Abstract
Large segmental duplications (SDs) constitute at least 3.6% of the human genome and have increased its size, complexity, and diversity. SDs can mediate ectopic sequence exchange resulting in gross chromosomal rearrangements that could contribute to speciation and disease. We have identified and evaluated a subset of human SDs that harbor an 88-member subfamily of olfactory receptor (OR)-like genes called the 7Es. At least 92% of these genes appear to be pseudogenes when compared to other OR genes. The 7E-containing SDs (7E SDs) have duplicated to at least 35 regions of the genome via intra- and interchromosomal duplication events. In contrast to many human SDs, the 7E SDs are not biased towards pericentromeric or subtelomeric regions. We find evidence for gene conversion among 7E genes and larger sequence exchange between 7E SDs, supporting the hypothesis that long, highly similar stretches of DNA facilitate ectopic interactions. The complex structure and history of the 7E SDs necessitates extension of the current model of large-scale DNA duplication. Despite their appearance as pseudogenes, some 7E genes exhibit a signature of purifying selection, and at least one 7E gene is expressed.
[Supplemental material is available online at www.genome.org.]
Large segmental duplications (SDs) are defined as duplicated blocks of genomic DNA that contain both interspersed high-copy repeat elements, such as Alus, and the intervening coding and intergenic sequences (IHG Sequencing Consortium 2001). A recent comprehensive survey by Bailey et al. found that SDs of ≥90% identity and ≥1 kb comprise at least 3.6% of the human genome (Bailey et al. 2001). They found SDs as large as 300 kb. Approximately 86% of these duplications appeared to involve the transfer of material within, rather than between, chromosomes (Bailey et al. 2001, 2002).
The mechanism by which SDs are generated has not been determined. The process is thought to involve replicative transposition or nonreciprocal recombination (Lundin 1993; Venter et al. 2001; Samonte and Eichler 2002). A possible clue to the duplicative mechanism is the observation that SDs are found more often in pericentromeric and subtelomeric regions than expected by chance (Bailey et al. 2001). One explanation for this finding is that these regions are less gene-dense than typical euchromatic regions, and insertion of a large segment of DNA is less likely to cause disruption of critical loci. However, SDs are found in euchromatic sequence as well as near genes in pericentromeric and subtelomeric regions, suggesting that multiple types of insertion sites for duplication events are tolerated in the genome (Hattori et al. 2000; Bailey et al. 2001).
Mounting evidence indicates that SDs mediate ectopic (i.e., homologous, but nonallelic) interaction of loci that can result in chromosomal rearrangements such as duplications, deletions, and inversions (Mazzarella and Schlessinger 1998). Some recurring SD-mediated rearrangements cause human disease, such as Velocardiofacial, Smith-Magenis, Prader-Willi, and Angelman syndromes (Ji et al. 2000; Emanuel and Shaikh 2001; Stankiewicz and Lupski 2002). The frequency of detrimental genomic rearrangements mediated by SDs is high, estimated at 0.7 per 1000 births, making the propensity of SDs to interact an important factor in human disease (Mazzarella and Schlessinger 1998).
Duplication of genomic segments containing genes can also be beneficial. This process can generate or expand the membership and diversity of gene families. After the duplication of a gene, selective pressure on one of the two copies is relieved only after it accumulates mutation that renders it nonfunctional. Once relieved of selective pressure, a gene may acquire further mutation, which, in some cases, gives it function distinct from the other copy (Hughes 2002; Kondrashov et al. 2002; Prince and Pickett 2002; Zhang et al. 2002). This model may explain the expansion of large gene families such as the olfactory receptors (ORs). ORs comprise the largest gene family in the human genome, with ∼900 members, and encode the proteins responsible for odorant binding and discrimination (Buck and Axel 1991; Glusman et al. 2001; Zozulya et al. 2001). New ORs generated by duplication and subsequent sequence divergence could increase the repertoire of perceived odorants and/or acquire new functions beyond olfaction.
Most OR genes have arisen by local duplication, but some, especially in humans, have duplicated interchromosomally (Trask et al. 1998; Brand-Arpon et al. 1999; Glusman et al. 2000b; Young et al. 2002). A subfamily of OR genes, called the 7Es (Glusman et al. 2000a), have expanded extensively in the human genome as part of large segmental duplications (Trask et al. 1998), such that 7Es account for ∼10% of all the human OR gene sequences (Glusman et al. 2001). The 7E SDs also account for ∼50% of the locations where ORs are found, demonstrating the significant contribution that 7E SDs have made to the genomic landscape of the human OR gene family (Trask et al. 1998; Glusman et al. 2001; Young et al. 2002). The 7E genes have been reported to be predominantly pseudogenes (Glusman et al. 2001) and therefore are unlikely to confer a selectively beneficial function. Moreover, there is evidence that 7E SDs can be disadvantageous, as they can mediate harmful genomic rearrangements (Giglio et al. 2001, 2002). So far, 7E SDs have been found at the break-points of multiple large intrachromosomal rearrangements of 8p causing mental handicap and a common translocation between 4p and 8p that leads to either Wolf-Hirschhorn syndrome or a variety of dysmorphic phenotypes (Giglio et al. 2001, 2002).
Using publicly available sequence databases and custom computational tools, we have identified human segmental duplications that contain 7E genes and evaluated their structure and genomic location. Our analyses provide insight into the dispersal of 7E genes in the genome via the 7E SDs, their subsequent ectopic interaction, and their potential for function.
RESULTS
Identification and Phylogenetic Analysis of 88 7E Genes in the Human Genome
We identified 7E gene sequences in the UCSC August 2001 human genome assembly by using the 112 unique 7E genes described in the Human Olfactory Receptor Data Exploratorium (HORDE) database (Glusman et al. 2001) (http://bioinformatics.weizman.ac.il/HORDE) as BLAT queries. This process yielded >350 ORs in the genome that matched a query gene with 60%–100% nucleotide identity and over ≥400 bp. Seventy of these OR sequences matched with ≥99% identity to HORDE 7E genes. An additional 15 genes not represented in the current HORDE database matched for ≥400 nucleotides with higher (≥80%) identity across their entire length to a member of the HORDE 7E family than to a member of any other HORDE OR gene family. These 15 genes are included in our analysis as members of the 7E family. Two HORDE 7E genes mapped to the UCSC August 2001 assembly (OR7E110P and OR7E98P) fell below our match criteria and are not included in our analyses, because their classification as members of the 7E family is ambiguous. Our final set of 88 7E genes includes three 7E genes in two finished BACs (AL360083 and AC073648) that are not included in the August 2001 assembly (details in Fig. 1 legend), but are mapped to a chromosomal location in later assemblies. Of the 88 genes in our set, 60 are wholly contained within finished sequence and therefore are expected to contain less than one error in 104 nucleotides.
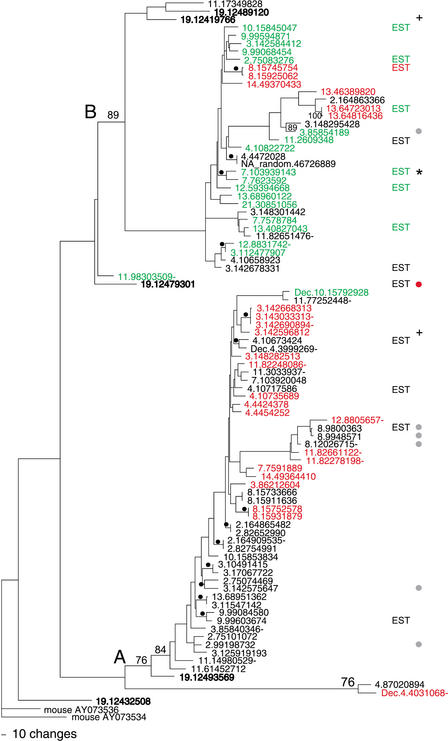
Figure 1.
A parsimony tree of the 88 7E nucleotide sequences. The two major clades of the tree are labeled (A) and (B). Bootstrap values (% of 1000 iterations) are indicated when >75% on the major branches and marked with black dots when ≥85% on the minor banches. The 7E genes are labeled by their position in the UCSC August 2001 assembly of human draft sequence (see Methods); Supplementary Table A gives the corresponding names assigned to the genes by Glusman et al. (2000a) and/or in HORDE. We also included in our set of 88 the sequences for three 7E genes that are found in two finished BACs (AL360083 and AC073648) included in later assemblies (these names of these genes carry the prefix Dec). Genes in bold type are those found in the ancestral locus on chromosome 19. Genes with names in red contain a common substitution resulting in a stop codon in TM6, and those in green contain a common frame shift leading to a stop codon in TM3. The gene marked by a red dot encodes an ORF containing seven TM regions and also encodes a methionine at the beginning of the first predicted extracellular region. Genes marked by a gray dot have ORFs that are predicted to encode six TM regions. Genes marked + have a Ks/Ka value ≥5 on average when compared to 75% of the other 7E genes. “EST” designates genes that match (≥98%) human ESTs. “EST*” designates a gene that matches spliced ESTs. Gene names followed by a dash are not part of 7E SDs.
