Abstract
Point mutation rates in exons (synonymous sites) and noncoding (introns and intergenic) regions are generally assumed to be the same. However, comparative sequence analyses of synonymous substitutions in exons (81 genes) and that of long intergenic fragments (141.3 kbp) of human and chimpanzee genomes reveal a 30%–60% higher mutation rate in exons than in noncoding DNA. We propose a differential CpG content hypothesis to explain this fundamental, and seemingly unintuitive, pattern. We find that the increased exonic rate is the result of the relative overabundance of synonymous sites involved in CpG dinucleotides, as the evolutionary divergence in non-CpG sites is similar in noncoding DNA and synonymous sites of exons. Expectations and predictions of our hypothesis are confirmed in comparisons involving more distantly related species, including human–orangutan, human–baboon, and human–macaque. Our results suggest an underlying mechanism for higher mutation rate in GC-rich genomic regions, predict nonlinear accumulation of mutations in pseudogenes over time, and provide a possible explanation for the observed higher diversity of single nucleotide polymorphisms (SNPs) in the synonymous sites of exons compared to the noncoding regions.
Point mutation is a major source of genetic variation and evolution. The rate at which these mutations arrive in a genome is routinely estimated in mutation accumulation experiments and in disease mutation analysis (Drake et al. 1998; Giannelli et al. 1999; Lynch et al. 1999; Denver et al. 2000). In comparative sequence analysis, mutation rates are inferred by estimating the rate at which neutral substitutions accumulate, because the rate of mutation and substitution is expected to be the same for neutral mutations (Kimura 1983; Kondrashov and Crow 1993; Nei and Kumar 2000). Pseudogenes, introns, intergenic regions, and synonymous sites in exons are largely free from selection in mammals and are therefore routinely used in comparative sequence analysis for this purpose (Wolfe et al. 1989; Li and Graur 1991; Keightley and Eyre-Walker 2000; Nachman and Crowell 2000; Zhao et al. 2000; Kumar and Subramanian 2002).
A survey of mutation rate estimates obtained from the synonymous sites of protein coding genes and noncoding DNA for the human and chimpanzee comparison reveal a significant rate difference in these two types of genomic regions. For instance, we find that the analyses of pseudogenes (Li and Tanimura 1987; Nachman and Crowell 2000; Martinez-Arias et al. 2001) and introns (Bergstrom et al. 1999; Chen et al. 2001) have produced much lower estimates of evolutionary divergence than those observed in synonymous sites in protein coding genes (Li and Tanimura 1987; Wolfe et al. 1989; Easteal 1991; Keightley and Eyre-Walker 2000; Duret et al. 2002; Kumar and Subramanian 2002). Also, intergenic DNA (excluding regulatory sites), which is expected to mutate at a rate similar to that for the coding DNA, also consistently shows much smaller evolutionary divergences than those obtained from coding sequences (Bohossian et al. 2000; Zhao et al. 2000; Chen et al. 2001; Mathews et al. 2001; Yu et al. 2001). This observation is not a random chance occurrence due to paucity of data, because these studies have involved direct comparisons of long intergenic regions (Bohossian et al. 2000; Zhao et al. 2000; Chen et al. 2001) and relatively large numbers of coding genes (Wolfe et al. 1989; Keightley and Eyre-Walker 2000; Duret et al. 2002; Kumar and Subramanian 2002). This difference between coding and noncoding regions in interspecies comparisons is also seen in within-species analysis of single nucleotide polymorphisms (SNPs) that show higher nucleotide diversity in the synonymous sites of exons compared to the noncoding DNA (Moriyama and Powell 1996; Cargill et al. 1999; Halushka et al. 1999; Zwick et al. 2000).
In order to systematically examine the extent and nature of mutation rate differences that have potentially given rise to the observed differences between synonymous sites of exons and noncoding DNA, we present an analysis of an extensive data set containing long stretches of intergenic DNA, introns, and pseudogenes as well as a large number of coding genes from human and other primates.
RESULTS
Patterns of Mutation Rate Difference
We estimated neutral evolutionary distances between human and chimpanzee genomes using the 141.3 kbp of intergenic DNA, 19 pseudogenes, 81 protein-coding genes, and introns from 48 of these protein-coding genes (Table 1; Methods). The synonymous divergence was significantly higher than the sequence divergence observed in different noncoding genomic regions (P < 0.002; Table 1). Similar patterns of difference are seen in the human–orangutan and human–baboon comparisons, suggesting that this phenomenon is common to hominids and old world monkeys (Table 1). It is unlikely that this difference can be explained by increased purifying selection in noncoding regions, as the data set analyzed contains rather long intergenic blocks (10–60 kb) from different human chromosomes and a large number of introns and pseudogenes. All of these vastly different genomic regions are unlikely to be under strong purifying selection with a similar intensity.
Table 1.
