Abstract
We previously described the whole-genome assembly program Arachne, presenting assemblies of simulated data for small to mid-sized genomes. Here we describe algorithmic adaptations to the program, allowing for assembly of mammalian-size genomes, and also improving the assembly of smaller genomes. Three principal changes were simultaneously made and applied to the assembly of the mouse genome, during a six-month period of development: (1) Supercontigs (scaffolds) were iteratively broken and rejoined using several criteria, yielding a 64-fold increase in length (N50), and apparent elimination of all global misjoins; (2) gaps between contigs in supercontigs were filled (partially or completely) by insertion of reads, as suggested by pairing within the supercontig, increasing the N50 contig length by 50%; (3) memory usage was reduced fourfold. The outcome of this mouse assembly and its analysis are described in (Mouse Genome Sequencing Consortium 2002).
In the whole-genome shotgun method of genome sequencing, as presently practiced, the entire genome is sheared to specified approximate sizes (generally chosen from the range 2–200 kb), yielding random fragments (called inserts), whose ends can then be determined, yielding sequence reads of size about 600 bp, given in pairs whose genomic separation is approximately known from the insert size (Sanger et al. 1977; Edwards et al. 1990). In principle, the genome can then be reconstructed in silico from these reads, using their overlap and pairing as glue, provided that the inserts gave sufficiently even representation of the genome, that an appropriate mix of insert lengths (short to long) was used, and that the DNA itself was sufficiently free of polymorphism (Sanger et al. 1982; Fleischmann et al. 1995; Myers et al. 2000; Aparicio et al. 2002; Yu et al. 2002).
This assembly process produces sequence units (called supercontigs or scaffolds), which under the best of circumstances can approximate chromosomes, but which in general are smaller, less contiguous, and have errors. Within a supercontig, contiguous segments (or contigs) are punctuated by gaps (captured by paired reads) whose sizes are approximately known (as a consequence of knowing the insert lengths). The resulting draft sequence may be an end goal or may be the starting point for production of finished sequence, through additional clone-based sequencing.
For whole-genome shotgun reads from large repetitive genomes such as those of mammals, assembly is a computational challenge (because a priori any read may overlap any other read), and an algorithmic challenge, because repetitive structures and defects in data make it easy to falsely join distant parts of the genome. Still, assemblies consisting of either a mixture of whole-genome shotgun and clone-based reads, or only whole-genome shotgun reads have been produced (human: Venter et al. 2001; mouse chromosome 16: Mural et al. 2002).
Presented with the publicly available mouse whole-genome data set, we and another group (Mullikin and Ning 2003) set out to produce high-quality draft sequence for the mouse genome. Our starting point was the work presented in (Batzoglou et al. 2002), wherein the Arachne software was used to assemble simulated reads from small to mid-sized genomes. In this paper we describe algorithmic enhancements to the program, made while assembling the mouse data set and yielding the mouse assembly described in (Mouse Genome Sequencing Consortium 2002).
These enhancements fall under three general headings: global assembly improvement, local assembly improvement, and computational performance improvement. These changes were made over a six-month period during which successively larger collections of mouse reads were made available to us and which we assembled, while modifying our algorithms. To expose the motivation for the solutions, we proceed where possible by explaining our successive attempts and their outcomes.
Global Assembly Improvement
We begin our analysis at the point where the mouse assembly had been run up to the end of the released Arachne code (Batzoglou et al. 2002). The N50 supercontig size was 265 kb (half the total bases in the supercontigs lay in supercontigs of size 265 kb or larger). By improving our algorithms, we hoped to raise this value to 1 Mb.
To that end, we set out to design code to look off the ends of each supercontig, in the hopes of finding genomically adjacent supercontigs with which it could be merged.
