Abstract
Accumulation and deposition of amyloid-β peptide (Aβ) in the brain is a primary cause of the pathogenesis of Alzheimer’s disease (AD). Aβ is generated from amyloid-β precursor protein (APP) through sequential cleavages first by β-secretase and then by γ-secretase. Inhibiting β-secretase activity is believed to be one of the most promising strategies for AD treatment. In the present study, we found that a resveratrol trimer, miyabenol C, isolated from stems and leaves of the small-leaf grape (Vitisthunbergii var. taiwaniana), can markedly reduce Aβ and sAPPβ levels in both cell cultures and the brain of AD model mice. Mechanistic studies revealed that miyabenol C affects neither protein levels of APP, the two major α-secretases ADAM10 and TACE, and the γ-secretase component Presenilin 1, nor γ-secretase-mediated Notch processing and TACE activity. In contrast, although miyabenol C has no effect on altering protein levels of the β-secretase BACE1, it can inhibit both in vitro and in vivo β-secretase activity. Together, our results indicate that miyabenol C is a prominent β-secretase inhibitor and lead compound for AD drug development.
Introduction
Alzheimer’s disease (AD) is a devastating neurodegenerative disorder characterized by impaired memory and cognition. One of the major pathological hallmarks of AD in the brain is senile plaques, which are composed of heterozygous amyloid-β (Aβ) peptides. Ample evidence indicates that accumulation of Aβ peptides in vulnerable brain regions plays a central role in AD pathogenesis: Aβ is neurotoxic and can trigger a cascade of neurodegenerative steps including the formation of senile plaques and neurofibrillary tangles, synaptic dysfunction, and eventual neuronal loss [1, 2]. Aβ is a proteolytic product of the amyloid-β precursor protein (APP) and is generated through sequential cleavages by enzymes called β- and γ-secretases. During this amyloidogenic processing, β-secretase first cleaves the type I transmembrane APP protein to generate an extracellular fragment known as sAPPβ and a membrane-associated carboxyl-terminal fragment known as APP β-CTF. APP β-CTF is then cleaved by γ-secretase to release Aβ. Alternatively, APP can be subjected to a non-amyloidogenic processing and cleaved by α-secretase within the Aβ domain. α-secretase-mediated cleavage precludes Aβ generation and generates an extracellular domain of APP known as sAPPα instead [3, 4].
β-cleavage of APP is the first and rate-limiting step in Aβ production. The transmembranous aspartic protease β-site APP cleaving enzyme 1 (BACE1) has been identified as the essential β-secretase in vivo[5–8]. The level and activity of BACE1 are found to be elevated in postmortem brain of sporadic AD patients [9–11], suggesting a causative role of BACE1 in AD. Although homozygous BACE1 knock-out mice develop certain phenotypic abnormalities including reduced body size, hyperactive behavior, decreased grip strength and elevated pain sensitivity [12–14], probably because the cleavage of other BACE1 substrates such as neuregulin 1 [13, 14] and β-subunits of voltage-gated sodium channels [15] is blocked, BACE1 heterozygous knockout mice do not develop such abnormal phenotypes and heterozygous knockout of BACE1 still can reduce Aβ deposition in AD mice [16, 17]. Therefore, moderate inhibition of β-secretase activity is considered as a promising strategy for AD intervention.
Natural products have been recognized as sources of new lead compounds for the treatment of various diseases including AD [18, 19]. The small-leaf grape Vitisthunbergii var. taiwaniana (VTT) is a wild grape native to Taiwan where, along with other species of Vitis spp., is used as a folk medicine [20]. The extracts or purified compounds from VTT have been reported to have anti-microbial [21], anti-inflammatory [22], anti-hypertensive [23] and neuroprotective activities [20]. In the present study, we isolated a resveratrol trimer, miyabenol C, from the stem and leaf extracts of VTT. Importantly, we demonstrate that miyabenol C is a prominent β-secretase inhibitor and can reduce Aβ levels both in vitro and in vivo, suggesting that miyabenol C may be a lead compound for AD drug development.
Materials and Methods
Isolation of miyabenol C from stem and leaf extracts of VTT
Dried stems and leaves of VTT (5.5 kg) were boiled and refluxed for 2.5 h with 60% of aqueous ethanol solution (10 L × 2 times). After filtration, the extracted solution was concentrated in vacuo. The extract (230.3 g) was then separated over a Diaion HP-20 column, using EtOH-H2O as mobile phase, into three fractions (A-C). Fraction B (60% EtOH-H2O eluent, 130.0 g) was chromatographed on silica gel column using stepwise gradient elution with CHCl3-MeOH (100:0∼0:100) to obtain 11 fractions (Fr. 1-11). Fr.5 (5.0 g) was subjected to silica gel column chromatography and eluted with CHCl3-MeOH (95:5, 9:1, 85:15, 0:100) to generate 8 sub-fractions (Fr.5-1 ∼ Fr.5-8). Fr. 5-5 (835 mg) was applied to ODS column chromatography and eluted with MeOH-H2O (3:7, 4:6 and 1:1). The elution of MeOH-H2O (4:6) was further repeated by RP-18 and purified through preparative HPLC (RestekPrinnacle DB C18, 5 μm, 250×20 mm) with elution of 45% aqueous methanol solution to generate a pure compound (65mg), whose structure was then determined by means of NMR and MS.
Cell culture and treatment
Maintenance of human neuroblastoma SH-SY5Y cells [24], mouse neuroblastoma N2a naive cells (N2aWT) [25] and N2a cells stably expressing human APP695 (N2a695) [26] was as previously reported [27, 28].
Purified miyabenol C and β-secretase inhibitor II (Millipore) were dissolved in DMSO (Sigma) as 20 mM stock solution. For treatment, cells grown to confluency were switched to be incubated in FBS-free media that were added with indicated concentrations of miyabenol C, β-secretase inhibitor II or DMSO for 10h.
