Abstract
Long noncoding RNAs (lncRNAs) are an emerging class of oncogenic molecules implicated in a diverse range of human malignancies. We recently identified SChLAP1 as a novel lncRNA that demonstrates outlier expression in a subset of prostate cancers, promotes tumor cell invasion and metastasis, and associates with lethal disease. Based on these findings, we sought to develop an RNA in situ hybridization (ISH) assay for SChLAP1 to 1) investigate the spectrum of SChLAP1 expression from benign prostatic tissue to metastatic castration-resistant prostate cancer and 2) to determine whether SChLAP1 expression by ISH is associated with outcome after radical prostatectomy in patients with clinically localized disease. The results from our current study demonstrate that SChLAP1 expression increases with prostate cancer progression, and high SChLAP1 expression by ISH is associated with poor outcome after radical prostatectomy in patients with clinically localized prostate cancer by both univariate (hazard ratio = 2.343, P = .005) and multivariate (hazard ratio = 1.99, P = .032) Cox regression analyses. This study highlights a potential clinical utility for SChLAP1 ISH as a novel tissue-based biomarker assay for outcome prognostication after radical prostatectomy.
Introduction
In the era of prostate-specific antigen (PSA) screening, prostate cancer has a varied clinical course. The majority of tumors are detected early and are usually cured by definitive treatment with radical prostatectomy or radiotherapy. However, even among those with definitively treated, clinically localized prostate cancer, subsets of patients will eventually progress and develop metastatic, castration-resistant prostate cancer (mCRPC)—a disease which is nearly always lethal. Standard pathologic evaluation of radical prostatectomy specimens provides basic risk stratification for prostate cancer progression; however, even these parameters may fail to accurately predict outcome of a proportion of high-risk tumors. With this in mind, considerable discovery efforts have focused on delineating tissue-based prognostic biomarkers for prostate cancer—with little overall success [1].
To date, the majority of biomarker efforts have focused on protein-coding genes, which comprise only a subset of all transcribed genes [2], [3]. Among the more than 90% of transcription that generates noncoding genes, long noncoding RNAs (lncRNAs) most closely resemble protein-coding genes in that they are transcribed by RNA polymerase II, polyadenylated, and associated with specific epigenetic signatures (i.e., H3K4me3 at the promoter and H3K36me3 throughout the gene length) [4], [5]. Although the precise molecular functions of lncRNAs remain poorly understood, an emerging body of evidence indicates that lncRNAs have essential oncogenic roles in a variety of tumor types, suggesting potential utility for clinical assays that detect lncRNA expression.
Using transcriptome sequencing, we recently identified a set of 121 novel lncRNAs that were differentially expressed in prostate cancer versus normal tissue or demonstrated outlier expression in a subset of prostate cancers [6]. SChLAP1, an lncRNA that is highly overexpressed in a subset of prostate cancers and associated with lethal disease, is involved in tumor cell invasion and metastasis [7]. In a limited number of samples, our preliminary data indicate that SChLAP1 expression levels can be detected in formalin-fixed, paraffin-embedded (FFPE) tissue sections by RNA in situ hybridization (ISH), suggesting potential utility of SChLAP1 as a tissue-based prostate cancer biomarker.
Based on these initial findings, we sought to determine SChLAP1 expression by ISH on FFPE tissue in a cohort of patients with clinically localized prostate cancer and lethal mCRPC. We identified a subset of clinically localized prostate cancer patients with high SChLAP1 expression; these patients are associated with high-risk clinicopathologic features (e.g., high Gleason score [GS], seminal vesicle invasion), decreased time to PSA recurrence after radical prostatectomy, and poor clinical outcome after univariate and multivariate Cox regression analyses. In addition, high SChLAP1 expression is detected in a significant proportion of patients with lethal mCRPC. Overall, our results demonstrate suitability of SChLAP1 ISH for the detection of aggressive prostate cancers and indicate that SChLAP1 is a promising tissue-based prognostic biomarker for prostate cancer.
Material and Methods
This study was approved by the University of Michigan (Ann Arbor) Health Sciences Institutional Review Board, and all patients provided written informed consent.
Tissue Microarray (TMA) Construction
TMAs comprised of surgical pathology material from 208 patients with clinically localized prostate cancer were constructed using tumor and benign tissue from radical prostatectomy specimens; all patients had undergone radical prostatectomy at the University of Michigan Health System as primary monotherapy (i.e., no neoadjuvant hormonal or radiation therapy). This radical prostatectomy series is part of the University of Michigan Prostate Cancer Specialized Program of Research Excellence Tissue Core. Three tissue cores (each 0.6 mm in diameter) were obtained from representative FFPE tissue blocks for each included patient sample. Detailed clinicopathologic data for this cohort (summarized in Table 1) are updated and maintained on a secure relational database.
Table 1.