Relative Rates of Evolutionary Divergence Between Synonymous Sites in Coding DNA and Different Types of Noncoding Regions
| Coding DNA (#genes, #bp) | Noncoding DNA (#segments, #bp) | Relative rate ratioa | |
| All sitesb | CpG sites excludedc | ||
| Human–Chimpanzee | |||
| Coding genes (81,14790) | Pseudogenes (19,21003) | 1.3 | 1.0 |
| Coding genes (81,14790) | Intergenic regions (4,141322) | 1.6 | 1.0 |
| Coding genes (48,8464)d | Intronic regions (48e,327344) | 1.4 | 1.0 |
| Human–Orangutan | |||
| Coding genes (38,5634) | Intergenic regions (1,9772) | 1.5 | 1.1 |
| Human–Baboon | |||
| Coding genes (31,6512) | Intergenic regions (1,37937) | 1.6 | 1.0 |
| Coding genes (25,5264)d | Intronic regions (25e,260186) | 1.4 | 0.9 |
Relative rate ratio is the ratio between the synonymous divergence at the fourfold-degenerate sites of the exons and the sequence divergence in the noncoding DNA.
Difference in evolutionary distances between synonymous sites of coding genes and noncoding DNA is significant at least at a 0.2% level using a Z-test.
Difference in evolutionary distances (excluding CpG sites) between the synonymous sites of coding genes and noncoding DNA is not significant at the 5% level in each comparison.
Neutral evolutionary divergence from the exons and introns of the same genes.
Count refers to the number of genes from which the introns were obtained.
The Differential CpG Content Hypothesis
We explored an alternative hypothesis to explain the observed difference in mutation rates, which is based on the known phenomenon of hypermutability of methylated CpG dinucleotides to TpG (Coulondre et al. 1978; Bird 1980; Britten et al. 1988; Ponger et al. 2001). As formulated, the differential-CpG-content hypothesis states that the difference in the CpG content is responsible for the observed mutation rate difference between exons and noncoding DNA. To test this hypothesis, we re-estimated the evolutionary divergence in exons, pseudogenes, introns, and intergenic sequences by excluding synonymous sites involved in the CpG dinucleotides in each case. The mutation rate difference among coding and noncoding DNA disappeared, as the ratio of synonymous distance to that observed in pseudogenes, introns, or intergenic sequences is now close to 1 (Table 1). This result is further confirmed in analyses of 48 genes from human–chimpanzee comparison and 25 genes from human–baboon comparison for which the exon and intron sequences were available for the same genes (Table 1).
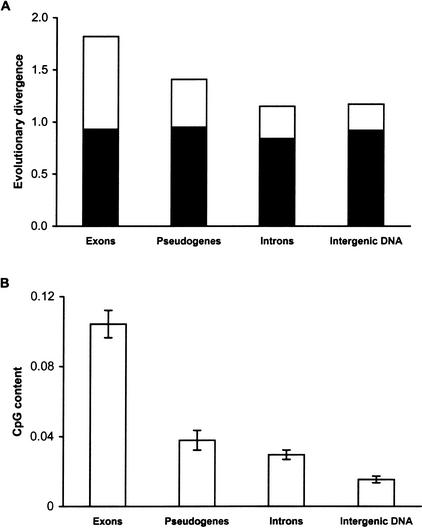
The differential contribution of CpG mutations to the overall mutation rate in noncoding DNA and the synonymous sites of exons is evident from Figure 1. The observed difference in evolutionary divergence correlates with the larger proportion of CpG dinucleotides in exons than those in the noncoding DNA (cf. open bars in Fig. 1A,B). This result holds true even when we normalize the CpG content by accounting for GC-content bias (data not shown). The finding that divergence at non-CpG sites (black bars in Fig. 1A) is almost the same in the synonymous sites of exons and in different types of noncoding regions indicates that the difference in CpG content, rather than selection, is largely responsible for the observed difference in the neutral evolutionary divergence in exons and noncoding DNA.
Figure 1.
(A) Evolutionary divergence per 100 sites and (B) CpG contents of synonymous sites in exons, pseudogenes, introns, and intergenic DNA for the human–chimpanzee comparison. The mean and the standard error estimates shown were obtained from 81 protein coding genes, 19 pseudogenes, introns of 48 genes, and four intergenic blocks. In (A), the total height of each column (including black and open bars) corresponds to the overall evolutionary divergence. The black portion in each column depicts divergence with CpG sites excluded, and therefore, the open bars show the contribution of mutations at CpG sites to the overall divergence. In (B), the CpG content is the proportion of sites involved in the CpG dinucleotide configuration. For noncoding DNA, all sites were included for the estimation of CpG content and evolutionary divergence, whereas for exons only the fourfold-degenerate sites were included (see Methods).
