomics ArrayScan VTI 1700 Plus. The relative distance traveled because of SGC7901 (G) and MKN45 (H) cell mobility was calculated with this instrument. In all panels, the results are representative of at least three independent experiments.
To investigate whether AK058003 might have a role in hypoxia-induced metastasis, we first determined whether AK058003 affected normoxic GC cell migration and invasion. In transwell assays with or without Matrigel, SGC7901 and MKN45 cells with stable AK058003 knockdown showed significantly decreased migration and invasion compared with control cells (Figure 3, E and F). Moreover, we examined cell motility in different groups under normoxia using high-content screening, which indicated that cell motility was reduced in AK058003 siRNA-lentivirus–treated cells compared with scrambled siRNA–treated cells (Figure 3, G and H). Taken together, these results indicate that although AK058003 did not affect GC cell growth, the lncRNA promoted migration and invasion.
Given that AK058003 can promote normoxic GC cell migration and invasion, we hypothesized that AK058003 might play a role in hypoxia-induced migration and invasion. To test this hypothesis, we first measured hypoxic GC cell migration and invasion. Hypoxia significantly increased the migration and invasion potential of SGC7901 and MKN45 cells (Figure 4, A–D), which is consistent with previous reports [4], [5]. After transfection with a siRNA-lentivirus to decrease endogenous AK058003 expression, the hypoxia-induced migration and invasion of SGC7901 and MKN45 cells were dramatically diminished (Figure 4, A–D). In addition, wound-healing assays showed that, under hypoxia, the cells more rapidly closed wounds (Figure 4, E–H). However, after AK058003 knockdown in GC cells, wound healing was not significantly promoted under hypoxia compared with the control cells (Figure 4, E–H).
Figure 4.
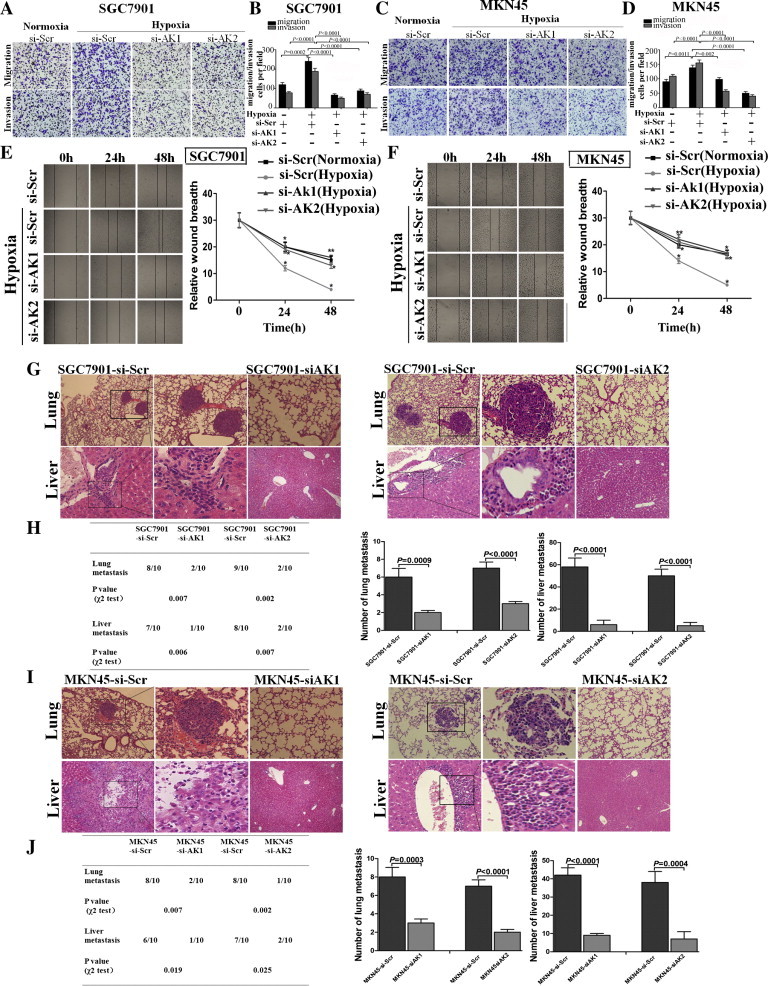
AK058003 mediates hypoxia-induced GC cell migration and invasion. (A, B) Transwell migration and invasion assays of SGC7901 cells were performed after transfection with AK058003 siRNA-lentivirus (si-AK1 and si-AK2) or a scrambled siRNA control (si-Scr) under normoxic or hypoxic conditions. (C, D) Transwell migration and invasion assays of MKN45 cells were performed after transfection with AK058003 siRNA-lentivirus (si-AK1 and si-AK2) or a scrambled siRNA control (si-Scr) under normoxic or hypoxic conditions. In all panels, the results are representative of at least three independent experiments. (E, F) Wound-healing assays were performed to evaluate the effect of AK058003 expression on cell migration. Cell culture plates with SGC7901 cells (E) that were transfected with AK058003 (si-AK1 and si-AK2) or a scrambled siRNA control (si-Scr). MKN45 cells (F) that were transfected with AK058003 (si-AK1 and si-AK2) or a scrambled siRNA control (si-Scr). Cells were wounded and incubated under normoxic or hypoxic conditions. Healing was determined at the indicated times. (H, J) The incidence of metastasis in mice (left panel) and the mean number of visible tumor nodules in the liver and lung (right panel) due to SGC7901-transfected cells (H) and MKN45-transfected cells (J) (n = 10 per group). (H) SGC7901 cells were transfected with the AK058003 siRNA-lentivirus (si-AK1 and si-AK2) or a scrambled siRNA control (si-Scr), and (J) MKN45 cells were transfected with the AK058003 siRNA-lentivirus (si-AK1 and si-AK2) or a scrambled siRNA control (si-Scr). The cells were then injected into nude mice via the tail vein for an in vivo metastasis assay, and the animals were sacrificed 6 weeks after injection. (G, I) Representative hematoxylin and eosin staining of lungs and livers isolated from mice injected with SGC7901-si-Scr or SGC7901-si-AK cells (G) and MKN45-si-Scr or MKN45-si-AK cells (I). The arrows indicate tumor foci in the lungs and livers. Magnification, × 100 (left) and × 200 (right).