Forty HORDE 7E genes are not mapped in the August 2001 assembly. Thirty-two of these 40 genes are GenBank entries of single sequencing reads from PCR products. These genes differ by 2%–4% from their best match in our set of 88 genes, possibly due to some combination of sequencing errors, artifacts, and allelic variation. Some of these genes might represent paralogues not yet in the current draft assembly. We excluded them from further analysis, because they lack flanking genomic sequence and map information. An additional eight 7E sequences in the HORDE database were identified in five unfinished BACs that are not included in the UCSC August 2001 assembly, or the most recent June 2002 assembly. We did not include these eight genes in our analysis, but when the sequence and assembly of these BACs becomes reliable, there may be opportunity to analyze at most two additional 7E clusters and three additional orphan 7E genes in the genome.
The 88 7E genes are 87%–99% identical to each other at the nucleotide level and 58%–99% identical at the amino acid level. The phylogenetic relationships of the 7E genes are represented in Figure 1 by a parsimony tree based on the alignment of their nucleotide sequences. They split into two phylogenetic clades (A and B in Fig. 1) that are ∼7% divergent at the nucleotide level on average. The A clade is slightly larger than the B clade, and contains 55% of the 7E genes. For approximately half of the 7E genes on branches supported by bootstrap values >85% (numbers and black dots in Fig. 1), the closest phylogenetic neighbor is located on a different chromosome, indicating that 7E genes are as likely to duplicate interchromosomally as intrachromosomally and/or undergo gene conversion with distant neighbors.
Protein-Coding Potential of 7E Genes
Only seven of the 88 7E sequences have predicted ORFs exceeding 300 amino acids, the typical length of functional OR genes (Glusman et al. 2001; Zozulya et al. 2001; Young et al. 2002; Zhang and Firestein 2002) (Fig. 1, gray and red dots). The single-exon ORF of one of these seven genes is predicted to encode seven transmembrane (TM) domains, as is typical for most intact OR genes (Fig. 1, red dot) (Sosinsky et al. 2000). An N-terminal glycosylation site, another common sequence element of ORs (Gat et al. 1994), is located in the first TM domain, 22 amino acids from the first methionine in this ORF. The other six ORFs (Fig. 1, gray dots) encode their first methionine at a position usually found within the first TM region of OR genes, a highly atypical location. These six genes also encode a putative N-terminal glycosylation site seven amino acids downstream of this methionine, but hydrophobicity plots of these six sequences predict six TM regions and place the N-terminus in the first intracellular region (data not shown). Alternatively, mRNA of these genes might include a 5′ coding exon(s), and an earlier starting methionine could be included through splicing, as has been observed in other OR genes (Walensky et al. 1998; Linardopoulou et al. 2001).
Of the remaining 81 genes with shorter predicted 7E ORFs, 24 contain the same nucleotide substitution that causes a premature stop codon in TM6 (Fig. 1, red names). Except this stop codon, 15 of the 24 genes would encode an ORF of 293–304 amino acids, albeit most without a methionine in the first extracellular domain, as is typical for ORs, unless it is donated by an upstream exon. These 24 genes are seen in both A and B clades of the tree, a feature we discuss below. Another 19 7E genes have a 1-bp insertion mutation that causes a frameshift and premature stop codon just upstream of the very common OR amino acid motif “MAYDRYVAIC” in TM3 (Fig. 1, green names). Seventeen of these genes have at least one other deleterious mutation further downstream. All the TM3-truncated genes except one are in clade B. The proteins encoded by the remaining 38 genes would be prematurely truncated because of a variety of mutations causing early stop codons.
We also determined the longest ORF for each of 30 nonhuman primate 7E gene sequences reported by Rouquier et al. (2000) and compared these to the 88 human 7E genes (not shown). The nonhuman sequences were obtained from seven hominid and New and Old World monkey species. The TM6 stop mutation is present in some, but not all, of the 7E genes reported for chimpanzee (1 of 7), orangutan (1 of 8), and gibbon (3 of 6). Thus, the TM6 mutation predates the last common ancestor of humans and gibbons. The TM3 frameshift (but not the TM3 stop mutation) was seen in three of the seven chimpanzee 7E sequences collected by Rouquier et al., but was not found in any of their other nonhuman primate 7E sequences.Because the sequences of nonhuman primates are far from complete, we cannot rule out the presence of the TM3 and TM6 stop mutations in more distantly related species.
The Ancestral 7E Locus Is in 19p13.2
Through comparative analysis of the mouse genome, we have determined that the entire set of 7E genes in humans descended from a single locus on chromosome 19p13.2, in general agreement with a previous analysis (Glusman et al. 2001). Pair-wise comparisons of each human 7E gene to all known mouse OR genes (Young et al. 2002) reveal that every human 7E gene is most similar at the nucleotide level (82%–89%) to one of two genes in the mouse (AY073534 and AY073536) than to any of the other ∼1500 mouse OR genes. These two genes are the only 7E-like genes in the public and Celera mouse genome sequence available as of August 2001. They map to mouse chromosome 9 in a location that is syntenic to the 7E locus on human chromosome 19. Both the mouse and the human chromosome 19 7E clusters are neighbored by orthologous non-7E OR genes on both sides and several orthologous zinc finger genes on one side (data not shown). The human chromosome 19 sequence contains five 7E genes and four OR genes from other subfamilies (1M, 7D, 7G, and 7H) (Fig. 2). The 7E genes on human chromosome 19 are found in both the A and B clades (Fig. 1, names in bold), and several are among the human genes phylogenetically closest to the mouse 7E-like genes. One of the five chromosome 19 7E genes encodes a full-length ORF (Fig. 1, 19.12479301, red dot). Both of the mouse 7E orthologs are expressed in mouse olfactory epithelium (J. Young, unpubl.) and are predicted to encode full-length ORFs of ≥310 amino acids.
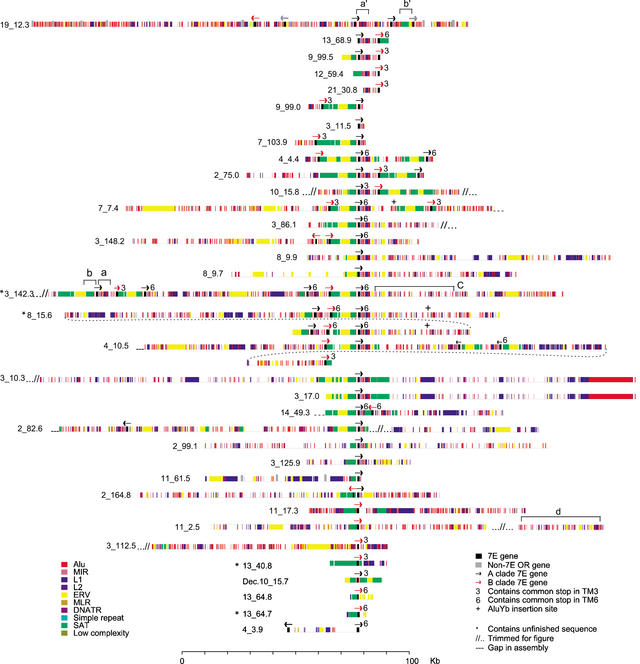
Figure 2.
The complex structure of 7E SDs. Each 7E SD is shown with its constituent repeat-element structure (colored bars) around 7E gene(s) (black bars). The arrows, which are not drawn to scale, represent the 5′ to 3′ transcriptional direction of each 7E gene; black and red arrows designate members of clade A and clade B, respectively. The labels to the left of each 7E SD give the chromosome and position in the UCSC August 2001 assembly at which the SD begins. The top line shows the putative ancestral locus on human chromosome 19 and includes non-SD material. The brackets a–d, a‘, and b‘ denote features discussed in the text. The four SDs that include unfinished sequence are marked with asterisks.
