Abstract
Background
Cell fate is fluid, and may be altered experimentally by the forced expression of master regulators mediating cell lineage. Such reprogramming has been achieved using viral vectors encoding transcription factors. We recently discovered that the viral vectors are more than passive vehicles for transcription factors, as they participate actively in the process of nuclear reprogramming to pluripotency by increasing epigenetic plasticity. Based on this recognition, we hypothesized that small molecule activators of toll-like receptor 3 (TLR3), together with external microenvironmental cues that drive EC specification, might be sufficient to induce transdifferentiation of fibroblasts into ECs (iECs).
Methods and Results
We show that TLR3 agonist Poly I:C, combined with exogenous EC growth factors, transdifferentiated human fibroblasts into ECs. These iECs were comparable to HMVEC in immunohistochemical, genetic and functional assays, including the ability to form capillary-like structures and to incorporate acetylated-LDL. Furthermore, iECs significantly improved limb perfusion and neovascularization in the murine ischemic hindlimb. Finally, using genetic knockdown studies, we find that the effective transdifferentiation of human fibroblasts to endothelial cells requires innate immune activation.
Conclusions
This study suggests that manipulation of innate immune signaling may be generally used to modify cell fate. As similar signaling pathways are activated by damage associated molecular patterns, epigenetic plasticity induced by innate immunity may play a fundamental role in transdifferentiation during wound healing and regeneration. Finally, this study is a first step toward development of a small molecule strategy for therapeutic transdifferentiation for vascular disease.
Keywords: transdifferentiation, innate immunity, induced endothelial cells, cellular plasticity, ischemia
Introduction
Shinya Yamanaka received the Nobel Prize for Medicine or Physiology in 2012 for his discovery that the forced expression of the transcription factors Oct4, Sox2, Klf4 and cMyc (“OSKM”) can induce nuclear reprogramming of somatic cells to induced pluripotent stem cells (iPSCs)1, 2. We recently discovered that the retroviral vectors used in nuclear reprogramming are more than mere vehicles for the transcription factors. Indeed, by activating innate immune signaling (through Toll-like receptor 3; TLR3) these viral vectors cause global changes in the expression and activity of epigenetic modifiers. Specifically, viral dsRNA stimulated TLR3-activated transcriptional pathways (mediated by NF-κB and IRF3) that caused global changes in epigenetic modifiers (e.g. downregulation of the histone de-acetylase [HDAC] family; upregulation of histone acetyltransferases [HATs]). The altered expression of epigenetic enzymes favored histone modifications that facilitated activation of endogenous genes in the core pluripotency network3. Finally, as demonstrated by gain- and loss-of-function studies, innate immune signaling also participated in the induction of pluripotency using other methods, including mmRNA-based reprogramming, and reprogramming of somatic cells containing the dox-inducible cassete encoding OKSM.
Dr. Yamanaka's discovery has stimulated others to search for sets of transcriptional factors that may induce transdifferentiation to another somatic cell type, also known as direct reprogramming. Indeed, several groups have converted fibroblasts to neurons4, cardiomyocytes5 or ECs6, 7 by overexpressing specific transcription factors. In each of these cases, transdifferentiation was induced using viral vectors encoding transcription factors. However, for clinical applications, it would be more desirable to develop strategies that avoid genetic manipulation. Accordingly, we sought to demonstrate that safe and functional ECs can be generated from fibroblasts using small molecules alone. Cognizant of the role of innate immunity in nuclear reprogramming, and its directed manipulation to increase epigenetic plasticity, we hypothesized that activation of TLR3, together with inductive growth cues, may be sufficient to induce the transdifferentiation of fibroblasts into ECs (“iECs”).
Methods
Cell Culture and iEC generation
BJ human newborn foreskin fibroblast cells (Stemgent), tail-tip fibroblasts isolated from 6-week old Tie2GFP knock-in and TLR3 knock-out mice were cultured and maintained in DMEM with 10% FBS and 1% penicillin/streptomycin (5% CO2, 37°C). Human Dermal Microvascular Endothelial Cells (HMVEC, Lonza) were cultured with endothelial cell growth medium-2MV (EGM-2, Lonza). For iEC generation, BJ fibroblasts were seeded on gelatin-coated plates and after overnight culture, cells were treated with Poly I:C (30ng/ml) and the medium changed to DMEM with 7.5% FBS and 7.5% knockout serum replacement (KSR). With daily treatments of Poly I:C for 7 days, the medium was gradually transitioned to contain 10% KSR. Following these treatments, cells were treated with transdifferentiation medium containing 20ng/ml bone morphogenetic protein-4 (BMP-4, Peprotech), 50ng/ml vascular endothelial growth factor (VEGF, Peprotech) and 20ng/ml basic fibroblast growth factor (bFGF, BD Bioscience) for another 7 days. Transdifferentiation medium was changed every 2 days. For maintenance of transdifferentiation, the cells were cultured for another 2 weeks in the presence of bFGF, VEGF and 0.1mM 8-bromoadenosine-3′:5′-cyclic monophosphate sodium salt (8-Br-cAMP, Sigma), with the medium changed every 2 days.
Fluorescence Activated Cell Sorting (FACS)
On day 28 of transdifferentiation, the iECs were purified using FACS. Cells were dissociated into single cells with Accutase (Life Technologies) for 5 minutes at 37°C, washed with 1× PBS containing 5% BSA and passed through a 70-μm cell strainer. Cells were then incubated with either Alexa Fluor 488-conjugated CD31 antibody (eBioscience) or PE-conjugated CD144 antibody (eBioscience) for 30 mins. Isotype-matched antibody served as negative control. The purified iECs were expanded in EGM-2 media and 10uM SB431542, a specific TGFβ receptor inhibitor that promotes ESC-derived endothelial cell growth and sheet formation6, 8.
RNA extraction and quantitative PCR
Using RNeasy Mini Kit (Qiagen), total RNA was extracted and quantitative polymerase chain reaction was performed using Taqman gene expression assays (Applied Biosystems). Genes associated with the EC phenotype were analyzed with the data normalized to β-actin housekeeping gene and expressed as relative fold changes using the ΔCt method of analysis.
Immunofluorescent staining
iECs were fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton X-100, blocked with 1% normal goat serum and stained for anti-human CD31 (R&D Systems), anti-human CD144 (R&D Systems), anti-human eNOS (BD Transduction Laboratories), anti-human von Willebrand factor (vWF, Abcam) and anti-KDR (R&D Systems) overnight at 4°C. After washes with PBS, the cells were treated with Alexa Fluor-488 or -594 secondary antibodies.