To further explore the role of AK058003 in tumor invasion and metastasis in vivo, SGC7901 and MKN45 cells with stable AK058003 repression by an AK058003 siRNA-lentivirus or a scrambled siRNA vector-transfected control cells were delivered into nude mice via tail vein injection. We found that the number and size of lung and liver metastatic nodules dramatically decreased in mice administered cells with low AK058003 expression compared with the scrambled siRNA controls (Figure 4, G–J). Taken together, these observations suggest that AK058003 is a positive metastatic regulator of GC.
SNCG an AK058003-Regulated Gene in GC
To explore the mechanism by which AK058003 promotes GC cell migration and invasion, we attempted to identify the lncRNA’s potential target genes. Recent studies have reported that lncRNAs may function by positively or negatively regulating the expression of their neighboring protein-coding genes [14], [16], [26], [27], [28].Thus, we retrieved the genomic locus information from the UCSC genome browser (http://genome.ucsc.edu/) and found that the tumor oncogene SNCG is located 8.6 kb downstream of AK058003, whereas another tumor suppressor gene, BMPR1A, is found 33.3 kb upstream of AK058003 (Figure 5A). To validate the association between the lncRNA AK058003 and BMPR1A or SNCG, we performed RT-PCR and western blotting using SGC7901 and MKN45 cells with AK058003 knockdown. BMPR1A expression was unchanged (data were not shown), whereas we observed significantly decreased SNCG levels in cells with low AK058003 expression compared with scrambled siRNA control cells (Figure 5, B–E) (This SNCG is a monomer; the molecular weight of the bands in the figure is approximately 16 kDa, predicted molecular weight: 13 kDa (Abcam, Cambridge, MA, Cat. #: ab52633)). Moreover, we showed that AK058003 suppressed SNCG mRNA and protein levels in GC cells and that there was a dose-dependent relationship between the expression of SNCG and AK058003 (Figure 5, B–E). These results suggest that SNCG is an AK058003-regulated gene in GC. We also found that SNCG was predominantly located in the cytoplasm of GC cells. Moreover, we found that SNCG expression was heterogeneous in tumor tissue; it was predominantly located in the center of the tumor but had almost negative expression in the stroma of GC tissues. There was little SNCG expression in paracancerous tissues. These results confirmed that SNCG protein levels in GC tissue were significantly higher than in paracancerous tissues. Additional analysis revealed that SNCG showed strong staining at primary sites from patients with metastatic GC compared with samples from patients with nonmetastatic GC (Figure 6A). Next, we analyzed the correlation between SNCG expression and the clinicopathologic parameters of GC patients. As shown in Supplementary Table 5, SNCG overexpression was associated with the depth of tumor invasion, the clinical tumor node metastasis stage, lymph node metastasis, and vascular invasion (P < .001 for all by the Mann-Whitney test). SNCG expression in GC patients did not correlate with age, sex, or cell differentiation.
Figure 5.
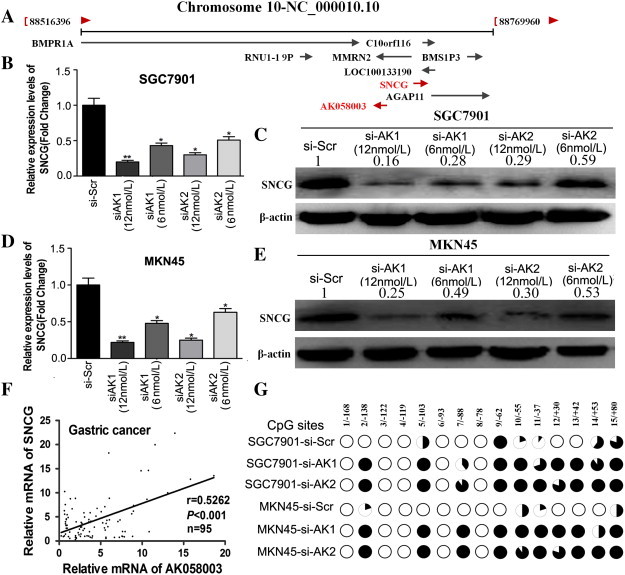
AK058003 upregulates SNCG in GC cells. (A) A diagram of the genes located around AK058003. (B–E) The mRNA (B, D) and protein (C, E) levels of SNCG were assessed through RT-PCR and Western blotting, respectively, in SGC7901 and MKN45 cells transfected with an AK058003 siRNA-lentivirus (si-AK1 and si-AK2) or a scrambled siRNA control (si-Scr) in a dose-dependent manner. (F) Correlation between the relative mRNA levels of AK058003 and SNCG in 95 GC tissue specimens (r = 0.5262, P < .001, Pearson’s correlation). (G) Methylation mapping of 15 CpG island in SNCG exon 1 region obtained from bisulfite sequencing in scrambled siRNA control MKN45 or SGC7901 cells, and MKN45 or SGC7901cells infected with AK058003 siRNA. CpG positions are indicated relative to the translation start codon, and each circle in the figure represents a single CpG site. For each cell line, the percentage methylation at a single CpG site is calculated from the sequencing results of 10 independent clones. Black circles, 100% methylated; white circles, 0% methylation.