Identification of 35 7E Segmental Duplications
We next collected genomic sequences to determine the extent of similarity between 7E-containing regions of the genome. We used an 11-kb sequence centered around a 7E gene on chromosome 3 (3.142575647 [OR7E130P], Fig. 1) to probe the August 2001 UCSC human genome assembly and the BAC sequences AL360083 and AC073648 mentioned above. We downloaded a total of ∼2 Mbp of sequence around each of 44 matches to this probe (≥1000 bp and ≥70% identical). Many of these 2-Mbp regions contained more than one 7E gene. Cursory examination also indicated that the 44 regions contain varying amounts of common sequence elements, which are not always in the same relative orientation. Additionally, these regions have independently acquired many new interspersed repeat elements, such as Alus, because of the original duplication events that formed them, making multiple alignments of even small spans of sequence difficult. Therefore, we adopted the technique of “fuguization,” that is, excising the repeat elements (Bailey et al. 2001), to decrease the computational time needed to compare the regions and identify common sequences. We applied a custom algorithm to process the output of cross_match analyses of the fuguized regions to identify paralogous segments among the 7E SDs. Any fuguized segment of ≥1 kb shared by two or more of the 44 regions was considered part of a 7E SD. The boundaries of the SDs were defined as the positions where a continuous match with high (≥70%) similarity to the sequence of any other 7E SD decreased markedly (typically from ≥70% to unalignable). Sequence segments at the same locus sharing sufficient similarity and length to other loci, but interrupted by >5 kb of low similarity, were divided into separate SDs. The 2-Mb window around each of the 44 matches to our probe was sufficient to contain each 7E SD.
We defined 35 7E-containing SDs within the 44 regions (Fig. 2). These 7E SDs range in size from 10 to 800 kb, with an average length of 113 kb. The remaining nine regions contained no 7E gene and shared only 3–5 kb of repeats with our probe (data not shown). Of the sequences in the 35 SDs, 94% was finished at the time of our analysis, and draft sequence is confined to only 4 SDs (Fig. 2). The 7E SDs contain 70 (79%) of the 88 7E genes identified in the genome assembly. The sequences surrounding the remaining 18 7E genes do not share paralogy with the 7E SDs beyond the genes themselves (these genes are noted in Fig. 1). The 7E SDs contain at least one, and as many as six (e.g., 3_142.3, Fig. 2), 7E genes. The genes are unevenly distributed within SDs, but the average amount of SD material per gene is ∼66 kb. Sixteen of the 17 SDs that contain multiple 7E genes contain representatives of both clades A and B (Figs. 1 and 2). Only one SD other than the ancestral locus on chromosome 19 contains ORs from a different family (11_61.5 contains a member of the 5F family, Fig. 2).
Portions of 23 non-OR genes are annotated in the UCSC Genome Browser to be within the boundaries of the regions defined as SDs. Although none of these genes is annotated at more than one locus, we found paralogues for each in one to five other 7E SDs, depending on the gene (not shown). The annotated genes are distributed among the SDs on chromosomes 2, 3, 7, 10, 11, and 13. The described functions of the non-OR genes vary widely and include a hypothetical zinc finger gene, an oxytocin receptor, a HERV-H protein, and an A-kinase anchoring protein (AKAP)-binding sperm roporin (Kimura et al. 1992; Lindeskog and Blomberg 1997; Carr et al. 2001). PC3–96, an autophagy-like protein, is the only non-OR gene that lies within 50 kb of a 7E gene (∼9 kb 5′ of the 7E gene at 3.125919193).
Locations of the 7E SDs Correlate Well With Locations Identified With FISH
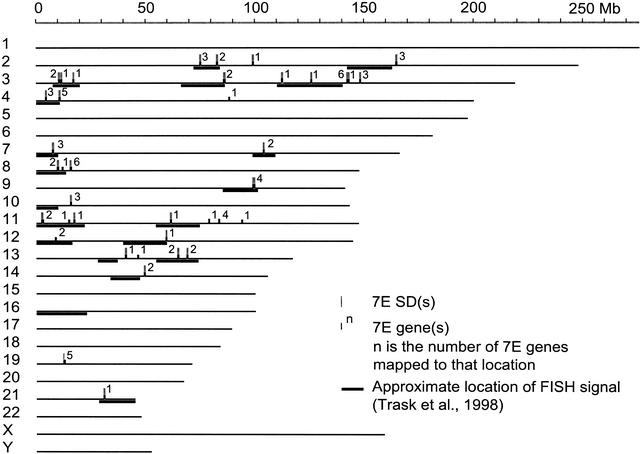
The 35 7E SDs are distributed across 12 human chromosomes (Fig. 3), indicating that the 7Es have been part of at least 11 interchromosomal duplicative transfers. The 7E genes not found in SDs are distributed on the same 12 chromosomes (Fig. 3). The 7E SDs are not biased for pericentromeric or subtelomeric regions, and no 7E SDs lie within 500 kb of subtelomeric or pericentromeric sequence motifs (data not shown).
Figure 3.
The 7E SDs (tall gray vertical bars) and 7E genes (short black vertical bars) in the human draft sequence assembly of August 2001 (http://genome.ucsc.edu/) and estimates of positions of cytogenetic bands showing cross-hybridization to 7E SD-containing clones by FISH (black horizontal bars) (Trask et al. 1998). Eighty-five 7E genes and 35 7E SDs are depicted, but because of their proximity to each other, not all sites are independently resolved. SDs are drawn to scale, but genes are not. The three 7E genes from BACs not included in the August 2001 assembly are not depicted in this figure. The number of 7E genes in each SD is indicated. The correlation between the UCSC map and the FISH results is imperfect, because FISH signals are mapped only with the precision of a chromosome band (∼6 Mb), and band boundaries are only approximately defined in the draft sequence (BAC Consortium 2001; Kent et al. 2002).
We compared the placement of 7E SDs to results published by Trask et al. (1998), who used fluorescence in situ hybridization (FISH) with probes containing portions of 7E SDs to survey the human genome for homology. FISH signals of varying intensities were found at 20 cytogenetically resolved locations on 13 chromosomes. Using UCSC's correlation of cytogenetic bands to genome sequence (BAC Consortium 2001; Kent et al. 2002), we compared the coordinates of the bands showing FISH signals to the locations of the 7E SDs in the August 2001 assembly. Almost all 7E SDs have corresponding FISH signals. Overall, 15 of the 20 FISH-positive locations overlap the sequence locations of 7E SDs, and all but one signal (on chromosome 16) are within 10 Mb of a 7E SD (within the precision with which the two maps are correlated).
The Structures of the 7E SDs Are Complex
Each 7E SD, except for a highly similar pair of SDs on chromosome 3, is a different complex mosaic of repetitive elements, 7E gene(s), and nonrepetitive sequence. On average, ∼50% of 7E SD sequence is occupied by interspersed repeat elements. Alu, satellite, LTR/ERV1, and L1 elements make up ∼70% of the repeat sequence, with component percentages of 18, 16, 14, and 21%, respectively. These densities of the first three repeat classes are notably higher than the human genome average (International Human Genome Sequencing Consortium 2001).
Two relatively common sequence patterns can be observed among 7E SDs. First, 30 7E genes are flanked on their 3′ side by a characteristic collection of Alu and L1 elements (red and blue pattern in bracket a, Fig. 2). Second, 31 genes are flanked on their 5′ side by a pairing of satellite (SATR1 and SATR2) and ERV elements (green and yellow pattern in bracket b, Fig. 2). Other repeat patterns are conserved in a smaller number of SDs, such as the interspersed Alu and MIR repeats seen on chromosomes 3_142.3 and 8_15.6 (Fig. 2, bracket c).
There is great variability in the elements contained in each 7E SD (Fig. 2). In some cases, very large segments have duplicated to generate 7E SDs. For example, ∼100 kb of the SDs 3_10.3 and 3_17.0 are nearly identical. Other 7E SDs show only partial or disrupted blocks of similarity to other SDs. The structural diversity among 7E SDs suggests that no specific sequence elements are necessary or important for duplication. Indeed, we found no common sequence or repeat elements just inside or just outside the break points of the SDs or in the regions directly flanking the common Alu/L1 and ERV/SAT patterns around many 7E genes.
The ancestral locus on human chromosome 19 contains rudiments of the common repeat patterns of ERV/SATR1/SATR2 and Alu/L1 elements seen in other 7E SDs (Fig. 2, brackets a‘ and b‘), but these are arranged differently than they are in all other 7E SDs. The mouse ancestral locus has no interspersed repeat patterns in common with any of the 7E SDs in humans.