Functional Assays
The functions of the generated iECs were characterized in angiogenic assays and compared to HMVECs and fibroblasts. Uptake of Ac-LDL: was evaluated by incubating cells with ac-LDL-594 at 1:200 dilution for 5 hours before washing the cells with PBS and then measuring the mean fluorescence of the cells in 10 high power fields. Vascular tube-like formation: The ability of the cells to form tube-like structures was assessed in vitro by seeding 104 cells in wells coated with matrigel in the presence of EGM-2 media containing 50ng/ml VEGF and incubated for 24 h. NO production: The ability of the cells to produce NO was assessed by measuring the concentration of NO in the culture medium using the NO detection kit (Molecular Probe, Carlsbad, CA) according to the manufacturer's instructions. The amount of nitrate was determined by converting it to nitrite, followed by the colorimetric determination of the total concentration of nitrite as a colored azo dye product of the Griess reaction that absorbed visible light at 540 nm using a microplate reader. In vivo matrigel angiogenesis assay: To assess the capacity of the iECs to form functional vessels in vivo, matrigel was mixed with 5 × 105 iECs in a final volume of 200μl and injected subcutaneously into the lower abdominal region of SCID mice. After 2 weeks, the matrigel plugs were dissected, fixed in 4% paraformaldehyde and stained for human CD31.
Assay for angiogenic cytokines
Human Angiogenesis Proteome Profiler™ antibody arrays (R&D Systems) were used to assess the various cytokines secreted by the iECs in hypoxic conditions. Briefly, iECs were grown in hypoxic condition (1% O2) for 8 hr and the conditioned media pooled, filtered and incubated with the antibody cocktail (1:1000) for 1 h at RT. The nitrocellulose membrane was then blocked and the sample/detection antibody cocktail mixture added to the membrane. Following overnight incubation at 4°C, the membrane was washed 3 times before incubation with Steptavidin HRP (1:2000) for 30 min at RT. The membranes were then washed prior to incubation with ECLplus (Amersham, Buckinghamshire, UK). The array data was quantified by densitometry using Image J software.
Chromatin Immunoprecipitation and ChIP-qPCR
qChIP was performed as previously described (Lim et al., 2009; Peng et al., 2009). For qChIP and qRT-PCR, error estimates are standard deviations. Recovery of genomic DNA as the percentage input was calculated as the ratio of copy numbers in the immunoprecipitate to the input control. Primers of CD31 promoter (Li et al., Arterioscler Thromb Vasc Biol. 2013;33(6):1366-75) were custom-designed and synthesized from Invitrogen.
Primer Sequence (5′ to 3′)
CD31 (F):CGAGACAGAGGGAGGGTCAA
CD31 (R):GGCCTGATATCTCCTCAGGAA
RNA-seq analysis
RNA-Seq–based expression analysis was carried out using RNA samples converted into individual cDNA libraries using Illumina (San Diego, CA). TruSeq methods employed single reads of 50 base-lengths sequenced at 20-30 million read depths using the Illumina HiSeq 2500 instrument. Trimmed sequences were generated as fastq outputs and analyzed based on the TopHat/Cufflinks pipeline based on reference annotations produced by the Ensembl Gencode project. Differential and significant gene expression analysis was carried out using gene-level FPKM (fragments per kilobase of gene locus summarized mRNA per million reads) expression levels. Genes were selected using the criteria of an absolute expression level greater than 3 fpkm in either HMVEC or iEC samples with at least two fold higher expression in iEC or HMVEC than fibroblasts. Gene lists relative enrichments for various functional associations was determined using ToppGene9.
Hindlimb Ischemia Model
To assess the therapeutic potential of iECs, we used a murine model of hindlimb ischemia (HLI)10, 11. Briefly, unilateral HLI was induced by ligating the femoral artery of aged-matched NOD-SCID male mice (Jackson Labs, 13 weeks old). The mice were randomized into several groups, each receiving either EGM-2 medium (Controls) or 106 iECs or 106 HMVECs intramuscularly (IM) in the gastrocnemius muscle (n=5 each group). To test the survival and localization of iECs, a similar study was carried out with IM administration of 106 transduced VE-cadherin-expressing iECs. The ischemia score index was modified from Brenes et al. Journal of Vascular Surgery. Volume 56, Issue 6, December 2012, Pages 1669 – 1679. The criteria used were: 0 - No changes; 1 - Discoloration/Necrosis of the nails; 2 - Discoloration/Necrosis of the toes; 3 - Foot Necrosis; 4 - Leg Necrosis (up to the gastrocnemius muscle); 5 - Autoamputation (Loss of Limb). There were two independent observers, blinded to the group assignment. Animal studies were approved by the Administrative Panel on Laboratory Animal Care. Total capillary density of hindlimb cross-sections was determined by staining the slides with rabbit anti-mouse CD31 (Abcam), followed by horseradish peroxidase-conjugated secondary antibody. Quantification of capillaries was performed on four different fields at magnification of 20×. The capillary density is normalized to the total number of cells in the field.
Laser Doppler Based Tissue Perfusion Measurement
Blood flow to the ischemic or normal (nonischemic) hindlimb was assessed using a PeriScan PIM3 laser Doppler system (Perimed AB, Sweden) as described previously11. Animals were prewarmed to a core temperature of 37.5°C and heart rate was constantly monitored for signs of stress from overheating. Hindlimb blood flow was measured pre- and postoperatively on selected days (0, 4, 7, 11, 14 and 18). Blood perfusion rate was measured via laser Doppler scans over a 3 minute cycle. The level of perfusion in the ischemic and normal hindlimbs was quantified using the mean pixel value within the region of interest, and the relative changes in hindlimb blood flow were expressed as the ratio of the left (ischemic) over right (normal) LDBP.
Short Hairpin RNA Design
Short hairpin RNA was obtained from Invivogen. Target sequences: TLR3 shRNA, GCTTGGCTTCCACAACTAGAA.
Statistical Analysis
Data shown are reported as mean ± standard error of mean (s.e.m.). Differences between groups were calculated by independent t test, 1-way ANOVA corrected with Bonferroni or Benjamini-Hochberg method, and also Mann-Whitney non-parametric test whether appropriate. In the RT-PCR analyses using independent samples from those used for RNAseq, correlations were evaluated with Spearman nonparametric correlation analysis, and multivariate regression was used for assessments of independence of correlations. Repeated-measures ANOVA followed by multiple comparisons with Bonferroni's method were used. Statistical significance was accepted at P<0.05.
Results
1. Direct reprogramming of fibroblasts to induced endothelial cells (iECs)
To determine if human fibroblasts could be converted to iECs by the by the combined activation of innate immunity and EC specification pathways, human foreskin fibroblasts (BJ) were treated with Poly I:C (30 ng/ml) and cultured in a mixture of fibroblast medium and defined growth medium containing knockout serum (induction medium; Fig. 1A). After culture for 7 days in this condition, the medium was changed to transdifferentiation medium, supplemented with bFGF (20ng/ml), VEGF (50ng/ml) and BMP4 (20ng/ml), which are known to promote endothelial lineage12, 13. To further increase the efficiency of endothelial transdifferentiation, we added 8-Br-cAMP (an agonist of cyclic AMP-dependent protein kinase to our protocol, as it enhances endothelial specification14. After 28 days of differentiation, the cells were dissociated and purified for EC-specific marker VE-cadherin or CD31 by Fluorescence-activated cell sorting (FACS). Approximately 2% of cells treated with induction and then transdifferentiation media expressed CD31 (Fig. 1B-C). By contrast, in the absence of induction by Poly IC, transdifferentiation media did not induce the expression of CD31 in any cells.
Figure 1.