Figure 6.
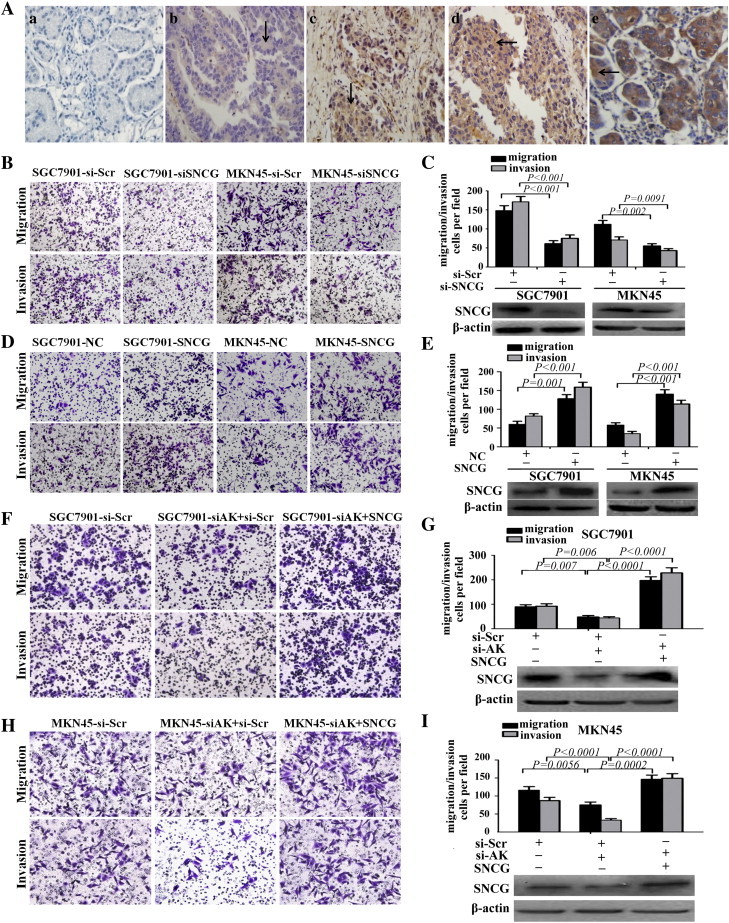
SNCG functions as a metastasis-related gene in GC and can promote AK058003-induced GC cell migration and invasion. (A) Immunohistochemical analysis of SNCG in metastatic and nonmetastatic GC: (a) noncancerous region of GC, (b) primary site of nonmetastatic GC, (c, d) primary site of metastatic GC, and (e) breast cancer as positive control. (B, C) Transwell migration and invasion assays of SGC7901 and MKN45 cells were performed after transfection with siRNA directed against SNCG or a scrambled siRNA control (si-Scr). SNCG protein levels were detected by Western blot analysis as well (at bottom of Figure C). (D, E) Transwell migration and invasion assays of SGC7901 and MKN45 cells were performed after transduction with SNCG or a negative control. SNCG protein levels were detected by Western blot analysis as well (at bottom of E). (F, G, H, I) Transwell migration and invasion assays of SGC7901-si-Scr or SGC7901-si-AK cells (F, G) and MKN45- si-Scr and MKN45-si-AK cells (H, I) were performed after transduction with a scrambled siRNA control (si-Scr) or SNCG. SNCG protein was detected by Western blot analysis as well (at bottom of G and I). In all panels, the results are representative of at least three independent experiments.
We further investigated the correlation between AK058003 and SNCG expression in 95 clinical GC tissues and found that AK058003 and SNCG expression levels were positively correlated (Figure 5F). These observations indicate that SNCG is frequently upregulated in GC samples, possibly because of AK058003 overexpression.
Next, to investigate the potential mechanism by which AK058003 contributes to malignant behaviors of GC cells, we focused on DNA methylation of SNCG because lncRNAs are known to be involved in epigenetic regulation and DNA hypomethylation of SNCG has been confirmed in laryngeal cancer, including GC. Then, we performed bisulfite sequencing of cloned alleles over the region of − 169 to 81 of the SNCG exon1 CpG islands, which has been demonstrated to be involved in GC [29]. The results showed that SNCG CpG islands were densely hypomethylated in control SGC7901-si-Scr and MKN45-si-Scr cells, but GC cells transduced with AK058003 siRNA showed more methylated CpG dinucleotides (Figure 5G). Taken together, these results indicate that knockdown of AK058003 can downregulate the expression of SNCG in GC cells by regulating SNCG DNA methylation, which illustrates that SNCG may act as a downstream target of AK058003 in GC cells.