Clinicopathologic Characteristics of 160 Patients with Clinically Localized Prostate Cancer Treated by Radical Prostatectomy.
| Age, y | ||
| ≤ 60 | 86 | (53.8%) |
| > 60 | 74 | (46.2%) |
| GS | ||
| < 7 | 41 | (25.6%) |
| = 7 | 109 | (68.1%) |
| > 7 | 10 | (6.3%) |
| Tumor size, cm | ||
| < 1 | 25 | (15.6%) |
| ≥ 1 | 135 | (84.4%) |
| AJCC T stage | ||
| pT2a | 19 | (11.9%) |
| pT2b | 103 | (64.4%) |
| pT3a | 31 | (19.4%) |
| pT3b | 6 | (3.8%) |
| pT4 | 1 | (0.6%) |
| Surgical margin | ||
| Negative | 114 | (71.2%) |
| Positive | 46 | (28.8%) |
| Preoperative PSA, ng/ml | ||
| < 4 | 22 | (13.8%) |
| 4-7 | 68 | (42.5%) |
| > 7 | 70 | (43.8%) |
| PSA recurrence | ||
| No | 109 | (68.1%) |
| Yes | 51 | (31.9%) |
Similarly, TMAs comprised of rapid autopsy material from 60 patients with lethal mCRPC were constructed; this material was obtained as part of the University of Michigan Prostate Cancer Specialized Program of Research Excellence Rapid Autopsy Program, as described previously [8]. All patients received multimodal therapy, including a combination of radical prostatectomy, hormone deprivation, radiation, and/or chemotherapy; detailed clinicopathologic data for a portion of this cohort have been reported previously [9]. As described above, three tissue cores (each 0.6 mm in diameter) were obtained from representative FFPE tissue blocks from all metastatic tumor sites, as well as primary tumor within the prostate (when present at the time of autopsy; i.e., no prior radical prostatectomy).
SChLAP1 ISH
SChLAP1 ISH was performed on thin (approximately 4 µm thick) TMA sections (Advanced Cell Diagnostics, Inc., Hayward, CA), as described previously [7]; in parallel, SChLAP1 ISH was performed on previously identified positive and negative control FFPE tissue sections, and all controls worked adequately (data not shown). All slides were examined for SChLAP1 ISH signals in morphologically intact cells and scored manually by a study pathologist (R.M.). Specific SChLAP1 ISH signal was identified as brown, punctate dots, and expression level was scored as follows: 0 = no staining or less than 1 dot per 10 cells, 1 = 1 to 3 dots per cell, 2 = 4 to 9 dots per cell (few or no dot clusters), 3 = 10 to 14 dots per cell (less than 10% in dot clusters), and 4 = greater than 15 dots per cell (more than 10% in dot clusters). For each evaluable tissue core, a cumulative ISH product score was calculated as the sum of the individual products of the expression level (0 to 4) and percentage of cells (0 to 100) (i.e., [A% × 0] + [B% × 1] + [C% × 2] + [D% × 3] + [E% × 4]; total range = 0 to 400). For each tissue sample, the ISH product score was averaged across evaluable TMA tissue cores.
Statistical Methods
All statistical analyses were performed using R (version 3.0.2). Mean SChLAP1 expression for benign prostatic glands, clinically localized prostate cancer, and mCRPC were compared using the Student’s t test and analysis of variance. The relationship between SChLAP1 expression (low versus high; see Results section below) and PSA recurrence in patients with clinically localized prostate cancer was examined using the “survival” package in R. Briefly, the PSA recurrence event time was calculated as the date of radical prostatectomy to the time of serum PSA recurrence. (Patients without PSA recurrence were censored on the date of last follow-up.) The probability of PSA recurrence-free survival was then calculated using the Kaplan-Meier product limit method, and the log-rank test was used to compare the survival curves between low– and high–SChLAP1 expression groups. To test for association between SChLAP1 expression and specific clinicopathologic features in clinically localized prostate cancer, Fisher exact test and the Student’s t test were used for categorical and continuous data, respectively. Finally, Cox proportional hazard univariate and multivariate regression models were used to calculate the hazard ratio and associated 95% confidence intervals for SChLAP1 expression and specific clinical variables (e.g., extraprostatic extension), and the statistical significance of Cox models covariate hazard ratios was determined by Wald test.