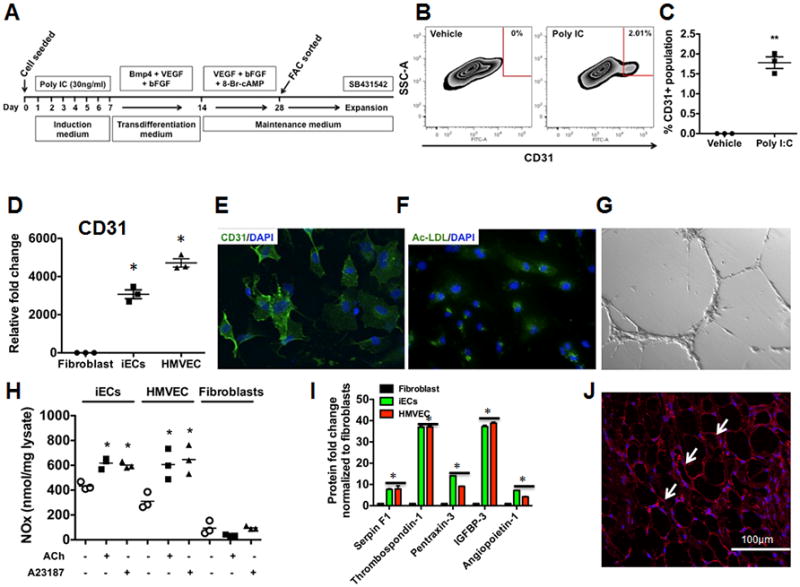
Direct reprogramming of human fibroblasts to functional endothelial cells via activation of innate immunity and microenvironmental cues. (A) Protocol for direct reprogramming of human BJ fibroblasts to endothelial cells. The figure outlines the time course and sequential treatments: Human fibroblasts were treated with Poly I:C (30ng/ml) for 7 days in induction medium containing DMEM/FBS and 7.5% knockout serum replacement (KSR). Following 7 days, the medium was changed to transdifferentiation medium containing DMEM/FBS and 10% KSR containing 20ng/ml BMP4, 50ng/ml VEGF and 20ng/ml bFGF. After another 7 days, the medium was replaced with endothelial growth medium (EGM-2) containing 50ng/ml VEGF, 20ng/ml bFGF and 0.1 mM 8-Br-cAMP for another 14 days. The medium was changed every 2-3 days. Cells were then FAC sorted with CD31 and then expanded in EGM-2 medium containing SB431542, a specific TGFβ receptor inhibitor. (B) Fluorescent activated cell sorting (FACs) plot data obtained using CD31+ antibody to quantify iECs. (left panel) - vehicle control; (right panel) - Poly I:C. (C) Dot plot depicting FACs plot data obtained using CD31+ antibody to quantify iECs. All data represented as mean ± S.E.M., (n=3) **p<0.005 vs Vehicle control, standard unpaired student t test. (D) RT-PCR data showing iECs express endothelial markers CD31 compared to Fibroblasts (n=3) *p<0.05, 1-way ANOVA corrected with Bonferroni method. (E) Immunofluorescence staining of iECs for endothelial markers CD31. (F-G) iECs take up acetylated LDL and form capillary-like networks on matrigel. (H) iECs are capable of producing NO in response to acetylcholine and Ca2+ ionophore A23187 (n=3) *p<0.05, 1-way ANOVA corrected with Bonferroni method. (I) Angiogenic cytokines are produced by iECs when subjected to hypoxia (n=3, done in duplicates) *p<0.05, 1-way ANOVA corrected with Bonferroni method. (J) iECs form blood capillaries (arrows) in subcutaneous matrigel plugs. The numerous spaces in the figure are the areas of matrigel that are bounded by the capillaries.
To further enhance the expansion of iECs, we added SB431542, a specific TGFβ receptor inhibitor that promotes ESC-derived endothelial cell growth and monolayer formation6, 8. After expansion, the iECs were sorted to 78% purity for CD31 (Fig. S1A). The iECs formed a typical “cobblestone” monolayer (Fig. S1B), and continued to express endothelial markers, by PCR including CD31, VE-cadherin, KDR, Von Willebrand factor (vWF) and eNOS (Fig. 1D and Fig. S1C-F). Similarly, immunofluorescence staining revealed that these iECs were positive for EC markers such as CD31, VE-cadherin and vWF (Fig.1E and S1G). These iECs were able to incorporate acetylated LDL and form networks of tubular structures on matrigel (Fig.1F-G and S1G-I). Furthermore, when stimulated with acetylcholine or Ca2+ ionophore A23187, these iECs were capable of generating NO as determined by Griess reaction (Fig. 1H). This NO production was comparable to that produced by HMVECs. This increase in NO production was inhibited by L-NAME, a NOS inhibitor, consistent with an active NOS pathway in the iECs (Fig. S1J).
To further confirm the function of our iECs, we assessed the cells in vitro for the expression of angiogenic cytokines under hypoxic conditions. Indeed, iECs expressed a majority of the angiogenic cytokines including Angiopoietin-1, Serpin F1, Thrombospondin-1, Pentraxin-3 and IGFBP-3 (Fig. 1I). In addition, these iECs, showed the capacity to form capillaries when injected subcutaneously in immunodeficient mice after placing them in matrigel and adding growth factor VEGF (Fig.1J). Furthermore, immunohistochemical images revealed red blood cells in the capillaries formed by the iECs, which indicates that the capillaries were conducting blood (Fig. S1K). By contrast, when human fibroblasts were subjected to the same in vivo conditions, they did not generate capillary structures.
To determine the specificity of the protocol, we repeated our induction/transdifferentiation protocol on tail-tip fibroblasts (TTFs) derived from Tie2GFP mice. In these mice, GFP is expressed under the direction of the endothelial-specific receptor tyrosine kinase (Tie2) promoter. To eliminate the possibility of any contaminating ECs in the starting culture of TTFs, the cells were FAC sorted to exclude any GFP positive cells (Fig. 2A). The GFP-negative TTFs were then subjected to the induction/transdifferentiation protocol and monitored daily by immunofluorescence for any GFP expression. On Day 19, we observed GFP expression in the TTFs undergoing this protocol, and by Day 28 we obtained approximately 4% GFP-positive cells (Fig. 2B-C) By contrast, when vehicle was substituted for Poly IC during the induction phase, no GFP+ cells were obtained with exposure to the PolyIC-deficient transdifferentiation media at Day 19 or 28 (Fig. 2C).
Figure 2.
