Abstract
Peripheral events in olfaction involve odorant binding proteins (OBPs) whose role in the recognition of different volatile chemicals is yet unclear. Here we report on the sensitive and quantitative measurement of the weak interactions associated with neutral enantiomers differentially binding to OBPs immobilized through a self-assembled monolayer to the gate of an organic bio-electronic transistor. The transduction is remarkably sensitive as the transistor output current is governed by the small capacitance of the protein layer undergoing minute changes as the ligand–protein complex is formed. Accurate determination of the free-energy balances and of the capacitance changes associated with the binding process allows derivation of the free-energy components as well as of the occurrence of conformational events associated with OBP ligand binding. Capacitance-modulated transistors open a new pathway for the study of ultra-weak molecular interactions in surface-bound protein–ligand complexes through an approach that combines bio-chemical and electronic thermodynamic parameters.
 Organic bioelectronic devices can be used to convert biological recognition events to a signal output. Here, the authors show how a water gated field-effect transistor modified with odorant binding proteins can be used to analyse the enantioselectivity of carvone binding and also estimate free-energy changes.
Organic bioelectronic devices can be used to convert biological recognition events to a signal output. Here, the authors show how a water gated field-effect transistor modified with odorant binding proteins can be used to analyse the enantioselectivity of carvone binding and also estimate free-energy changes.
Olfactory transduction is initiated when small volatile compounds (odorant molecules) in air interact with membrane-bound olfactory receptor proteins in olfactory sensory neurons1,2 after partitioning through the aqueous compartment of nasal mucus of vertebrates3 or the sensillum lymph in the antennae of insects. In the mucus or in the lymph small soluble odorant binding proteins (OBPs) are found, which are secreted in high concentrations (millimolar range)4,5. The binding properties are characterized by a broad selectivity to a range of odorant ligands with dissociation constants (K) in the micromolar range4. The physiological role of OBPs in the odour perception process is still unclear. They may shuttle odorant molecules from air to the olfactory receptors through the mucus or the lymph, or have a role in the odorant clearance mechanism after the transduction of the olfactory signal6,7. Recent evidence indicates direct involvement of OBPs in the recognition of different volatile chemicals in insects8,9,10,11.
OBPs are extremely stable to temperature, organic solvents and proteolytic digestion. They can be expressed in bacterial systems at low cost, are easily purified12 and the binding properties can be tailored through mutagenesis, making them attractive for a number of biotechnological applications. Vertebrate OBPs are characterized by a hydrophobic β-barrel cavity13, where the ligand is hosted and the monomeric porcine OBPs (pOBPs)14 form a useful model system to study binding affinities to odorant compounds. While pOBPs are weakly negatively charged in pure water15, (S)-(+)- and (R)-(−)-carvone enantiomers are odorant molecules that bear a dipole moment16. Understanding of the nature of the ligand–protein interactions at a molecular level is limited, and little is yet known about the interaction of chiral molecules with OBPs.
OBPs have been used as recognition elements in different transducing systems but the reported detection limits do not go below nanomolar level. However, lately bio-detection at very low concentrations has become possible using field-effect back gate17,18,19 and water-gated organic field-effect transistor (WGOFET)20 sensors21,22. Particularly relevant is the case of a back-gate OFET that can provide differential detection of carvone enantiomers at the tens of micromolar concentration level19. WGOFETs are operated at very low voltages23 and can be fabricated at low cost on flexible substrates24. In a bio-FET the gate electrode is generally modified with an electro-active bio-recognition element, and on exposure to the target molecule a threshold voltage (VT)25 shift that scales with the logarithm of the analyte concentration26,27 has been so far generally measured. The scaling of VT with the concentration is a general property of FET transduction28 since the reaction electrochemical Gibbs free energy (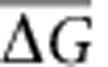 )29 changes the electrochemical potential of the gate metal electrons,
)29 changes the electrochemical potential of the gate metal electrons, 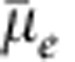 . In an ideal trap-free OFET, VT defines the gate bias needed to offset the mismatch between the gate metal and the semiconductor electrons electrochemical potential
. In an ideal trap-free OFET, VT defines the gate bias needed to offset the mismatch between the gate metal and the semiconductor electrons electrochemical potential 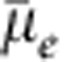 . When the reaction takes place at the gate, the
. When the reaction takes place at the gate, the 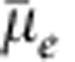 is changed and VT shifts. As
is changed and VT shifts. As  is equal to μ+nFV (μ=chemical potential, F=Faraday’s constant, n=moles of charges and V=electrostatic potential), it is associated with strong long-range coulomb interactions (10–100 kJ mol−1). This is why FETs generally provide sensitive detection in reactions that involve charged species, but are less successful for quantitative detection of neutral species30. The Gibbs free energy (G) of interactions involving neutral species coinvolves the system energy and entropy, ignoring the effect of net charges or monopoles31. G can be associated with the interaction energy which accounts for weak shorter-range interactions such as dipole–dipole or dispersive interactions (~2 kJ mol−1).
is equal to μ+nFV (μ=chemical potential, F=Faraday’s constant, n=moles of charges and V=electrostatic potential), it is associated with strong long-range coulomb interactions (10–100 kJ mol−1). This is why FETs generally provide sensitive detection in reactions that involve charged species, but are less successful for quantitative detection of neutral species30. The Gibbs free energy (G) of interactions involving neutral species coinvolves the system energy and entropy, ignoring the effect of net charges or monopoles31. G can be associated with the interaction energy which accounts for weak shorter-range interactions such as dipole–dipole or dispersive interactions (~2 kJ mol−1).
This article reports on the binding of (S)-(+)- and (R)-(−)-carvone enantiomers to a pOBP-mutant-F88W, detected by means of a WGOFET. With this approach we achieve the sensitive evaluation of energies as low as 1.1±0.5 kJ mol−1, associated with neutral enantiomers differentially binding to a pOBP immobilized through a self-assembled monolayer (SAM). The WGOFET output current is governed by the protein nano-layer capacitance where subtle changes can be transduced, with a high signal-to-noise ratio, due to the transistor current gain. Analysis of the free-energy balance and of the degree of capacitance decreases, provides strong support for the hypothesis that on S-(+)-carvone ligand binding the pOBP undergoes a conformational change that is not seen with the R-(−) enantiomer.
Results
Dissociation constants of pOBPs in solution
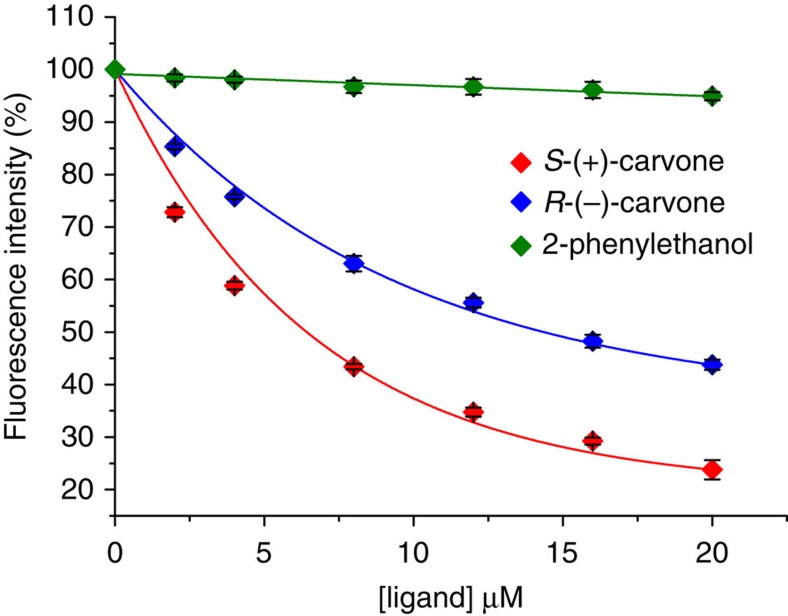
A competitive fluorescent binding assay performed on pOBPs in solution, returned for pOBP-carvone enantiomer complexes (pOBP-C) dissociation constants KSol+ of 0.50±0.01 μM and KSol− of 1.22±0.05 μM, for the (S)-(+)- and (R)-(−)-carvone, respectively. The data are reported in Fig. 1 and the details of the dissociation constants extraction are reported in the Method section. These data prove the pOBP-mutant-F88W to be chiral active. The binding is relatively selective, and control experiments indicate that 2-phenylethanol binds very weakly to pOBP (KSol~40 μM) (Fig. 1), while wasp-OBP is not capable of differentiation between carvone enantiomers (Supplementary Fig. 1).
Figure 1. Competitive binding curves for carvone enantiomers and 2-phenylethanol.
The affinity of binding of the porcine pOBP was measured using the 1-AMA fluorescence probe. The fluorescent signal is reported as a function of the ligand concentration. A decrease in fluorescence intensity of 1-AMA with increasing concentrations of binding ligands is seen. There is also clear difference in affinities between the carvone and the 2-phenylethanol ligands. Moreover, the decrease in fluorescence intensity of 1-AMA for two enantiomers differs significantly, confirming the differential selectivity of carvone enantiomers by pOBP-F88W.