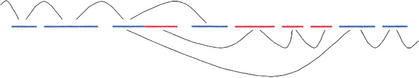
Manual examination of the data revealed many cases where it was clear in principle how to merge, but many of the cases were complicated by the presence of multiple supercontigs that linked off the end of a given supercontig (Fig. 1). To specify precisely how such cases should be handled, we posed the following mathematical problem (J, see Methods): Off a particular end of a given supercontig, consider all supercontigs it links to, and which, based on pairing separations, should lie off the end; for each ordering of these supercontigs, find the nonoverlapping positioning of them which minimizes ‘stretching’ of links; mark the order as acceptable if the stretching is modest, at most 2.5 standard deviations; determine whether among the acceptable orders, there is only one supercontig, nearest the given one; if so, merge the given supercontig with this nearest neighbor.
Figure 1.
Joining of supercontigs. Three supercontigs (a, b, c) are seen off the end of supercontig s. There are two or more read pair links from s to each of them. Each has an optimal position relative to s, determined by the insert lengths corresponding to the read pairs. However, each insert length has a standard deviation associated to it, and so the positions of a, b, and c relative to s also have standard deviations. Supposing that we allow each of them to slide from their optimal positions by up to 2.5 standard deviations, but that we do not allow overlap between any of the supercontigs, is there more than one possible order for the supercontigs? Among the possible orders, does a always appear first (after s)? If so, we join supercontig s to supercontig a.
This technique facilitated some mergers, but other potential mergers were inhibited by misassemblies. This suggested that to optimize supercontigs, we needed to break supercontigs at suspicious points, rejoin them as described (J), and repeat, iteratively.
Multiple breaking schemes were required because there are diverse sources of assembly error. Genomic repeats are the most important source, but there are others, which act in combination with repeats to create “hazardous” sections of the genome to assemble. For example, there are regions of the genome whose assembly coverage is low or of low quality, either for random reasons, or because of the nature of the genome itself, as reflected in cloning bias or sequencing difficulty. Misassemblies arise from errors in linking (themselves arising from chimeric clones, from intraplate contamination, from labeling errors, from duplicated sequencing of clones). Misassemblies also arise from chimeric reads (from chimeric clones or bleed-through of contaminating sequence). Finally, misassemblies can arise from undiagnosed algorithmic defects in the assembly process.
Given this complex background, we did not expect a single supercontig breaking approach to suffice, and so we developed several methods, not all of which are correlated with particular sources of assembly error. We describe some of these methods now, and another later.
Negative Breaking
These methods detect regions of the assembly which are weakly held together. A simple case (which we call method B1) is where we break supercontigs between contigs if the template coverage across the gap is only one. Evidently this situation arises now because earlier in the assembly process, we merged supercontigs connected by only one link. Nevertheless, in aggregate, it was not a mistake to do this initially, as some were reinforced subsequently by longer links. As a generalization, in method B2, we break supercontigs at gaps whose covering templates fall nearly on top of each other. This subsumes the case where the same clone was end-sequenced more than once, giving rise to the appearance of multiple links. Next, in method B3, we break supercontigs at places within a contig where only sequence holds together the supercontig (no links hold it together; see Methods). In the most glaring cases, we found that the supercontig was held together by only a thread: a single poor, short join between two reads. However, the same criterion was designed to identify subtler mistakes, in which templates might falsely overlap by about a read length. Further, after assembling mouse, we devised criteria to identify disguised instances of the same phenomenon, exemplified in Figure 2.
Figure 2.
A disguised instance where sequence join alone holds together a supercontig. A long supercontig (blue) from one part of the genome subsumes a small foreign inset (red) from a completely different part of the genome, held together by a single point of attachment within a contig (bicolor): in fact only a sequence join ties blue to red. This was not recognized in the version of the code which produced the released mouse assembly (Mouse Genome Sequencing Consortium 2002). Resolution: break at the bicolor juncture, move the red sequence to where it links in another supercontig.
Breaking Off Ends
The ends of supercontigs are enriched for misassembly. In method B4, we removed 10 kb of sequence from the ends of long supercontigs, breaking mid-contig. No specific evidence of error was required; we expect that most of these supercontig ends were in fact correct.
In combination, these methods proved effective. After applying them (iteratively breaking and rejoining), the N50 supercontig size rose to 11 Mb, surpassing our initial expectation of 1 Mb, but possibly aided by false global joins. To assess this possibility, we aligned the mouse genetic markers (Dietrich et al. 1996) to the assembly. Because genetic markers are the most reliable source of long-range ordering, and are not used by Arachne, this was an independent test for assembly integrity. Yet from the alignments of the genetic markers to the assembly, we could see instances where supercontigs crossed chromosomes, proof of global misassembly.