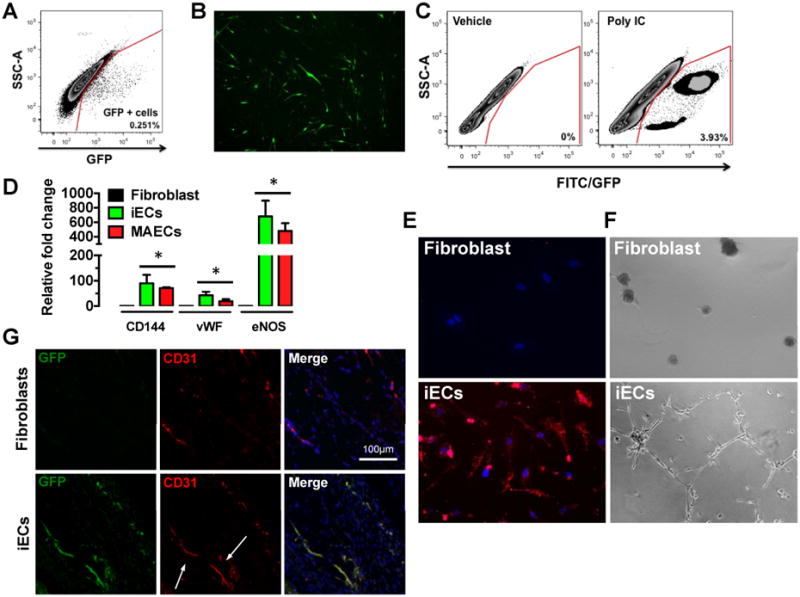
Direct reprogramming of mouse tail-tip fibroblasts to functional iECs. (A) FACs plot obtained by sorting GFP positive and GFP negative cells from tail-tip (TT) fibroblasts. GFP positive cells were eliminated and GFP negative cells were collected by FACs. (B) Representative image of tail-tip fibroblasts expressing GFP at day 19 of the transdifferentiation protocol. (C) FACs plot obtained by sorting GFP positive cells at day 28 following the transdifferentiation protocol to quantify iECs. (left panel) - vehicle control; (right panel) - Poly I:C. (D) Real-Time RT-PCR for endothelial markers CD144, vWF and eNOS in miECs mouse and aortic endothelial cells (MAECs), which are comparable (n=3, done in duplicates) *p<0.05, 1-way ANOVA corrected with Bonferroni method. (E) miECs incorporate acetylated LDL compared to tail-tip fibroblasts. (F) miECs form capillary-like network compared to TT fibroblasts when seeded on matrigel. (G) miECs form blood capillaries in subcutaneous matrigel plugs as shown by CD31 staining.
Furthermore, we confirmed the molecular and functional characteristics of these mouse iECs (miECs) after passaging the cells. Based on gene expression analysis, the miECs resembled murine aortic endothelial cells in their expression of endothelial markers including VE-cadherin, vWF and eNOS (Fig. 2D). Importantly, these cells showed the capacity to form tubular networks when placed on matrigel and could incorporate acetylated LDL (Fig. 2E-F). When placed in matrigel and injected subcutaneously in NOD-SCID mice, the miECs were capable of forming capillaries (Fig. 2G).
2. iECs are heterogeneous
We have previously shown that endothelial cells derived from iPSCs (hiPSC-ECs) expressed various markers associated with arterial, venous and lymphatic ECs and thereby represent a heterogeneous population of ECs (Rufaihah et al, 2013). To determine whether iECs derived directly from fibroblasts exhibited all three subtypes, we employed similar subtype specific markers for lymphatic, venous and arterial subtypes of ECs. We found that iECs expressed markers for all three subtypes of ECs with high levels for arterial markers (Ephrin B2, Notch 1 and Notch 4) and lower levels for venous (CoupTF-II, EphB4) and lymphatic markers (podoplanin, Prox-1 and LYVE-1; Fig. S2A). These results suggest that transdifferentiated iECs are comprised of a heterogeneous population of ECs derived from all three subtypes. Furthermore, we provide gene expression data of cells that have undergone the transdifferentiation protocol at day 28 immediately prior to FAC sorting (Fig. S2B). At this time point, there is no increase in endodermal and ectodermal markers, nor is there expression of pluripotency markers such as Nanog and Oct4. Only the mesodermal (endothelial) markers PECAM1 and DES are increased. Thus, our transdifferentiation protocol appears to be fairly specific for inducing a transdifferentiation within the mesodermal lineage, from fibroblast to endothelial cell, without an intermediate pluripotent or progenitor cell.
3. Histone modifications during direct reprogramming to ECs
To further characterize the epigenetic changes during the induction/transdifferentiation protocol, we performed chromatin immunoprecipitation followed by PCR analysis (ChIP-PCR) to detect trimethylation of histone H3 at lysine 4 (H3K4me3), comparing iECs with parental fibroblasts and HMVECs. This epigenetic modification marks transcriptionally active genes. As expected, we observed a significant increase of H3K4me3 in the promoter regions of CD31, when compared to parental fibroblasts (Fig. S3A). Similarly, we assessed histone H3 at lysine 27 (H3K27me3), an epigenetic modification that marks transcriptionally silenced genes, in the same promoter regions of CD31. A concomitant decrease of H3K27me3 was observed at the CD31 promoter regions when compared to fibroblasts (Fig. S3B). These epigenetic modifications are consistent with transdifferentiation of fibroblasts to endothelial lineage.
4. Transcriptional profiling of induced-ECs
To investigate whether iECs are similar to HMVECs with respect to their global transcriptome, we performed RNA-Seq-based expression analysis. RNA samples from independently induced cultures were converted into individual cDNA libraries using the Illumina TruSeq method, and then subjected to single-end 50 base-sequence analysis at 20-30 million read depths using the Illumina HiSeq 2500 instrument. Trimmed sequences were then generated as fastq outputs and analyzed via the TopHat/Cufflinks pipeline15-17 based on reference annotations produced by the Ensembl Gencode project18-20. Gene expression analysis was carried out using gene-level FPKM (fragments per kilobase of gene locus-summarized mRNA per million reads) expression levels to identify genes that were differentially expressed in iECs and HMVEC versus fibroblasts. The selection criteria for the heat map were either 1.) those genes that were expressed at a level >3 FPKM in HMVECS and at least two-fold greater expression than in fibroblasts; or 2.) those genes that were expressed at a level >3 FPKM in fibroblasts and at least two-fold greater expression than in HMVECs (Fig. 3). The iECs were highly similar to HMVECs in the up- and down-regulated genes that they shared versus fibroblasts. Indeed, the odds that the overlap of HMVEC and iEC genes was due to chance alone was <10-100. Importantly, of the 1085 EC-upregulated genes (by comparison to fibroblasts), 172 are known to be functionally significant for angiogenesis or other endothelial functions (supplementary table 1 and 2).
Figure 3.