Directionality of the CpG Mutations
The depletion of CpG dinucleotides in vertebrate genomes has been attributed to the directional hypermutability of these dinucleotides to TpG or CpA (Bird 1980; Cooper and Krawczak 1989; Karlin and Mrazek 1997; Hendrich et al. 1999). We examined this directionality of the CpG mutations (Bird 1980) in an analysis of introns and synonymous sites of 22 genes from human and chimpanzee. For these genes, baboon sequences were used to identify the direction of the change at the CpG sites in the human lineage and in the chimpanzee lineage. Results show that for a total of 4600 sites that were potentially CpG in the ancestral sequence (based on the most parsimonious reconstructions), 5.72% experienced CpG → TpG/CpA changes. In contrast, only 0.82% of ancestrally TpG or CpA show a change to CpG. This roughly estimated ∼sevenfold difference in forward and reverse mutation rates is in agreement with previous studies on disease mutations and comparative sequence analysis, which have shown CpG mutations to be 5–20 times more frequent than other types of point mutations (Batzer et al. 1990; Yang et al. 1996; Krawczak et al. 1998; Giannelli et al. 1999; Ebersberger et al. 2002).
Pseudogenes As the Missing Link Between Coding and Noncoding DNA
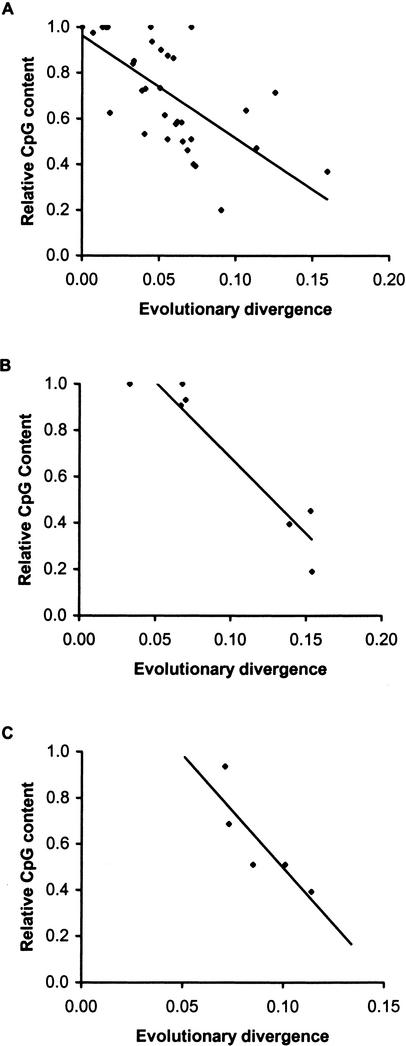
We can also track the evolutionary dynamics of CpG depletion by comparing pseudogenes with their functional counterparts (e.g., Sved and Bird 1990). This provides a glimpse into the process of decay of CpG dinucleotides, as recently emerged pseudogenes are expected to show much higher CpG content than the older pseudogenes. An analysis of 39 pseudogenes from the human genome indeed shows that the overall evolutionary divergence correlates negatively with the ratio of CpG content between the pseudogenes and their functional counterparts (R2 = −0.52, P < 0.01; Fig. 2A). This clear pattern of CpG decay over time is further demonstrated in the analysis of the pseudogenes created at different times from the same functional genes (Fig. 2B,C).
Figure 2.
Relationship between evolutionary divergence and the ratio of CpG contents in pseudogenes and their functional counterparts. (A) Analysis of 39 human pseudogenes [R2 = 0.52; P < 0.01], (B) seven pseudogenes created from the same Arginino succinate synthetase gene [R2 = 0.90; P < 0.01], and (C) five pseudogenes of the same Beta tubulin Q gene [R2 = 0.73; P < 0.05]. The relative CpG content is the ratio of the overall CpG content in the pseudogene divided by overall CpG content of the functional gene.
A comparison of Figure 1A and B also shows that the evolutionary divergence as well as the CpG content of pseudogenes are at an average intermediate compared to those at synonymous sites and noncoding DNA. This result is now explained by the finding that the pseudogenes created recently from the functional coding genes have higher CpG content than introns and intergenic regions (Fig. 2). However, as pseudogenes become older their CpG content declines until it reaches the level present in the surrounding noncoding DNA. This also means that the average rate of mutation per site in a pseudogene will decrease with time, because the number of CpG dinucleotides will decrease over time. Therefore, there are no linear mutation clocks for pseudogene evolution.