Results and Discussion
SChLAP1 Expression Increases with Prostate Cancer Progression
To determine its association with prostate cancer progression, we examined SChLAP1 expression by ISH in TMA cohorts of patients with clinically localized prostate cancer or lethal mCRPC. Benign prostate glands, clinically localized prostate cancer, and mCRPC demonstrated a spectrum of SChLAP1 expression by ISH (Figure 1). When present, SChLAP1 staining was predominantly nuclear (Figure 1). Overall, there were significant differences in SChLAP1 expression between benign prostatic glands, clinically localized prostate cancer, and lethal mCRPC (P < .001; Figure 2A). Benign prostatic glands were available for evaluation in 74 patients with clinically localized prostate cancer and, overall, showed absent to low SChLAP1 expression (mean ISH product score = 13.8; range = 0 to 100). Out of a total of 208 patients with clinically localized prostate cancer, tissue from 160 patients (76.9%) was available for SChLAP1 expression evaluation (Table 1). Of these, 58 (36.3%) showed no SChLAP1 expression (ISH product score = 0), whereas the remaining 102 (63.7%) demonstrated a wide spectrum of SChLAP1 expression (overall mean ISH product score = 44.5; range = 0 to 337; Figure 1). Relative to benign prostatic glands, SChLAP1 expression was significantly increased in clinically localized prostate cancer (P < .001; Figure 2A). In addition, high SChLAP1 expression was associated with an increased proportion of clinically localized tumors with high GS (≥ 8; Figure 2B). Rapid autopsy material from a total of 28 patients with lethal mCRPC was available for SChLAP1 expression evaluation, and a large proportion of the patients (15 cases, 53.6%) demonstrated high SChLAP1 expression at one or more tissue sites. Relative to benign prostatic glands (P < .001) and clinically localized prostate cancer (P < .001), SChLAP1 expression was significantly increased in lethal mCRPC (mean ISH product score = 136.4; range = 0 to 370; Figure 2A). Overall, these data indicate that SChLAP1 expression is associated with prostate cancer progression.
Figure 1.
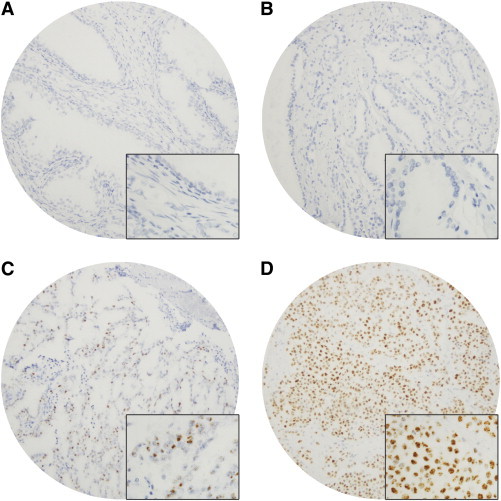
Spectrum of SChLAP1 expression in benign prostatic glands, clinically localized prostate cancer, and lethal mCRPC by ISH. SChLAP1 expression in (A) benign prostatic glands, (B) low- and (C) high-GS clinically localized prostate cancer, and (D) lethal mCRPC. SChLAP1 expression varies from negative to low in benign prostatic glands and low-grade, clinically localized prostate cancers to high expression in a subset of high-grade, clinically localized prostate cancers and lethal mCRPC. Magnification, × 100. Inset in (A), (B), (C), and (D) = magnification, × 400.
Figure 2.

SChLAP1 expression increases with prostate cancer progression. (A) Histogram representation of mean SChLAP1 ISH product score for benign prostatic glands (Benign), clinically localized prostate cancer (PCA), and lethal mCRPC (METS) in a large TMA cohort. Error bars represent standard deviation. SChLAP1 expression is significantly associated with prostate cancer progression, from benign glands to clinically localized prostate cancer to mCRPC (P < .001). (B) Histogram representation of proportion of clinically localized prostate cancer with negative (ISH product score = 0), low (ISH product score > 0 and < 100), or high (ISH product score ≥ 100) SChLAP1 expression, stratified by GS. High SChLAP1 expression is associated with increasing GS in clinically localized prostate cancer.
High SChLAP1 Expression in Clinically Localized Prostate Cancer Is Associated with High-Risk Clinicopathologic Features and Poor Clinical Outcome
To investigate the relationship between SChLAP1 expression and PSA recurrence after radical prostatectomy, we performed logistic regression with the ISH product score and plotted the data as a receiver operating characteristic curve (area under the curve = 0.668; Figure 3A). In this model, the threshold which maximizes the sum of sensitivity and specificity corresponds to a SChLAP1 ISH product score of 100.9. Hence, we used an ISH product score of 100 (rounded down from 100.9 for practicality and convenience) to differentiate patients with low and high SChLAP1 expression in our clinically localized prostate cancer cohort. Comparing these groups revealed that high SChLAP1 expression is associated with significantly decreased time to PSA recurrence after radical prostatectomy (P = .004; Figure 3B). Next, we evaluated the association between SChLAP1 expression by ISH and standard clinicopathologic parameters in our TMA cohort of clinically localized prostate cancer. In these patients, high SChLAP1 expression is associated with several high-risk clinicopathologic features, including high GS (≥ 8) and seminal vesicle invasion (P < .05; Table 2 and Figure 3C). Finally, by both univariate (hazard ratio = 2.343, P = .005) and multivariate (hazard ratio = 1.99, P = .032) Cox regression analyses, high SChLAP1 expression was associated with poor clinical outcome in clinically localized prostate cancer (Figure 3C, Tables 3 and 4). We performed permutation tests to validate our selection of the cutoff of ISH scores and found very little inflation of type-I errors, and all the findings still hold significant based on the permutation tests.
Figure 3.