The pOBP-WGOFET functional mechanism
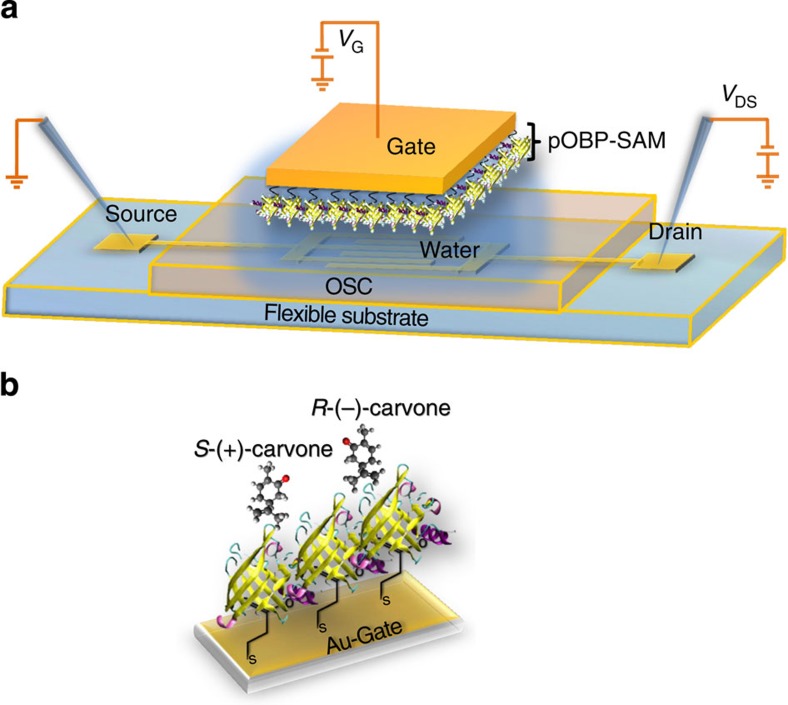
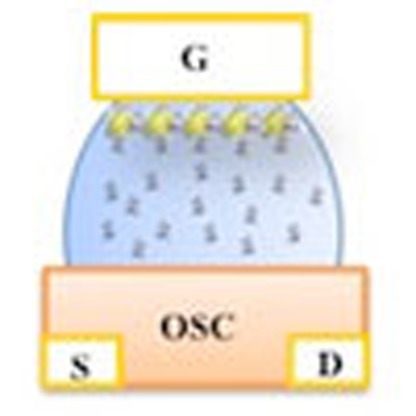
In Fig. 2a, a schematic of the pOBP-WGOFET organic bio-electronic32 structure is reported, showing source (S) and drain (D) contacts covered by a spin-coated poly[2,5-bis(3-tetradecylthiophen-2-yl)thieno[3,2-b]thiophene] (PBTTT-C14) p-type organic semiconductor (OSC) deposited on a flexible substrate. A droplet of water is placed on the OSC hydrophobic surface and electrical contact to the gate (G) is made through a gold plate whose water exposed surface is functionalized with a very compact pOBP-SAM, schematically shown in Fig. 2b. An extensive surface characterization of the pOBP-SAM is reported in the Supplementary Information. The data in Supplementary Figs 2 and 3 assess the surface coverage of the gold electrode by evaluating the electrochemical hindrance. In Supplementary Figs 4–6, the SAM surface chemical composition is studied by means of X-ray photoelectron spectroscopy analysis, while in Supplementary Fig. 7 the pOBP-SAM morphology is evaluated by scanning electron microscopy.
Figure 2. Water-gated bio-organic transistor comprising a pOBP-SAM.
In a, the WGOFET device schematic structure is shown. The source (S) and drain (D) interdigitated contacts are defined on a flexible foil. The S–D patterned substrate is covered by the p-type, PBTTT-C14 and a 3 μl droplet of water lies on the OSC surface. A bio-functionalized Au-plate hangs in contact with the water droplet, acting as gate (G). The pOBP protein structure is sketched in b along with the SAM on the gate surface. pOBP is a monomer of 157 amino acid residues (molecular mass of ~19 kDa) with a height of 38.04 Å and a base of 25.70 × 26.40 Å.
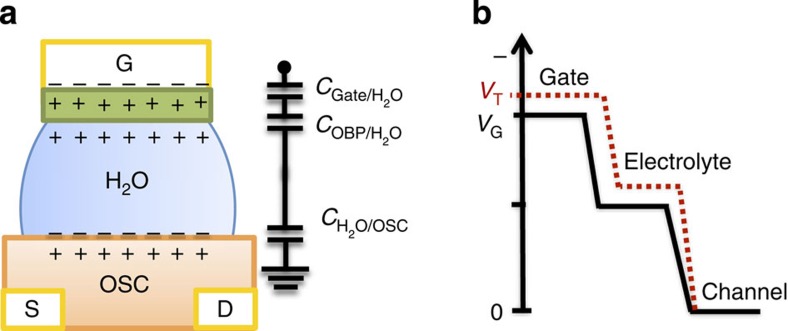
A p-type FET is operated by applying negative VDS and VGS biases to the drain and the gate contacts while the source contact is grounded (Fig. 2a). Due to self-ionization, pure water becomes an ionic conductor and, on application of VG, the ions redistribute so that H+ ions face the negatively biased gate plate while OH− align at the OSC surface (Fig. 3a). High capacitance electrical double layers at the gate/water, pOBP-SAM/water and water/OSC interfaces eventually form giving rise to the potential drop profile33 reported in Fig. 3b. The Au-gate/water and the water/OSC interfaces capacitances per unit area are reported20 to be 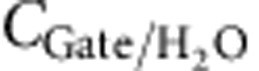 ~40 μF cm−2 and
~40 μF cm−2 and 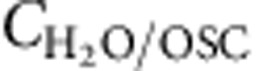 ~1–5 μF cm−2, respectively. The pOBP-SAM capacitance per unit area (COBP) is modelled as a planar capacitor, C=ε0εrd−1, with ε0 and εr being the vacuum and the relative permittivity while d is the distance between capacitor plates. Taking εr=3, typical for a protein system34, and d as the height of the pOBP-SAM (4.22 nm), a COBP=0.63 μF cm−2 is estimated. The COBP can be approximated to the whole pOBP-WGOFET gating-system capacitance as this is the smallest in the series (Fig. 3a). This general occurrence is solely connected with the differences in the dielectric constants of a protein-SAM and an OSC compared with water. The actual value of COBP depends, in turn, on several microscopic quantities and parameters.
~1–5 μF cm−2, respectively. The pOBP-SAM capacitance per unit area (COBP) is modelled as a planar capacitor, C=ε0εrd−1, with ε0 and εr being the vacuum and the relative permittivity while d is the distance between capacitor plates. Taking εr=3, typical for a protein system34, and d as the height of the pOBP-SAM (4.22 nm), a COBP=0.63 μF cm−2 is estimated. The COBP can be approximated to the whole pOBP-WGOFET gating-system capacitance as this is the smallest in the series (Fig. 3a). This general occurrence is solely connected with the differences in the dielectric constants of a protein-SAM and an OSC compared with water. The actual value of COBP depends, in turn, on several microscopic quantities and parameters.
Figure 3. Water-gating mechanism.
In a, the functioning mechanism of a water-gated field-effect transistor is schematically described along with the capacitors forming the gating systems that are in series. In b, the profile of the gate potential drops is reported.
The measurements of the electronic responses
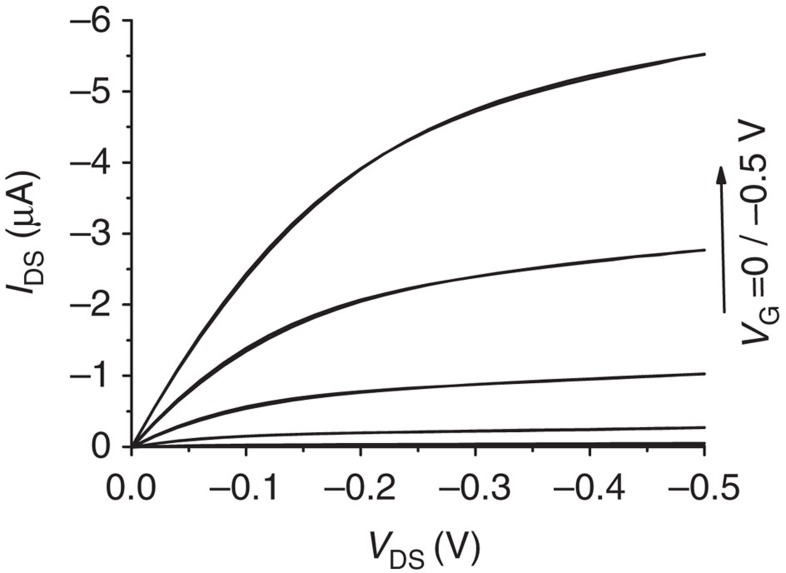
The applied negative VG bias, through capacitive coupling between the gating system and the OSC, induces positive charges in the OSC generating a two-dimensional channel of holes35,36 (Fig. 3a). As VG is applied, the conditions for charge accumulation are set but the actual IDS current flow does not start (under an applied VDS bias) until VG equals VT and the energy barrier, originated by the gate electrode and the OSC 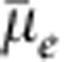 mismatch, is levelled. Beyond VT, the further injected charges can drift through the channel with a given mobility μFET and output characteristics such as those reported in Fig. 4 are measured. Each applied gate bias (0≤VG≤−0.5 V) sets the IDS maximum intensity with the curves exhibiting the expected linear (at lower VDS) and saturation regimes while measured at remarkably low biases (VDS≤−0.5 V). The IDS current flowing in the FET channel in the saturation regime is given by25:
mismatch, is levelled. Beyond VT, the further injected charges can drift through the channel with a given mobility μFET and output characteristics such as those reported in Fig. 4 are measured. Each applied gate bias (0≤VG≤−0.5 V) sets the IDS maximum intensity with the curves exhibiting the expected linear (at lower VDS) and saturation regimes while measured at remarkably low biases (VDS≤−0.5 V). The IDS current flowing in the FET channel in the saturation regime is given by25:
Figure 4. pOBP-SAM WGOFET IDS output characteristics.