Phylogenetic Dynamics of the 7E Duplications
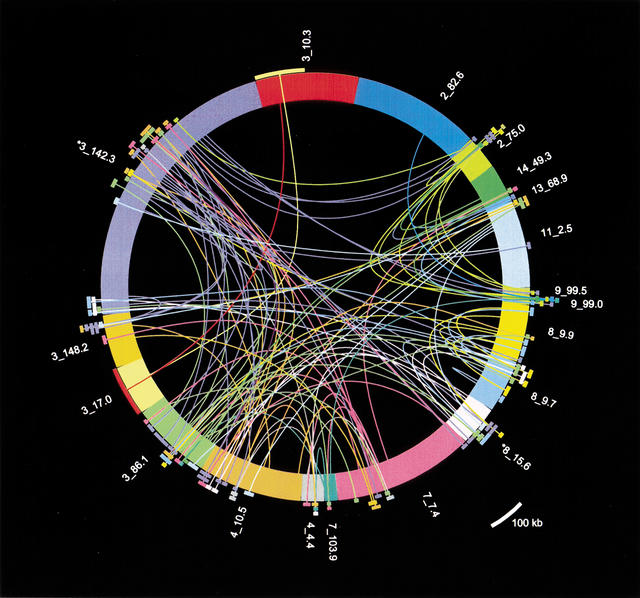
The structural complexity of the 7E SDs demonstrates that the SD family was generated from the ancestral locus through duplication and exchange of multiple constituent sequence segments. To evaluate this complex history of the 7E SDs, we compared all 7E SDs, including repeats, to each other and determined the most similar sequence match of any segment. For this analysis, we considered only the best match of any SD segment of ≥90% in identity and ≥10 kb in length. Portions of 19 7E SDs matched another 7E SD with these criteria, and these relationships are shown in Figure 4.
Figure 4.
A plot of 19 7E SD sequences that contain at least one segment of ≥10 kb that matches another 7E SD at ≥82% nucleotide identity. Although many 7E segments are homologous to multiple other locations in 7E SDs, only the most similar match for each segment is depicted here for simplicity. The 7E SDs are plotted as the differently colored thick arcs making up the circle, with length proportional to SD length (see Fig. 2). Each curved line connects the center of a 7E SD segment to its most highly homologous segment in the set of 35 SDs and terminates in an external arc of the same color as the originating 7E SD. The length of the terminating arc is proportional to the length of the matching segment. We show both lines for bidirectional matches, because not all best matches are reciprocal. The radial positions of the terminating arcs are varied only to allow viewing of the overlapping matches. Asterisks mark the two SDs that include unfinished sequence.
The analysis shown in Figure 4 shows considerable exchange activity among 7E SDs. Two patterns in particular provide some understanding of 7E SD dynamics. First, multiple 7E SDs can spawn duplications and/or exchange sequence with other loci. Such loci are the best match for many other SD segments and have multiple, differently colored lines emanating from them in Figure 4. For example, portions of the SD 2_75.0 are the best match for a single location on chromosome 13 and for multiple locations on chromosomes 3 and 9 and are locally duplicated in 2_75.0. If only one 7E SD was actively duplicating/exchanging, only one SD would exhibit this networked pattern, but several such examples are evident in Figure 4. Second, exchange activity can be seen from the fact that neighboring segments of an SD match best to different SDs. For example, the five turquoise lines leading from neighboring segments on 11_2.5 are connected to (i.e., these segments have the highest percent identity with) segments in SDs 3_148.2, 3_86.1, 4_10.5 and 8_15.6. Sequences near these patches in SD 11_2.5 also have homology to many other SDs (including sequence near the best matches on chromosomes 3, 4, 8, and 11), but the lengths of homology are not greater than 10 kb and thus are not depicted in Figure 4. The 7E SD at 11_2.5 could have been created de novo through independent duplicative transfer of segments from chromosomes 3, 4, and 8 to the 11_2.5 locus, which would also cause this pattern of lines and arcs. However, it is extremely unlikely that these segments would accumulate in 11_2.5 in the exact arrangement in which they are found in other SDs (Fig. 2, 11_2.5, bracket d, and similar patterns in 3_148.2, 3_86.1, 4_10.5, and 8_15.6). A more parsimonious explanation for this pattern is that SD sequences on 11_2.5 have undergone ectopic exchange with 7E SDs on chromosomes 3, 4, and 8 (and perhaps others) in the past.
We also examined Alu insertions in the 7E SDs in an attempt to roughly date duplication activity. The Alu elements in the common block patterns near the 7E genes (bracket a, Fig. 2) are all members of the Sc and Sx subfamilies. These AluS subfamilies were active ∼30 million years ago in the ancestral human genome after its divergence from rodents (Kapitonov and Jurka 1996; Kumar and Hedges 1998), indicating that most 7E SD expansion took place after this time. The 7E SDs also contain many copies of the even older AluJ class at common positions. Only three AluYb8 elements, which are among the most recently active Alus (Carroll et al. 2001), are observed in the 7E SDs (Fig. 2). Two are at identical sites in two duplicated segments of SD 8_15.6, which implies that these segments were the result of a very recent duplication event, and the third appears to represent an independent insertion into SD 7_7.4.
The 7E Genes Exhibit Evidence of Gene Conversion
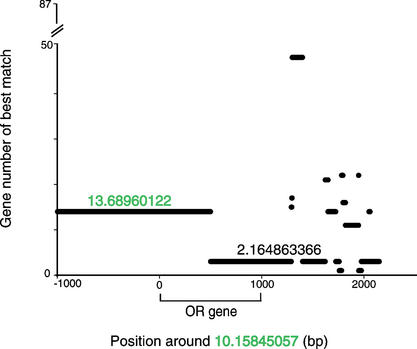
Given the propensity of 7E SDs to interact with each other, we tested for gene conversion between 7E ORs. We used GeneConv (Sawyer 1989) to compare all 7E genes to each other and compute the statistical likelihood of gene conversion (see Methods). Forty-five percent of 7E genes showed significant evidence (P < 0.0001) of involvement in a gene conversion event. The 7E genes in SD 3_142.3 are the most active genes, showing evidence of gene conversion with 28 other 7E genes (data not shown). This SD also demonstrated substantial exchange activity in the analysis shown in Figure 4. Notably, we see evidence of gene conversion between 7E genes containing the mutations causing the TM3 or TM6 stop codons and genes without such mutations. In the example depicted graphically in Figure 5, the first portion of 10.15845047 (OR7E68P) and 5′ flanking sequence are most similar (at 94% nucleotide identity) to 13.68960122 (OR7E33P), both of which contain the TM3 stop codon, while the remainder of 10.15845047 and 3′ flanking sequence is most similar (97%) to 2.164863366 (OR7E107P), which does not have the mutation. The transition of similarity from one sequence to another is abrupt, supporting the idea that 10.15845047 experienced a gene conversion event.
Figure 5.
One example of gene conversion among the 7E genes. The x-axis gives position around the 7E gene at 10.15845047. The gene is located from position 0 to 1000. Each of the 87 other 7E genes is assigned an identifying number from 1 to 87 (y-axis). A 30-bp window was moved in 1-bp steps across the sequence around 10.15845047 (x-axis) and the best match of sequence in the window to any other 7E gene region was recorded (y-axis). This plot illustrates a gene conversion event involving 10.15845047. The average percent nucleotide identity between the first portion of 10.15845057 sequence and 13.68960122 is 94%. The average nucleotide identity between the 3′ portion of 10.15845057 and 2.164863366 is 97%. The transition between these regions is abrupt. Where the chromosome-10 sequence matches chromosome 13 best, it is only ∼80% identical to chromosome 2, and where it matches chromosome 2 best, it is ∼91% identical to chromosome 13. Both 10.15845047 and 13.689601222 contain the common TM3 frameshift and premature stop, but 2.164863366 does not.
Some 7E Genes Exhibit a Signature of Selection, and at Least One Is Expressed
The majority of 7E genes exhibit the signature of mild purifying selection, and some exhibit the signature of strong purifying selection. We aligned the nucleotide sequences corresponding to their longest predicted ORFs of the 88 7E genes and observed an average ratio of synonymous (Ks) substitutions to nonsynonymous (Ka) substitutions between pairs of 7E genes of ∼1.7. As a Ks/Ka ratio of ∼1 is expected under a neutral-mutation model, a ratio of 1.7 suggests that multiple 7E genes have been under purifying selective pressure at some point in evolution. As a comparison, the average Ks/Ka for ∼350 mouse ORs that are known to be expressed (J. Young, unpubl.) is ∼4. Thus, although the human 7E genes show some purifying selection, it is not as strong as the selection acting on expressed OR genes in the mouse. Notably, two of the human 7E genes have average Ks/Ka ratios of ≥5 when compared to ∼75% of other 7E genes (and ≥2 when compared to the other 25%) (Fig. 1).