DISCUSSION
It is now clear that there is a significant difference in the evolutionary divergence between the noncoding and the synonymous sites of the coding DNA. This difference correlates with differences in the content of the hypermutable CpG dinucleotides in these two types of genomic DNA. This is a causal relationship, as the removal of CpG dinucleotides from data eliminates the difference. The higher CpG content in codon positions is already well appreciated as the observed versus expected (O/E) ratio of CpG contents in the 1st–2nd, 2nd–3rd, and 3rd–1st positions is known to be higher than that for the genomic sequences (Sved and Bird 1990; Caccio et al. 1997; Jabbari and Bernardi 1998; Halushka et al. 1999). In the data set of introns and exons of 48 genes from human and chimpanzee comparison, we find that the noncoding DNA shows an O/E ratio of 0.23, whereas the synonymous sites in exons show an O/E ratio of 0.40. This is an almost twofold difference and is likely caused by the fact that one of the nucleotides of the CpG dinucleotides involving a fourfold-degenerate site is under purifying selection, as they are always from the 1st or 2nd codon position. This increases the probability of occurrence and maintenance of CpG dinucleotides in exons compared to noncoding DNA, where both nucleotides in the CpG dinucleotides are free to undergo neutral nucleotide substitution. This results in a higher average mutational input per synonymous site, as synonymous sites are largely free to evolve irrespective of the selection on the adjacent codon positions.
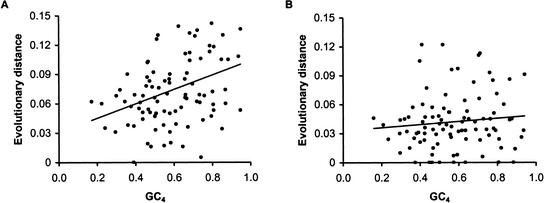
The knowledge of CpG content-mediated difference in mutation rates also provides new insights into reasons behind a number of previous observations. For instance, a significant positive correlation has been reported between the GC content of the 3rd codon position and the synonymous substitution in primates (Bielawski et al. 2000). This is often thought to be due to differences in rates of mutations among regions with differing base compositions (Smith et al. 2002; Yi et al. 2002). However, GC-rich regions are expected to contain more CpG dinucleotides than GC-poor regions. It is therefore possible that differences in CpG content in GC-poor and GC-rich regions are a major factor in the observed synonymous distance difference. This is indeed the case, as revealed in an analysis of 93 genes from our human–macaque comparison (Fig. 3A,B). The positive correlation between GC content and the synonymous distance becomes rather small when synonymous sites involved in CpG dinucleotides are excluded. These results are consistent with the reports of CpG-mediated correlation between the heterozygosity and GC content of the human SNPs (Sachidanandam et al. 2001).
Figure 3.
(A) Relationship between evolutionary divergence and GC content, GC4, at synonymous sites of 93 genes from the human–macaque comparison; R2 = 0.125, P < 0.01. (B) Relationship between evolutionary divergence and GC4 after excluding fourfold-degenerate sites involved in CpG dinucleotides; R2 = 0.01, P > 0.05.
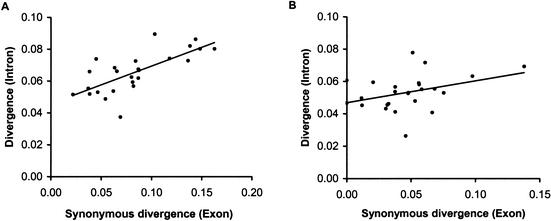
Furthermore, there have been reports of positive correlation between evolutionary distances observed at synonymous sites and the noncoding DNA for the human–mouse genome comparison (Hardison et al. 2003). This positive correlation (R2 = 0.48, P < 0.01) is also reflected in our analysis of 25 genes of human and baboon (Fig. 4A). However after the exclusion of CpG sites, this correlation reduces significantly (Fig. 4B; R2 = 0.12), suggesting that the CpG mutations mediate this relationship in these primate genomes. In fact, the R2 value for this relationship drops down to 0.04 if we remove just a single outlier (Fig. 4B). Thus, the actual rates of mutation at non-CpG sites, which comprise >95% of noncoding DNA and >90% of synonymous sites, are quite similar in GC-rich and GC-poor regions. This result is in contrast to some of the previous studies of human–mouse genome comparisons (e.g., Waterston et al. 2002; Hardison et al. 2003), which presumably removed the influence of CpG mutations when estimating neutral distances. However, when the time of species divergence is very large, as is the case for human and mouse (90–110 million years ago, see Kumar and Hedges 1998; Hedges and Kumar 2003; Springer et al. 2003), the identification of CpG sites as well as the assessment of CpG mutations on overall divergence becomes unreliable (see also Methods). Therefore, it is important to use closely related species to examine the mutation rate variation across the genome.
Figure 4.
(A) Relationship between synonymous divergence in exons and evolutionary divergence in introns; R2 = 0.511, P < 0.01. (B) Relationship between synonymous divergence in exons and evolutionary divergence in introns after removing CpG sites; R2 = 0.137, P > 0.05. With the rightmost outlier excluded, R2 = 0.06, P > 0.05.