The current–voltage curves are measured in the common-source mode with VDS scanned between 0 and −0.5 V. The gate bias, VG, varies in the same range in steps of −0.1 V.
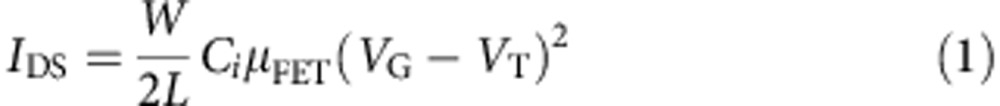 |
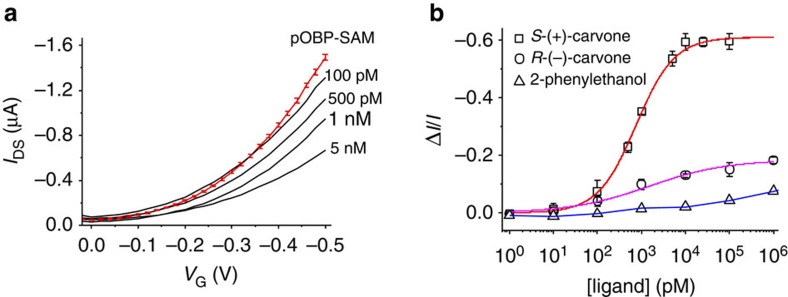
where Ci≈COBP is the capacitance per unit area. In Fig. 5a, the pOBP-WGOFET transfer-characteristics (IDS versus VG at VDS=−0.5 V) are reported for a device gated with a pristine pOBP-SAM (baseline, red curve) along with the traces measured after exposing the pOBP-SAM to progressively higher concentrations of (S)-(+)carvone (signal, black curves). Apparently, a significant degree of current decrease can be measured even at very low ligand concentrations. In the Supplementary Fig. 8, the gate leakage currents (IG) for the data of Fig. 5a are reported for comparison. These leakage currents are about 3 orders of magnitude lower than IDS and do not display a consistent trend with the concentration of the ligand, confirming the need of the FET gain to gather useful responses. From the transfer curve the COBP and μFET product, as well as the VT values are extracted17. μFET can be as high as (1.1±0.2) × 10−1 cm2 V−1 s−1, showing a very good level of performance for the pOBP-SAM WGOFET37. The fractional current decrease upon binding was evaluated from the transfer curves as ΔI/I=[(I−I0)I0−1] with I and I0 being IDS values at VG=−0.5 V for the signal and for the base-line curves, respectively. ΔI/I was taken as the WGOFET electronic response as this normalizes the device-to-device variation in FET biosensors38, resulting in high response reproducibility.
Figure 5. Electronic chiral differential detection of odorant molecules.
In a, the device IDS versus VG transfer characteristics are reported both for a pristine pOBP-SAM gate (red curve) and for a gate exposed to concentrations of (S)-(+)-carvone in the 100 pM—5 nM range (black curves). These curves were measured after the device stabilization had been performed and an average relative error of 5% is measured for the base-line curve. In b, the binding curves gathering the ΔI/I data points measured with a WGOFET whose pOBP-SAM gate is exposed to the (R)-(−)- and (S)-(+)-carvone as well as the 2-phenylethanol ligands, in the 1 to 106 pM range, are reported. The data are plotted as the ΔI/I average values measured on three different devices for each curve.
Enantiomer binding to a pOBP-SAM
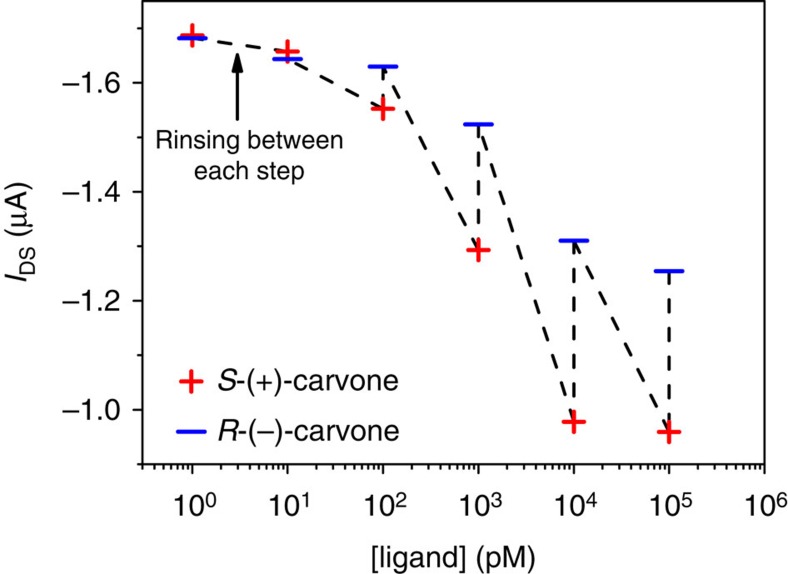
The binding curves of the (S)-(+)-carvone, (R)-(−)-carvone and 2-phenylethanol ligands to the pOBP-SAM are reported in Fig. 5b as ΔI/I versus ligand concentration. The curves are distinguishable down to few tens of picomolar concentrations, showing that chiral differential detection can be achieved at extremely low concentrations with a WGOFET. A control experiment, reported in Fig. 6, shows the IDS values measured using exactly the same pOBP-SAM gold gate alternating the exposure to the (S)-(+)-carvone and (R)-(−)-carvone solutions. Between two subsequent exposures the Au-gate plate was rinsed thoroughly with water. The (+) and (−) symbols in Fig. 6 mark the IDS values measured at different concentrations of the two enantiomers. The differential detection effect is clearly visible also in this experiment, already in the 10–100 pM range, though to a lower extent compared with the results of Fig. 5b. The S-(+)-carvone traces return a maximum fractional decrease of ~60%, this being very similar to the data reported in Fig. 5b. A much higher response at saturation is seen for the R-(−)-carvone. This can be explained considering that the memory effect impacts more on the weaker binding species.
Figure 6. Chiral differential detection of carvone enantiomers performed with the same gate electrode.
The IDS values measured with a WGOFET bearing the very same pOBP-SAM gate exposed, alternatively, to one of the two carvone enantiomer solutions. Between two subsequent exposures, the gate is rinsed thoroughly. The (+) and (−) symbols mark the IDS current values after exposure to the same concentration of either one of the two carvone enantiomers, in the 1 to 106 pM concentration range.
The fitting of the (S)-(+)-carvone binding curve (red solid line in Fig. 5b) is performed using Langmuir’s isotherm Y=bMAX(X/(KFET+X)), where Y is the ΔI/I electronic response proportional, through bmax, to the degree of saturation. X is the ligand concentration and KFET is the dissociation constant. The fitting of the (R)-(−)-carvone (magenta solid curve in Fig. 5b) was calculated using Hill’s binding model, also used to model the data relevant to the 2-phenylethanol (blue curve). The Hill’s equation is Y=bMAX(Xα/(KFET+Xα)), with α giving the degree of cooperativity (α=1 for non-cooperative, α>1 and α<1 for positive and negative cooperativity, respectively). The fit of the S-(+)-carvone data to a Langmuir’s non-cooperative binding isotherm39 returns a dissociation constant KFET+=0.81±0.05 nM and a plateau response at a saturation of (ΔI/I)+=0.62±0.01, while the limit-of-detection is 50 pM and the limit-of-quantification is 150 pM. The detection of carvone enantiomers here performed is about 6 orders of magnitude better than the determinations performed with a back-gate device19. A comparison between the two systems is not straightforward as they hold different device structures and different recognition elements. However, it is a fact that in the WGOFET a direct interaction between the bio-recognition element layer and an electronic interface, is created. The R-(−)-carvone data could only be accounted for by a Hill’s binding isotherm38 that, through the α coefficient, models the extent of cooperativity of a given binding site. The best-fit parameters, KFET−=20±20 nM, plateau response at saturation of (ΔI/I)−=0.17±0.02 and α=0.5±0.1, suggest an anti-cooperative binding for R-(−)-carvone. Hill’s equation successfully accounts also for the 2-phenylethanol binding, exhibiting an anti-cooperative behaviour too (α=0.3±0.1) but with a much higher KFET of 0.7±0.33 μM. The fitting of the S-(+)-carvone to the Hill’s isotherm, performed as control, returns a KFET=0.4±0.2 nM and an α=1.1±0.1, consistent with the Langmuir’s fit. The errors are taken as 1 s.d., as for all data presented.
An enantio-selectivity factor (ESF), taken as the ratio between the slopes in the binding curves linear branch19 as high as 6.3 is measured here. The binding of the two carvone enantiomers to the pOBP-SAM being characterized by markedly different levels of cooperativity and an exceptionally high ESF, provides evidence that different interactions indeed occur. Chiral detection is a research topic of great relevance40,41 and, to our knowledge, there has been no previous report showing both differential detection at low picomolar concentrations together with an ESF >6 (refs 42, 43).