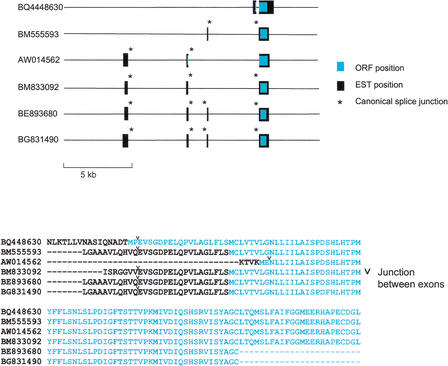
We searched the NCBI human EST database to see if any 7E genes have been observed to be expressed. We found 89 ESTs that matched a 7E gene with ≥98% identity. These ESTs correspond to 14 7E genes, marked on Figure 1 by “EST”. Most of these ESTs show no evidence of splicing when aligned with the genomic sequence and could represent genomic contaminants in the cDNA libraries analyzed. However, six ESTs match one of these 14 genes, 7.103939143 (OR7E38P), over sufficient length to give evidence of 5′ splicing (Fig. 6), suggesting that these six ESTs are true 7E transcripts. These ESTs came from skin, retinal epithelial, and germ-line tissues, as well as mammary tumor. These EST matches represent five splice forms, containing two to four exons. We observed canonical splice-site signals at 15 of the 24 of the intron–exon boundaries. The longest predicted ORF for two ESTs (Fig. 6, BQ4448630 and AW014562) encodes a putative translation start codon in a 5′ exon. The ORFs of the remaining four ESTs encode their first methionine in what would be TM1 of a typical OR protein (Fig. 6). All of the proteins encoded by the 7.103939143 transcripts are predicted to terminate in TM3 due to the frame-shift mutation that this gene shares with 18 other 7E genes (Fig. 1, green names). Four of the six ESTs also show this mutation in their sequences (Fig. 6).
Figure 6.
Spliced ESTs for one 7E gene and alignment of predicted amino acid sequences. The 7E gene from chromosome 7 (7.103939143 OR7E38P) matches six ESTs with 98.5 to 99.9% nucleotide identity. These ESTs exhibit 5′ splicing and demonstrate five splice forms for this gene. The positions of the matches to the genomic sequence are indicated by black bars. Turquoise bars show the position of the predicted ORF for each EST. The corresponding predicted amino acid sequences are shown in alignment below. Splice junctions marked by an “*” have the canonical sequence AG…GT.
DISCUSSION
Assembly and Coverage of 7E SDs
We have identified 7E SDs in 35 locations in the human genome. These SDs account for 70 of the 88 7E genes we have identified in the UCSC August 2001 assembly and in two additional finished BACs not included in that assembly. Eighteen 7Es are not part of SDs. Additional 7E genes may exist in the as-yet unsequenced or unassembled portions of the human genome. The HORDE database includes 40 7E genes that do not have coordinates in the August 2001 assembly. Most of these sequences are short PCR products, which may or may not represent additional paralogues. Although future refinements of the genome sequence may affect the detailed relationships between 7E genes (e.g., the relative positions of taxa on the minor branches in phylogenetic trees), our main conclusions are supported by strong bootstrap values. The positions of the 7E SDs in the assembled draft sequence correlate well with earlier FISH results obtained using 7E-containing probes (Trask et al. 1998), thereby validating the chromosomal localization of most 7E SDs in the draft sequence. FISH showed that a 7E SD-containing probe cross-hybridized to one location where we did not find 7E genes or SDs in the current assembly (Fig. 3, chromosome 16). 7E genes corresponding to this location may emerge as more sequence is accumulated and assembled. The 7E locus on chromosome 19 failed to produce a detectable FISH signal, likely because this locus has only short, noncontiguous stretches of similarity to components of other 7E SDs.
Human SDs May Be Generated by Multiple Mechanisms
Segmental duplication is one of the many evolutionary processes shaping genomes, but this process is not well understood (Ji et al. 2000; Bailey et al. 2001; Emanuel and Shaikh 2001; International Human Genome Sequencing Consortium 2001; Samonte and Eichler 2002). Although 40% of all human segmental duplications are found in pericentromeric and subtelomeric regions (Bailey et al. 2001), none of the 7E SDs is found within 500 kb of these areas, indicating that the 7E SDs might utilize duplication mechanisms differing from many other human SDs. Additionally, the 7E SDs have duplicated interchromosomally as much as intrachromosomally. This pattern contrasts with the entire human set of SDs, of which 86% duplicated intrachromosomally (Bailey et al. 2001). The 7E SDs also differ from classic retrotransposition events in that they lack any simple conserved sequence motif at or near their breakpoints (Grindley 1978; Johnsrud et al. 1978), although a more robust search might identify degenerate common motifs.
7E SDs Extend the Model of Segmental Duplication
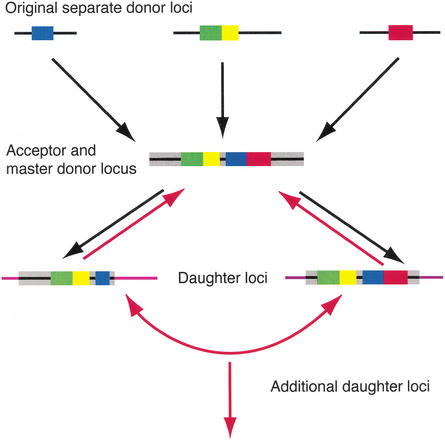
The current working model for SD generation presented by Samonte and Eichler is a hierarchical process of large replicative DNA transposition (Fig. 7, black arrows) (Samonte and Eichler 2002). In this model, segments of DNA from various locations in the genome, called “original separate donor loci,” duplicate to a single locus, called the acceptor locus. This acceptor locus can become a master donor locus and can transfer material, again through duplicative transposition, to other locations. Although much of our data fit this model, the model must be extended to accommodate all of our results. The 7E SD on chromosome 19 appears to be the oldest human 7E locus, because of its syntenic relationship with the only 7E locus in mouse and the close phylogenetic relationship between the mouse 7E and human chromosome 19 genes. The chromosome 19 locus could be equated with the original donor locus in the model as it contains 7E genes from both the A and B clades and neighboring DNA segments that appear to have duplicated in a shuffled order to other loci. At one or more of these loci, additional material was acquired and carried along in subsequent duplications.
Figure 7.
Model of segmental duplication extended from the model proposed by Samonte and Eichler (2002). The model begins with donor loci separated in the genome and proceeds sequentially in time down through the development of the master donor and daughter loci. Black arrows indicate Samonte and Eichler’s formulation of the model. Red lines indicate additional steps added to extend the model to accommodate data from the 7E SDs (see text).
By tracking the best match of each duplication (Fig. 4), we conclude that there is no master 7E donor locus, as the basic model would predict. Rather, several 7E SD loci have donated sequence, either through duplication or exchange, to multiple other locations. We observe significant exchange activity between homologous regions of the 7E SDs, including ectopic interaction among the nongene segments of the 7E SDs and gene conversion between the 7E genes (Figs. 4, 5). Samonte and Eichler’s model of duplication can be extended to accommodate the 7E SD data by allowing for duplication of, and exchange between, 7E SDs (Fig. 7, red arrows).
A Common Mutation in the 7E Genes Was Transmitted Through Gene Conversion
Two characteristic mutations, leading to premature stops in either TM3 or TM6, are common to 20% and 28% of 7E genes, respectively. The presence of these common mutations on both major branches of the tree, but only in some of the taxa in each branch, is intriguing. The ancestral locus for the 7E genes on human chromosome 19 contains five 7E genes representing both clades of the 7E phylogenetic tree, but none contains the mutations causing either the TM3 or TM6 stop codon. Therefore, these mutations must have occurred after the ancestral locus spawned the first 7E duplication and were then propagated in subsequent duplications and/or exchanges. One possible model for the presence of the TM6 stop codon on both branches of the tree is as follows (a similar model might hold for the propagation of the TM3 frameshift as well). At some time during hominid evolution, at least two original 7E genes, which had already diverged to seed the A and B clades, began to duplicate, most likely as a pair. Pairs of A and B genes are frequently seen together in the SDs (Fig. 2). This process continued to some extent to populate the 7E gene family with A and B genes lacking the mutation. At some time, genes of both the A and B subfamilies acquired the mutation causing the TM6 stop codon and subsequently duplicated to further populate the 7E gene family. Sequences from Rouquier et al (2000) indicate that this mutation predated the last common ancestor of humans and gibbons. It is highly improbable that both an A and B gene would mutate independently to the same nucleotide at the same position (Kimura 1969). Neither the TM6 nor the TM3 mutation is part of a highly mutable motif such as a CpG or mononucleotide run. We therefore propose that the mutation occurred once in either an A or B gene and was transferred between the two clades by one or more gene conversion events.
The high frequency of gene conversion observed among the extant 7E genes indicates that this mechanism is plausible. The fact that the A and B genes currently segregate into two phylogenetic clades suggests that the gene conversion events taking place between A and B genes involved only small segments surrounding the mutation relative to the total length of the gene. This gene conversion might have occurred between neighboring A and B genes in one SD. Additional 7E genes containing the mutation were likely spawned from this SD or its progeny.