Finally, the demonstration of significant differences in evolutionary rates in exonic and noncoding DNA due to CpG-content difference provides a potential explanation for the observed 30%–40% higher diversity in the human SNPs in the fourfold-degenerate sites of exons compared to noncoding DNA (Cargill et al. 1999; Halushka et al. 1999). This difference is similar to that observed in the interspecies comparisons (Table 1). It was also reflected in an analysis of 6109 human SNPs, which we mapped to fourfold-degenerate sites in the reference human sequence (August 7, 2002 release of dbSNP from NCBI was used). We found that 39.5% of all SNPs at the fourfold-degenerate sites were of CpG ↔ TpG/CpA types, which is close to the proportion of variable sites with CpG ↔ TpG/CpA difference in the interspecies comparisons (39.0%–48.4%). Therefore, although the synonymous sites of exons and noncoding DNA may be used for estimating coalescence times of population polymorphisms (and alleles), they will require the use of different mutation rates unless the CpG mutations are excluded.
METHODS
Data Mining and Assembly
Protein coding genes used in this study were obtained from the HOVERGEN database, release 36 (Duret et al. 1994) and from the GenBank. Phylogenetic trees of 8627 HOVERGEN gene families were constructed from amino acid sequence alignments using the neighbor-joining (NJ) method in MEGA2 (Kumar et al. 2001). The cDNA sequence alignments for orthologous sequence sets were then generated using amino acid sequence alignments as guides. NJ trees were scanned automatically, followed by manual inspection, to identify orthologous sequence sets. We enforced strict orthology definitions by considering sequences to be orthologous only if no gene duplication events were detected since their divergence from the most recent common ancestor. Gene duplication events were identified by comparing the gene family tree with the species tree from NCBI (see also Zmasek and Eddy 2001). All gene families containing fewer than three sequences were excluded. This produced a data set consisting of 33, 38, 93, and six orthologous pairs of sequences for human–chimpanzee, human–orangutan, human–macaque, and human–baboon comparisons, respectively. We also obtained a set of 19 autosomal pseudogenes through searching the literature (Nachman and Crowell 2000; Chen et al. 2001). In addition, orthologous noncoding intergenic segments belonging to human, chimpanzee, and orangutan genomes were obtained (Bohossian et al. 2000; Zhao et al. 2000). To compare the evolutionary divergence at the neutral sites of exons and introns of the same gene, we obtained (from GenBank) 48 genes for the human–chimpanzee comparison and 25 genes for the human–baboon comparison. When not available, the intron–exon boundaries of chimpanzee and baboon genes were identified by using the corresponding human exons in the BLASTZ software (Schwartz et al. 2003). For intergenic sequence comparisons, five large contigs (human: AC002066, AC002080, chimpanzee: AC087253, AC087512, baboon: AC084730) were obtained from GenBank. All repeat elements in these contigs were identified using RepeatMasker (http://ftp.genome.washington.edu/cgi-bin/RepeatMasker) and removed prior to the sequence alignment. CpG islands in the upstream regions are usually unmethylated and are not hypermutable (Bird 1999). Therefore, they were also excluded from further analyses. (However, our results remained the same whether or not they were excluded). Also, the coding genes known from the annotated human genome were removed, along with the 5-kb flanking regions immediately upstream and downstream of each gene to exclude potential regulatory regions. This resulted in intergenic regions with sizes of 52.6 kbp (AC002066 and AC087512) and 65.0 kbp (AC002080 and AC087253) for the human–chimpanzee comparison, and 38.0 kbp (AC002066 and AC084730) for the human–baboon comparison. All noncoding sequences were aligned using BLASTZ.
For analysis of CpG depletion in pseudogenes, we used 19 pseudogenes for the human–chimpanzee comparison. In addition, we obtained 116 human pseudogenes from the GenBank. Using local BLAST with 31,289 human cDNA (obtained from ftp://ftp.ncbi.nih.gov/genomes/H_sapiens/), we found the functional counterparts of these pseudogenes, which had an e-value of 0. We restricted our analysis to the pseudo-functional gene pairs for which evolutionary distance was small (d < 0.2). Our final data set contains 39 pseudogene-functional gene pairs.
Estimation of Evolutionary Divergence
Evolutionary divergences were computed using only the fourfold-degenerate sites for coding sequences and all sites for pseudogenes, introns, and intergenic regions. We took a stringent approach in identifying fourfold-degenerate sites by selecting only those sites that have remained fourfold-degenerate in both the species compared. A genomic synonymous distance estimate was obtained by concatenating all coding genes for a given species pair. Evolutionary distances were estimated for pseudogenes and introns by concatenating all the respective sequences. Correction for multiple hits was done using the Tamura-Nei method, which accounts for transition/transversion and base composition biases (Tamura and Nei 1993). CpG content was estimated as the number of sites involved in the CpG configuration divided by the total number of sites. For exons, only the fourfold-degenerate sites involved in the CpG configuration (G of the CpG dinucleotide in the second and third codon positions and C of the CpG dinucleotide in the third and first [adjacent] codon positions) were used. For noncoding DNA (including pseudogenes and introns) and the coding genes used for the CpG depletion study (Fig. 2), all sites involved in CpG dinucleotides were included. In the pseudogene-functional gene comparisons, we excluded all CpG sites when estimating the evolutionary divergence, in order to avoid introducing spurious negative correlation.