Cell viability test
Cell viability was measured using the Cell Counting Kit-8 (CCK-8) assay (Dojindo), following the manufacturer’s protocol.
Western blot and antibodies
Conditioned media from treated cells were assayed for sAPPα and sAPPβ by Western blot. Treated cells were lysed in ice-cold lysis buffer (10mM Tris pH7.5, 150mM NaCl, 1mM EDTA, 1% NP-40, 1× Protease Inhibitor Cocktail), and equal protein amounts of cell lysates were analyzed by Western blot. Antibodies used for Western blot included: 369 (against the carboxyl terminus of APP, 1:1000) and ab14 (against PS1 amino-terminal fragment, 1:1000) developed in our laboratory; 6E10 (against sAPPα and β-CTF, 1:1000) and anti-sAPPβ antibodies (1:500) from Covance; anti-α-tubulin (1:10000) and anti-ADAM10 (1:1000) antibodies from Millipore; anti-ADAM17 (1:1000) antibody from Abcam; and anti-c-Myc (1:1000) and anti-NICD (1:1000) antibodies from CST. The mouse monoclonal antibody 3D5 (against BACE1, 1:1000) was a kind gift from Dr. Robert Vassar at Northwestern University.
NotchΔE cDNA construct and cell based γ-secretase activity assay
NotchΔE fragment contains the transmembrane and intracellular domains of Notch1 and is an immediate substrate of γ-secretase for generating Notch intracellular domain (NICD). For γ-secretase activity assay, N2aWT cells were transiently transfected with the NotchΔE plasmid. After splitting equally, cells were treated with DMSO, the γ-secretase inhibitor compound E (Millipore) or miyabenol C for 10h. Cell lysates were analyzed by Western blot for NotchΔE and NICD levels. Cellular γ-secretase activity was estimated by the generation of NICD.
α-secretase activity assay
The activity of α-secretase in cells was measured by using InnoZyme TACE Activity Kit (Millipore), following the manufacturer’s protocols.
β-secretase activity assays
Cell-based β-secretase activity and cell-free BACE1 activity were measured by using commercial Kits from Millipore and Sigma, respectively, following the manufacturers’ protocols.
Aβ40 and Aβ42 ELISA assays
Human Aβ40 and Aβ42 in conditioned media from treated N2a695 cells were assayed by sandwich ELISA, following a previously described protocol [29].
Ethics statement
All animal procedures were in accordance with the National Institute of Health Guidelines for the Care and Use of Laboratory Animals and were approved by the Animal Ethics Committee of Xiamen University (IACUC #: XMULAC20120012).
Miyabenol C treatment of APP/PS1 mice and sample analysis
C57BL6 mice co-expressing the Swedish mutant APP and the exon-9 deletion mutant PS1 (APP/PS1) were provided by Nanjing Biomedical Research Institute of Nanjing University, China. For treatment, 12-month-old APP/PS1 transgenic mice were anesthetized with sodium pentobarbital (50μg/g) and placed in a stereotaxic apparatus before intracerebroventricular injection of vehicle (45% DMSO in artificial cerebrospinal fluid: 148 mM NaCl, 3 mM KCl, 1.4 mM CaCl2, 0.75 mM MgCl2, 0.8 mM Na2HPO4, 0.2 mM NaH2PO4) or miyabenol C (0.6μg/g). Vehicle and miyabenol C solution were injected at 4μL into the lateral ventricle using a 5μL-blunt needle equipped with asyringe pump (KD Scientific). The stereotaxic coordinates for the lateral ventricle were AP 0.5 (0.5 mm posterior to bregma), L 1 (1 mm left from mid-sagittal line) and H 2.2 (2.2 mm below bregma). Sixty seconds after insertion of the needle, vehicle or miyabenol C solution were injected at a constant flow rate of 0.4μL/min. The injection needle was kept in place for an additional 10 min to prevent reflux of fluid.
Three days after treatment, mice were anesthetized and euthanized by transcardial perfusion with ice-cold physiological saline. Brain cortex and hippocampus were dissected and lysed for Western blot analysis of APP and sAPPβ. Alternatively, samples were weighed and sequentially extracted into TBSX- (25 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% Triton X-100) soluble and GuHCl-soluble fractions for Aβ sandwich ELISAs. This separation was carried out following a previously validated method [30] with some modification. Briefly, samples were placed in 2mL ice-cold glass dounce homogenizer containing TBSX homogenization buffer. Samples were homogenized on ice, transferred into pre-chilled 1.5 mL polyallomer ultracentrifuge tubes, and centrifuged at 100,000×g for 1 h at 4°C. The supernatant was collected as TBSX-soluble fraction. The pellet was re-suspended in 5M GuHCl, mixed by rotation at room temperature for 6 h, and centrifuged at 16,000×g for 30 min. The supernatant was collected as GuHCl soluble fraction.
Statistical analysis
The statistical analysis was carried out by using GraphPad Prism Software 5.0. Data were analyzed by Student’s t-test or One-way analysis of variance (ANOVA) followed by Tukey’s post-hoc test, and presented as the mean ± standard deviation.