SNCG Mediates AK058003-induced Migration and Invasion of GC Cells
To further explore the biological function of SNCG in GC, siRNAs targeting SNCG were designed and transfected into SGC7901 and MKN45 cells. These siRNAs significantly decreased SNCG expression (Figure 6C). The migration and invasion of the two cell lines were significantly decreased by SNCG siRNA but not by the scrambled siRNA control (Figure 6, B and C). To support these findings, we overexpressed SNCG in SGC7901 and MKN45 cells, which significantly increased cell migration and invasion (Figure 6, D and E).
If SNCG is indeed a functional gene of AK058003 in GC, then SNCG restoration in AK058003-downregulated GC cells should abrogate the effects of AK058003. To test this hypothesis, we introduced an SNCG expression vector into SGC7901 and MKN45 GC cells that had reduced AK058003 expression, restoring SNCG protein level (Figure 6, F–I). After SNCG restoration, the migration and invasion of SGC7901 and MKN45 cells that were previously inhibited by si-AK058003 significantly increased. These results indicate that SNCG is a functional gene of AK058003 in GC cells.
SNCG Regulation by AK058003 Mediates Hypoxia-Induced GC Metastasis
To determine whether SNCG is involved in AK058003-induced hypoxic GC cell metastasis, we first detected SNCG expression under hypoxia. The results showed that SNCG expression in GC cells increased under hypoxia compared with normoxia (Figure 7, A and B; upper panel). When AK058003 expression was downregulated in SGC7901 and MKN45 cells, the hypoxia-induced SNCG increase was abrogated (Figure 7, A and B; lower panel), indicating that SNCG overexpression was mediated by AK058003 upregulation under hypoxia. Moreover, transwell assays indicated that inhibiting SNCG expression promoted hypoxia-induced migration and invasion (Figure 7, C–F). To determine the function of SNCG in AK058003-induced GC metastasis under hypoxia, an SNCG expression vector and a scramble control were cotransfected into SGC7901 and MKN45 cells with stable AK058003 suppression. In transwell experiments, the impairment of the effects of siRNA-mediated AK058003 on hypoxia-induced GC cell migration and invasion was partially relieved by SNCG but not by the negative control (Figure 7, G–J). Taken together, these results indicate that SNCG expression is increased by hypoxia and that SNCG upregulation by AK058003 mediates hypoxia-induced GC metastasis.
Figure 7.
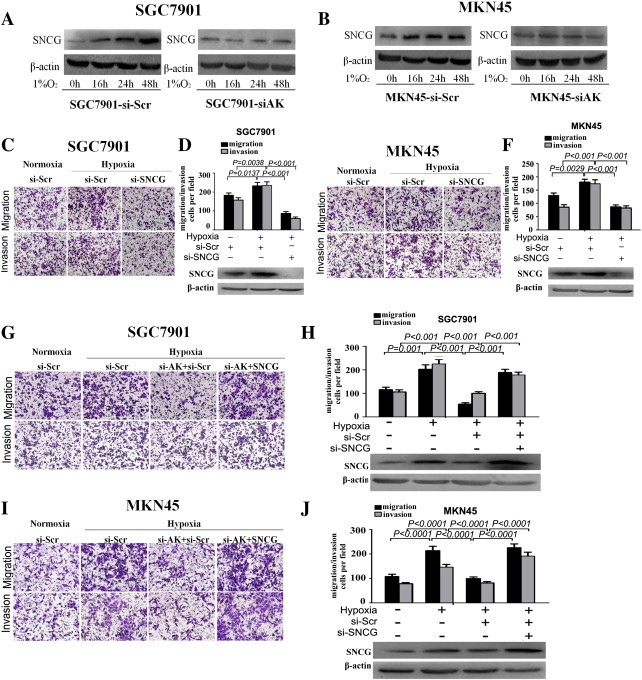
Restoration of SNCG significantly increased the GC cell migration and invasiveness inhibited by AK058003 knockdown under hypoxia. (A, B) SNCG protein levels in SGC7901 and MKN45 cells. The cells were transfected with an AK058003 siRNA-lentivirus (si-AK) or a scrambled siRNA control (si-Scr) and then exposed to hypoxia after transfection. The cells were then collected and subjected to Western blot analysis at specific time points, as indicated. β-Actin served as an internal control. (C, D, E, F) Transwell migration and invasion assays of SGC7901-si-Scr and SGC7901-si-SNCG cells (C, D) and MKN45-si-Scr and MKN45-si-SNCG cells (E, F) were performed under normoxic or hypoxic conditions. SNCG protein levels were determined by Western blot assays as well (at bottom of D and F). (G, H, I, J) Transwell migration and invasion assays of SGC7901 (G, H) and MKN45 (I, J) cells were performed after transfection with an AK058003 siRNA-lentivirus (si-AK), SNCG expression vector, or a scrambled siRNA control (si-Scr) under normoxic or hypoxic conditions. SNCG protein levels were determined through Western blot assays as well (at bottom of H and J). In all panels, the results are representative of at least three independent experiments.