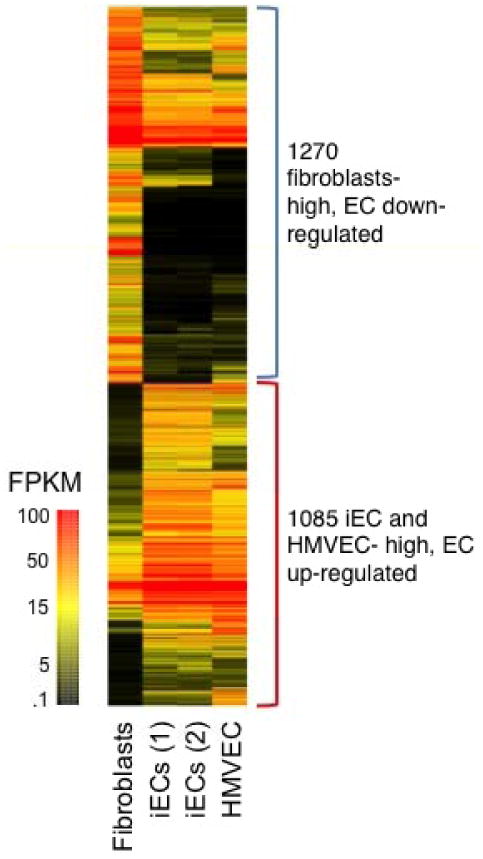
Transcriptional profiling of iECs. Heat map of genes differentially expressed in iECs and HMVEC versus fibroblasts. The selection criteria for gene transcripts shown in the heat map were either 1.) those genes that were expressed at a level >3 FPKM in HMVECS and at least two-fold greater expression than in fibroblasts; or 2.) those genes that were expressed at a level >3 FPKM in fibroblasts and at least two-fold greater expression than in HMVECs as selected from the initially filtered list of ENSEMBL-modeled genes with FPKM >3 (13,181 transcripts), and that exhibited Benjamini-Hochberg adjusted p<0.05 in a Welch ANOVA testing fibroblasts versus iEC versus HMVEC (740 expected, Welch: 3,459 observed, Students: 7466 observed). The pool of 2× differential genes were hierarchically clustered with equal weighting of both absolute and differential expression levels and colored as shown based on absolute FPKM expression levels as calculated via the Topphat Cufflinks pipeline using ENSEMBL Gencode locus definitions.
5. Therapeutic potential of iECs in a model of peripheral arterial disease
To functionally characterize our iECs and to determine their capacity for vascular regeneration, we induced hindlimb ischemia by ligating the femoral artery of NOD SCID mice. For non-invasive tracking in vivo, the iECs were transduced with a lentiviral vector carrying the ubiquitin promoter driving firefly luciferase (Fluc). Upon transplantation into the ischemic limb, the iECs could be detected in the ischemic limb noninvasively by BLI up to Day 7 (data not shown). Consistent with our previous studies21, we saw a decline in BLI that became undetectable by Day 10. Thus, cells were injected again on Day 10. The mice were assigned to receive injection (into the gastrocnemius muscle) of either iECs, human ECs (HMVECs) or endothelial growth medium (EGM-2). To assess hindlimb blood flow, laser Doppler perfusion was performed at Days 0, 4, 7, 11, 14 and 18 post-injection (Fig. S4). The hindlimb perfusion ratio (ischemic/control hindlimb perfusion values) was improved in the iEC-treated mice compared to the EGM-2-treated mice (Fig. 4A-B).
Figure 4.
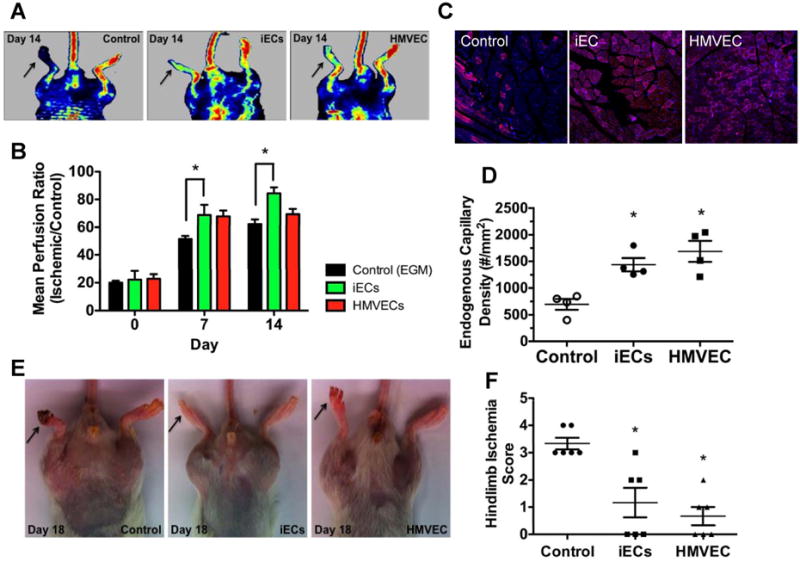
Blood flow and capillary density is improved in ischemic hindlimbs by iEC transplantation. (A) Representative images of laser Doppler perfusion imaging. (B) Summarized data of perfusion ratio (value of the ischemic limb divided by that of non-ischemic limb) at day 0, 7 and 14 post-treatment (n=5 each group, *P<0.05, Repeated-measures ANOVA followed by multiple comparisons with Bonferroni's method). (C) Immunofluorescence CD31 staining of ischemic tissues from mice treated with iECs, vehicle or HMVEC. (D) Quantification of total capillary density in the ischemic limbs (n=3, *P<0.05 – Control vs iECs; #P<0.05 – Control vs HMVECs, 1-way ANOVA corrected with Bonferroni method). (E) Representative images of ischemic limbs at day 18 from iECs, vehicle and HMVEC treated animals. (F) Hindlimb ischemia score obtained by two blinded observers (n=3, *P<0.05 – Control vs iECs; #P<0.05 – Control vs HMVECs, 1-way ANOVA corrected with Bonferroni method).
To confirm the laser Doppler data, sections of ischemic hindlimbs at day 18 were stained with mouse CD31 antibody to assess capillary density by immunofluorescence. Capillary density was significantly greater in the iEC group compared to the control group (Fig. 4C-D and Fig S1L). Furthermore, we assessed the severity of the ischemic tissue damage by assigning a score (by observers blinded to the treatment groups) to the iEC-, HMVEC- and EGM-2-treated animals. As seen in Fig. 4E-F, by comparison to media control, injection of iECs improved the clinical score. Taken together, these results indicate that, similar to HMVECS, the iECs promote expansion of the endogenous microvasculature, improve tissue perfusion, and reduce tissue injury.
6. Innate immunity enables efficient transdifferentiation of fibroblasts to ECs
To determine whether TLR3 signaling was necessary for efficient transdifferentiation of human fibroblasts, we repeated the induction/transdifferentiation protocol in BJ fibroblasts previously treated with scrambled shRNA, or with shRNA to knockdown (KD) the expression of TLR3. Following treatment with the induction/transdifferentiation protocol described above, cells were dissociated after 28 days, and FAC sorted for VE-cadherin. Notably TLR3 KD reduced iEC generation by comparison to scramble treated cells (Fig. 5A-B). In addition to reducing iEC generation, TLR3 KD impaired the function of iECs. The iECs generated from TLR3 KD cells had a significant deficiency in their capacity to take up acetylated LDL (Fig. 5C-D). Furthermore, the ability of TLR3KD iECs had a markedly impaired ability to form networks of tubular structures on matrigel (Fig. 5E-F). The functional differences in iECs generated from the TLR3 KD cells were associated with some differences in gene and protein expression for EC markers when compared to scrambled control (Fig. S5A-F). To gain more insights into the molecular mechanism underlying the role of innate immunity in transdifferentiation, we performed RNA-seq on the iECs generated from scramble and TLR3 KD fibroblasts. Many of the genes associated with endothelial function that were upregulated with transdifferentiation, were not upregulated during transdifferentiation in the TLR3 KD cells. Specifically, 385 genes associated with endothelial biology (Fig. S5G) and 59 genes associated with angiogenesis (Fig. 5G), which were up-regulated in the scramble-iECs were not so in the TLR3 KD-iECs. Importantly, a variety of genes whose expression was activated in iEC was prevented by anti-TLR3 but unaffected by the scrambled vector. For example, angiogenesis essential genes GJA4, HOXA1, HOXA3, HOXB3, HEY2, GBX2, and SEMA3A were all prevented from being activated in iEC treated with TLR3 KD but not so in the scrambled-treated iEC (Fig. S5H).