Results
Isolation and identification of miyabenol C
We isolated a series of compounds from stems and leaves of VTT (unpublished data). One of the compounds was purified in a yellow and solid form. This compound was subjected to MS and NMR and the spectral data were:
ESI-MS (Bruker FTMS): m/z 679 [M-H]−. 1H-NMR (BrukerAvance spectrometer, DMSO-d6, 400MHz) δ: 7.18 (2H, d, J = 8.8Hz, H-2a, H-6a), 7.13 (2H, d, J = 8.4Hz, H-2c, H-6c), 6.90 (1H, d, J = 16.4Hz, H-7c), 6.84 (2H, d, J = 8.8Hz, H-3a, H-5a), 6.75 (2H, d, J = 8.4Hz, H-3c, H-5c), 6.62 (1H, d, J = 16.4Hz, H-8c), 6.58 (2H, d, J = 8.8Hz, H-3b, H-5b), 6.52 (2H, d, J = 8.8Hz, H-2b, H-6b), 6.32 I (1H, d, J = 2.0Hz, H-12b), 6.24 (1H, d, J = 2.0Hz, H-12a), 6.19 (2H, d, J = 2.0Hz, H-10a, H-14a), 6.09 (1H, d, J = 2.0Hz, H-14b), 5.39 (1H, d, J = 5.2Hz, H-7a), 5.22 (1H, d, J = 1.2Hz, H-7b), 4.64 (1H, d, J = 5.2Hz, H-8a), 4.33 (1H, d, J = 1.2Hz, H-8b). 13C-NMR (BrukerAvance spectrometer, DMSO-d6,100MHz) δ: 162.3 (C-11b), 160.7 (H-11c), 160.3 (C-13b), 160.3 (C-13a), 160.3 (C-11a), 159.5 (C-13c), 158.2 (C-4c), 157.6 (C-4b), 157.0 (C-4a), 147.6 (C-9a), 143.5 (C-9b), 135.9 (C-9c), 133.4(C-1a), 133.3 (C-1b), 131.2 (C-7c), 129.3 (C-1c), 128.7 (C-2c, C-6c), 127.8 ( C-2a, C-6a), 127.4 (C-2b, C-6b), 122.8 (C-8c), 121.4 (C-10c), 118.6 (C-10b), 116.7 (C-3c, C-5c), 116.5 (C-3a, C-5a), 115.7 (C-3b, C-5b), 107.7 (C-14b), 107 (C-10a, C-14a), 104.4 (C-14c), 102.6 (C-12a), 97.2 (C-12c), 96.4 (C-12b), 94.6 (C-7a), 92.3 (C-7b), 57.2 (C-8a), 51.1 (C-8b). By comparison, we found that the spectral data were identical to that of miyabenol C (Fig. 1A), a previously reported resveratrol trimer [31], indicating that the compound we isolated is miyabenol C.
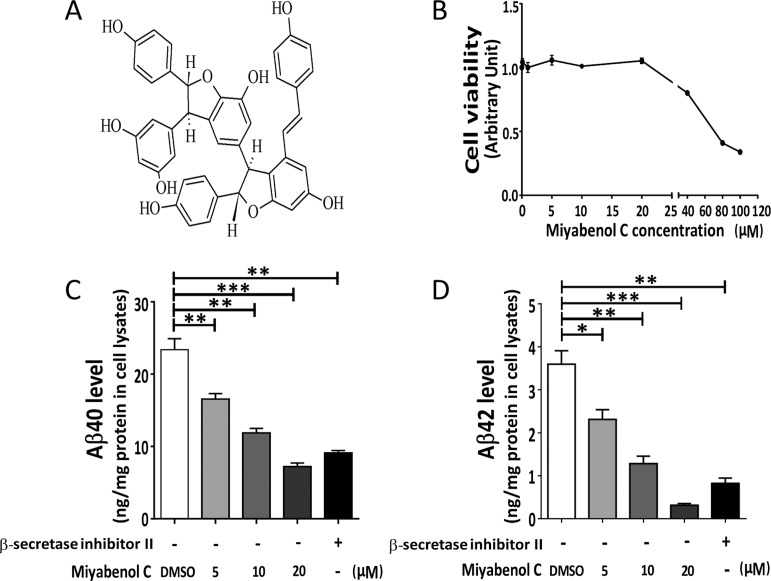
Figure 1. Miyabenol C treatment inhibits Aβ secretion.
(A) Chemical structure of miyabenol C. (B) N2a695 cells were treated with indicated doses of miyabenol C for 10h. Cell viability was measured using CCK-8 assay, n = 3. Error bars indicate standard deviation. (C, D) N2a695 cells were treated with DMSO (negative control), β-secretase inhibitor II (positive control, 2μM) or indicated doses of miyabenol C for 10h. Extracellular Aβ40 (C) and Aβ42 (D) levels were quantified by ELISA (One-way ANOVA followed by Tukey’s post hoc test, n = 3, *: p< 0.05, **: p< 0.01, ***: p< 0.001).
Miyabenol C treatment reduces Aβ secretion
To study whether miyabenol C can affect Aβ generation, we first measured its cytotoxicity. N2a695 cells were treated with increasing doses of miyabenol C (0, 0.1, 1, 5, 10, 20, 40, 80, 100μM) for 10h and cell viability was evaluated by CCK-8 assay. The results showed that miyabenol C had no cytotoxicity at lower doses (5–20μM) but showed a dose-dependent cytotoxicity at higher doses (40–100μM) (Fig. 1B). Therefore, we used doses of 0–20μM in the following studies. When N2a695 cells were treated with miyabenol C (5, 10, 20μM) for 10 h, levels of Aβ40 (Fig. 1C) and Aβ42 (Fig. 1D) in conditioned media were markedly decreased in a dose-dependent manner.
Miyabenol C does not affect γ-secretase activity
Because γ-secretase-mediated cleavage of APP is the final step for Aβ generation, it is possible that miyabenol C may inhibit γ-secretase activity and reduce Aβ production. To study this possibility, we transfected N2aWT cells with NotchΔE that can be directly cleaved by γ-secretase for NICD generation [32, 33]. Cells were then treated with miyabenol C. The generation of NICD was measured by Western blot and the results showed that miyabenol C had no effect on inhibiting NICD generation, whereas the γ-secretase inhibitor compound E dramatically blocked NICD generation (Fig. 2). In addition, miyabenol C treatment did not affect the protein levels of PS1, a major γ-secretase component (Fig. 2). These results suggest that miyabenol C does not inhibit γ-secretase activity.
Figure 2. Miyabenol C does not inhibit γ-secretase activity.