Note that the existing methods to correct for multiple hits in evolutionary distances estimation do not account for the CpG substitutions, which occur at a much higher rate than other mutations and with a different pattern. Use of Tamura-Nei method (Tamura and Nei 1993) is expected to lead to underestimation in this case. However in our analysis we have used only closely related species. In such cases the choice of a simpler substitution model is known to produce minimal underestimation (see Nei and Kumar 2000). For the current scenario, we examined the extent of underestimation by conducting a computer simulation in which two identical sequences with a prespecified CpG content were made to evolve over time, and the evolutionary divergence between sequences was estimated using the Tamura-Nei distance. CpG substitutions were made to occur at 10 times the rate of all other substitutions, and the total actual number of substitutions was recorded. The estimated numbers of substitutions obtained using Tamura-Nei (1993) distance were compared to the actual number of substitutions at difference levels of sequence divergence for the sequence length of 10,000. Simulations were conducted with different GC contents (25%, 50%, and 75%). We found that for sequence divergences considered here (<0.1 per site), the Tamura-Nei distances were less than 10% underestimates of the true distance (in each case they were slight underestimates; but they were large for divergences >0.1 per site). This means that the difference in rates between exons and other genomic regions reported here are conservative estimates, as the sequence divergence from exons with higher CpG content will be underestimated more than that for the CpG-poor noncoding regions.
WEB SITE REFERENCES
http://ftp.genome.washington.edu/cgi-bin/RepeatMasker; RepeatMasker Web server.
http://www.nisc.nih.gov; NIH intramural sequencing center.
ftp://ftp.ncbi.nih.gov/genomes/H_sapiens/; Human genomic sequences at National Center for Biotechnology Information.
Acknowledgments
We thank Philip Hedrick, Koichiro Tamura, Michael Rosenberg, Robert Friedman, Alan Filipski, Rekha Iyer, and Patrick Kolb for helpful discussions and comments. We also thank Alan Filipski for help with computer simulations, Michael Rosenberg for analysis of SNP data, Balaji Ramanujam for some data retrieval, and David Cutler, Phil Green, and an anonymous reviewer for insightful comments. Many of the genomic contigs used in this study were generated by the NIH Intramural Sequencing Center (http://www.nisc.nih.gov). This research was supported by research grants from the NIH, NSF, and the Burroughs-Wellcome Fund to S.K.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
E-MAIL s.kumar@asu.edu; FAX(480) 965-2519.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1152803.
REFERENCES
- 1.Batzer M.A., Kilroy, G.E., Richard, P.E., Shaikh, T.H., Desselle, T.D., Hoppens, C.L., and Deininger, P.L. 1990. Structure and variability of recently inserted Alu family members. Nucleic Acids Res. 18: 6793-6798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bergstrom T.F., Erlandsson, R., Engkvist, H., Josefsson, A., Erlich, H.A., and Gyllensten, U. 1999. Phylogenetic history of hominoid DRB loci and alleles inferred from intron sequences. Immunol. Rev. 167: 351-365. [DOI] [PubMed] [Google Scholar]
- 3.Bielawski J.P., Dunn, K.A., and Yang, Z. 2000. Rates of nucleotide substitution and mammalian nuclear gene evolution. Approximate and maximum-likelihood methods lead to different conclusions. Genetics 156: 1299-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bird A. 1999. DNA methylation de novo. Science 286: 2287-2288. [DOI] [PubMed] [Google Scholar]
- 5.Bird A.P. 1980. DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res. 8: 1499-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bohossian H.B., Skaletsky, H., and Page, D.C. 2000. Unexpectedly similar rates of nucleotide substitution found in male and female hominids. Nature 406: 622-625. [DOI] [PubMed] [Google Scholar]
- 7.Britten R.J., Baron, W.F., Stout, D.B., and Davidson, E.H. 1988. Sources and evolution of human Alu repeated sequences. Proc. Natl. Acad. Sci. 85: 4770-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caccio S., Jabbari, K., Matassi, G., Guermonprez, F., Desgres, J., and Bernardi, G. 1997. Methylation patterns in the isochores of vertebrate genomes. Gene 205: 119-124. [DOI] [PubMed] [Google Scholar]
- 9.Cargill M., Altshuler, D., Ireland, J., Sklar, P., Ardlie, K., Patil, N., Shaw, N., Lane, C.R., Lim, E.P., Kalyanaraman, N., et al. 1999. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat. Genet. 22: 231-238. [DOI] [PubMed] [Google Scholar]