Discussion
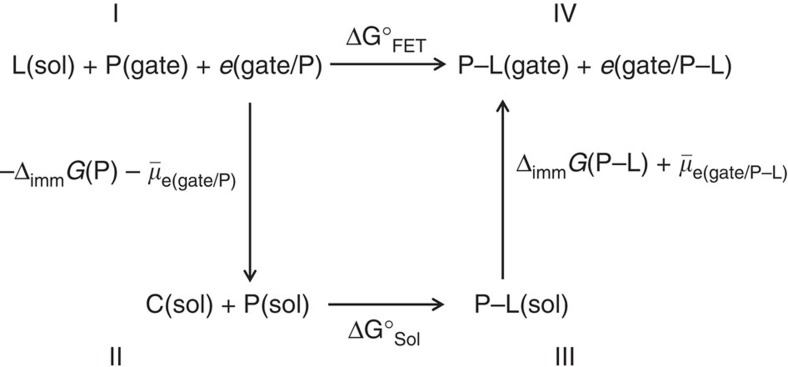
All the dissociation constants evaluated with the WGOFET (KFET) are at least 3 orders of magnitude lower than those measured in solution (KSol), although the scale of affinity is preserved. A rationale for the differences in K values involves the surface work (W) associated with a ligand binding to a layer of orderly immobilized receptors44. The change in the metal-gate electron free energy, ΔEF, also needs to be accounted for. This is opposite to the molar free energy associated to the VT shift, so that ΔEF=−nFΔVT. Since ΔVT is the difference between the VT at zero ligand concentration (no binding site occupied) and at saturation (all the binding sites occupied) and F being a molar quantity, ΔEF is the electrostatic contribution to the molar free energy of the pOBP-C complex formation. The binding equilibrium between a ligand and the pOBP can be written as 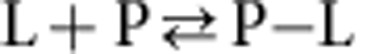 , where L is the ligand (carvone enantiomers in the present case) that binds to the pOBP protein (P) to give the P–L complex. As the pOBP-SAM is anchored to a gold-gate surface, the equilibrium involves a protein layer/water interface rather than a protein dissolved in solution. The overall interfacial effect can be described considering the thermodynamic cycle reported in Fig. 7. The cycle goes from the immobilized protein P (state I) to the immobilized complex P–L (state IV) involving two intermediate steps. The first step (I→II) concerns the release of the protein from the surface into the solution. The free-energy change associated encompasses the surface work needed to release the protein, that is opposite to the work required to immobilize the protein, ΔimmG(P), and the change in the gold-gate electron electrochemical potential when the protein layer is removed
, where L is the ligand (carvone enantiomers in the present case) that binds to the pOBP protein (P) to give the P–L complex. As the pOBP-SAM is anchored to a gold-gate surface, the equilibrium involves a protein layer/water interface rather than a protein dissolved in solution. The overall interfacial effect can be described considering the thermodynamic cycle reported in Fig. 7. The cycle goes from the immobilized protein P (state I) to the immobilized complex P–L (state IV) involving two intermediate steps. The first step (I→II) concerns the release of the protein from the surface into the solution. The free-energy change associated encompasses the surface work needed to release the protein, that is opposite to the work required to immobilize the protein, ΔimmG(P), and the change in the gold-gate electron electrochemical potential when the protein layer is removed 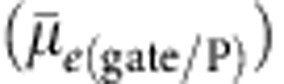 . The second step (II→III) is the binding of L to P taking place in solution with a free-energy change ΔG°Sol=RTln(KSol). The third step (III→IV) is the immobilization of the complex P–L to the gold surface with a free-energy change including the surface work of immobilization ΔimmG(P-L) and the change in the gold-gate electron electrochemical potential when passing from bare gold to a surface coated with the immobilized P–L layer
. The second step (II→III) is the binding of L to P taking place in solution with a free-energy change ΔG°Sol=RTln(KSol). The third step (III→IV) is the immobilization of the complex P–L to the gold surface with a free-energy change including the surface work of immobilization ΔimmG(P-L) and the change in the gold-gate electron electrochemical potential when passing from bare gold to a surface coated with the immobilized P–L layer 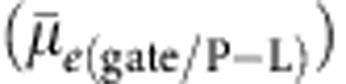 . The fourth step (IV→I) closes the cycle and corresponds to−ΔG°FET=RTln(KFET). Following the cycle, the binding standard molar Gibbs free energy probed with the WGOFET, ΔG°FET, is given by:
. The fourth step (IV→I) closes the cycle and corresponds to−ΔG°FET=RTln(KFET). Following the cycle, the binding standard molar Gibbs free energy probed with the WGOFET, ΔG°FET, is given by:
Figure 7. The thermodynamic cycle.
It describes the surface and electrostatic effects on the binding equilibrium of proteins immobilized on a gold surface.
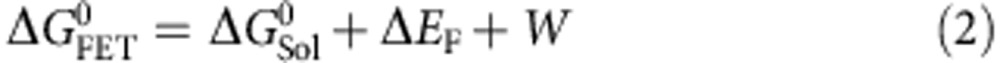 |
where W=ΔimmG(P–L)—ΔimmG(P) is the binding surface work term and 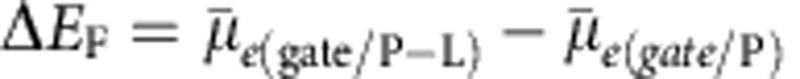 is the change in the gold electron electrochemical potential
is the change in the gold electron electrochemical potential 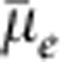 as the gold gate passes from being coated with the P–L to the P layer. ΔVT=−(ΔEF/nF) is the difference between VT at 0.1 μM and zero carvone concentrations. The binding Gibbs energy on a gold gate is split into a molecular interaction and a surface work contribution. The molecular interaction term is associated with the ligand–protein recognition energy in solution. The surface work contribution describes the work spent to accommodate the ligand into a protein bound to a surface. This term (ΔEF+W) includes the conformational rearrangements as well as the change of the gold-gate electron energy.
as the gold gate passes from being coated with the P–L to the P layer. ΔVT=−(ΔEF/nF) is the difference between VT at 0.1 μM and zero carvone concentrations. The binding Gibbs energy on a gold gate is split into a molecular interaction and a surface work contribution. The molecular interaction term is associated with the ligand–protein recognition energy in solution. The surface work contribution describes the work spent to accommodate the ligand into a protein bound to a surface. This term (ΔEF+W) includes the conformational rearrangements as well as the change of the gold-gate electron energy.
The binding energy in solution ΔG°Sol=RTln(KSol) is defined as the difference in standard chemical energy between the complex P–L and the reagent partners P and L. The ΔG°FET term can be described considering the change in Gibbs free energy between states IV and I. In this case the ligand binding causes the change of physical-chemistry properties of the electrode (including not only the gold plate but also the pOBP-SAM) such as the interfacial tension and the electrons electrochemical potential. The Gibbs energy equation for the system moving from state I to state IV (keeping T and P constant) is44:
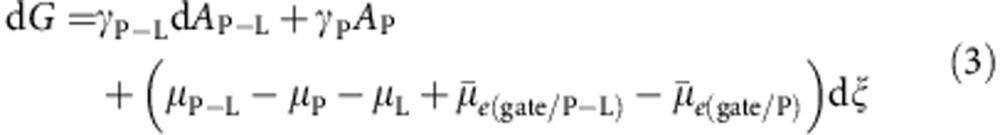 |
where μP, μL and μP–L are the chemical potentials of the protein, the ligand and the complex, respectively; ξ is the extent of the reaction variable; AP–L and AP are the surface areas of the γP–L and γP surface tensions, respectively. The dAP–L area increase for the γP–L interfacial phase, corresponds to an area decrease −dAP of the γP interfacial phase. Thus, by using the relation dA=d AP–L=−dAP, equation 3 becomes:
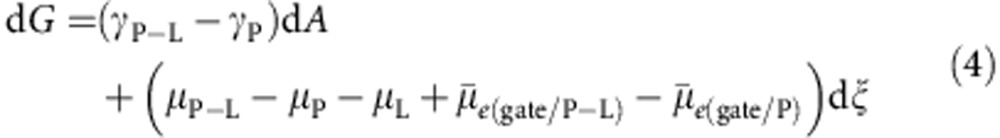 |
The change of the molar Gibbs energy of the reaction is then obtained by taking the partial derivative with respect to ξ:
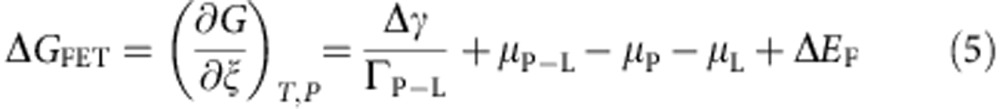 |
where, Δγ=(γP–L−γP) and ΓP–L=dξ/dAP–L describe how the reaction proceeds when the surface is occupied by the complex P–L, that is, it is the surface density of the complex P–L that is constant. By writing the chemical potential as a function of the activity ai of the i-th species and recalling that at equilibrium ΔGFET=0, equation 5 becomes:
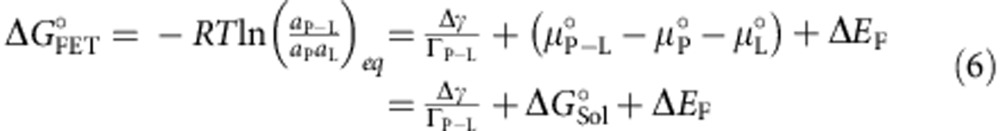 |
where the activities are at equilibrium and thus the ratio aPaL/aP−L equals the dissociation constant. By identifying the surface binding work W with Δγ/ΓP–L, equation 2 is fully recovered. The comparison between equations 2 and 6 indicates that RTln(KFET) is actually the ΔG°FET encompassing the molecular recognition in solution, the surface work and the electronic contribution.