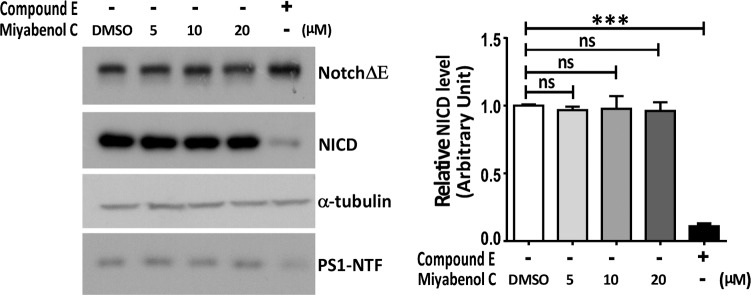
N2aWT cells were first transfected with NotchΔE. After splitting equally, cells were treated with DMSO (negative control), the γ-secretase inhibitor compound E (positive control, 0.5μM) or indicated doses of miyabenol C for 10h. Equal protein amounts of cell lysates were subjected to SDS-PAGE and Western blot. NICD levels were quantified by densitometry using image J for comparison (One-way ANOVA followed by Tukey’s post hoc test, n = 3, ns: p> 0.05, ***: p< 0.001).
Miyabenol C inhibits APP β-cleavage and promotes APP α-cleavage
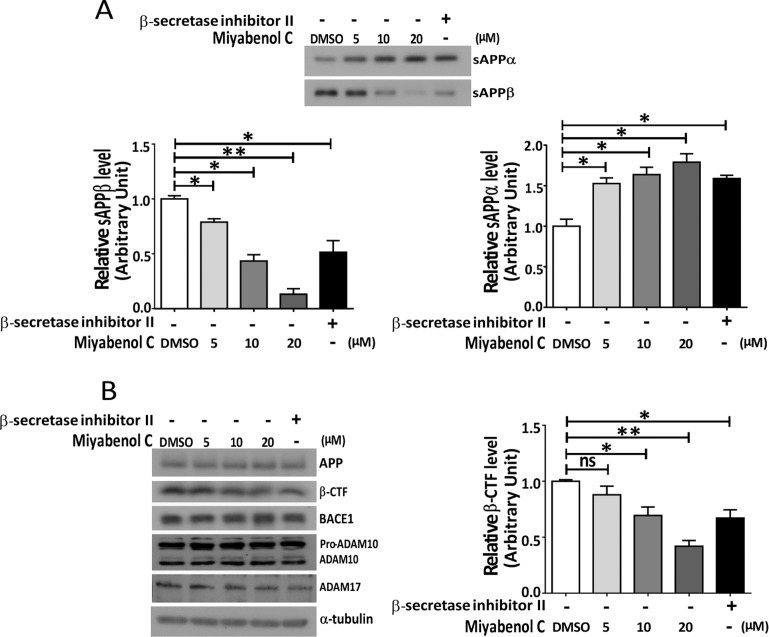
Because β-secretase-mediated APP processing is the first step leading to Aβ generation, we studied whether miyabenol C affects β-secretase. In N2a695 cells, miyabenol C treatment dose-dependently decreased the secreted level of sAPPβ, an amino-terminal fragment of APP generated by β-secretase cleavage (Fig. 3A). Consistently, the level of APP β-CTF (a carboxyl-terminal fragment of APP generated by β-secretase cleavage) was also decreased upon miyabenol C treatment (Fig. 3B). These results suggest that miyabenol C inhibits β-cleavage of APP. In addition, miyabenol C treatments increased the level of secreted sAPPα, the major extracellular fragment of APP released by α-secretase cleavage (Fig. 3A). Moreover, we found that miyabenol C treatment did not affect protein levels of APP, the major β-secretase BACE1 and two major α-secretases ADAM10 and ADAM17 (i.e. Tumor necrosis factor alpha converting enzyme, TACE) [34–36] (Fig. 3B). These results suggest that miyabenol C reduces APP amyloidogenic processing not through affecting α- and β-secretase protein levels.
Figure 3. Miyabenol C treatment reduces APP β-CTF and sAPPβ levels and increases sAPPα levels.
N2a695 cells were treated with DMSO (negative control), β-secretase inhibitor II (2μM) or indicated doses of miyabenol C for 10h. (A) Conditioned media and (B) cellular lysates were subjected to SDS-PAGE and Western blot. sAPPα, sAPPβ and β-CTF levels were quantified by densitometry using image J for comparison (One-way ANOVA followed by Tukey’s post hoc test, n = 3, ns: p>0.05, *: p<0.05, **: p<0.01).
Miyabenol C inhibits β- but not α-secretase activity
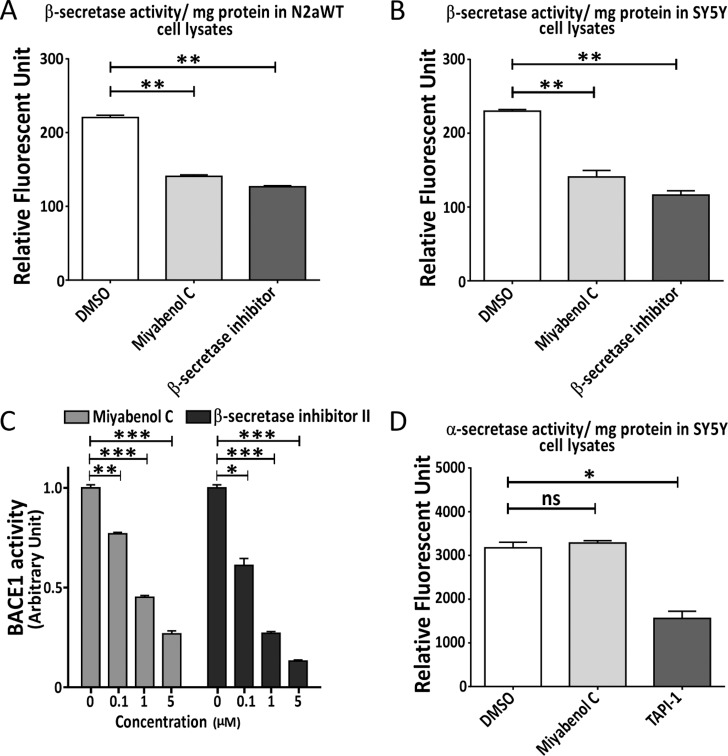
Since miyabenol C inhibits β-processing of APP without affecting BACE1 levels, miyabenol C probably directly inhibits BACE1 activity. To ascertain this possibility, we carried out a cell-based assay to measure the β-secretase activity. The results revealed that miyabenol C indeed dramatically inhibited β-secretase activity in both N2aWT and SY5Y cells (Fig. 4A, B). Moreover, miyabenol C markedly inhibited BACE1 acitivity in vitro, and its effect was comparable to that of β-secretase inhibitor II in vitro (Fig. 4C). Since there is another possibility that miyabenol C inhibits APP β-processing and Aβ generation through promoting α-secretase activity, we also assayed the activity of TACE, a major α-secretase in SY5Y cells treated with miyabenol C. We found that miyabenol C did not affect TACE activity (Fig. 4D), suggesting that miyabenol C does not affect α-secretase activity.