Discussion
Hypoxia is an important microenvironmental factor in nearly every solid tumor, including GC, and it is related to an increased risk of metastasis and invasion [30]. The contribution of hypoxia-induced signaling pathways to malignancy is of great interest [31], [32]. Our previous studies found that many protein-coding genes associated with hypoxia correlation with GC metastasis and invasion [4], [5], [6]. However, the exact mechanism in this process has not been thoroughly elucidated. Recently, mounting evidence suggests that the expression of many lncRNAs is altered in different types of human cancer [33]. Fu Yang et al. demonstrated that a hypoxic microenvironment suppresses lncRNA-LET, which was associated with HepatoCellular Carcinoma metastasis [10]. In another study, Fan Yang et al. found that lincRNA-p21 is a hypoxia-responsive lncRNA and is essential for hypoxia-enhanced glycolysis [34]. However, whether lncRNA is also involved in the hypoxic GC remains uncharacterized. In this study, as a first attempt to investigate whether lncRNA expression is altered under hypoxic conditions, we first searched for potential hypoxia-responsive lncRNAs using lncRNA microarrays between hypoxia-induced GC cells and normoxia-induced GC cells. The results showed that hypoxia alters lncRNA expression and indicate that hypoxic GC may have specific lncRNA profiles. We then selected six lncRNAs from the differentially expressed lncRNAs with a fold change > 3 and performed a quantitative RT-PCR to examine these lncRNAs’ expression levels in GC cells under both normoxic and hypoxic conditions. Of these six lncRNAs, only AK058003, which is a 1197-bp transcript and is located in the chromosome 10q22 on the forward strand, was strongly induced by hypoxia in three GC cells and upregulated in GC samples. In addition, lncRNA-AK058003 was induced by hypoxia at a time-dependent manner in different GC cells. These data indicate the specificity of lncRNA-AK058003 in hypoxic induction. Moreover, we found that AK058003 mediated hypoxia-induced GC migration and invasion in vitro and in vivo. To our knowledge, this study is the first report demonstrating that an lncRNA enhances GC cell migration and invasion under hypoxia. Thus, our results suggest that lncRNA-AK058003 may not only act as a tumor hypoxia marker or adjust cells to hypoxic stress in tumors but may also have a biological role in tumor malignancy and metastasis.
However, the mechanism of lncRNAs in hypoxic cancer has not been thoroughly elucidated. Several reports have demonstrated that the expression levels of certain lncRNAs are correlated with the nearest protein-coding genes and that these lncRNAs may have either a positive [26], [35] or negative [14], [16] effect on gene expression. Accordingly, we were interested in the genes surrounding AK058003. We then retrieved the genomic locus information and found that the SNCG gene, which is a prometastatic oncogene, is only approximately 8.6 kb downstream from the AK058003 gene.
SNCG was first discovered by direct differential cDNA sequencing in breast cancer samples [36] and can promote breast cancer metastasis and invasion [37]. Certain studies have revealed a strong correlation between SNCG expression in primary tumors and distant metastasis in many cancer types, including liver, esophageal, colon, gastric, lung, prostate, and cervical cancers [38]. Furthermore, SNCG expression may be related to the tumor microenvironment [39]. In our study, we found that SNCG expression is increased in GC samples, particularly in metastatic tissues, and promotes GC metastasis. After analyzing the relationship between SNCG expression and the clinicopathologic factors of GC patients, we found a significant association between SNCG expression and the depth of tumor invasion, clinical tumor node metastasis stage, lymph node metastasis, and vascular invasion. Moreover, we observed that SNCG was also involved in hypoxia-induced GC metastasis. Furthermore, we found that the SNCG mRNA levels were positively correlated with the expression of AK058003 in 95 pairs of clinical GC samples. Accordingly, we have found that AK058003 knockdown could downregulate SNCG expression at the mRNA and protein levels in a dose-dependent manner. These data demonstrated that SNCG downregulation is at least partly caused by AK058003 downregulation in GC.
However, the mechanism of AK058003 regulation of SNCG is still unknown. Surgucheva et al. demonstrated that mature miRs repress protein expression primarily through base pairing of a seed region with the 3’-UTR of SNCG [40]. Furthermore, several studies reported that SNCG expression is activated by demethylation of the SNCG CpG island in the development of cancer, including GC [29], [41], [42]. In another study, H. Liu et al. found that cigarette smoke induces the demethylation and subsequent expression of SNCG gene in lung cancer cells through downregulation of DNA methyltransferase 3B [43]. Recently, increasing evidence confirmed that lncRNA can regulate DNA methylation of protein-coding genes during the development of disease [22], [35], [44], [45]. For instance, in a recent paper published in Nature, Di Ruscio and colleagues reported that the lncRNA-ecCEBPA interacts with DNA methyltransferase 1, resulting in prevention of CEBPA gene methylation and robust CEBPA mRNA production [35]. Wei Li et al. demonstrated that linc-POU3F3 promotes cell viability and proliferation in esophageal squamous cell carcinoma cell levels and altered linc-POU3F3 levels could drive the methylation of the POU3F3 gene, which is located in the downstream of 4 kb from the linc-POU3F3 [46]. Because SNCG gene is subject to silencing by DNA methylation, we assessed whether depletion of AK058003 resulted in changes in DNA methylation of SNCG. Interestingly, using Bisulfite sequencing PCR assays, we observed a robust increase in levels of DNA methylation of SNCG following the depletion of AK058003.
Furthermore, we have observed that SNCG overexpression was mediated by AK058003 upregulation under hypoxia and that the SNCG upregulation by AK058003 mediates hypoxia-induced GC metastasis. This finding indicated that hypoxia/AK058003/SNCG is a new signaling pathway involved in hypoxia-induced GC metastasis and invasion.