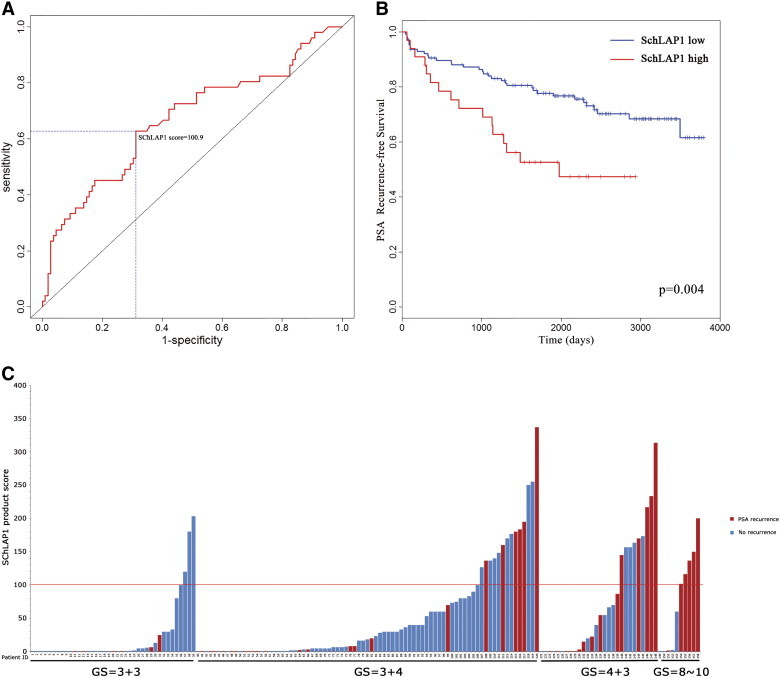
High SChLAP1 expression in clinically localized prostate cancer is associated with decreased time to PSA recurrence after radical prostatectomy. (A) Receiver operating characteristic curve for logistic regression of SChLAP1 expression and PSA recurrence after radical prostatectomy (area under the curve = 0.668). The threshold which maximizes the sum of sensitivity and specificity corresponds to a SChLAP1 ISH product score of 100.9. (B) Kaplan-Meier survival curve for PSA recurrence in clinically localized prostate cancer patients after radical prostatectomy. High SChLAP1 expression is associated with decreased time to PSA recurrence. (C) Graphical representation of SChLAP1 expression for each individual patient with clinically localized prostate cancer, stratified by GS, with associated PSA recurrence status. High SChLAP1 expression is associated with high GS (≥ 8) and PSA recurrence.
Table 2.
Association of SChLAP1 Expression with Clinicopathologic Parameters in a Cohort of Patients with Clinically Localized Prostate Cancer Treated by Radical Prostatectomy.
| SChLAP1 ISH Score < 100 (n = 127) | SChLAP1 ISH Score ≥ 100 (n = 33) | P Value | |
|---|---|---|---|
| Age, y | |||
| ≤ 60 | 68 (53.5%) | 18 (54.5%) | 1.000 |
| > 60 | 59 (46.5%) | 15 (45.5%) | |
| Race | |||
| Black | 14 (11.0%) | 4 (12.1%) | .294 |
| White | 104 (81.9%) | 24 (72.7%) | |
| Other/unknown | 9 (7.1%) | 5 (15.2%) | |
| Preoperative PSA, ng/ml | |||
| ≤ 7 | 70 (55.1%) | 20 (60.6%) | .694 |
| > 7 | 57 (44.9%) | 13 (39.4%) | |
| DRE | |||
| T1 | 87 (68.5%) | 18 (54.5%) | .152 |
| T2 | 40 (31.5%) | 15 (45.5%) | |
| Gland weight, g | |||
| ≤ 50 | 79 (62.2%) | 27 (81.8%) | .039 |
| > 50 | 48 (37.8%) | 6 (18.2%) | |
| Tumor size, cm | |||
| ≤ 1.5 | 69 (54.3%) | 17 (51.5%) | .846 |
| > 1.5 | 58 (45.7%) | 16 (48.5%) | |
| Multifocal | |||
| No | 29 (22.8%) | 8 (26.7%) | .639 |
| Yes | 98 (77.2%) | 22 (73.3%) | |
| GS | |||
| < 7 | 37 (29.1%) | 4 (12.1%) | .017 |
| = 7 | 85 (66.9%) | 24 (72.7%) | |
| > 7 | 5 (3.9%) | 5 (15.2%) | |
| Surgical margin | |||
| Negative | 92 (72.4%) | 22 (66.7%) | .523 |
| Positive | 35 (27.5%) | 11 (33.3%) | |
| EPE | |||
| Negative | 102 (80.3%) | 21 (63.6%) | .063 |
| Positive | 25 (19.7%) | 12 (36.4%) | |
| SVI | |||
| Negative | 125 (98.4%) | 28 (84.8%) | .004 |
| Positive | 2 (1.6%) | 5 (15.2%) | |
| AJCC N stage | |||
| pNX or pN0 | 115 (90.6%) | 27 (90.0%) | 1.000 |
| pN1 | 12 (9.4%) | 3 (10.0%) | |
| PSA recurrence | |||
| Negative | 92 (72.4%) | 17 (51.5%) | .035 |
| Positive | 35 (27.6%) | 16 (48.5%) | |
| Path1992 | |||
| ≤ 3 | 102 (80.3%) | 20 (60.6%) | .023 |
| > 3 | 25 (19.7%) | 13 (30.4%) | |
| Path1997 | |||
| < 3 | 102 (80.3%) | 20 (60.6%) | .023 |
| ≥ 3 | 25 (19.7%) | 13 (39.4%) |
DRE, digital rectal examination; EPE, extraprostatic extension; SVI, seminal vesicle invasion.