Figure 4. Miyabenol C inhibits β-secretase activity.
(A) N2aWT and (B) SH-SY5Y cells were treated with DMSO (negative control), miyabenol C (10μM) or a β-secretase inhibitor (positive control, 2μM, provided in the kit) for 10h. Cell lysates were assayed for β-secretase activity by using a commercial kit from Millipore and subjected to comparison. (C) Indicated amounts of miyabenol C and β-secretase inhibitor II were incubated with BACE1 and its substrate provided by a commercial kit from Sigma, for 2h at 37°C. BACE1 activity was measured for comparison. (D) SY5Y cells were treated with DMSO (negative control), miyabenol C (10μM) or the α-secretase inhibitor TAPI-1 (positive control, 10μM) for 10h. Cell lysates were assayed for the α-secretase (TACE) activity for comparison (One-way ANOVA followed by Tukey’s post hoc test, n = 3, ns: p> 0.05, *: p< 0.05, **: p< 0.01, ***: p< 0.001).
Miyabenol C treatment reduces sAPPβ and soluble Aβ levels in the brains of APP/PS1 transgenic AD mice
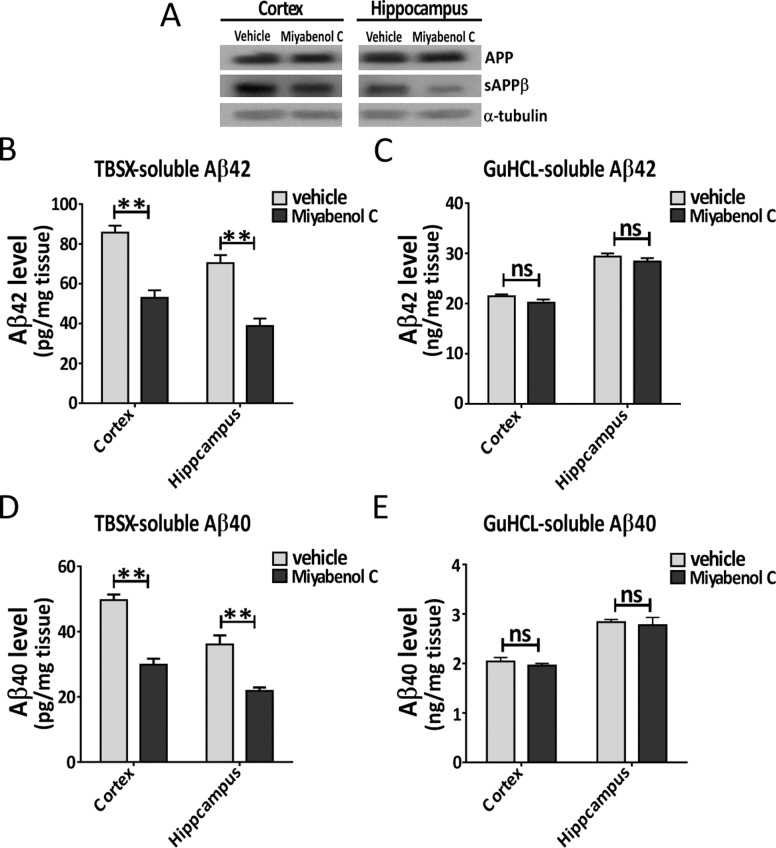
We also examined the in vivo inhibitory effects of miyabenol C on β-secretase activity using APP/PS1 transgenic mice, a transgenic mouse model of AD that expresses APP Swedish mutant and PS1 exon-9 deletion mutant. We injected miyabenol C or vehicle control into the lateral ventricle of 12 month-old APP/PS1 mice for a short period of time (72h). Mice were then sacrificed for analyses. We found that mice that received miyabenol C treatment had markedly reduced sAPPβ levels in both cortex and hippocampus when compared with mice treated with vehicle (Fig. 5A). Moreover, miyabenol C treatment significantly reduced levels of Aβ42 and Aβ40 in TBSX-soluble fractions (Fig. 5B, D). However, neither Aβ42 nor Aβ40 levels in the TBSX-insoluble fractions were affected by miyabenol C treatment (Fig. 5C, E).
Figure 5. Short term miyabenol C treatment reduces sAPPβ and soluble Aβ levels in APP/PS1 AD mouse brain.
APP/PS1 AD mice (12 month old) were treated with miyabenol C (0.6μg/g) or vehicle (45% DMSO in artificial cerebrospinal fluid) through intracerebroventricular injection (4μl) for 72h. (A) Cortex and hippocampus of treated mice were dissected and lysed. Equal protein amounts of lysates were analyzed by Western blot. Levels of TBSX-soluble Aβ42 (B) and Aβ40 (D), as well as TBSX-insoluble (re-dissolved in GuHCl) Aβ42 (C) and Aβ40 (E) in the cortex and hippocampus of treated mice were measured by ELISA assays (Student’s t test, n = 6, ns: p>0.05, **: p<0.01).