In conclusion, our results indicated that lncRNA-AK058003 expression is frequently increased in hypoxic GC and promotes hypoxia-induced GC metastasis and invasion. SNCG, a metastasis-associated gene in cancer that is involved in hypoxia-induced GC metastasis, was identified as a functional gene that was regulated by AK058003 through DNA demethylation. SNCG upregulation by AK058003 mediates hypoxia-induced GC cell migration and invasion. Further study will explore the exact mechanism of SNCG DNA demethylation that is induced by AK058003. In summary, this finding suggests that the hypoxia/AK058003/SNCG pathway may contribute to the development of new anticancer therapeutics directed against hypoxic tumor targets.
The following are the Supplementary data related to this article.
The expression profiles of lncRNAs and mRNAs were compared between hypoxia-induced and normoxia-induced GC cells. The box plot is a convenient way to quickly visualize the distribution of the lncRNA (A) and mRNA (B) data set profiles. After normalization, the distribution of log2 ratios among six samples was nearly the same. Meanwhile, volcano plots are useful tools for visualizing differential expression between two different conditions. The vertical lines correspond to 1.5-fold upregulation and downregulation, and the horizontal line represents a P value of .05. Thus, the red points in the plot represent the differentially expressed lncRNAs (C) and mRNAs (D) with statistical significance.
The relative expression of lncRNAs in hypoxia-induced and normoxia-induced GC cells by real-time PCR. We used real-time PCR to validate the differential expression of six randomly chosen lncRNAs (three upregulated and three downregulated) in hypoxia-induced (24 hours, 1% O2 and 94% N2) and normoxia-induced GC cells. The relative expression rate of each lncRNA was compared between hypoxic and normoxic GC cells. The data are shown as the mean ± standard deviation of at least three independent experiments. *, P< 0.05; **, P< 0.01.
AK058003 did not affect the proliferation of GC cells in vitro. After silencing AK058003, cell proliferation was determined by (A, B) 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide and (C, D) colony formation assays. The data are reported as the mean ± SD for three independent experiments, and the results showed that the groups did not significantly differ.
Oligonucleotide Sequences Used in This Study
The Clinicopathologic Features of GC Tissues Used in Our Study
Array Data of All 84 Upregulated lncRNAs in the Hypoxia-Induced GC Cells Compared with the Normoxia
Array Data of All 70 Downregulated lncRNAs in the Hypoxia-Induced GC Cells Compared with the Normoxia
Relationship Between Clinicopathologic Parameters and SNCG Expression in GC
Supporting Materials and Methods
Acknowledgments
The authors thank the Xijing Hospital (Xi’an, China) for specimens.
Footnotes
Conflict of Interest: No potential conflicts of interest were disclosed.
Financial Support: This work was supported by the National Nature Science Foundation of China (81272349 and 81372608).
Contributor Information
Helong Zhang, Email: cnxazhl@163.com.
Lili Liu, Email: lily123fmmu@aliyun.com.
References
- 1.Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gupta GP, Massague J. Cancer metastasis: building a framework. Cell. 2006;127:679–695. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 3.Majmundar AJ, Wong WJ, Simon MC. Hypoxia-inducible factors and the response to hypoxic stress. Mol Cell. 2010;40:294–309. doi: 10.1016/j.molcel.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu L, Sun L, Zhao P, Yao L, Jin H, Liang S, Wang Y, Zhang D, Pang Y, Shi Y. Hypoxia promotes metastasis in human gastric cancer by up-regulating the 67-kDa laminin receptor. Cancer Sci. 2010;101:1653–1660. doi: 10.1111/j.1349-7006.2010.01592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y, Li Z, Zhang H, Jin H, Sun L, Dong H, Xu M, Zhao P, Zhang B, Wang J. HIF-1alpha and HIF-2alpha correlate with migration and invasion in gastric cancer. Cancer Biol Ther. 2010;10:376–382. doi: 10.4161/cbt.10.4.12441. [DOI] [PubMed] [Google Scholar]
- 6.Liu L, Zhang H, Sun L, Gao Y, Jin H, Liang S, Wang Y, Dong M, Shi Y, Li Z. ERK/MAPK activation involves hypoxia-induced MGr1-Ag/37LRP expression and contributes to apoptosis resistance in gastric cancer. Int J Cancer. 2010;127:820–829. doi: 10.1002/ijc.25098. [DOI] [PubMed] [Google Scholar]
- 7.Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang H, Zhong Y, Xie H, Lai X, Xu M, Nie Y, Liu S, Wan YJ. Induction of the liver cancer-down-regulated long noncoding RNA uc002mbe.2 mediates trichostatin-induced apoptosis of liver cancer cells. Biochem Pharmacol. 2013;85:1761–1769. doi: 10.1016/j.bcp.2013.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matouk IJ, Mezan S, Mizrahi A, Ohana P, Abu-Lail R, Fellig Y, Degroot N, Galun E, Hochberg A. The oncofetal H19 RNA connection: hypoxia, p53 and cancer. Biochim Biophys Acta. 2010;1803:443–451. doi: 10.1016/j.bbamcr.2010.01.010. [DOI] [PubMed] [Google Scholar]