- 10.Chen F.C., Vallender, E.J., Wang, H., Tzeng, C.S., and Li, W.H. 2001. Genomic divergence between human and chimpanzee estimated from large-scale alignments of genomic sequences. J. Hered. 92: 481-489. [DOI] [PubMed] [Google Scholar]
- 11.Cooper D.N. and Krawczak, M. 1989. Cytosine methylation and the fate of CpG dinucleotides in vertebrate genomes. Hum. Genet. 83: 181-188. [DOI] [PubMed] [Google Scholar]
- 12.Coulondre C., Miller, J.H., Farabaugh, P.J., and Gilbert, W. 1978. Molecular basis of base substitution hotspots in Escherichia coli. Nature 274: 775-780. [DOI] [PubMed] [Google Scholar]
- 13.Denver D.R., Morris, K., Lynch, M., Vassilieva, L.L., and Thomas, W.K. 2000. High direct estimate of the mutation rate in the mitochondrial genome of Caenorhabditis elegans. Science 289: 2342-2344. [DOI] [PubMed] [Google Scholar]
- 14.Drake J.W., Charlesworth, B., Charlesworth, D., and Crow, J.F. 1998. Rates of spontaneous mutation. Genetics 148: 1667-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duret L., Mouchiroud, D., and Gouy, M. 1994. HOVERGEN: A database of homologous vertebrate genes. Nucleic Acids Res. 22: 2360-2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duret L., Semon, M., Piganeau, G., Mouchiroud, D., and Galtier, N. 2002. Vanishing GC-rich isochores in mammalian genomes. Genetics 162: 1837-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Easteal S. 1991. The relative rate of cDNA evolution in primates. Mol. Biol. Evol. 8: 115-127. [DOI] [PubMed] [Google Scholar]
- 18.Ebersberger I., Metzler, D., Schwarz, C., and Paabo, S. 2002. Genome-wide comparison of DNA sequences between humans and chimpanzees. Am. J. Hum. Genet. 70: 1490-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giannelli F., Anagnostopoulos, T., and Green, P.M. 1999. Mutation rates in humans. II. Sporadic mutation-specific rates and rate of detrimental human mutations inferred from hemophilia B. Am. J. Hum. Genet. 65: 1580-1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Halushka M.K., Fan, J.B., Bentley, K., Hsie, L., Shen, N., Weder, A., Cooper, R., Lipshutz, R., and Chakravarti, A. 1999. Patterns of single-nucleotide polymorphisms in candidate genes for blood-pressure homeostasis. Nat. Genet. 22: 239-247. [DOI] [PubMed] [Google Scholar]
- 21.Hardison R.C., Roskin, K.M., Yang, S., Diekhans, M., Kent, W.J., Weber, R., Elnitski, L., Li, J., O'Connor, M., Kolbe, D., et al. 2003. Covariation in frequencies of substitution, deletion, transposition and recombination during eutherian evolution. Genome Res. 13: 13-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hedges, S.B. and Kumar, S. 2003. Genomic clocks and evolutionary timescales. Trends Genet. (in press). [DOI] [PubMed]
- 23.Hendrich B., Hardeland, U., Ng, H.H., Jiricny, J., and Bird, A. 1999. The thymine glycosylase MBD4 can bind to the product of deamination at methylated CpG sites. Nature 401: 301-304. [DOI] [PubMed] [Google Scholar]
- 24.Jabbari K. and Bernardi, G. 1998. CpG doublets, CpG islands and Alu repeats in long human DNA sequences from different isochore families. Gene 224: 123-127. [DOI] [PubMed] [Google Scholar]
- 25.Karlin S. and Mrazek, J. 1997. Compositional differences within and between eukaryotic genomes. Proc. Natl. Acad. Sci. 94: 10227-10232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keightley P.D. and Eyre-Walker, A. 2000. Deleterious mutations and the evolution of sex. Science 290: 331-333. [DOI] [PubMed] [Google Scholar]
- 27.Kimura M., 1983. The neutral theory of molecular evolution. Cambridge University Press, Cambridge, UK.
- 28.Kondrashov A.S. and Crow, J.F. 1993. A molecular approach to estimating the human deleterious mutation rate. Hum. Mutat. 2: 229-234. [DOI] [PubMed] [Google Scholar]
- 29.Krawczak M., Ball, E.V., and Cooper, D.N. 1998. Neighboring-nucleotide effects on the rates of germ-line single-base-pair substitution in human genes. Am. J. Hum. Genet. 63: 474-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar S. and Hedges, B. 1998. A molecular timescale for vertebrate evolution. Nature 392: 917-919. [DOI] [PubMed] [Google Scholar]
- 31.Kumar S. and Subramanian, S. 2002. Mutation rates in mammalian genomes. Proc. Natl. Acad. Sci. 99: 803-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumar S., Tamura, K., Jakobsen, I.B., and Nei, M. 2001. MEGA2: Molecular evolutionary genetics analysis software. Bioinformatics 17: 1-2. [DOI] [PubMed] [Google Scholar]
- 33.Li W.-H. and Graur, D., 1991. Fundamentals of molecular evolution. Sinauer Associates, Sunderland, MA.