Discussion
Natural products provide excellent sources of new therapeutic compounds for disease intervention. Resveratrol and its derivatives are rich in grapes and possess strong anti-oxidant functions. Although resveratrol and its derivatives are considered to have therapeutic potential in AD [37, 38], detailed mechanism underlying their protective effects has yet to be determined.
Miyabenol C is a resveratrol trimer and has been found in plants including various Vitis species [39, 40], Caragana species [41–44], Carex species [45, 46], Parthenocissus quinquefolia [47], and Sophora davidii [48]. Previous studies have shown that miyabenol C possesses various functions, such as being anti-proliferative and pro-apoptotic in tumor cells [45, 49], inhibiting the activity of protein kinas C [44, 50], antagonizing human 5-hydroxytryptamine (5-HT) receptor [51], having estrogenic activity through binding to estrogen receptor [52, 53], and having ecdysteroid antagonistic activity in Drosophila cell bioassays [46]. However, information on the role of miyabenol C in AD is scarce, with only one recent study showing that miyabenol C has marginal effect on inhibiting Aβ fibril formation [39].
In the present study, we isolated miyabenol C from the small-leaf grape (Vitisthunbergii var. taiwaniana). Importantly, we found that miyabenol C treatment can dramatically reduce Aβ secretion (Fig. 1) and increase sAPPα release (Fig. 3) in cell culture. Since miyabenol C does not affect total APP levels (Fig. 3B), we studied whether miyabenol C affects α-, β- or γ-secretases. Miyabenol C treatment did not affect the protein level of PS1, a major component of the γ-secretase complex, and γ-secretase-mediated Notch processing for NICD generation, suggesting that miyabenol C does not affect γ-secretase. In addition, miyabenol C treatment did not affect protein levels of two major α-secretases, ADAM10 and ADAM17/TACE, and cell-based TACE activity, implying that miyabenol C does not affect α-secretase. In contrast, although miyabenol C treatment did not affect protein levels of the essential β-secretase BACE1, it reduced cell-based β-secretase activity and in vitro BACE1 activity, as well as both sAPPβ and APP β–CTF levels, indicating that miyabenol C reduces Aβ generation through inhibiting β-secretase activity. Consistently, we found that short-term (72h) treatment with miyabenol C in the brain of APP/PS1 mice markedly reduced the levels of both sAPPβ and TBSX-soluble Aβ that represent newly generated portion through β-cleavage, whereas miyabenol C treatment did not affect the levels of TBSX-insoluble Aβ that are from aggregated species (Fig. 5).
Together, our results demonstrate that miyabenol C is a prominent β-secretase inhibitor and can reduce Aβ generation both in vitro and in vivo. Since β-secretase-mediated APP processing is a rate-limiting step for Aβ generation and inhibition of β-secretase activity is a promising strategy for AD intervention, miyabenol C may be a lead compound for AD drug development.
Acknowledgments
We thank Dr. Robert Vassar at Northwestern University for providing the BACE1 antibody 3D5 and Dr. Chi-Luan Wen at Taiwan Seed Improvement and Propagation Station for providing Plant material. This work was supported by National Natural Science Foundation of China (Nos. 81202419, 81225008, 81161120496, 91332112, and 91332114), Xiamen Science and Technology Key program grant (No. 3502Z20100006), and Fundamental Research Funds for the Central Universities of China.
Data Availability
All relevant data are within the paper.
Funding Statement
This work was supported by National Natural Science Foundation of China (Nos. 81202419, 81225008, 81161120496, 91332112, and 91332114), Xiamen Science and Technology Key program grant (No. 3502Z20100006), and Fundamental Research Funds for the Central Universities of China. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Hardy JA, Higgins GA (1992) Alzheimer’s disease: the amyloid cascade hypothesis. Science 256: 184–185. [DOI] [PubMed] [Google Scholar]
- 2. Hardy J, Selkoe DJ (2002) The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science 297: 353–356. [DOI] [PubMed] [Google Scholar]
- 3. Zheng H, Koo EH (2011) Biology and pathophysiology of the amyloid precursor protein. Mol Neurodegener 6: 27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhang YW, Thompson R, Zhang H, Xu H (2011) APP processing in Alzheimer’s disease. Mol Brain 4: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hussain I, Powell D, Howlett DR, Tew DG, Meek TD, et al. (1999) Identification of a novel aspartic protease (Asp 2) as beta-secretase. Molecular and cellular neurosciences 14: 419–427. [DOI] [PubMed] [Google Scholar]
- 6. Vassar R, Bennett BD, Babu-Khan S, Kahn S, Mendiaz EA, et al. (1999) Beta-secretase cleavage of Alzheimer’s amyloid precursor protein by the transmembrane aspartic protease BACE. Science 286: 735–741. [DOI] [PubMed] [Google Scholar]
- 7. Lin X, Koelsch G, Wu S, Downs D, Dashti A, et al. (2000) Human aspartic protease memapsin 2 cleaves the beta-secretase site of beta-amyloid precursor protein. Proc Natl Acad Sci U S A 97: 1456–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sinha S, Anderson JP, Barbour R, Basi GS, Caccavello R, et al. (1999) Purification and cloning of amyloid precursor protein beta-secretase from human brain. Nature 402: 537–540. [DOI] [PubMed] [Google Scholar]