Note: There were 3 patients missing AJCC N stage and multifocal records; hence, we used 157 patients for testing AJCC N stage and multifocal, but 160 patients for other parameters.
Table 3.
Univariate Cox Regression Analysis.
| Covariate | Hazard Ratio | 95% CI | P Value | |
|---|---|---|---|---|
| SChLAP1 ISH score | ||||
| (≥ 100 vs < 100) | 2.343 | 1.285 | 4.270 | .005 |
| Age, y | ||||
| (> 60 vs ≤ 60) | 1.715 | 0.984 | 2.988 | .057 |
| Preoperative PSA, ng/ml | ||||
| (> 7 vs ≤ 7) | 2.244 | 1.279 | 3.939 | .005 |
| Tumor size, cm | 1.879 | 1.075 | 3.284 | .027 |
| Gland weight, g | ||||
| Gland weight (< 50 vs ≥ 50) | 1.499 | 0.858 | 2.621 | .155 |
| GS | ||||
| (7 vs<7) | 2.558 | 1.136 | 5.758 | .023 |
| (> 7 vs<7) | 5.477 | 1.830 | 16.394 | .002 |
| (≥ 7 vs < 7) | 2.757 | 1.235 | 6.155 | .013 |
| Surgical margin (positive vs negative) | 1.991 | 1.142 | 3.471 | .015 |
| EPE (positive vs negative) | 5.235 | 3.009 | 9.109 | < .001 |
| SVI (positive vs negative) | 8.670 | 3.791 | 19.850 | < .001 |
| AJCC N stage (pN1 vs pN0 or pNX) | 0.921 | 0.330 | 2.569 | .875 |
| DRE (positive vs negative) | 1.644 | 0.944 | 2.862 | .079 |
| Race (white vs black) | 1.156 | 0.457 | 2.925 | .759 |
| Race (other/unknown vs black) | 0.751 | 0.180 | 3.145 | .696 |
| Multifocal (yes vs no) | 1.203 | 0.441 | 1.568 | .568 |
| Path1992 | 2.230 | 1.759 | 2.826 | < .001 |
| Path1997 | 3.379 | 2.389 | 4.781 | < .001 |
Table 4.
Multivariate Cox Regression Analysis.
| Covariate | Hazard Ratio | 95% CI | P Value | |
|---|---|---|---|---|
| SChLAP1 ISH score (≥ 100 vs < 100) | 1.99 | 1.06 | 3.73 | .032 |
| Preoperative PSA | 1.05 | 1.004 | 1.100 | .034 |
| GS (≥ 7 vs < 7) | 1.72 | 0.75 | 3.95 | .202 |
| Surgical margin (positive vs negative) | 1.09 | 0.57 | 2.08 | .785 |
| EPE (positive vs negative) | 3.76 | 2.04 | 6.93 | < .001 |
| SVI (positive vs negative) | 1.47 | 0.47 | 4.56 | .505 |
CI, confidence interval.
To our knowledge, this is the first report to characterize SChLAP1 expression by ISH in a hospital-based cohort of American men treated for clinically localized prostate cancer, as well as a rapid autopsy cohort of patients with lethal mCRPC. We have used a novel ISH assay to assess SChLAP1 expression on routine surgical pathology prostate cancer specimens. We note several major findings from this work. First, we confirm that SChLAP1 is predominantly a nuclear RNA transcript (Figure 1), which supports in vitro studies of SChLAP1 in prostate cancer cells and preliminary in situ data [7]. Second, we find that SChLAP1 expression is enriched in metastatic samples, suggesting that expression of this lncRNA may be preferentially selected for during prostate cancer progression. Third, we find that, in clinically localized prostate cancer, SChLAP1 expression is enriched in samples from tumors with high GSs (≥ 8) compared to tumors with lower GSs, which also suggests an association with aggressive disease. Most importantly, however, we find that SChLAP1 expression is highly predictive of disease recurrence after prostatectomy and that this observation remains significant even after adjusting for all major clinicopathologic covariates (including GS and seminal vesicle invasion, among others).