Figure 5.
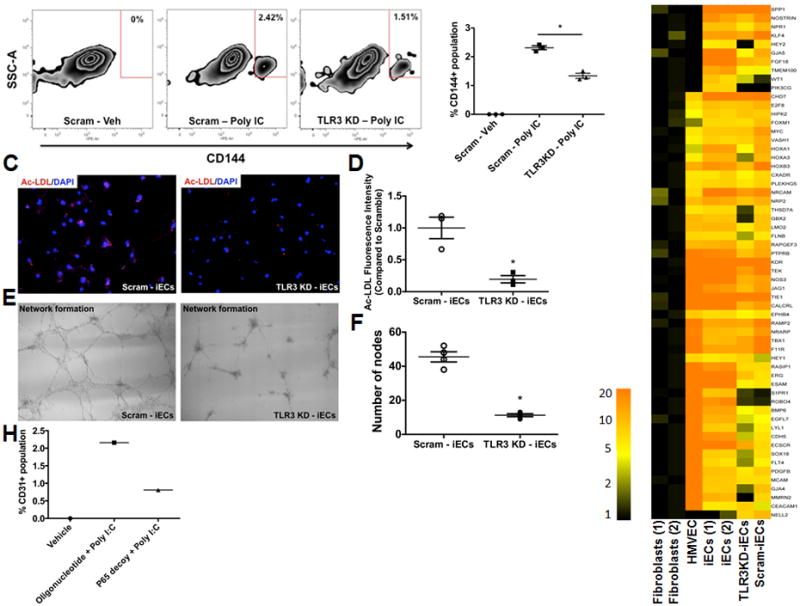
TLR3 signaling is required for efficient transdifferentiation of human fibroblasts to functional endothelial cells. (A) Fluorescent activated cell sorting (FACs) plot data obtained using CD144+ antibody to quantify iECs: (left panel) – scramble cells treated with vehicle control; (middle panel) – scramble cells treated with Poly I:C and (right panel) – TLR3-KD cells treated with Poly I:C. (B) Dot plot depicting FACs plot data obtained using CD144+ antibody to quantify iECs. All data represented as mean ± S.E.M, (n=3) *p<0.05, standard unpaired student t test, 1-way ANOVA corrected with Bonferroni method. (C) Representative images of iECs generated from Scramble treated cells (left panel) or TLR3-KD cells (right panel) showing reduced capacity to incorporate acetylated LDL by TLR3-KD cells. (D) Dot plot showing a significant decrease in the acetylated-LDL fluorescent intensity in TLR3-KD cells compared to scramble (n=3) *p<0.05, standard unpaired student t test and Mann-Whitney non-parametric test. (E) Representative images of iECs generated from scramble treated cells (left panel) or TLR3-KD cells (middle panel) showing failure to form capillary-like networks on matrigel by TLR3-KD cells. (F) Dot plot (right panel) showing a significant decrease in the number of nodes in TLR3-KD cells compared to Scramble (n=3) *p<0.05, standard unpaired student t test and Mann-Whitney non-parametric test. (G) Heat map of genes differentially expressed in iECs generated from scramble- or TLR3-KD fibroblasts showing 59 genes associated with angiogenesis reversed in the TLR3 KD-iECs when compared to scramble-treated iECs. (H) Dot plot depicting FACs plot data obtained using CD31+ antibody to quantify iECs: (left bar) – vehicle treated; (middle bar) control oligonucleotide treated with Poly I:C; (right bar) – P65 decoy (p65i) treated with Poly I:C.
To further elucidate the elements of TLR3 signaling involved in transdifferentiation, we inhibited the action of downstream effector NF-κB using a decoy oligonucleotide. NF-κB is a transcriptional effector of TLR3 activation22, 23 and interacts with CBP/p300 to positively regulate gene expression. The number of iECs generated by the induction/transdifferentiation protocol (Fig. 5H), was markedly reduced by the addition of p65 decoy. A similar result was observed when we repeated our transdifferentiation studies using tail-tip fibroblasts of TLR3 knockout or WT littermates. There was a 50% reduction in the miECs generated by TLR3KO fibroblasts when compared to WT fibroblasts (Fig. S6).
Discussion
The salient observations of this study are: 1) A small molecule agonist of TLR3, combined with endothelial growth factors, is sufficient to transdifferentiate human fibroblasts to iECs that are morphologically and immunohistochemically similar to human microvascular endothelial cells (HMVECs). 2) The iECs share a highly similar transcriptional profile to HMVECs, and when administered to the ischemic hindlimb, are as effective as HMVECs in improving limb perfusion and reducing tissue injury; and 3) The generation of effective iECs using this small molecule methodology is dependent upon innate immune signaling. When fibroblasts were NOT exposed to the induction medium (i.e. containing Poly I:C), endothelial growth medium (transdifferentiation medium) had no effect. Furthermore, TLR3 or NFKB knockdown each reduced the generation of iECs using this protocol.
Previously, we showed that activation of innate immunity increased epigenetic plasticity so as to enhance the action of cell-permeant transcriptional factors to induce the gene network required for pluripotency. In the current paper, we used the combination of immunity-induced epigenetic plasticity and outside-in signaling to induce transdifferentiation of fibroblasts to endothelial cells. The fact that epigenetic plasticity, combined with outside-in signaling, is sufficient for transdifferentiation to a therapeutic cell type, is a conceptual departure and a technical advance. Previous studies have demonstrated the conversion of fibroblasts into angioblast-like cells24 or endothelial cells25, 26. However, the reported methods include either the use of transcription factors encoding OSKM24, 26 or OK25. To our knowledge, this is also the first time that transdifferentiation to a therapeutic cell type has been accomplished with a small molecule approach alone. It is likely that modifications of this small molecule approach may be used to generate other somatic cells of therapeutic interest.
Generation of functional human ECs could be beneficial for various vascular disorders including wound healing and ischemic syndromes. Our previous studies have shown that ECs can be derived from pluripotent stem cells such as embryonic stem cells11 or induced pluripotent stem cells21. These ESC-ECs and iPSC-ECs are effective at increasing microvascular density and improving perfusion when administered to ischemic tissue. These effects appear to be largely mediated by paracrine effects of the cells, rather than a direct contribution to the capillary bed21. However, generating human iPSCs, and deriving endothelial cells from them, is cumbersome. For therapeutic purposes, such iPSC-ECs must be free of pluripotent cells that could induce teratoma formation. Accordingly, direct reprogramming to ECs is an attractive alternative approach and indeed, several groups have been successful in generating functional ECs directly from human fibroblasts6, 7. However, these groups employed viral vectors encoding transcription factors, which raises concerns regarding integration of viral DNA into the reprogrammed cells.