- 9. Yang LB, Lindholm K, Yan R, Citron M, Xia W, et al. (2003) Elevated beta-secretase expression and enzymatic activity detected in sporadic Alzheimer disease. Nat Med 9: 3–4. [DOI] [PubMed] [Google Scholar]
- 10. Johnston JA, Liu WW, Todd SA, Coulson DT, Murphy S, et al. (2005) Expression and activity of beta-site amyloid precursor protein cleaving enzyme in Alzheimer’s disease. Biochem Soc Trans 33: 1096–1100. [DOI] [PubMed] [Google Scholar]
- 11. Li R, Lindholm K, Yang LB, Yue X, Citron M, et al. (2004) Amyloid beta peptide load is correlated with increased beta-secretase activity in sporadic Alzheimer’s disease patients. Proc Natl Acad Sci U S A 101: 3632–3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dominguez D, Tournoy J, Hartmann D, Huth T, Cryns K, et al. (2005) Phenotypic and biochemical analyses of BACE1- and BACE2-deficient mice. J Biol Chem 280: 30797–30806. [DOI] [PubMed] [Google Scholar]
- 13. Hu X, Hicks CW, He W, Wong P, Macklin WB, et al. (2006) Bace1 modulates myelination in the central and peripheral nervous system. Nat Neurosci 9: 1520–1525. [DOI] [PubMed] [Google Scholar]
- 14. Willem M, Garratt AN, Novak B, Citron M, Kaufmann S, et al. (2006) Control of peripheral nerve myelination by the beta-secretase BACE1. Science 314: 664–666. [DOI] [PubMed] [Google Scholar]
- 15. Wong HK, Sakurai T, Oyama F, Kaneko K, Wada K, et al. (2005) beta Subunits of voltage-gated sodium channels are novel substrates of beta-site amyloid precursor protein-cleaving enzyme (BACE1) and gamma-secretase. J Biol Chem 280: 23009–23017. [DOI] [PubMed] [Google Scholar]
- 16. Laird FM, Cai H, Savonenko AV, Farah MH, He K, et al. (2005) BACE1, a major determinant of selective vulnerability of the brain to amyloid-beta amyloidogenesis, is essential for cognitive, emotional, and synaptic functions. J Neurosci 25: 11693–11709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McConlogue L, Buttini M, Anderson JP, Brigham EF, Chen KS, et al. (2007) Partial reduction of BACE1 has dramatic effects on Alzheimer plaque and synaptic pathology in APP Transgenic Mice. J Biol Chem 282: 26326–26334. [DOI] [PubMed] [Google Scholar]
- 18. Williams P, Sorribas A, Howes MJ (2011) Natural products as a source of Alzheimer’s drug leads. Nat Prod Rep 28: 48–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ebrahimi A, Schluesener H (2012) Natural polyphenols against neurodegenerative disorders: potentials and pitfalls. Ageing Res Rev 11: 329–345. 10.1016/j.arr.2012.01.006 [DOI] [PubMed] [Google Scholar]
- 20. Wang CK, Chen LG, Wen CL, Hou WC, Hung LF, et al. (2010) Neuroprotective activity of Vitis thunbergii var. taiwaniana extracts in vitro and in vivo. J Med Food 13: 170–178. 10.1089/jmf.2009.1162 [DOI] [PubMed] [Google Scholar]
- 21. Dodson SE, Andersen OM, Karmali V, Fritz JJ, Cheng D, et al. (2008) Loss of LR11/SORLA enhances early pathology in a mouse model of amyloidosis: evidence for a proximal role in Alzheimer’s disease. J Neurosci 28: 12877–12886. 10.1523/JNEUROSCI.4582-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang KT, Chen LG, Tseng SH, Huang JS, Hsieh MS, et al. (2011) Anti-inflammatory effects of resveratrol and oligostilbenes from Vitis thunbergii var. taiwaniana against lipopolysaccharide-induced arthritis. J Agric Food Chem 59: 3649–3656. 10.1021/jf104718g [DOI] [PubMed] [Google Scholar]
- 23. Liu CC, Kanekiyo T, Xu H, Bu G (2013) Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy. Nat Rev Neurol 9: 106–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Biedler JL, Roffler-Tarlov S, Schachner M, Freedman LS (1978) Multiple neurotransmitter synthesis by human neuroblastoma cell lines and clones. Cancer Res 38: 3751–3757. [PubMed] [Google Scholar]
- 25. Olmsted JB, Carlson K, Klebe R, Ruddle F, Rosenbaum J (1970) Isolation of microtubule protein from cultured mouse neuroblastoma cells. Proc Natl Acad Sci U S A 65: 129–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Thinakaran G, Teplow DB, Siman R, Greenberg B, Sisodia SS (1996) Metabolism of the “Swedish” amyloid precursor protein variant in neuro2a (N2a) cells. Evidence that cleavage at the “beta-secretase” site occurs in the golgi apparatus. J Biol Chem 271: 9390–9397. [DOI] [PubMed] [Google Scholar]
- 27. Zhao Y, Wang Y, Yang J, Wang X, Zhang X, et al. (2012) Sorting nexin 12 interacts with BACE1 and regulates BACE1-mediated APP processing. Mol Neurodegener 7: 30 10.1186/1750-1326-7-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang YW, Liu S, Zhang X, Li WB, Chen Y, et al. (2009) A Functional mouse retroposed gene Rps23r1 reduces Alzheimer’s beta-amyloid levels and tau phosphorylation. Neuron 64: 328–340. 10.1016/j.neuron.2009.08.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Das P, Verbeeck C, Minter L, Chakrabarty P, Felsenstein K, et al. (2012) Transient pharmacologic lowering of Abeta production prior to deposition results in sustained reduction of amyloid plaque pathology. Mol Neurodegener 7: 39 10.1186/1750-1326-7-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Youmans KL, Leung S, Zhang J, Maus E, Baysac K, et al. (2011) Amyloid-beta42 alters apolipoprotein E solubility in brains of mice with five familial AD mutations. J Neurosci Methods 196: 51–59. 10.1016/j.jneumeth.2010.12.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ono M, Ito Y, Kinjo J, Yahara S, Nohara T, et al. (1995) Four new glycosides of stilbene trimer from Foeniculi fructus (fruit of Foeniculum vulgare MILLER). Chem Pharm Bull 43: 868–871. [Google Scholar]