From the KFET+/− values extracted from the data in Fig. 5b, 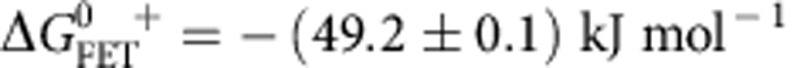 and
and 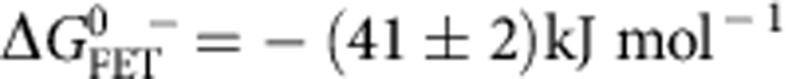 are derived. For the pOBP in solution the figures are:
are derived. For the pOBP in solution the figures are: 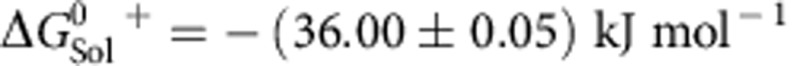 and
and 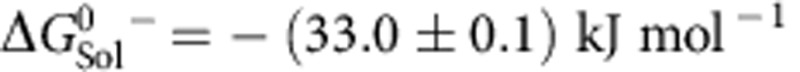 . The
. The 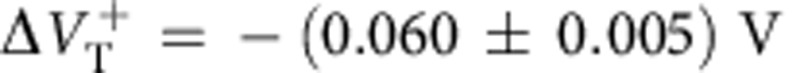 for the S-(+)- and
for the S-(+)- and 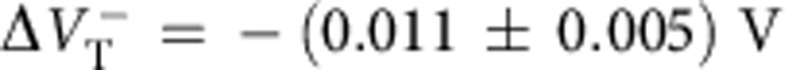 for the R-(−)- enantiomers, result in
for the R-(−)- enantiomers, result in 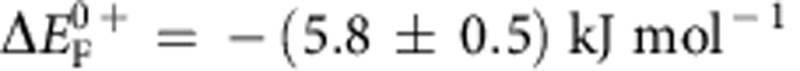 and
and 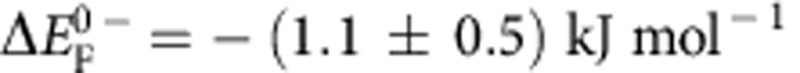 , respectively. All the ΔG° molar energies estimated so far are shown in Table 1, along with the (ΔG0+−ΔG0−) chiral differential values. As
, respectively. All the ΔG° molar energies estimated so far are shown in Table 1, along with the (ΔG0+−ΔG0−) chiral differential values. As 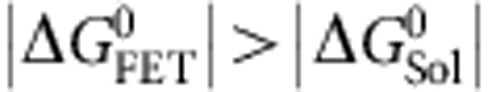 and they are both negative, the binding at the pOBP-SAM is favoured compared with pOBP in solution. This occurrence likely reflects the lower entropy change associated with ligands binding to the orderly immobilized proteins (|ΔSFET|<|ΔSSol|, both being negative contributions). The surface work, W, accounts for a fraction of the differences in ΔG°FET and ΔG°Sol (see equation 2), and since W holds indistinguishable values for the two enantiomers (Table 1) the complex immobilization work would be independent from the chiral interaction that involves mainly the pOBP outer cavity. Accordingly, Table 1 shows that the chiral differential binding free energy
and they are both negative, the binding at the pOBP-SAM is favoured compared with pOBP in solution. This occurrence likely reflects the lower entropy change associated with ligands binding to the orderly immobilized proteins (|ΔSFET|<|ΔSSol|, both being negative contributions). The surface work, W, accounts for a fraction of the differences in ΔG°FET and ΔG°Sol (see equation 2), and since W holds indistinguishable values for the two enantiomers (Table 1) the complex immobilization work would be independent from the chiral interaction that involves mainly the pOBP outer cavity. Accordingly, Table 1 shows that the chiral differential binding free energy 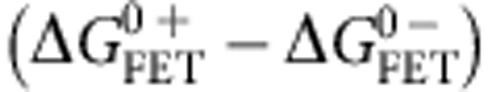 for the immobilized pOBP compares very well to the sum of the
for the immobilized pOBP compares very well to the sum of the 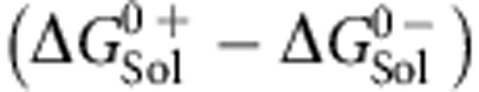 and
and 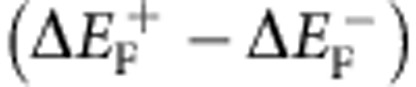 of independently estimated terms. Such an agreement is remarkable considering that the
of independently estimated terms. Such an agreement is remarkable considering that the 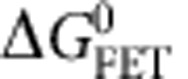 and the
and the 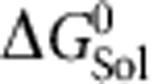 data are measured under different experimental conditions. Indeed, while with the WGOFET the binding of a ligand to the empty cavity takes place, in the fluorescent assay it is the displacement of a bound reporter ligand that is studied.
data are measured under different experimental conditions. Indeed, while with the WGOFET the binding of a ligand to the empty cavity takes place, in the fluorescent assay it is the displacement of a bound reporter ligand that is studied.
Table 1. Standard Gibbs free energies for the S-(+) and R-(−) carvones binding to the pOBP.
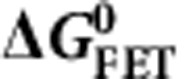 (kJ mol−1)
(kJ mol−1)
|
 (kJ mol−1)
(kJ mol−1)
|
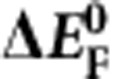 (kJ mol−1)
(kJ mol−1)
|
W (kJ mol−1) | |
|---|---|---|---|---|
| S-(+) | −(49.2±0.1) | −(36.00±0.05) | −(5.8±0.5) | −(7.2±0.5) |
| R-(−) | −(41±2) | −(33.0±0.1) | −(1.1±0.5) | −(7.9±2) |
| ΔG0+−ΔG0− | −(7±2) | −(3.0±0.2) | −(4.7±1) |
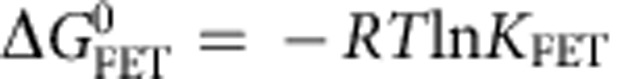 is estimated from the WGOFET binding curves of Fig. 5b;
is estimated from the WGOFET binding curves of Fig. 5b; 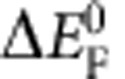 is the molar electrostatic energy evaluated from the WGOFET VT shift;
is the molar electrostatic energy evaluated from the WGOFET VT shift; 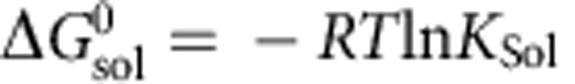 is the binding free energy of the pOBP dissolved in solution;
is the binding free energy of the pOBP dissolved in solution; 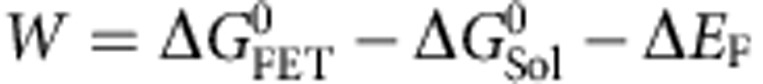 is the surface binding work.
is the surface binding work.