- 10.Yang F, Huo XS, Yuan SX, Zhang L, Zhou WP, Wang F, Sun SH. Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Mol Cell. 2013;49:1083–1096. doi: 10.1016/j.molcel.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 11.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsai MC, Spitale RC, Chang HY. Long intergenic noncoding RNAs: new links in cancer progression. Cancer Res. 2011;71:3–7. doi: 10.1158/0008-5472.CAN-10-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tano K, Akimitsu N. Long non-coding RNAs in cancer progression. Front Genet. 2012;3:219. doi: 10.3389/fgene.2012.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Du Y, Kong G, You X, Zhang S, Zhang T, Gao Y, Ye L, Zhang X. Elevation of highly up-regulated in liver cancer (HULC) by hepatitis B virus X protein promotes hepatoma cell proliferation via down-regulating p18. J Biol Chem. 2012;287:26302–26311. doi: 10.1074/jbc.M112.342113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng J, Bi C, Clark BS, Mady R, Shah P, Kohtz JD. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 2006;20:1470–1484. doi: 10.1101/gad.1416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, Cui H. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berghoff EG, Clark MF, Chen S, Cajigas I, Leib DE, Kohtz JD. Evf2 (Dlx6as) lncRNA regulates ultraconserved enhancer methylation and the differential transcriptional control of adjacent genes. Development. 2013;140:4407–4416. doi: 10.1242/dev.099390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mattick JS, Taft RJ, Faulkner GJ. A global view of genomic information—moving beyond the gene and the master regulator. Trends Genet. 2010;26:21–28. doi: 10.1016/j.tig.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 20.Wang X, Arai S, Song X, Reichart D, Du K, Pascual G, Tempst P, Rosenfeld MG, Glass CK, Kurokawa R. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454:126–130. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kornienko AE, Guenzl PM, Barlow DP, Pauler FM. Gene regulation by the act of long non-coding RNA transcription. BMC Biol. 2013;11:59. doi: 10.1186/1741-7007-11-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li D, Feng J, Wu T, Wang Y, Sun Y, Ren J, Liu M. Long intergenic noncoding RNA HOTAIR is overexpressed and regulates PTEN methylation in laryngeal squamous cell carcinoma. Am J Pathol. 2013;182:64–70. doi: 10.1016/j.ajpath.2012.08.042. [DOI] [PubMed] [Google Scholar]
- 23.Surguchov A. Synucleins: are they two-edged swords? J Neurosci Res. 2013;91:161–166. doi: 10.1002/jnr.23149. [DOI] [PubMed] [Google Scholar]
- 24.Ahmad M, Attoub S, Singh MN, Martin FL, El-Agnaf OM. Gamma-synuclein and the progression of cancer. FASEB J. 2007;21:3419–3430. doi: 10.1096/fj.07-8379rev. [DOI] [PubMed] [Google Scholar]
- 25.Li JP, Liu LH, Li J, Chen Y, Jiang XW, Ouyang YR, Liu YQ, Zhong H, Li H, Xiao T. Microarray expression profile of long noncoding RNAs in human osteosarcoma. Biochem Biophys Res Commun. 2013;433:200–206. doi: 10.1016/j.bbrc.2013.02.083. [DOI] [PubMed] [Google Scholar]
- 26.Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Orom UA, Derrien T, Guigo R, Shiekhattar R. Long noncoding RNAs as enhancers of gene expression. Cold Spring Harb Symp Quant Biol. 2010;75:325–331. doi: 10.1101/sqb.2010.75.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Orom UA, Shiekhattar R. Noncoding RNAs and enhancers: complications of a long-distance relationship. Trends Genet. 2011;27:433–439. doi: 10.1016/j.tig.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yanagawa N, Tamura G, Honda T, Endoh M, Nishizuka S, Motoyama T. Demethylation of the synuclein gamma gene CpG island in primary gastric cancers and gastric cancer cell lines. Clin Cancer Res. 2004;10:2447–2451. doi: 10.1158/1078-0432.ccr-03-0107. [DOI] [PubMed] [Google Scholar]
- 30.Tsai YP, Wu KJ. Hypoxia-regulated target genes implicated in tumor metastasis. J Biomed Sci. 2012;19:102. doi: 10.1186/1423-0127-19-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ying Q, Liang L, Guo W, Zha R, Tian Q, Huang S, Yao J, Ding J, Bao M, Ge C. Hypoxia-inducible microRNA-210 augments the metastatic potential of tumor cells by targeting vacuole membrane protein 1 in hepatocellular carcinoma. Hepatology. 2011;54:2064–2075. doi: 10.1002/hep.24614. [DOI] [PubMed] [Google Scholar]
- 32.Ghosh G, Subramanian IV, Adhikari N, Zhang X, Joshi HP, Basi D, Chandrashekhar YS, Hall JL, Roy S, Zeng Y. Hypoxia-induced microRNA-424 expression in human endothelial cells regulates HIF-alpha isoforms and promotes angiogenesis. J Clin Invest. 2010;120:4141–4154. doi: 10.1172/JCI42980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reis EM, Verjovski-Almeida S. Perspectives of long non-coding RNAs in cancer diagnostics. Front Genet. 2012;3:32. doi: 10.3389/fgene.2012.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang F, Zhang H, Mei Y, Wu M. Reciprocal regulation of HIF-1alpha and LincRNA-p21 modulates the Warburg effect. Mol Cell. 2014;53:88–100. doi: 10.1016/j.molcel.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 35.Di Ruscio A, Ebralidze AK, Benoukraf T, Amabile G, Goff LA, Terragni J, Figueroa ME, De Figueiredo Pontes LL, Alberich-Jorda M, Zhang P. DNMT1-interacting RNAs block gene-specific DNA methylation. Nature. 2013;503:371–376. doi: 10.1038/nature12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ji H, Liu YE, Jia T, Wang M, Liu J, Xiao G, Joseph BK, Rosen C, Shi YE. Identification of a breast cancer-specific gene, BCSG1, by direct differential cDNA sequencing. Cancer Res. 1997;57:759–764. [PubMed] [Google Scholar]