- 32. Li X, Liu Y, Zheng Q, Yao G, Cheng P, et al. (2013) Ferritin light chain interacts with PEN-2 and affects gamma-secretase activity. Neurosci Lett 548: 90–94. 10.1016/j.neulet.2013.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sorensen EB, Conner SD (2010) gamma-secretase-dependent cleavage initiates notch signaling from the plasma membrane. Traffic 11: 1234–1245. 10.1111/j.1600-0854.2010.01090.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lammich S, Kojro E, Postina R, Gilbert S, Pfeiffer R, et al. (1999) Constitutive and regulated alpha-secretase cleavage of Alzheimer’s amyloid precursor protein by a disintegrin metalloprotease. Proc Natl Acad Sci U S A 96: 3922–3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Buxbaum JD, Liu KN, Luo Y, Slack JL, Stocking KL, et al. (1998) Evidence that tumor necrosis factor alpha converting enzyme is involved in regulated alpha-secretase cleavage of the Alzheimer amyloid protein precursor. J Biol Chem 273: 27765–27767. [DOI] [PubMed] [Google Scholar]
- 36. Zhang H, Ma Q, Zhang YW, Xu H (2012) Proteolytic processing of Alzheimer’s beta-amyloid precursor protein. J Neurochem 120 Suppl 1: 9–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vingtdeux V, Dreses-Werringloer U, Zhao H, Davies P, Marambaud P (2008) Therapeutic potential of resveratrol in Alzheimer’s disease. BMC Neurosci 9 Suppl 2: S6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Baur JA, Sinclair DA (2006) Therapeutic potential of resveratrol: the in vivo evidence. Nat Rev Drug Discov 5: 493–506. [DOI] [PubMed] [Google Scholar]
- 39. Chaher N, Arraki K, Dillinseger E, Temsamani H, Bernillon S, et al. (2014) Bioactive stilbenes from Vitis vinifera grapevine shoots extracts. J Sci Food Agric 94: 951–954. 10.1002/jsfa.6341 [DOI] [PubMed] [Google Scholar]
- 40. Lambert C, Richard T, Renouf E, Bisson J, Waffo-Teguo P, et al. (2013) Comparative analyses of stilbenoids in canes of major Vitis vinifera L. cultivars. J Agric Food Chem 61: 11392–11399. 10.1021/jf403716y [DOI] [PubMed] [Google Scholar]
- 41. Jin Q, Han XH, Hong SS, Lee C, Choe S, et al. (2012) Antioxidative oligostilbenes from Caragana sinica. Bioorg Med Chem Lett 22: 973–976. 10.1016/j.bmcl.2011.12.012 [DOI] [PubMed] [Google Scholar]
- 42. Liu HX, Lin WH, Yang JS (2004) Oligomeric stilbenes from the root of Caragana stenophylla. Chem Pharm Bull (Tokyo) 52: 1339–1341. [DOI] [PubMed] [Google Scholar]
- 43. Chen G, Luo H, Ye J, Hu C (2001) Identification and determination of oligomeric stilbenes in the roots of Caragana species by capillary electrophoresis. Planta Med 67: 665–668. [DOI] [PubMed] [Google Scholar]
- 44. Kulanthaivel P, Janzen WP, Ballas LM, Jiang JB, Hu CQ, et al. (1995) Naturally occurring protein kinase C inhibitors; II. Isolation of oligomeric stilbenes from Caragana sinica. Planta Med 61: 41–44. [DOI] [PubMed] [Google Scholar]
- 45. Gonzalez-Sarrias A, Gromek S, Niesen D, Seeram NP, Henry GE (2011) Resveratrol oligomers isolated from Carex species inhibit growth of human colon tumorigenic cells mediated by cell cycle arrest. J Agric Food Chem 59: 8632–8638. 10.1021/jf201561e [DOI] [PubMed] [Google Scholar]
- 46. Meng Y, Bourne PC, Whiting P, Sik V, Dinan L (2001) Identification and ecdysteroid antagonist activity of three oligostilbenes from the seeds of Carex pendula (Cyperaceae). Phytochemistry 57: 393–400. [DOI] [PubMed] [Google Scholar]
- 47. Yang JB, Wang AG, Ji TF, Su YL (2014) Two new oligostilbenes from the stem of Parthenocissus quinquefolia. J Asian Nat Prod Res 16: 275–280. 10.1080/10286020.2013.877451 [DOI] [PubMed] [Google Scholar]
- 48. Tanaka T, Ito T, Iinuma M, Ohyama M, Ichise M, et al. (2000) Stilbene oligomers in roots of Sophora davidii. Phytochemistry 53: 1009–1014. [DOI] [PubMed] [Google Scholar]
- 49. Barjot C, Tournaire M, Castagnino C, Vigor C, Vercauteren J, et al. (2007) Evaluation of antitumor effects of two vine stalk oligomers of resveratrol on a panel of lymphoid and myeloid cell lines: comparison with resveratrol. Life Sci 81: 1565–1574. [DOI] [PubMed] [Google Scholar]
- 50. Xu G, Zhang LP, Chen LF, Hu CQ (1994) [Inhibition of protein kinase C by stilbenoids]. Yao Xue Xue Bao 29: 818–822. [PubMed] [Google Scholar]
- 51. Kim DH, Kim SH, Kim HJ, Jin C, Chung KC, et al. (2010) Stilbene derivatives as human 5-HT(6) receptor antagonists from the root of Caragana sinica. Biol Pharm Bull 33: 2024–2028. [DOI] [PubMed] [Google Scholar]
- 52. Tian CY, Gao HD, Zhang YG, Xu G, Hu CQ, et al. (2003) [The binding sites of estrogen receptor for miyabenol C and kobophenol A]. Sheng Wu Hua Xue Yu Sheng Wu Wu Li Xue Bao (Shanghai) 35: 77–81. [PubMed] [Google Scholar]
- 53. Tian CY, Hu CQ, Xu G, Song HY (2002) Assessment of estrogenic activity of natural compounds using improved E-screen assay. Acta Pharmacol Sin 23: 572–576. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.