Although the genetic map could be used to evaluate the assembly, substantial spacing between markers prohibited its use in editing the assembly. Therefore we devised a supercontig breaking criterion which might remedy the defects uncovered by the genetic map, without using the map itself.
Positive Breaking
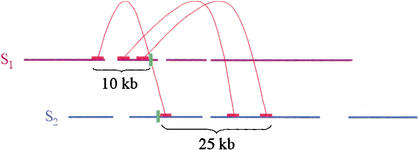
We found examples where there were multiple consistent links from the middle of one supercontig to another. These supercontigs were adequately held together [as evidenced by inapplicability of (B1–4)], but strong evidence supported an alternative structure. A specific criterion was devised to address this. In method B5 (described further in Methods), we found correlated clusters of links between pairs of supercontigs, which could not be explained without breaking them. Then we broke the supercontigs, immediately after the last link, mid-contig. This criterion depended on heuristics: The minimum number of links in a cluster, and especially, the minimum acceptable spread of the cluster, where the spread is the minimum (over the two participating supercontigs) of the total span in bases of the link end reads on a given supercontig (Fig. 3). For mouse, we did not experiment, but used five as the minimum number of links, and 50 kb as the minimum acceptable spread. Notably, it is clear that these values are inappropriate for assemblies where there is less long-range coverage, and it is clear that in such cases, long-range correct assembly may be impossible. For mouse, we were fortunate to have a rich data set, replete with long-range linking.
Figure 3.
Positive breaking of supercontigs. Three correlated links are seen between supercontigs S1 and S2. The spread of the connection between S1 and S2 is, in this case, the lesser of 10 kb and 25 kb, which is 10 kb. Because the positive breaking algorithm as applied to mouse required five links with spread at least 50 kb, this connection would not have been sufficient to break the supercontigs. If it were, the respective supercontigs would have been broken at the exact ends of reads (green bars).
After positive breaking in the mouse assembly, we aligned again to the genetic markers, finding no evidence of global misjoins. This and continued breaking and rejoining raised the N50 supercontig value from 11 Mb to 17 Mb, again without evidence of global error. We note that the only way we used the genetic map during the assembly was to suggest the method of positive breaking.
Local Assembly Improvement
Once one has globally valid long-range supercontigs, the assembly problem is local. One can scan through small regions of a supercontig, improving each region. In particular, we describe here our work to shrink and where possible close as many gaps as possible, or in short, to patch gaps.
Gap patching is possible because some read overlaps were undetected or rejected in prior assembly stages: because of repetitive sequence, or because the overlaps were short, or because the reads themselves contained defects, such as low-quality bases, isolated errors at high-quality bases, or untrimmed vector. Some of these unexploited overlaps are wrong. To safely patch gaps, we first formed a pool of reads for which there was linking evidence to support placement of the read in the gap. This pool contained the partners of reads already placed in the neighborhood of the gap. Further, when we found two paired reads, with neither placed in the neighborhood but with both members having good overlaps to reads in the neighborhood (including those in the pool), we added them to the pool; this process was iterated several times. We then restricted the pool by requiring that placement of its reads within the gap was consistent with pairing separations.
We then attempted to close (or partially close) the gap, using the given pool of reads. For this purpose, we recomputed the alignments between all involved sequences, choosing not to rely on the precomputed alignments (from much earlier in the assembly process), to circumvent omissions in that set. We accepted alignments as short as 15 bases, so long as they were perfect and consisted of nonrepetitive sequence (as gauged by abundance of 12-mers in the reads). Then we tried to walk from one side of the gap to the other, starting at both sides. In the end, progress would be made when we extended on either side, or (ideally) closed the gap.