As such, our work contributes to the growing body of literature suggesting that lncRNAs are major drivers of cancer biology and, therefore, clinically important molecular entities. For example, the lncRNA HOTAIR has been studied extensively in a wide variety of tumors and has been shown to be an important risk factor in breast, colorectal, and other cancers [10], [11]. Interestingly, however, HOTAIR is not significantly associated with prostate cancer [6]. Expression of the lncRNA MALAT1 has also been associated with poor patient outcomes in lung and hepatocellular cancer [12], [13]. In addition, the lncRNA PCAT1, previously described by our group [6], has been associated with poor clinical outcome in colorectal cancer [14]. Our recent work suggests that PCAT1 confers a BRCA-deficient phenotype via impaired DNA repair in prostate cancer, potentially contributing to tumorigenesis and disease progression [15]. Ongoing work on other lncRNAs as potential cancer biomarkers continues to be an area of high interest for the cancer biology community [16], [17], [18], [19].
Our own studies have demonstrated that SChLAP1 expression is a critical driver of aggressive prostate cancer biology. We have previously shown that SChLAP1 promotes the metastatic phenotype of prostate cancer cells by interfering with the function of the SWI/SNF tumor suppressor complex, leading to global dysregulation of oncogenic gene expression signatures [7]. SChLAP1 expression in prostate cancer cells is essential for metastasis in mouse models and for hematogenous spread of cancer cells in chick embryo experiments. In retrospective multi-institutional studies, we have also shown that SChLAP1 expression is an independent predictor of metastasis for patients with clinically localized prostate cancer [20].
To date, we are unaware of any other lncRNA ISH assay that is currently in clinical development for determining cancer outcome. In this regard, the current study is unique. Clinically, our observation that high SChLAP1 expression by ISH is an independent risk factor for disease recurrence has important implications for the management of early-stage disease. Indeed, although we observe an enrichment of SChLAP1 expression in tumors with high GSs (≥ 8), we also observe a fraction of low-stage, low-grade tumors with high SChLAP1 expression. We are optimistic that SChLAP1 ISH may be able to identify low-grade prostate cancer with a high risk of recurrence.
Furthermore, our ISH assay has several important advantages for clinical translation. First, although not the focus of the current study, we can perform ISH assays on patient biopsy samples. Thus, in the future, we may be able to determine SChLAP1 expression in different prostate cancer foci before definitive therapy. Second, ISH assays are able to assess SChLAP1 expression even when the tumor content is very low. This is an advantage over RNA-based polymerase chain reaction or microarray analyses, where low tumor content leads to poor assay performance because of dilution of tumor RNA by stromal and benign gland RNA.
Given these attributes, we ultimately envision that an ISH assay for SChLAP1 could be combined with serum PSA and GS for risk stratification early in disease management and, therefore, potentially impact clinical decision making for patients (i.e., active clinical surveillance versus definitive therapy). Additionally, in the post–radical prostatectomy population, patients with high SChLAP1 expression may benefit from more rigorous clinical surveillance or, potentially, adjuvant therapy. Although these speculations remain to be proven, we are hopeful that further studies demonstrate that detection of SChLAP1 expression improves management and risk stratification of prostate cancer patients.
Next, we note that SChLAP1 joins several other RNA-based assays which have become prominent over the past 5 years. In particular, urine assays for the lncRNA PCA3 and the TMPRSS2-ERG gene fusion have shown improvements in the diagnosis of prostate cancer [21]. The PCA3 assay, in particular, has a high sensitivity for prostate cancer detection, whereas the TMPRSS2-ERG fusion, which is present in approximately 50% of tumors [22], has a very high specificity for prostate cancer. We envision that a SChLAP1-based assay (i.e., ISH) could complement these tests because neither PCA3 nor TMPRSS2-ERG has been shown as a definitive strong prognostic early-stage biomarker [23], [24], [25], [26]. Thus, although SChLAP1 has a poor sensitivity for cancer overall, its utility as a strong prognostic test could complement the PCA3 and TMPRSS2-ERG tests to identify patients at a high risk for disease recurrence.
Finally, this study has limitations. First, we analyzed a relatively small number of patients in a single cohort from a single institution. Larger, multicohort evaluations will be needed to confirm our findings. Second, further study regarding the relationship between SChLAP1, serum PSA, and GS will be needed to establish more specific implementation of SChLAP1 ISH as an assay in the clinical decision-making algorithm for routine patient care. Third, in this study we use the primary outcome of biochemical recurrence as a measure for patient survival. Therefore, additional independent studies focusing on larger cohorts using PSA recurrence, as well as prostate cancer–specific death, as an end point may further define the overall clinical utility of the SChLAP1 ISH assay. Lastly, integration of SChLAP1 ISH with established multifactor clinical nomograms, such as the Cancer of the Prostate Risk Assessment (CAPRA-S) score [27], [28], will be important to assess whether SChLAP1 staining improves the performance of these risk-stratification algorithms.
In summary, we have developed and optimized a novel RNA ISH approach to detect SChLAP1 expression in routine surgical pathology prostate cancer specimens. We demonstrate that approximately 16% of clinically localized prostate cancers in a hospital-based cohort of American men exhibit high SChLAP1 expression, which is significantly associated with disease recurrence after prostatectomy. Furthermore, we observe high levels of SChLAP1 expression in mCRPC, which lend strong evidence to the idea that high SChLAP1 expression represents an aggressive molecular subtype of prostate cancer with a susceptibility to evolve into the castration-resistant metastatic state. Although the current study remains an initial evaluation of SChLAP1 ISH, we believe that its clinical application as a prognostic assay for prostate cancer is very promising and warrants additional, large-scale evaluation to further define its role as a potential clinical test.