Binding curves in Fig. 5b also indicate that the WGOFET response itself is markedly different for the two enantiomers. At saturation, the response to S-(+)-carvone is in excess of 60% while it drops to <20% for the R-(−)-carvone. From equation 1 (at constant μFET) the response can be written as:
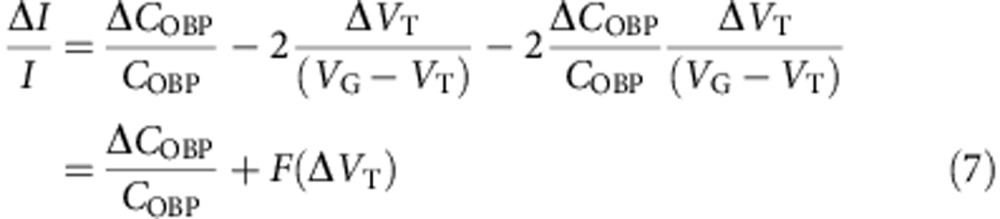 |
where only the second order terms in ΔVT/(VG–VT) were disregarded. In principle, if |VT|≪|VG| and |ΔVT|≪|VG| equation 7 becomes:
 |
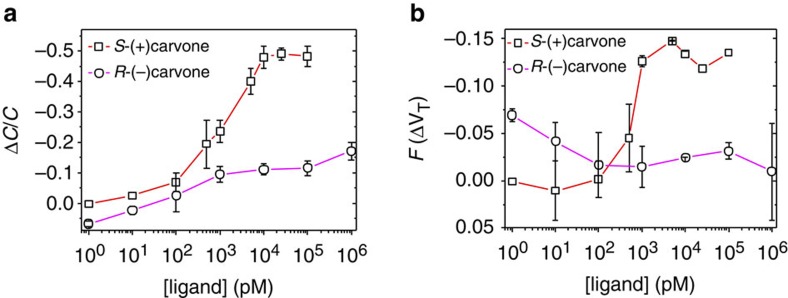
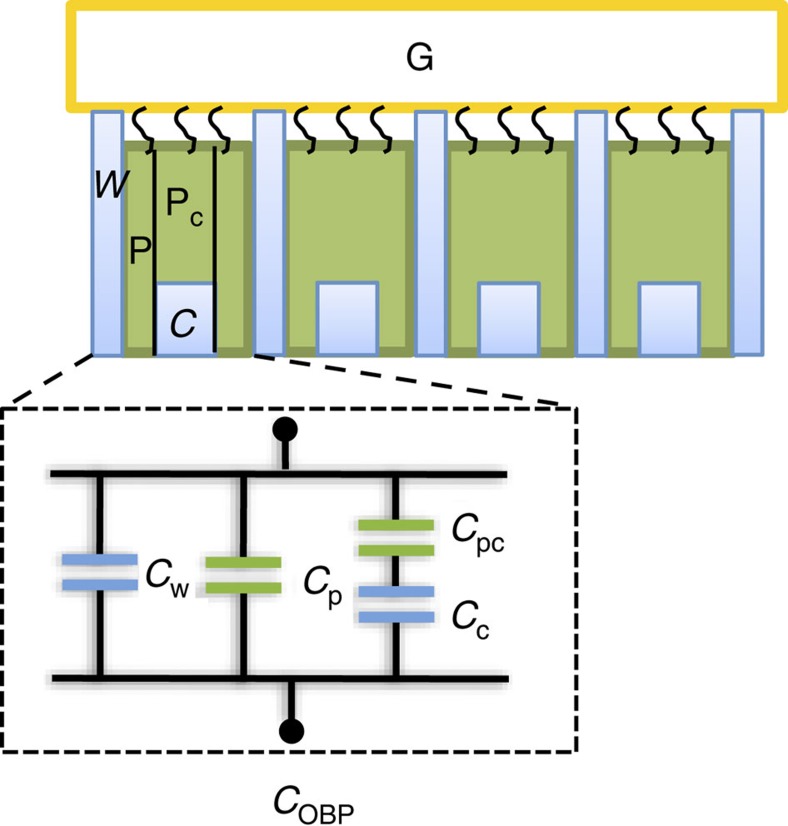
In the present case at saturation, F(ΔVT)≅0.1, while ΔCOBP/COBP≅0.5, as it can be seen in Fig. 8a,b respectively. So, the WGOFET output current is mostly affected by the COBP term. The WGOFET chiral large differential responses are thus associated with subtle changes occurring in the protein upon ligand binding, which lower the pOBP-SAM capacitance to a large extent, as it can be explained through the simple model reported in Fig. 9. The dielectric properties of a protein in the pOBP-SAM layer are described as being characterized by regions with lower (green) or higher (light-blue) dielectric constants. The green parts in Fig. 9 describe the plain protein regions (εr=3) while the light-blue ones are the regions characterized by the presence of water (εr=80). Indeed the presence of water in pOBP has been already proven45. In this model, repeated units are featured that represent the proteins, each one with its cavity filled with water or with the odorant molecule. Each pOBP comprises two different protein regions (P, PC) and the elicited cavity (C). The cavity region is modelled with a capacitor, CC, characterized by a εr=80 in the absence of the ligand as in this case water has been shown to be present in the cavity. The cavity εr becomes equal to 3 when it is filled with the carvone. The protein regions, all characterized by the same εr=3, can be modelled with a plane plates capacitor having a distance between the plates, d, as high as the whole pOBP-SAM (P) or with a capacitor with a smaller d due to the presence of the cavity (PC). These two regions hold capacitances CP and CPC, respectively. Contiguous pOBPs are separated by a high dielectric percolative path, W, addressed as the ‘water channel’ holding a capacitance CW. The overall COBP can be represented by the equivalent circuit, formed by the CP, CC, CPC and CW elements, reported also in Fig. 9. The capacitors CC and CPC in series are in parallel with the capacitors associated with the water channel (CW) and the plain protein (CP), respectively.
Figure 8. Capacitance and threshold voltage contribution to current fractional changes.
In a, the fractional changes of the capacitance, and in b, the F(ΔVT) term data as extracted from the source–drain current measured on exposure to the (R)-(−)- and (S)-(+)-carvone ligands in the 1 to 106 pM range.
Figure 9. Capacitive-modulated transduction of protein–ligand interactions.
The pOBP-SAM layer is detailed as an array of surface immobilized proteins. For the capacitance model each pOBP comprises two plain protein regions (P, PC) and a cavity (C). The cavity holds a capacitance CC, while the protein regions can either be almost as high as the whole pOBP-SAM or be smaller as in the presence of the cavity. These two regions hold capacitances CP and CPC, respectively. Contiguous pOBPs are separated by a high dielectric channel, W, holding a capacitance CW. COBP can be represented by the equivalent circuit, formed by the CP, CC, CPC and CW capacitances arranged as reported in the bottom of this panel.
Each different element (P, C and W) in the model contributes to the equivalent capacitance per unit surface with weighting factors αp for the protein regions, αc for the cavity and αw for the water channel. These weighting factors correspond also to the ratio between the free surface occupied by a given element and the entire unit, hence: αp+αc+αw=1. Assuming that the cavity occupies approximately one-third of the total protein-SAM (d=4.22 nm), the equivalent capacitance per unit surface is:
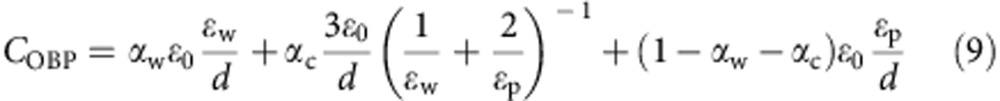 |
assuming εw=80 and εp=3, equation 9 becomes:
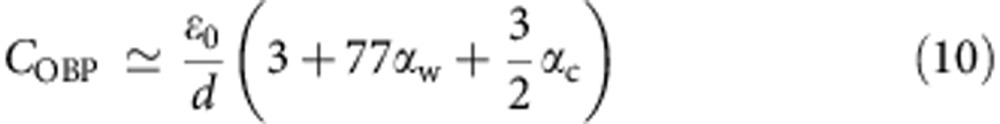 |
In general terms, the same formula holds also when the ligand (S)-(+)-carvone or (R)-(−)-carvone is bound. Assuming that the cavity is completely filled by a medium with the same dielectric constant of the protein, we obtain:
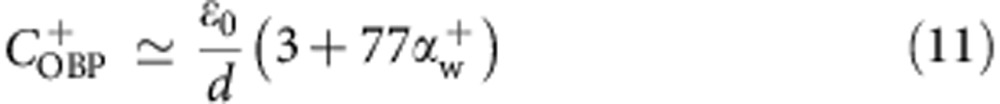 |
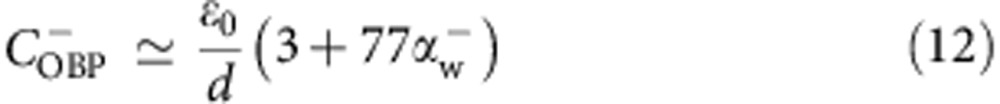 |
The measured ratios ΔCOBP/COBP are therefore:
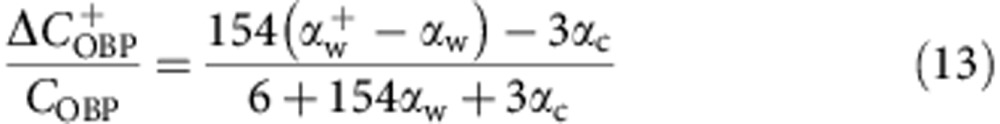 |
 |
These last two equations were used to fit the measured chiral responses ΔC+/C=−0.485 and ΔC−/C=−0.17.
The model presented contains four unknown parameters, hence some hypotheses are needed to reduce the number of degrees of freedom. From the equations, it is apparent that the crucial parameter is the weighting factor of the water channel that affects the equivalent capacitance with a high numerical coefficient. Since the experimental data show a remarkable difference between the two responses measured for (S)-(+)-carvone and (R)-(−)-carvone, the fitting had to be carried out assuming that the water channel disappears completely upon binding of the (S)-(+)-carvone, while it is largely unaffected when (R)-(−)-carvone binds to the hydrophobic OBP cavity. With these hypotheses (corresponding to 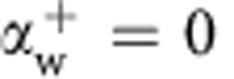 and
and 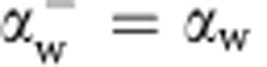 ), we can estimate the unknown parameters as:
), we can estimate the unknown parameters as:
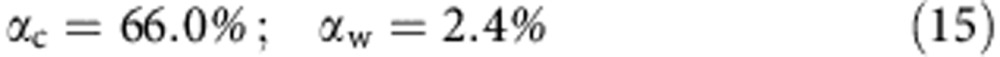 |
As the high percolative and low εr paths are in parallel, the overall capacitance is dominated by the water channel capacitance. The surface occupied by the percolative channel is a mere 2.4% of the total surface of the protein exposed to the solvent (~7 nm2) and hence it may well-accommodate a few water molecules. Any event that interrupts the high dielectric pathway can drastically lower the layer capacitance as this becomes suddenly dominated by the low capacitance path of the protein. The measured responses can be fully accounted for by assuming that this water channel is indeed interrupted upon binding of the S-(+)-carvone, while it is unaffected upon binding of the R-(−)-carvone, proving, though indirectly, the existence of a probably very subtle conformational change in the pOBP on chiral interaction that dramatically impacts on the complex dielectric properties and is efficiently transduced by the WGOFET.
In conclusion, a minute change in protein layer capacitance is shown to efficiently modulate a water-gated OFET, allowing for chiral differential detection with large enantiomeric discrimination factor and accurate estimation of interaction energies as low as few kilojoules per mole. This is a unique tool with general applicability that allows neutral ligand detection in the picomolar concentration range. The FET binding curves modelling provide information on the electrochemical free energies derived from the FET dissociation constants while the electrostatic component is isolated from the threshold voltage shifts. These can be combined with the chemical free energies gathered from the complex formation in solution, overall providing a comprehensive picture of the energy balances for a surface-bound pOBP-carvone complex undergoing chiral interactions. The computation of the relative decrease in capacitance on binding, associated to the FET binding curves maximum relative current changes, provides strong support to the hypothesis of pOBP undergoing a conformational change on binding of the S-(+)-carvone. This study shows also that an ultra-sensitive detection system can be achieved with an organic bio-electronic device fabricated on a flexible substrate with low cost, printing compatible technology.