- 34.Li W.-H. and Tanimura, M. 1987. The molecular clock runs more slowly in man than in apes and monkeys. Nature 326: 93-96. [DOI] [PubMed] [Google Scholar]
- 35.Lynch M., Blanchard, J., Houle, D., Kibota, T., Schultz, S., Vassilieva, L., and Willis, J. 1999. Perspective: Spontaneous deleterious mutation. Evolution 53: 645-663. [DOI] [PubMed] [Google Scholar]
- 36.Martinez-Arias R., Calafell, F., Mateu, E., Comas, D., Andres, A., and Bertranpetit, J. 2001. Sequence variability of a human pseudogene. Genome Res. 11: 1071-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mathews D.J., Kashuk, C., Brightwell, G., Eichler, E.E., and Chakravarti, A. 2001. Sequence variation within the fragile X locus. Genome Res. 11: 1382-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moriyama E.N. and Powell, J.R. 1996. Intraspecific nuclear DNA variation in Drosophila. Mol. Biol. Evol. 13: 261-277. [DOI] [PubMed] [Google Scholar]
- 39.Nachman M.W. and Crowell, S.L. 2000. Estimate of the mutation rate per nucleotide in humans. Genetics 156: 297-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nei M. and Kumar, S., 2000. Molecular evolution and phylogenetics. Oxford University Press, New York, NY.
- 41.Ponger L., Duret, L., and Mouchiroud, D. 2001. Determinants of CpG islands: Expression in early embryo and isochore structure. Genome Res. 11: 1854-1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sachidanandam R., Weissman, D., Schmidt, S.C., Kakol, J.M., Stein, L.D., Marth, G., Sherry, S., Mullikin, J.C., Mortimore, B.J., Willey, D.L., et al. 2001. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature 409: 928-933. [DOI] [PubMed] [Google Scholar]
- 43.Schwartz S., Kent, W.J., Smit, A., Zhang, Z., Baertsch, R., Hardison, R.C., Haussler, D., and Miller, W. 2003. Human–mouse alignments with BLASTZ. Genome Res. 13: 103-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith N.G., Webster, M.T., and Ellegren, H. 2002. Deterministic mutation rate variation in the human genome. Genome Res. 12: 1350-1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Springer M.S., Murphy, W.J., Eizirik, E., and O'Brien, S.J. 2003. Placental mammal diversification and the Cretaceous-Tertiary boundary. Proc. Natl. Acad. Sci. 100: 1056-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sved J. and Bird, A. 1990. The expected equilibrium of the CpG dinucleotide in vertebrate genomes under a mutation model. Proc. Natl. Acad. Sci. 87: 4692-4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tamura K. and Nei, M. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10: 512-526. [DOI] [PubMed] [Google Scholar]
- 48.Waterston R.H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J.F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520-562. [DOI] [PubMed] [Google Scholar]
- 49.Wolfe K.H., Sharp, P.M., and Li, W.-H. 1989. Mutation rates differ among regions of the mammalian genome. Nature 337: 283-285. [DOI] [PubMed] [Google Scholar]
- 50.Yang A.S., Gonzalgo, M.L., Zingg, J.M., Millar, R.P., Buckley, J.D., and Jones, P.A. 1996. The rate of CpG mutation in Alu repetitive elements within the p53 tumor suppressor gene in the primate germline. J. Mol. Biol. 258: 240-250. [DOI] [PubMed] [Google Scholar]
- 51.Yi S., Ellsworth, D.L., and Li, W.H. 2002. Slow molecular clocks in old world monkeys, apes, and humans. Mol. Biol. Evol. 19: 2191-2198. [DOI] [PubMed] [Google Scholar]
- 52.Yu N., Zhao, Z., Fu, Y.X., Sambuughin, N., Ramsay, M., Jenkins, T., Leskinen, E., Patthy, L., Jorde, L.B., Kuromori, T., et al. 2001. Global patterns of human DNA sequence variation in a 10-kb region on chromosome 1. Mol. Biol. Evol. 18: 214-222. [DOI] [PubMed] [Google Scholar]
- 53.Zhao Z., Jin, L., Fu, Y.X., Ramsay, M., Jenkins, T., Leskinen, E., Pamilo, P., Trexler, M., Patthy, L., Jorde, L.B., et al. 2000. Worldwide DNA sequence variation in a 10-kilobase noncoding region on human chromosome 22. Proc. Natl. Acad. Sci. 97: 11354-11358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zmasek C.M. and Eddy, S.R. 2001. A simple algorithm to infer gene duplication and speciation events on a gene tree. Bioinformatics 17: 821-828. [DOI] [PubMed] [Google Scholar]
- 55.Zwick M.E., Cutler, D.J., and Chakravarti, A. 2000. Patterns of genetic variation in Mendelian and complex traits. Annu. Rev. Genomics Hum. Genet. 1: 387-407. [DOI] [PubMed] [Google Scholar]