We hypothesized that transdifferentiation might be achieved using small molecules exclusively. Our strategy was to activate innate immunity to induce epigenetic plasticity, and to provide microenvironmental cues to promote endothelial lineage. We used the TLR3 agonist poly I:C, which has been approved for preclinical use and has potential for use as a clinical grade adjuvant27, 28. In addition, we used growth factors (VEGF, bFGF, BMP4), 8Br-cAMP, and the TGFβ inhibitor SB431542. This empirical formulation is based on literature indicating the role of these growth factors in endothelial development, endothelial proliferation, and prevention of endothelial-to-mesenchyme transformation6, 14, 29. Though our yield of iECs was modest (∼2%), our yield is not substantially different from those reported by others using viral constructs encoding transcriptional factors. We intend to perform high throughput screening of small molecules that can increase this yield.
With respect to the other cells that may be generated by this protocol, we used markers for mesodermal, endodermal and ectodermal lines, eg. CD45, CD14, CD41, Tau, Tuj1, runx2, α-actinin, α-MHC and brachyury. We do not observe any increase in these markers over the course of the 28 days during our transdifferentiation protocol. Furthermore, we were not able to detect any c-kit or CD34 positive cells (which includes the subset of “endothelial progenitor cells”) during the process of transdifferentiation during temporal studies of gene expression (Figure S7). Importantly, we performed qPCR for EC markers for the entire 28 days of the transdifferentiation protocol. For each day, RNA was collected after treating human fibroblasts through the various stages of treatment. We then checked for EC markers, specifically, CD31, CD144, KDR and eNOS (Fig. S8). Intriguingly, KDR is the first of these to increase in expression at about 14d (KDR is known to be an earlier marker in endothelial development), followed by CD144, CD31, and lastly eNOS. The data suggests that, during the transdifferentiation, there is an epigenetic resistance that is greater for some endothelial genes than others. The mechanism(s) of such epigenetic resistance may be a very interesting topic for further study.
Previous studies have assigned detrimental roles to TLRs in the progression of atherosclerosis. However, Cole et al. found that deficiency of TLR3 accelerated the onset of atherosclerosis in hypercholesterolemic ApoE-/- mice. This finding suggests a protective role for TLR signaling in the vessel wall.30 Taken together, our work and the observations of Cole and colleagues might indicate that some amount of immune activation is necessary for endothelial regeneration and repair.
The iECs we generated did not incorporate into the murine microvasculature. Nevertheless, we observed a significant increase in capillary density and an improvement in limb blood flow. We therefore assume that the effect of the injected human cells in the murine model of hindlimb ischemia, is due to a paracrine effect, rather than stable integration of the human cells into the murine limb vasculature.
We confirmed the role of innate immune activation in transdifferentiation using shRNA knockdown of TLR3. The knockdown of TLR3 reduced the yield of iECs. It is not surprising that we were still able to generate some iECs in this fibroblast line, as the TLR knockdown is incomplete. Furthermore, Poly I:C can also activate innate immunity through the alternate RIG-1 pathway31, 32. In the current investigation, we used the TLR3 agonist polyI:C to activate innate immune signaling and epigenetic plasticity. However, we have unpublished evidence indicating that activation of other receptors involved in innate immune signaling, such as TLR4 and Rig1, are also capable of inducing epigenetic plasticity. We speculate that any activation of innate immunity, triggered by pathogen-associated molecular patterns (PAMPs) or damage-associated molecular patterns (DAMPs) induces global changes in epigenetic modifiers to increase epigenetic plasticity. We further speculate that this response permits cells to respond rapidly to microenvironmental cues, with a phenotypic plasticity that facilitates cellular response to pathogens or injury. We call this process “transflammation”.
Of great interest, the iECs derived from the knockdown lines were incompletely reprogrammed, as assessed by functional studies. Their ability to incorporate acetylated LDL, and to form a tubular network in matrigel, were markedly inhibited. These findings would be consistent with an incomplete activation of epigenetic plasticity and impaired reprogramming in the TLR3 knockdown lines. The role of innate immunity was further confirmed by our studies using the decoy oligonucleotide against p65. In these studies, the decoy oligonucleotide also suppressed the generation of iECs. Taken together, our observations are consistent with the hypothesis that activation of innate immunity induces a state of epigenetic plasticity where external cues can guide transdifferentiation. This concept may be useful in its application to the derivation of other cell types by transdifferentiation. In addition, our work provides a new avenue for therapeutic transdifferentiation using small molecules in the absence of viral vectors or transcription factors, an approach that may be more feasible in vivo.
Supplementary Material
Acknowledgments
We are grateful to Dr. Carolyn Lutzko and the Cincinnati Cell Characterization Core (CCCC) for their assistance in running the RNA-seq experiments.
Funding Sources: This work was supported by grants to Dr. Cooke from National Institutes of Health (U01HL100397, RC2HL103400). Dr. Sayed was supported by a NIH postdoctoral fellowship (HL098049-01A1), American Heart Association Scientist Development Grant (AHASDG) (13SDG17340025) and NIH PCBC Pilot grant (SR00003169/5U01HL099997). Dr. Wong was supported by a NIH PCBC Jump Start Award (PCBC_JS_2012/1_02) and American Heart Association Scientist Development Grant (AHASDG) (13SDG15800004)
Footnotes
Disclosures: Drs Cooke, Sayed and Lee are inventors of the intellectual property, assigned to Stanford University, which was generated by this research.