Methods
pOBP expression
F88WH(6) is a pOBP where the wild-type phenylalanine at residue 88 was substituted by tryptophan and a histidine tag (six residues) was attached to the N terminus of the protein. F88WH(6) pOBP was expressed in a bacterial system using the protocol described by Wei et al.46 A pET5b vector containing the pOBP F88WH(6) sequence was used to transform Escherichia coli BL21(DE3) cells. Bacteria colonies were grown overnight in 10 ml Luria-Bertani/Miller broth containing 100 mg l−1 of ampicillin. The culture was diluted 1:100 with fresh medium and grown at 37 °C until a bacterial optical density at λ=600 nm of 0.7 was reached. At this stage, protein expression was induced by adding isopropylthio-D-galactoside (IPTG) to a final concentration of 0.4 mM. After 2 h at 37 °C, the cells were harvested by centrifugation, resuspended in 50 mM Tris-HCl pH 7.4 and lysed by sonication. After centrifugation, the protein was found to be present in the supernatant. Protein purification was performed by using combinations of chromatographic steps, anionic-exchange resins, followed by gel filtration. Hydrophobic ligands, present in the broth used to grow the bacteria and entering into the protein binding pocket, were removed by a delipidation process at pH 4.5 (ref. 47). The protein was then dialysed against 50 mM sodium phosphate buffer, pH 7.4. The pOBP is characterized by a hydrophobic β-barrel cavity, whose inner surface is ~500 Å2 (ref. 13). The proteins, which have an isoelectric point of 4.55 (ref. 48) are negatively charged in deionized water. The (R)-(−)- and (S)-(+)-carvone enantiomers are perceived as spearmint or caraway odours with human threshold for detection of 30 and 420 nM (ref. 49), respectively. The weight ratio between pOBP and carvone is ~1.5 × 102 while the volume ratio between the β-barrel cavity and the carvone molecule is ~1.5–3.
Competitive fluorescent binding assay
Emission fluorescence spectra were recorded using a Perkin Elmer LS55 instrument at room temperature, with a 1 cm light path quartz cuvette and 5 nm slits for both excitation and emission. The affinity of binding of pOBP was measured using the fluorescence probe 1-AMA as previously reported50. About 1 μM pOBP-F88W solution in 10 mM phosphate buffer, pH 7.4, was titrated with aliquots of 1 mM 1-AMA in methanol to final concentrations of 0.25–8 μM. The probe was excited at 375 nm and the fluorescence spectra were recorded between 400 to 570 nm, monitoring the signals between 480 and 490 nm. The dissociation constant (K1-AMA) for the OBP/1-AMA complex was calculated using the equation y=(Bmax × x)/(K1-AMA+x), where y is the degree of saturation, Bmax is the number of maximum binding sites, and K1-AMA is the dissociation constant50. The calculation was run using the software Sigma Plot and a single binding site was considered for the OBP.
The affinity of pOBP-F88W towards S-(+)-carvone and R-(−)-carvone and 2-phenylethanol was measured in competitive binding assays, using both pOBP and probe at 1 μM as the final concentration. The ligand solutions, with concentrations ranging from 2 to 20 μM, were prepared in 10 mM phosphate buffer pH 7.4 and used for the binding experiments. Dissociation constants of competitors were calculated from the corresponding IC50 values, using the equation: KSol=[IC50]/1+[1-AMA]/K1-AMA, where [1-AMA] is the free concentration of the fluorescence probe and K1-AMA is the dissociation constant of the complex OBP/1-AMA.
For Polistes dominula OBP1 (Wasp-OBP), the same method was employed using N-phenyl-1-naphthylamine (1-NPN) as fluorescent probe51. The probe was excited at 295 nm and emission spectra were recorded between 337–450 nm.
pOBP-SAM WGOFET device fabrication
PBTTT-C14 was dissolved (7 mg ml−1) in a mixture of 1,2-dichlorobenzene and chloroform (9:1). Gold source (S) and drain (D) interdigitated electrodes were photo-lithographically defined on a flexible substrate. The distance between two differently biased fingers is the channel length (L), while the perimeter of each set of equipotential fingers is the channel width (W); they are 5 μm and 104 μm, respectively. The substrate with patterned electrodes was spin coated with a PBTTT-C14 solution at 7,000 r.p.m. for 60 s and annealed at 120 °C for 10 min. A gold plate with an area of ~3 10−2 cm2 served as the gate (G) electrode. The SAM functionalizing protocol52 involved a 50 mM solution of 3-mercaptopropionic acid (3MPA) in ethanol containing 5% acetic acid. N2 was bubbled through the 3MPA solution for at least 10 min to remove dissolved oxygen. The gold electrodes were immersed into the 3MPA solution and kept in the dark under N2 for 18 h at 22 °C. The 3MPA SAM obtained had a height of 0.42 nm (ref. 52). For the bio-functionalization protocol, the SAM was activated by immersion into a 100 mM 1-ethyl-3-dimethylaminopropylcarbodiimide hydrochloride and 200 mM N-hydroxysuccinimide aqueous solution for 1 h at 25 °C. Finally, the pOBPs (0.7 mg ml−1 in 20 mM Na phosphate buffer, pH 8.0) were left to immobilize for 2 h at 25 °C. Electrochemical inspection of the Au-electrode showed that after the bio-functionalization the electrode surface was passivated (Supplementary Fig. 2 and Supplementary Fig. 3). X-ray photoelectron spectroscopy analysis showed that the appearance of nitrogen and sulfur peaks is only seen on the bio-functionalized gate (Supplementary Fig. 4, Supplementary Fig. 5 and Supplementary Fig. 6). Also the scanning electron microscope micrographs of the pOBP-SAM gold surface showed a very compact and smooth morphology clearly different from that of the Au-surface (Supplementary Fig. 7a,b).
WGOFET measurement of pOBP-carvone binding curves
The pOBP-SAM gate was brought into contact with a previously dispensed 3 μl of deionized water droplet covering the OSC (Fig. 1a). The electrical characterization of the WGOFET was carried out by a semiconductor parameter analyzer. For the output characteristics, the drain current (IDS) was measured as a function of the drain voltage VDS at gate voltages VGS, ranging between 0 and −0.5 V, in steps of −0.1 V. The curves were measured in the forward and reverse mode. For the transfer characteristics IDS was measured as a function of VGS (+0.02 to −0.5 V) at VDS=−0.5 V. The device current was stabilized by cycling the measurement of the transfer curve until the overlap of subsequent traces was obtained. This process leads to the filling of the OSC low-mobility trap states leading to a stable VT value17. After stabilization, the pOBP-SAM gate was incubated for 5 min in a carvone solution and the transfer characteristic was recorded. The ΔI/I is the electronic response at a given concentration and the relevant dose–response curve is obtained by plotting these data points at all investigated concentrations as the average values over three replicates on different devices, with the relative error taken as 1 s.d.
Author contributions
M.Y.M. and M.M. performed the FET measurements while E.T. worked on the expression and purification of the OBPs and the complex formation measurements in solution. G.L. carried out the theoretical modelling of the capacitance while G.P. worked on the thermodynamics of the FET system. K.P. supervised all the work connected with the pOBP production and binding in solution. L.T. supervised the work performed on the FET and wrote the manuscript that was revised by all the authors.
Additional information
How to cite this article: Mulla, M. Y. et al. Capacitance-modulated transistor detects odorant binding protein chiral interactions. Nat. Commun. 6:6010 doi: 10.1038/ncomms7010 (2015).
Supplementary Material
Supplementary Figures 1-8, and Supplementary Reference
Acknowledgments
We thank Martin Grell and Gaetano Scamarcio for useful discussions. Nicoletta Ditaranto is acknowledged for performing the X-ray photoelectron spectroscopy surface analysis and Cinzia Di Franco and Maria Vittoria Santacroce for the morphological characterization. The ‘Gas sensor on flexible substrates for wireless applications−FlexSmell’ project, FP7-People-ITN-2008 under Grant Agreement 238454 and the PON project ‘Laboratorio per lo Sviluppo Integrato delle Scienze e delle Tecnologie dei Materiali Avanzati e per dispositivi innovativi-LABORATORIO SISTEMA’ and the PRIN 2010-2011 ‘Nanostructured Soft Matter’ by the Italian MIUR (the Ministry of Education, Universities and Research) are acknowledged for partial financial support of this work.