Potential Functional Consequences of 7E SDs
The 7E SDs are interesting in four respects. First, the expansion of the 7E SDs is potentially disadvantageous. Inserting large segments of DNA into the genome can have profound effects on the local gene structure and therefore the fitness of the organism (Pravtcheva and Wise 1995). Second, the presence of these highly similar large segments makes some of these chromosomal locations susceptible to illicit recombination and resulting rearrangements. A recently published curated set of 169 regions flanked by highly similar segmental duplications that are strong candidates for causing intrachromosomal rearrangements and disease includes seven 7E SDs (Bailey et al. 2002). Indeed, Giglio and colleagues have documented the involvement of 7E SDs on chromosome 4 and 8 in recurrent, disease-causing rearrangements (Giglio et al. 2001, 2002). Third, few non-OR genes reside in the 7E SDs. Finally, all but one 7E gene encode proteins that are shorter than typical ORs.
If the 7E genes are functionally important, the generation and dispersal of SDs could have some selective advantage. Several lines of evidence suggest that at least some of the 7Es were functional during their dispersal. The 7E genes have the signature of mild purifying selection, and at least two 7E genes show signs of strong purifying selection. Furthermore, ESTs in the public database match 14 7E genes, of which two are predicted to encode full-length OR proteins (Fig. 1). Multiple alternatively spliced transcripts were found for one 7E gene (Fig. 5). Notably, this gene contains the common TM3 frameshift mutation resulting in a premature stop codon.
Although mutated 7E genes might have been carried along in large duplications without contributing to the functional gene repertoire of the organism, it is possible that some 7E genes are not true pseudogenes. Several studies have shown that mRNAs can be recoded in mammals such that they encode proteins that do not directly correspond to the genomic sequence (Powell et al. 1987; Sommer et al. 1991; Higuchi et al. 1993; Navaratnam et al. 1995; Baranov et al. 2002). RNA recoding can even rescue frameshifted transcripts (Benne et al. 1986; Feagin et al. 1988). Alternatively, the 7E genes may encode proteins that function with fewer than seven TM-spanning segments. Transmembrane segments 2 through 5 are sufficient for the function of mutant cytokine receptors (normally 7-TM proteins) (Ling et al. 1999). Our discovery that some 7E genes, even those encoding proteins with premature stop codons, are expressed or carry the signature of purifying selection suggests the possibility that 7E transcripts code for functional proteins. Future studies are warranted to determine the spectrum of tissues and the form in which 7E genes are expressed as proteins. Our findings also add further impetus to analyze the functional consequences of other large segmental duplications in the human genome.
METHODS
7E Gene Identification
We identified 88 7E genes using the HORDE (Glusman et al. 2000a) set of 112 unique 7E genes as queries to search the UCSC August 2001 assembly (Kent et al. 2002) of human genomic sequence. The HORDE database was updated on October 20, 2002, to include 127 7E genes (http://bioinformatics.weizmann.ac.il/HORDE). However, 15 7E genes in the HORDE database appear to be duplicate entries (i.e., they are represented in the HORDE database twice, but have only one map location in the UCSC August 2001 assembly). Any sequence in the UCSC database that matched a HORDE 7E gene for ≥400 bp and ≥50% identity was acquired. This query yielded ∼350 sequences. We then compared this set of UCSC genes to the entire HORDE set of OR sequences. Any sequence from the UCSC set that matched a HORDE 7E gene with higher similarity than any other HORDE gene for ≥400 bp was called a 7E gene. We also searched the unassembled or “random” or unassigned sequence of each chromosome in the whole genome for 7E-like gene sequences. Only one 7E gene was found in the “random” sequence (NA_random.46726889). We included this gene in our analysis of the 7E genes, but not of the 7E SDs.
7E SD Identification
Using an 11-kb unmasked segment surrounding a 7E gene (UCSC chr3:142570647–142581647), we probed the human genome with BLAT (http://genome.ucsc.edu) to identify regions with identity ≥70% and ≥1 kb. This query identified 44 locations with matching sequence. We downloaded each matching sequence plus 2 Mb of surrounding sequence from the August 2001 assembly to define the boundaries of the 7E SDs. Overlapping regions were merged. The process of identifying the 7E SDs within these regions consisted of several steps. First, we located all common repeat elements (default settings, RepeatMasker) and excised them (using ExciseRepeats, T. Newman unpubl.), in a process called “fuguization” (Bailey et al. 2001). This step reduced the size of the sequences by ∼50%, making the regions small enough to compare in reasonable computational time. Next we used cross_match (-masklevel 101) (http://www.phrap.org/) to compare all fuguized sequences against each other. We used only matches of ≥70% identity and ≥1 kb. We further refined identification of paralogous segments with an algorithm ParaReg (T. Newman, unpubl.), which considers two matches to share a recent common ancestor if their homology spans a site where a repeat was excised from both sequences at the same position. Homologous sequences with mismatching repeat-excision sites would indicate more distant common ancestry and/or different repeat-insertion activity in the two sequences. This analysis identified a large set of sequences sharing various amounts similarity. The size of these matching segments ranged from ∼1 to 150 kb, excluding repeats. Finally, working outwards from each 7E gene, we looked for the position where identity to any other region dropped sharply (typically from ≥70% to unalignable). We then reinserted the repeat elements at their original positions.
Thirty nonhuman primate sequences described by Rouquier et al. (2000) were obtained from GenBank. The 30 genes are distributed among seven species as follows: Pongo pygmaeus (orangutan), eight; Pan troglodytes (chimpanzee), seven; Gorilla gorilla, three; Hylobates lar (gibbon), six; Saimiri scieureus (squirrel monkey, a New World monkey), three; Callithrix jacchus (common marmoset, a New World monkey), one, and Papio hamadryas (baboon, an Old World monkey), two.
Nomenclature
We refer here to all 7E genes, except three, by their location in the UCSC August 2001 draft assembly of the human genome to facilitate correlation of phylogenetic relationships and genomic locations. The number before the period is the chromosome; the number after the period is the position at which the gene begins. The three remaining genes are named Dec.4.4031068, Dec.4.3999269 and Dec.10.15792928, giving their coordinates on their respective chromosomes in the December 2001 assembly. Supplementary Table A (available at www.genome.org and www.fhcrc.org/labs/trask) links each of these coordinate-specifying names with the names assigned to these genes by Glusman et al. (2000a) and/or in HORDE, which follow the type OR7E1, OR7E2, etc. The 7E SDs are named starting with the chromosome number, followed by an underscore, and the position of the beginning of the SD in the August 2001 assembly in megabasepairs.
Phylogenetic Analysis of the 7E SDs
We performed pairwise comparisons between 7E SDs using cross_match (default parameters, except -init_gap -1, -gap_ext -1) and retained matches ≥10 kb in length. Many of these segments matched sequences from more than one other 7E SD. We developed a C++ program (BestMatch, T. Newman, unpubl.) to identify the most similar match for each 7E SD or portion thereof. This code also calculates and generates Postscript code to display the 7E SDs and show the best matching segment pairs (Fig. 4).
Phylogenetic Analysis of the 7E Genes
We aligned the 7E nucleotide sequences using CLUSTALW (http://www.ebi.ac.uk/clustalw/), refined the alignment by hand, and then used PAUP (Swofford 2000) to produce a phylogram from the alignment using a parsimony scoring method and an unweighted pairgroup method with arithmetic mean (UPGMA) to search the tree space. We also evaluated trees constructed using distance and maximum likelihood scoring algorithms and neighbor-joining and various heuristic search algorithms. The overall topologies of the trees were consistent among these methods. Bootstrap values for the parsimony UPGMA tree shown were generated over 1000 iterations.
Tests for Gene Conversion
We analyzed all 7E genes using GeneConv (http://www.math.wustl.edu/∼sawyer/geneconv) to identify gene conversion events (Sawyer 1989). GeneConv implements an algorithm that compares an alignment of nucleotide sequences, identifies the locations of the paralogous differences among the sequences, and scores sequences that appear to have differences found at the same position in both sequences in a contiguous stretch, separated by regions that contain no shared differences. The length and number of these stretches of shared differences is compared to an expected probability of length and number of shared differences and used to calculate a P-value. A low P-value indicates a high probability of past gene conversion.
Selection Pressure Acting on the 7E Genes
We used the aligned amino acid sequences and corresponding nucleotide sequences from the predicted ORFs of 7E genes as input to the GCG package Diverge and computed Ks and Ka values between every sequence pair (http://www.accelrys.com/products/gcg_wisconsin_package/). Diverge calculates the number of synonymous or nonsynonymous changes per synonymous or nonsynonymous site, respectively, using the algorithm first developed by Li et al. (1985). Diverge also accounts for the possible saturation of mutations in very diverged sequences (Li 1993; Pamilo and Bianchi 1993). We automated the analysis of Diverge output (GetSyn, T. Newman, unpubl.) to obtain the distribution of Ks/Ka ratios for 7E genes and identify genes with high Ks/Ka ratios. Hydrophobicity plots of predicted protein sequences were performed using the DAS program (http://www.sbc.su.sel/∼miklos/DAS/).