References
- 1.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 3.Lee J, Sayed N, Hunter A, Au KF, Wong WH, Mocarski ES, Pera RR, Yakubov E, Cooke JP. Activation of innate immunity is required for efficient nuclear reprogramming. Cell. 2012;151:547–558. doi: 10.1016/j.cell.2012.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Südhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ginsberg M, James D, Ding BS, Nolan D, Geng F, Butler JM, Schachterle W, Pulijaal VR, Mathew S, Chasen ST, Xiang J, Rosenwaks Z, Shido K, Elemento O, Rabbany SY, Rafii S. Efficient direct reprogramming of mature amniotic cells into endothelial cells by ets factors and TGFbeta suppression. Cell. 2012;151:559–75. doi: 10.1016/j.cell.2012.09.032. Epub 2012 Oct 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Margariti A, Winkler B, Karamariti E, Zampetaki A, Tsai Tn, Baban D, Ragoussis J, Huang Y, Han JDJ, Zeng L, Hu Y, Xu Q. Direct reprogramming of fibroblasts into endothelial cells capable of angiogenesis and reendothelialization in tissue-engineered vessels. Proc Natl Acad Sci U S A. 2012;109:13793–13798. doi: 10.1073/pnas.1205526109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Watabe T, Nishihara A, Mishima K, Yamashita J, Shimizu K, Miyazawa K, Nishikawa SI, Miyazono K. TGFBeta receptor kinase inhibitor enhances growth and integrity of embryonic stem cell-derived endothelial cells. J Cell Biol. 2003;163:1303–1311. doi: 10.1083/jcb.200305147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen J, Bardes EE, Aronow BJ, Jegga AG. Toppgene suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37:W305–311. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Niiyama H, Huang NF, Rollins MD, Cooke JP. Murine model of hindlimb ischemia. J Vis Exp. 2009;(23) doi: 10.3791/1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang NF, Niiyama H, Peter C, De A, Natkunam Y, Fleissner F, Li Z, Rollins MD, Wu JC, Gambhir SS, Cooke JP. Embryonic stem cell-derived endothelial cells engraft into the ischemic hindlimb and restore perfusion. Arterioscler Thromb Vasc Biol. 2010;30:984–991. doi: 10.1161/ATVBAHA.110.202796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu SJ, Feng Q, Caballero S, Chen Y, Moore MAS, Grant MB, Lanza R. Generation of functional hemangioblasts from human embryonic stem cells. Nat Meth. 2007;4:501–509. doi: 10.1038/nmeth1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chicha L, Feki A, Boni A, Irion O, Hovatta O, Jaconi M. Human pluripotent stem cells differentiated in fully defined medium generate hematopoietic cd34- and cd34+ progenitors with distinct characteristics. PLoS ONE. 2011;6:e14733. doi: 10.1371/journal.pone.0014733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamamizu K, Kawasaki K, Katayama S, Watabe T, Yamashita JK. Enhancement of vascular progenitor potential by protein kinase a through dual induction of flk-1 and neuropilin-1. Blood. 2009;114:3707–3716. doi: 10.1182/blood-2008-12-195750. [DOI] [PubMed] [Google Scholar]
- 15.Trapnell C, Pachter L, Salzberg SL. Tophat: Discovering splice junctions with rna-seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by rna-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotech. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roberts A, Trapnell C, Donaghey J, Rinn JL, Pachter L. Improving rna-seq expression estimates by correcting for fragment bias. Genome Biol. 2011;12:R22. doi: 10.1186/gb-2011-12-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flicek P, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, Fitzgerald S, Gil L, Gordon L, Hendrix M, Hourlier T, Johnson N, Kahari AK, Keefe D, Keenan S, Kinsella R, Komorowska M, Koscielny G, Kulesha E, Larsson P, Longden I, McLaren W, Muffato M, Overduin B, Pignatelli M, Pritchard B, Riat HS, Ritchie GR, Ruffier M, Schuster M, Sobral D, Tang YA, Taylor K, Trevanion S, Vandrovcova J, White S, Wilson M, Wilder SP, Aken BL, Birney E, Cunningham F, Dunham I, Durbin R, Fernandez-Suarez XM, Harrow J, Herrero J, Hubbard TJ, Parker A, Proctor G, Spudich G, Vogel J, Yates A, Zadissa A, Searle SM. Ensembl 2012. Nucleic Acids Res. 2012;40:D84–90. doi: 10.1093/nar/gkr991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, Barnes I, Bignell A, Boychenko V, Hunt T, Kay M, Mukherjee G, Rajan J, Despacio-Reyes G, Saunders G, Steward C, Harte R, Lin M, Howald C, Tanzer A, Derrien T, Chrast J, Walters N, Balasubramanian S, Pei B, Tress M, Rodriguez JM, Ezkurdia I, van Baren J, Brent M, Haussler D, Kellis M, Valencia A, Reymond A, Gerstein M, Guigo R, Hubbard TJ. Gencode: The reference human genome annotation for the encode project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan X, Ruan Y, Lassmann T, Carninci P, Brown JB, Lipovich L, Gonzalez JM, Thomas M, Davis CA, Shiekhattar R, Gingeras TR, Hubbard TJ, Notredame C, Harrow J, Guigo R. The gencode v7 catalog of human long noncoding rnas: Analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rufaihah AJ, Huang NF, Jamé S, Lee JC, Nguyen HN, Byers B, De A, Okogbaa J, Rollins M, Reijo-Pera R, Gambhir SS, Cooke JP. Endothelial cells derived from human ipscs increase capillary density and improve perfusion in a mouse model of peripheral arterial disease. Arterioscler Thromb Vasc Biol. 2011;31:e72–e79. doi: 10.1161/ATVBAHA.111.230938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hayden MS, West AP, Ghosh S. Nf-kappab and the immune response. Oncogene. 2006;25:6758–6780. doi: 10.1038/sj.onc.1209943. [DOI] [PubMed] [Google Scholar]
- 23.Hayden MS, Ghosh S. Signaling to nf-kappab. Genes Dev. 2004;18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- 24.Kurian L, Sancho-Martinez I, Nivet E, Aguirre A, Moon K, Pendaries C, Volle-Challier C, Bono F, Herbert JM, Pulecio J, Xia Y, Li M, Montserrat N, Ruiz S, Dubova I, Rodriguez C, Denli AM, Boscolo FS, Thiagarajan RD, Gage FH, Loring JF, Laurent LC, Izpisua Belmonte JC. Conversion of human fibroblasts to angioblast-like progenitor cells. Nat Meth. 2013;10:77–83. doi: 10.1038/nmeth.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li J, Huang NF, Zou J, Laurent TJ, Lee JC, Okogbaa J, Cooke JP, Ding S. Conversion of human fibroblasts to functional endothelial cells by defined factors. Arterioscler Thromb Vasc Biol. 2013;33:1366–1375. doi: 10.1161/ATVBAHA.112.301167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Margariti A, Winkler B, Karamariti E, Zampetaki A, Tsai TN, Baban D, Ragoussis J, Huang Y, Han JD, Zeng L, Hu Y, Xu Q. Direct reprogramming of fibroblasts into endothelial cells capable of angiogenesis and reendothelialization in tissue-engineered vessels. Proc Natl Acad Sci U S A. 2012;109:13793–13798. doi: 10.1073/pnas.1205526109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jasani B, Navabi H, Adams M. Ampligen: A potential toll-like 3 receptor adjuvant for immunotherapy of cancer. Vaccine. 2009;27:3401–3404. doi: 10.1016/j.vaccine.2009.01.071. [DOI] [PubMed] [Google Scholar]
- 28.Navabi H, Jasani B, Reece A, Clayton A, Tabi Z, Donninger C, Mason M, Adams M. A clinical grade poly i:C-analogue (ampligen) promotes optimal dc maturation and th1-type t cell responses of healthy donors and cancer patients in vitro. Vaccine. 2009;27:107–115. doi: 10.1016/j.vaccine.2008.10.024. [DOI] [PubMed] [Google Scholar]
- 29.Coultas L, Chawengsaksophak K, Rossant J. Endothelial cells and vegf in vascular development. Nature. 2005;438:937–945. doi: 10.1038/nature04479. [DOI] [PubMed] [Google Scholar]
- 30.Cole JE, Navin TJ, Cross AJ, Goddard ME, Alexopoulou L, Mitra AT, Davies AH, Flavell RA, Feldmann M, Monaco C. Unexpected protective role for toll-like receptor 3 in the arterial wall. Proc Natl Acad Sci U S A. 2011;108:2372–2377. doi: 10.1073/pnas.1018515108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saito T, Owen DM, Jiang F, Marcotrigiano J, Gale M., Jr Innate immunity induced by composition-dependent rig-i recognition of hepatitis c virus rna. Nature. 2008;454:523–527. doi: 10.1038/nature07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kawai T, Akira S. Toll-like receptor and rig-1-like receptor signaling. Ann N Y Acad Sci. 2008;1143:1–20. doi: 10.1196/annals.1443.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