Footnotes
Funding: This work was supported in part by the Prostate Cancer Foundation (R.M.., F.Y.F, J.R.P. and A.M.C.), the National Institutes of Health Prostate Cancer Specialized Program of Research Excellence P50CA69568 to A.M.C., and the Early Detection Research Network (UO1 CA113913 to A.M.C. and U01CA113913 to J.W.). A.M.C. is also supported by the American Cancer Society,Alfred A. Taubman Medical Institute, and The Howard Hughes Medical Institute. F.Y.F. is supported by the Evans Foundation.
Disclosures: R.M. serves as a consultant to GenomeDx Biosciences (Vancouver, BC, Canada). F.Y.F. Advisory board: GenomeDx Biosciences, Nanostring. The University of Michigan has filed a patent on lncRNAs in prostate cancer, including SChLAP1, in which A.M.C., J.R.P., and M.K.I. are named as coinventors. Wafergen Inc. has a nonexclusive license for the detection of lncRNAs including SChLAP1. GenomeDx Biosciences Inc. has a license for the detection SChLAP1 for the molecular analysis of clinical prostate cancer tissue samples. All other authors have no disclosures.
Contributor Information
Rohit Mehra, Email: mrohit@med.umich.edu.
Arul M. Chinnaiyan, Email: arul@med.umich.edu.
References
- 1.Udager A.M., Alva A., Mehra R. Current and proposed molecular diagnostics in a genitourinary service line laboratory at a tertiary clinical institution. Cancer J. 2014;20:29–42. doi: 10.1097/PPO.0000000000000017. [DOI] [PubMed] [Google Scholar]
- 2.Birney E., Stamatoyannopoulos J.A., Dutta A., Guigo R., Gingeras T.R., Margulies E.H., Weng Z., Snyder M., Dermitzakis E.T., Thurman R.E. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kapranov P., Cheng J., Dike S., Nix D.A., Duttagupta R., Willingham A.T., Stadler P.F., Hertel J., Hackermuller J., Hofacker I.L. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 4.Guttman M., Amit I., Garber M., French C., Lin M.F., Feldser D., Huarte M., Zuk O., Carey B.W., Cassady J.P. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prensner J.R., Chinnaiyan A.M. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prensner J.R., Iyer M.K., Balbin O.A., Dhanasekaran S.M., Cao Q., Brenner J.C., Laxman B., Asangani I.A., Grasso C.S., Kominsky H.D. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol. 2011;29:742–749. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prensner J.R., Iyer M.K., Sahu A., Asangani I.A., Cao Q., Patel L., Vergara I.A., Davicioni E., Erho N., Ghadessi M. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat Genet. 2013;45:1392–1398. doi: 10.1038/ng.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rubin M.A., Putzi M., Mucci N., Smith D.C., Wojno K., Korenchuk S., Pienta K.J. Rapid ("warm") autopsy study for procurement of metastatic prostate cancer. Clin Cancer Res. 2000;6:1038–1045. [PubMed] [Google Scholar]
- 9.Shah R.B., Mehra R., Chinnaiyan A.M., Shen R., Ghosh D., Zhou M., Macvicar G.R., Varambally S., Harwood J., Bismar T.A. Androgen-independent prostate cancer is a heterogeneous group of diseases: lessons from a rapid autopsy program. Cancer Res. 2004;64:9209–9216. doi: 10.1158/0008-5472.CAN-04-2442. [DOI] [PubMed] [Google Scholar]
- 10.Gupta R.A., Shah N., Wang K.C., Kim J., Horlings H.M., Wong D.J., Tsai M.C., Hung T., Argani P., Rinn J.L. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kogo R., Shimamura T., Mimori K., Kawahara K., Imoto S., Sudo T., Tanaka F., Shibata K., Suzuki A., Komune S. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71:6320–6326. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- 12.Lai M.C., Yang Z., Zhou L., Zhu Q.Q., Xie H.Y., Zhang F., Wu L.M., Chen L.M., Zheng S.S. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med Oncol. 2012;29:1810–1816. doi: 10.1007/s12032-011-0004-z. [DOI] [PubMed] [Google Scholar]
- 13.Schmidt L.H., Spieker T., Koschmieder S., Schaffers S., Humberg J., Jungen D., Bulk E., Hascher A., Wittmer D., Marra A. The long noncoding MALAT-1 RNA indicates a poor prognosis in non-small cell lung cancer and induces migration and tumor growth. J Thorac Oncol. 2011;6:1984–1992. doi: 10.1097/JTO.0b013e3182307eac. [DOI] [PubMed] [Google Scholar]