Conversion of FISH Locations to Sequence Coordinates
We approximated the locations in the UCSC assembly of the FISH signals recorded by Trask et al. (1998) by using band positions provided in the UCSC genome browser (Kent et al. 2002). Kent et al.'s band boundaries were estimated by reconciling the cytogenetic band locations of ≥7600 BAC clones with the positions of unique sequence tags derived from the clones in the UCSC assembly (BAC Consortium 2001).
Analysis of Telomeric and Pericentromeric Markers
A set of telomeric and pericentromeric markers was used as a query to search sequence extending 500 kb on either side of each 7E SD using cross_match (parameters -minmatch 10 and -minscore 15). This search query consisted of five pericentromere-associated repeat sequences (α, β, γ, and CER [Common Eliminated Region] satellites and CAGGG repeats) (Willard 1990; Eichler 1999; Horvath et al. 2000) and two telomere-associated repeat sequences (TAR [accession no. M55752] and [TTAGGG]13).
WEB SITE REFERENCES
http://bioinformatics.weizmann.ac.il/HORDE/; Human Olfactory Receptor Data Exploratorium.
http://genome.ucsc.edu/; UCSC Genome Bioinformatics.
http://www.math.wustl.edu/∼sawyer/geneconv/; GeneConv Molecular Biology Computer Program.
http://www.ebi.ac.uk/clustalw/; European Bioinformatics Institute Clustalw Program.
http://www.accelrys.com/products/gcg_wisconsin_package/; GCG Wisconsin Package.
http://www.sbc.su.sei/∼miklos/DAS/; Transmembrane Prediction Server.
http://www.phrap.org/; Phred/Phrap/Consed System Home Page.
Acknowledgments
We thank Janet Young for detailed comments on earlier drafts of the manuscript, and Elena Linardopoulou and Eleanor Williams for helpful discussions. This work was supported by NIH Grants RO1 DC04209 and T32 HG00035.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
EMAIL btrask@fhcrc.org; FAX 206-667-4023.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.769003.
REFERENCES
- 1.BAC Resource Consortium 2001. Integration of cytogenetic landmarks into the draft sequence of the human genome. Nature 409: 953-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bailey J.A., Yavor, A.M., Massa, H.F., Trask, B.J., and Eichler, E.E. 2001. Segmental duplications: Organization and impact within the current human genome project assembly. Genome Res. 11: 1005-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bailey J.A., Gu, Z., Clark, R.A., Reinert, K., Samonte, R.V., Schwartz, S., Adams, M.D., Myers, E.W., Li, P.W., and Eichler, E.E. 2002. Recent segmental duplications in the human genome. Science 297: 1003-1007. [DOI] [PubMed] [Google Scholar]
- 4.Baranov P.V., Gesteland, R.F., and Atkins, J.F. 2002. Recoding: Translational bifurcations in gene expression. Gene 286: 187-201. [DOI] [PubMed] [Google Scholar]
- 5.Benne R., Van den Burg, J., Brakenhoff, J.P., Sloof, P., Van Boom, J.H., and Tromp, M.C. 1986. Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell 46: 819-826. [DOI] [PubMed] [Google Scholar]
- 6.Brand-Arpon V., Rouquier, S., Massa, H., de Jong, P.J., Ferraz, C., Ioannou, P.A., Demaille, J.G., Trask, B.J., and Giorgi, D. 1999. A genomic region encompassing a cluster of olfactory receptor genes and a myosin light chain kinase (MYLK) gene is duplicated on human chromosome regions 3q13–q21 and 3p13. Genomics 56: 98-110. [DOI] [PubMed] [Google Scholar]
- 7.Buck L. and Axel, R. 1991. A novel multigene family may encode odorant receptors: A molecular basis for odor recognition. Cell 65: 175-187. [DOI] [PubMed] [Google Scholar]
- 8.Carr D.W., Fujita, A., Stentz, C.L., Liberty, G.A., Olson, G.E., and Narumiya, S. 2001. Identification of sperm-specific proteins that interact with A-kinase anchoring proteins in a manner similar to the type II regulatory subunit of PKA. J. Biol. Chem. 276: 17332-17338. [DOI] [PubMed] [Google Scholar]
- 9.Carroll M.L., Roy-Engel, A.M., Nguyen, S.V., Salem, A.H., Vogel, E., Vincent, B., Myers, J., Ahmad, Z., Nguyen, L., Sammarco, M., et al. 2001. Large-scale analysis of the Alu Ya5 and Yb8 subfamilies and their contribution to human genomic diversity. J. Mol. Biol. 311: 17-40. [DOI] [PubMed] [Google Scholar]
- 10.Eichler E.E. 1999. Repetitive conundrums of centromere structure and function. Hum. Mol. Genet. 8: 151-155. [DOI] [PubMed] [Google Scholar]
- 11.Emanuel B.S. and Shaikh, T.H. 2001. Segmental duplications: An “expanding” role in genomic instability and disease. Nat. Rev. Genet. 2: 791-800. [DOI] [PubMed] [Google Scholar]
- 12.Feagin J.E., Abraham, J.M., and Stuart, K. 1988. Extensive editing of the cytochrome c oxidase III transcript in Trypanosoma brucei. Cell 53: 413-422. [DOI] [PubMed] [Google Scholar]
- 13.Gat U., Nekrasova, E., Lancet, D., and Natochin, M. 1994. Olfactory receptor proteins. Expression, characterization and partial purification. Eur. J. Biochem. 225: 1157-1168. [DOI] [PubMed] [Google Scholar]
- 14.Giglio S., Broman, K.W., Matsumoto, N., Calvari, V., Gimelli, G., Neumann, T., Ohashi, H., Voullaire, L., Larizza, D., Giorda, R., et al. 2001. Olfactory receptor-gene clusters, genomic-inversion polymorphisms, and common chromosome rearrangements. Am. J. Hum. Genet. 68: 874-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giglio S., Calvari, V., Gregato, G., Gimelli, G., Camanini, S., Giorda, R., Ragusa, A., Guerneri, S., Selicorni, A., Stumm, M., et al. 2002. Heterozygous submicroscopic inversions involving olfactory receptor-gene clusters mediate the recurrent t(4;8)(p16;p23) translocation. Am. J. Hum. Genet. 71: 276-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glusman G., Bahar, A., Sharon, D., Pilpel, Y., White, J., and Lancet, D. 2000a. The olfactory receptor gene superfamily: Data mining, classification, and nomenclature. Mamm. Genome 11: 1016-1023. [DOI] [PubMed] [Google Scholar]
- 17.Glusman G., Sosinsky, A., Ben-Asher, E., Avidan, N., Sonkin, D., Bahar, A., Rosenthal, A., Clifton, S., Roe, B., Ferraz, C., et al. 2000b. Sequence, structure, and evolution of a complete human olfactory receptor gene cluster. Genomics 63: 227-245. [DOI] [PubMed] [Google Scholar]
- 18.Glusman G., Yanai, I., Rubin, I., and Lancet, D. 2001. The complete human olfactory subgenome. Genome Res. 11: 685-702. [DOI] [PubMed] [Google Scholar]
- 19.Grindley N.D. 1978. IS1 insertion generates duplication of a nine base pair sequence at its target site. Cell 13: 419-426. [DOI] [PubMed] [Google Scholar]
- 20.Hattori M., Fujiyama, A., Taylor, T.D., Watanabe, H., Yada, T., Park, H.S., Toyoda, A., Ishii, K., Totoki, Y., Choi, D.K., et al. 2000. The DNA sequence of human chromosome 21. Nature 405: 311-319. [DOI] [PubMed] [Google Scholar]
- 21.Higuchi M., Single, F.N., Kohler, M., Sommer, B., Sprengel, R., and Seeburg, P.H. 1993. RNA editing of AMPA receptor subunit GluR-B: A base-paired intron–exon structure determines position and efficiency. Cell 75: 1361-1370. [DOI] [PubMed] [Google Scholar]
- 22.Horvath J.E., Schwartz, S., and Eichler, E.E. 2000. The mosaic structure of human pericentromeric DNA: A strategy for characterizing complex regions of the human genome. Genome Res. 10: 839-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hughes A.L. 2002. Adaptive evolution after gene duplication. Trends Genet. 18: 433-434. [DOI] [PubMed] [Google Scholar]