References
- Meierhenrich U. J., Golebiowski J., Fernandez X. & Cabrol-Bass D. The molecular basis of olfactory chemoreception. Angew. Chem. Int. Ed. 43, 6410–6412 (2004). [DOI] [PubMed] [Google Scholar]
- Buck L. & Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell 65, 175–187 (1991). [DOI] [PubMed] [Google Scholar]
- Pevsner F., Sklar P. B., Hwang P. M. & Snyder S. H. Odorant-binding protein. sequence analysis and localization suggest an odorant transport function. Chem. Senses 1, 227–242 (1989). [Google Scholar]
- Tegoni M. et al. Mammalian odorant binding proteins. Biochim. Biophys. Acta 1482, 229–240 (2000). [DOI] [PubMed] [Google Scholar]
- Pelosi P. Odorant-binding proteins. Crit. Rev. Biochem. Mol. Biol. 29, 199–228 (1994). [DOI] [PubMed] [Google Scholar]
- Bignetti E. et al. Specificity of an immunoaffinity column for odorant-binding protein from bovine nasal mucosa. Chem. Senses 12, 601–608 (1987). [Google Scholar]
- Pevsner J., Hou V., Snowman A. M. & Snyder S. H. Odorant-binding protein. Characterization of ligand binding. J. Biol. Chem. 265, 6118–6125 (1990). [PubMed] [Google Scholar]
- Xu P., Atkinson R., Jones D. N. M. & Smith D. P. Drosophila OBP LUSH is required for activity of pheromone-sensitive neurons. Neuron 45, 193–200 (2005). [DOI] [PubMed] [Google Scholar]
- Matsuo T., Sugaya S., Yasukawa J., Aigaki T. & Fuyama Y. Odorant-binding proteins OBP57d and OBP57e affect taste perception and host-plant preference in Drosophila Sechellia. PLoS Biol. 5, e118 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup S., Williams T. I. & Anholt R. R. H. Functional dissection of odorant binding protein genes in Drosophila melanogaster. Genes Brain Behav. 10, 648–657 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y. et al. Crystal and solution structures of an odorant-binding protein from the southern house mosquito complexed with an oviposition pheromone. Proc. Natl Acad. Sci. USA 107, 19102–19107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelosi P., Mastrogiacomo R., Iovinella I., Tuccori E. & Persaud K. Structure and biotechnological applications of odorant-binding proteins. Appl. Microbiol. Biotechnol. 98, 61–70 (2014). [DOI] [PubMed] [Google Scholar]
- Spinelli S. et al. the structure of the monomeric porcine odorant binding protein sheds light on the domain swapping mechanism. Biochemistry 37, 7913–7918 (1998). [DOI] [PubMed] [Google Scholar]
- Vincent F. et al. Complexes of porcine odorant binding protein with odorant molecules belonging to different chemical classes. J. Mol. Biol. 300, 127–139 (2000). [DOI] [PubMed] [Google Scholar]
- Paolini S. et al. Amino acid sequence, post-translational modifications, binding and labelling of porcine odorant-binding protein. Chem. Senses 23, 689–698 (1998). [DOI] [PubMed] [Google Scholar]
- Lambert J., Compton R. N. & Crawford T. D. The optical activity of carvone: a theoretical and experimental investigation. J. Chem. Phys. 136, 114512 (2012). [DOI] [PubMed] [Google Scholar]
- Torsi L., Magliulo M., Manoli K. & Palazzo G. Organic field-effect transistor sensors: a tutorial review. Chem. Soc. Rev. 42, 8612–8628 (2013). [DOI] [PubMed] [Google Scholar]
- Angione M. D. et al. Interfacial electronic effects in functional biolayers integrated into organic field-effect transistors. Proc. Natl Acad. Sci USA 109, 6429–6434 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torsi L. et al. A sensitivity-enhanced field-effect chiral sensor. Nat. Mater. 7, 412–417 (2008). [DOI] [PubMed] [Google Scholar]
- Kergoat L. et al. A water-gate organic field-effect transistor. Adv. Mater. 22, 2565–2569 (2010). [DOI] [PubMed] [Google Scholar]
- Kim T. H. et al. Single-carbon-atomic-resolution detection of odorant molecules using a human olfactory receptor-based bioelectronic nose. Adv. Mater. 21, 91–94 (2009). [Google Scholar]
- Yoon H. et al. Polypyrrole nanotubes conjugated with human olfactory receptors: high-performance transducer for FET-type bioelectronic nose. Angew. Chem. Int. Ed. 48, 2755–2758 (2009). [DOI] [PubMed] [Google Scholar]
- Magliulo M. et al. Sensors: electrolyte-gated organic field-effect transistor sensors based on supported biotinylated phospholipid bilayer. Adv. Mater. 25, 1958–1958 (2013). [DOI] [PubMed] [Google Scholar]
- Dumitru L. M. et al. Plain poly(acrylic acid) gated organic field-effect transistors on a flexible substrate. ACS Appl. Mater. Interfaces 5, 10819–10823 (2013). [DOI] [PubMed] [Google Scholar]
- Sze S. M. & Kwok K. N. G. Physics of Semiconductor Devices 3rd edn293Wiley (2007). [Google Scholar]
- Casalini S., Leonardi F., Cramer T. & Biscarini F. Organic field-effect transistor for label-free dopamine sensing. Org. Electron. 14, 156–163 (2013). [Google Scholar]
- Lai S. et al. Ultralow voltage, OTFT-based sensor for label-free DNA detection. Adv. Mater. 25, 103–107 (2013). [DOI] [PubMed] [Google Scholar]
- Bergveld P. Development, operation, and application of the ion-sensitive field-effect transistor as a tool for electrophysiology. IEEE Trans. Biomed. Eng. 19, 342–351 (1972). [DOI] [PubMed] [Google Scholar]
- Bard A. J. & Faulkner L. R. Electrochemical Methods: fundamentals and Applications 2nd Edition 79Wiley (2001). [Google Scholar]
- Janata J. & Josowicz M. Chemical modulation of work function as a transduction mechanism for chemical sensors. Acc. Chem. Res. 31, 241–248 (1998). [Google Scholar]
- Evans D. F. & Wennestrom H. inThe Colloidal Domain: where Physics, Chemistry, Biology, and Technology Meet Ch. 3VCH (1994). [Google Scholar]
- Rivnay J., Owens R. M. & Malliaras G. G. The rise of organic bioelectronics. Chem. Mater. 26, 679–685 (2014). [Google Scholar]
- Bernards D. A. et al. Enzymatic sensing with organic electrochemical transistors. J. Mater. Chem. 18, 116–120 (2008). [Google Scholar]
- Kukic P. et al. Protein dielectric constants determined from NMR chemical shift perturbations. J. Am. Chem. Soc. 135, 16968–16976 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodabalapur A., Torsi L. & Katz H. E. Organic transistors: two-dimensional transport and improved electrical characteristics. Science 268, 270–271 (1995). [DOI] [PubMed] [Google Scholar]
- Laiho A., Herlogsson L., Forchheimer R., Crispin X. & Berggren M. Controlling the dimensionality of charge transport in organic thin-film transistors. Proc. Natl Acad. Sci USA 108, 15069–15073 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porrazzo R., Bellani S., Luzio A., Lanzarini E., Caironi M. & Antognazza M. R. Improving mobility and electrochemical stability of a water gated polymer field-effect transistor. Org. Electron. 15, 2126–2134 (2014). [Google Scholar]
- Ishikawa F. N. et al. A calibration method for nanowire biosensors to suppress device-to-device variation. ACS Nano 3, 3969–3976 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill K. A. & Bromberg S. inMolecular Driving Forces : Statistical Thermodynamics in Chemistry & Biology Ch. 27–28Garland Science (2003). [Google Scholar]
- Wattanakit C. et al. Enantioselective recognition at mesoporous chiral metal surfaces. Nat. Commun. 5, 3325 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernites R. B., Venkata S. K., Tiu B. D. B., Yago A. C. C. & Advincula R. C. Nanostructured, molecularly imprinted, and template-patterned polythiophenes for chiral sensing and differentiation. Small 8, 1669–1674 (2012). [DOI] [PubMed] [Google Scholar]
- Guo L. et al. Discrimination of enantiomers based on LSPR biosensors fabricated with weak enantioselective and non selective receptors. Biosens. Bioelectron. 47, 199–205 (2013). [DOI] [PubMed] [Google Scholar]
- Xu L. et al. Highly selective recognition and ultrasensitive quantification of enantiomers. J. Mater. Chem. B 1, 4478–4483 (2013). [DOI] [PubMed] [Google Scholar]
- Oliviero G., Federici S., Colombi P. & Bergese P. On the difference of equilibrium constants of DNA hybridization in bulk solution and at the solid-solution interface. J. Mol. Recognit. 24, 182–187 (2011). [DOI] [PubMed] [Google Scholar]
- Stepanenko O. V. et al. Hydrophobic interactions and ionic networks play an important role in thermal stability and denaturation mechanism of the porcine odorant-binding protein. Proteins 71, 35–44 (2008). [DOI] [PubMed] [Google Scholar]
- Wei Y., Brandazza A. & Pelosi P. Binding of polycyclic aromatic hydrocarbons to mutants of odorant-binding protein: a first step towards biosensors for environmental monitoring. Biochim. Biophys. Acta 1784, 666–671 (2008). [DOI] [PubMed] [Google Scholar]
- Lautenschlager C., Leal W. S. & Clardy J. Coil-to-helix transition and ligand release of Bombyx mori pheromone-binding protein. Biochem. Biophys. Res. Commun. 335, 1044–1050 (2005). [DOI] [PubMed] [Google Scholar]
- ProtParam tool from ExPASy Tools http://web.expasy.org/protparam/.
- Pelosi P. & Viti R. Specific anosmia to l-carvone: the minty primary odour. Chem. Senses 3, 331–337 (1978). [Google Scholar]
- Meillour P. N. et al. Phenylalanine 35 and tyrosine 82 are involved in the uptake and release of ligand by porcine odorant-binding protein. Biochim. Biophys. Acta 1794, 1142–1150 (2009). [DOI] [PubMed] [Google Scholar]
- Zhang S. et al. Binding characterization of recombinant odorant-binding proteins from the parasitic wasp, microplitis mediator (Hymenoptera: Braconidae). J. Chem. Ecol. 37, 189–194 (2011). [DOI] [PubMed] [Google Scholar]
- Techane S., Baer D. R. & Castner D. G. Simulation and modeling of self-assembled monolayers of carboxylic acid thiols on flat and nanoparticle gold surfaces. Anal. Chem. 83, 6704–6712 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures 1-8, and Supplementary Reference