- 37.Jiang Y, Liu YE, Goldberg ID, Shi YE. Gamma synuclein, a novel heat-shock protein-associated chaperone, stimulates ligand-dependent estrogen receptor alpha signaling and mammary tumorigenesis. Cancer Res. 2004;64:4539–4546. doi: 10.1158/0008-5472.CAN-03-3650. [DOI] [PubMed] [Google Scholar]
- 38.Liu H, Liu W, Wu Y, Zhou Y, Xue R, Luo C, Wang L, Zhao W, Jiang JD, Liu J. Loss of epigenetic control of synuclein-gamma gene as a molecular indicator of metastasis in a wide range of human cancers. Cancer Res. 2005;65:7635–7643. doi: 10.1158/0008-5472.CAN-05-1089. [DOI] [PubMed] [Google Scholar]
- 39.Hu H, Sun L, Guo C, Liu Q, Zhou Z, Peng L, Pan J, Yu L, Lou J, Yang Z. Tumor cell-microenvironment interaction models coupled with clinical validation reveal CCL2 and SNCG as two predictors of colorectal cancer hepatic metastasis. Clin Cancer Res. 2009;15:5485–5493. doi: 10.1158/1078-0432.CCR-08-2491. [DOI] [PubMed] [Google Scholar]
- 40.Surgucheva I, Gunewardena S, Rao HS, Surguchov A. Cell-specific post-transcriptional regulation of gamma-synuclein gene by micro-RNAs. PLoS One. 2013;8:e73786. doi: 10.1371/journal.pone.0073786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gupta A, Godwin AK, Vanderveer L, Lu A, Liu J. Hypomethylation of the synuclein gamma gene CpG island promotes its aberrant expression in breast carcinoma and ovarian carcinoma. Cancer Res. 2003;63:664–673. [PubMed] [Google Scholar]
- 42.Czekierdowski A, Czekierdowska S, Wielgos M, Smolen A, Kaminski P, Kotarski J. The role of CpG islands hypomethylation and abnormal expression of neuronal protein synuclein-gamma (SNCG) in ovarian cancer. Neuro Endocrinol Lett. 2006;27:381–386. [PubMed] [Google Scholar]
- 43.Liu H, Zhou Y, Boggs SE, Belinsky SA, Liu J. Cigarette smoke induces demethylation of prometastatic oncogene synuclein-gamma in lung cancer cells by downregulation of DNMT3B. Oncogene. 2007;26:5900–5910. doi: 10.1038/sj.onc.1210400. [DOI] [PubMed] [Google Scholar]
- 44.Szafranski P, Dharmadhikari AV, Brosens E, Gurha P, Kolodziejska KE, Zhishuo O, Dittwald P, Majewski T, Mohan KN, Chen B. Small noncoding differentially methylated copy-number variants, including lncRNA genes, cause a lethal lung developmental disorder. Genome Res. 2013;23:23–33. doi: 10.1101/gr.141887.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lai F, Shiekhattar R. Where long noncoding RNAs meet DNA methylation. Cell Res. 2014;24:263–264. doi: 10.1038/cr.2014.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li W, Zheng J, Deng J, You Y, Wu H, Li N, Lu J, Zhou Y. Increased levels of the long intergenic non-protein coding RNA POU3F3 promote DNA methylation in esophageal squamous cell carcinoma cells. Gastroenterology. 2014;146:1714–1726. doi: 10.1053/j.gastro.2014.03.002. [e1715] [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The expression profiles of lncRNAs and mRNAs were compared between hypoxia-induced and normoxia-induced GC cells. The box plot is a convenient way to quickly visualize the distribution of the lncRNA (A) and mRNA (B) data set profiles. After normalization, the distribution of log2 ratios among six samples was nearly the same. Meanwhile, volcano plots are useful tools for visualizing differential expression between two different conditions. The vertical lines correspond to 1.5-fold upregulation and downregulation, and the horizontal line represents a P value of .05. Thus, the red points in the plot represent the differentially expressed lncRNAs (C) and mRNAs (D) with statistical significance.
The relative expression of lncRNAs in hypoxia-induced and normoxia-induced GC cells by real-time PCR. We used real-time PCR to validate the differential expression of six randomly chosen lncRNAs (three upregulated and three downregulated) in hypoxia-induced (24 hours, 1% O2 and 94% N2) and normoxia-induced GC cells. The relative expression rate of each lncRNA was compared between hypoxic and normoxic GC cells. The data are shown as the mean ± standard deviation of at least three independent experiments. *, P< 0.05; **, P< 0.01.
AK058003 did not affect the proliferation of GC cells in vitro. After silencing AK058003, cell proliferation was determined by (A, B) 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide and (C, D) colony formation assays. The data are reported as the mean ± SD for three independent experiments, and the results showed that the groups did not significantly differ.
Oligonucleotide Sequences Used in This Study
The Clinicopathologic Features of GC Tissues Used in Our Study
Array Data of All 84 Upregulated lncRNAs in the Hypoxia-Induced GC Cells Compared with the Normoxia
Array Data of All 70 Downregulated lncRNAs in the Hypoxia-Induced GC Cells Compared with the Normoxia
Relationship Between Clinicopathologic Parameters and SNCG Expression in GC
Supporting Materials and Methods