- 24.International Human Genome Sequencing Consortium 2001. Initial sequencing and analysis of the human genome. Nature 409: 860-921. [DOI] [PubMed] [Google Scholar]
- 25.Ji Y., Eichler, E.E., Schwartz, S., and Nicholls, R.D. 2000. Structure of chromosomal duplicons and their role in mediating human genomic disorders. Genome Res. 10: 597-610. [DOI] [PubMed] [Google Scholar]
- 26.Johnsrud L., Calos, M.P., and Miller, J.H. 1978. The transposon Tn9 generates a 9 bp repeated sequence during integration. Cell 15: 1209-1219. [DOI] [PubMed] [Google Scholar]
- 27.Kapitonov V. and Jurka, J. 1996. The age of Alu subfamilies. J. Mol. Evol. 42: 59-65. [DOI] [PubMed] [Google Scholar]
- 28.Kent W.J., Sugnet, C.W., Furey, T.S., Roskin, K.M., Pringle, T.H., Zahler, A.M., and Haussler, D. 2002. The human genome browser at UCSC. Genome Res. 12: 996-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kimura M. 1969. The rate of molecular evolution considered from the standpoint of population genetics. Proc. Natl. Acad. Sci. 63: 1181-1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kimura T., Tanizawa, O., Mori, K., Brownstein, M.J., and Okayama, H. 1992. Structure and expression of a human oxytocin receptor. Nature 356: 526-529. [DOI] [PubMed] [Google Scholar]
- 31.Kondrashov F.A., Rogozin, I.B., Wolf, Y.I., and Koonin, E.V. 2002. Selection in the evolution of gene duplications. Genome Biol. 3: RESEARCH0008.0001-0008.0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumar S. and Hedges, S.B. 1998. A molecular timescale for vertebrate evolution. Nature 392: 917-920. [DOI] [PubMed] [Google Scholar]
- 33.Li W.H. 1993. Unbiased estimation of the rates of synonymous and nonsynonymous substitution. J. Mol. Evol. 36: 96-99. [DOI] [PubMed] [Google Scholar]
- 34.Li W.H., Wu, C.I., and Luo, C.C. 1985. A new method for estimating synonymous and nonsynonymous rates of nucleotide substitution considering the relative likelihood of nucleotide and codon changes. Mol. Biol. Evol. 2: 150-174. [DOI] [PubMed] [Google Scholar]
- 35.Linardopoulou E., Mefford, H.C., Nguyen, O.T., Friedman, C., van den Engh, G., Farwell, D.G., Coltrera, M., and Trask, B.J. 2001. Transcriptional activity of multiple copies of a subtelomerically located olfactory receptor gene that is polymorphic in number and location. Hum. Mol. Genet. 10: 2373-2383. [DOI] [PubMed] [Google Scholar]
- 36.Lindeskog M. and Blomberg, J. 1997. Spliced human endogenous retroviral HERV-H env transcripts in T-cell leukaemia cell lines and normal leukocytes: Alternative splicing pattern of HERV-H transcripts. J. Gen. Virol. 78: 2575-2585. [DOI] [PubMed] [Google Scholar]
- 37.Ling K., Wang, P., Zhao, J., Wu, Z.J., Cheng, Y.L., Wu, Z.J., Hu, W., Ma, L., and Pei, G. 1999. Five-transmembrane domains appear sufficient for a G protein-coupled receptor: Functional five-transmembrane domain chemokine receptors. Proc. Natl. Acad. Sci. 96: 7922-7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lundin L.G. 1993. Evolution of the vertebrate genome as reflected in paralogous chromosomal regions in man and the house mouse. Genomics 16: 1-19. [DOI] [PubMed] [Google Scholar]
- 39.Mazzarella R. and Schlessinger, D. 1998. Pathological consequences of sequence duplications in the human genome. Genome Res. 8: 1007-1021. [DOI] [PubMed] [Google Scholar]
- 40.Navaratnam N., Bhattacharya, S., Fujino, T., Patel, D., Jarmuz, A.L., and Scott, J. 1995. Evolutionary origins of apoB mRNA editing: Catalysis by a cytidine deaminase that has acquired a novel RNA-binding motif at its active site. Cell 81: 187-195. [DOI] [PubMed] [Google Scholar]
- 41.Pamilo P. and Bianchi, N.O. 1993. Evolution of the Zfx and Zfy genes: Rates and interdependence between the genes. Mol. Biol. Evol. 10: 271-281. [DOI] [PubMed] [Google Scholar]
- 42.Powell L.M., Wallis, S.C., Pease, R.J., Edwards, Y.H., Knott, T.J., and Scott, J. 1987. A novel form of tissue-specific RNA processing produces apolipoprotein- B48 in intestine. Cell 50: 831-840. [DOI] [PubMed] [Google Scholar]
- 43.Pravtcheva D.D. and Wise, T.L. 1995. A postimplantation lethal mutation induced by transgene insertion on mouse chromosome 8. Genomics 30: 529-544. [DOI] [PubMed] [Google Scholar]
- 44.Prince V.E. and Pickett, F.B. 2002. Splitting pairs: The diverging fates of duplicated genes. Nat. Rev. Genet. 3: 827-837. [DOI] [PubMed] [Google Scholar]
- 45.Rouquier S., Blancher, A., and Giorgi, D. 2000. The olfactory receptor gene repertoire in primates and mouse: Evidence for reduction of the functional fraction in primates. Proc. Natl. Acad. Sci. 97: 2870-2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Samonte R.V. and Eichler, E.E. 2002. Segmental duplications and the evolution of the primate genome. Nat. Rev. Genet. 3: 65-72. [DOI] [PubMed] [Google Scholar]
- 47.Sawyer S. 1989. Statistical tests for detecting gene conversion. Mol. Biol. Evol. 6: 526-538. [DOI] [PubMed] [Google Scholar]
- 48.Sommer B., Kohler, M., Sprengel, R., and Seeburg, P.H. 1991. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell 67: 11-19. [DOI] [PubMed] [Google Scholar]
- 49.Sosinsky A., Glusman, G., and Lancet, D. 2000. The genomic structure of human olfactory receptor genes. Genomics 70: 49-61. [DOI] [PubMed] [Google Scholar]
- 50.Stankiewicz P. and Lupski, J.R. 2002. Genome architecture, rearrangements and genomic disorders. Trends Genet. 18: 74-82. [DOI] [PubMed] [Google Scholar]
- 51.Swofford D.L., 2000. PAUP*. Phylogenetic analysis using parsimony (*and other methods). Sinauer Associates, Sunderland, MA.
- 52.Trask B.J., Massa, H., Brand-Arpon, V., Chan, K., Friedman, C., Nguyen, O.T., Eichler, E., van den Engh, G., Rouquier, S., Shizuya, H., et al. 1998. Large multi-chromosomal duplications encompass many members of the olfactory receptor gene family in the human genome. Hum. Mol. Genet. 7: 2007-2020. [DOI] [PubMed] [Google Scholar]
- 53.Venter J.C., Adams, M.D., Myers, E.W., Li, P.W., Mural, R.J., Sutton, G.G., Smith, H.O., Yandell, M., Evans, C.A., Holt, R.A., et al. 2001. The sequence of the human genome. Science 291: 1304-1351. [DOI] [PubMed] [Google Scholar]
- 54.Walensky L.D., Ruat, M., Bakin, R.E., Blackshaw, S., Ronnett, G.V., and Snyder, S.H. 1998. Two novel odorant receptor families expressed in spermatids undergo 5′-splicing. J. Biol. Chem. 273: 9378-9387. [DOI] [PubMed] [Google Scholar]
- 55.Willard H.F. 1990. Centromeres of mammalian chromosomes. Trends Genet. 6: 410-416. [DOI] [PubMed] [Google Scholar]
- 56.Young J.M., Friedman, C., Williams, E.M., Ross, J.A., Tonnes-Priddy, L., and Trask, B.J. 2002. Different evolutionary processes shaped the mouse and human olfactory receptor gene families. Hum. Mol. Genet. 11: 535-546. [DOI] [PubMed] [Google Scholar]
- 57.Zhang J., Zhang, Y.P., and Rosenberg, H.F. 2002. Adaptive evolution of a duplicated pancreatic ribonuclease gene in a leaf-eating monkey. Nat. Genet. 30: 411-415. [DOI] [PubMed] [Google Scholar]
- 58.Zhang X. and Firestein, S. 2002. The olfactory receptor gene superfamily of the mouse. Nat. Neurosci. 5: 124-133. [DOI] [PubMed] [Google Scholar]
- 59.Zozulya S., Echeverri, F., and Nguyen, T. 2001. The human olfactory receptor repertoire. Genome Biol. 2: RESEARCH0018.1-0018.12. [DOI] [PMC free article] [PubMed] [Google Scholar]