- 14.Ge X., Chen Y., Liao X., Liu D., Li F., Ruan H., Jia W. Overexpression of long noncoding RNA PCAT-1 is a novel biomarker of poor prognosis in patients with colorectal cancer. Med Oncol. 2013;30:588. doi: 10.1007/s12032-013-0588-6. [DOI] [PubMed] [Google Scholar]
- 15.Prensner J.R., Chen W., Iyer M.K., Cao Q., Ma T., Han S., Sahu A., Malik R., Wilder-Romans K., Navone N. PCAT-1, a long noncoding RNA, regulates BRCA2 and controls homologous recombination in cancer. Cancer Res. 2014;74:1651–1660. doi: 10.1158/0008-5472.CAN-13-3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rinn J.L., Chang H.Y. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ronnau C.G., Verhaegh G.W., Luna-Velez M.V., Schalken J.A. Noncoding RNAs as novel biomarkers in prostate cancer. Biomed Res Int. 2014;2014:591703. doi: 10.1155/2014/591703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wapinski O., Chang H.Y. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 19.Malik R., Patel L., Prensner J.R., Shi Y., Iyer M.K., Subramaniyan S., Carley A., Niknafs Y.S., Sahu A., Han S. The lncRNA PCAT29 inhibits oncogenic phenotypes in prostate cancer. Mol Cancer Res. 2014;12:1081–1087. doi: 10.1158/1541-7786.MCR-14-0257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prensner J.R., Zhao S., Erho N., Schipper M., Iyer M.K., Dhanasekaran S.M., Magi-Galluzzi C., Mehra R., Sahu A., Siddiqui J., Davicioni E., Den R.B., Dicker A.P., Karnes R.J., Wei J.T., Klein E.A., Jenkins R.B., Chinnaiyan A.M., Feng F.Y. RNA biomarkers associated with metastatic progression in prostate cancer: a multi-institutional high-throughput analysis of SChLAP1. Lancet Oncology. Dec. 2014;15:1469–1480. doi: 10.1016/S1470-2045(14)71113-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tomlins S.A., Aubin S.M., Siddiqui J., Lonigro R.J., Sefton-Miller L., Miick S., Williamsen S., Hodge P., Meinke J., Blase A. Urine TMPRSS2:ERG fusion transcript stratifies prostate cancer risk in men with elevated serum PSA. Sci Transl Med. 2011;3:94ra72. doi: 10.1126/scitranslmed.3001970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mehra R., Tomlins S.A., Shen R., Nadeem O., Wang L., Wei J.T., Pienta K.J., Ghosh D., Rubin M.A., Chinnaiyan A.M. Comprehensive assessment of TMPRSS2 and ETS family gene aberrations in clinically localized prostate cancer. Mod Pathol. 2007;20:538–544. doi: 10.1038/modpathol.3800769. [DOI] [PubMed] [Google Scholar]
- 23.Prensner J.R., Rubin M.A., Wei J.T., Chinnaiyan A.M. Beyond PSA: the next generation of prostate cancer biomarkers. Sci Transl Med. 2012;4:127rv123. doi: 10.1126/scitranslmed.3003180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakanishi H., Groskopf J., Fritsche H.A., Bhadkamkar V., Blase A., Kumar S.V., Davis J.W., Troncoso P., Rittenhouse H., Babaian R.J. PCA3 molecular urine assay correlates with prostate cancer tumor volume: implication in selecting candidates for active surveillance. J Urol. 2008;179:1804–1809. doi: 10.1016/j.juro.2008.01.013. [discussion 1809–1810] [DOI] [PubMed] [Google Scholar]
- 25.Whitman E.J., Groskopf J., Ali A., Chen Y., Blase A., Furusato B., Petrovics G., Ibrahim M., Elsamanoudi S., Cullen J. PCA3 score before radical prostatectomy predicts extracapsular extension and tumor volume. J Urol. 2008;180:1975–1978. doi: 10.1016/j.juro.2008.07.060. [discussion 1978-1979] [DOI] [PubMed] [Google Scholar]
- 26.Aubin S.M., Reid J., Sarno M.J., Blase A., Aussie J., Rittenhouse H., Rittmaster R., Andriole G.L., Groskopf J. PCA3 molecular urine test for predicting repeat prostate biopsy outcome in populations at risk: validation in the placebo arm of the dutasteride REDUCE trial. J Urol. 2010;184:1947–1952. doi: 10.1016/j.juro.2010.06.098. [DOI] [PubMed] [Google Scholar]
- 27.Cooperberg M.R., Hilton J.F., Carroll P.R. The CAPRA-S score: a straightforward tool for improved prediction of outcomes after radical prostatectomy. Cancer. 2011;117:5039–5046. doi: 10.1002/cncr.26169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Punnen S., Freedland S.J., Presti J.C., Jr., Aronson W.J., Terris M.K., Kane C.J., Amling C.L., Carroll P.R., Cooperberg M.R. Multi-institutional validation of the CAPRA-S score to predict disease recurrence and mortality after radical prostatectomy. Eur Urol. 2014;65:1171–1177. doi: 10.1016/j.eururo.2013.03.058. [DOI] [PubMed] [Google Scholar]