This procedure raised the N50 contig size from 16.3 kb to 24.8 kb, but as a side effect placed some reads more than once in the assembly. This could happen when both reads in a read pair had been placed in the assembly, but in different scaffolds. Usually one scaffold was very large, and one very small, with the small scaffold duplicating sequence within the large scaffold: The small scaffold could be discarded. Similar heuristics allowed us to remove almost all multiply placed reads. Still, 0.014% of the reads were multiply placed in the chromosomally anchored part of the final assembly. Some of these multiple placements are associated with misassemblies (of a type which in principle could be identified by breaking method B3, above), and we believe that these can be removed by adjusting the existing algorithms. However, we expect that some cases will remain where a read can go in more than one place, and it is not possible to determine which is right without obtaining further data.
In the process of closing gaps, we discovered certain types of problems which had to be solved earlier in the assembly. These were cases where a sequence read had incorrect sequence attached at its beginning or end, incorrectly terminating a contig. Where such sequence was attached at the beginning of the read, it was often a short fragment of vector which for some reason had not been trimmed from the read. Where such sequence was attached at the end of a read, it sometimes arose in the following coincidental situation. Two different sequences were originally present in adjacent wells of a plate, but via some unknown mechanism, sequence was transferred from one well to its neighbor, contaminating it at a low level. In turn, the dominant sequence in this neighboring well had sequence (perhaps, e.g., a large number of consecutive Gs) which would cause premature termination in the sequencing reaction. The net effect of these combined phenomena was to create a chimeric trace, arising from sequence native to the well, up to the point of its premature termination, followed by trace arising from the contaminating sequence, at lower magnitude.
To remove residual vector fragments from reads, we identified 10-mers overrepresented in the first 20 bases of reads (as compared to their frequency in the entire reads). We then used these suspect 10-mers as a guide in trimming the beginnings of the reads. To deal with cross-well contamination, we empirically modeled its signature (as manifested in bases, quality scores), explicitly looked for physically adjacent reads consistent with this signature, and trimmed them accordingly. Although this worked (as measured by a small test set), we also observed reads with “junk” at their ends, not explained by cross-well contamination, and which we were unable to systematically eliminate.
Computational Performance Improvement
For the mouse assembly, we had roughly ten times as many sequence reads as we had had on any previous Arachne assembly. Consequently, we devoted about two-thirds of our effort to reducing memory usage and runtime, rather than to algorithmic development aimed at assembly improvement.
Most of the computation was performed using a single processor equipped with 32 GB memory (which could not be increased without great expense; see Methods.) On the other hand, several processors were attached to the memory, which we could hope to multithread across. Some progress was made in this direction, hindered primarily by the number of independent executable modules in Arachne (69 for our mouse assembly, up from 17 for simulations reported in Batzoglou et al. 2002), many of which would have to be multithreaded to yield substantial runtime reduction. For the mouse assembly, we multithreaded a single module, which dealt with the initial handling of data (trimming, etc.), thereby reducing total runtime by a day or two.
Instead, our main computational strategy to reduce runtime was to locate computational hotspots and remediate them as best we could. For example, we found that the compiler for our language (C++) would inefficiently store very large vectors of smaller objects (such as the vector of all sequence reads), by performing a separate memory allocation for each of the objects. This resulted in runtime which behaved roughly quadratically as a function of vector size, rather than roughly linearly, as we expected from prior experience with Arachne. Accordingly we designed structures which stored the data using a single memory allocation, while looking like a vector to the compiler. This also reduced memory usage. As another related example, we found that by storing large data structures contiguously, we could quickly read them into memory. In combination, about a dozen such optimizations made it possible for us to assemble the mouse genome in about 15 days.
The main reason we had difficulty reducing memory usage was that many of Arachne's modules were designed so that at any time, all of the data could be accessed. For example, on demand, the error correction code accesses a read it wants, the associated quality scores, and the alignments of it with other reads. In assemblies of smaller genomes, we had exploited this capability to optimize assembly quality. Now with the larger mouse data set at hand, we wanted to preserve gains in assembly quality, while living within the confines of our allotted memory (32 GB).
This was accomplished by economizing in the storage of large data structures (discussed above), and by localizing the computation. We did this by reordering the reads (after computing all read-read alignments; see Methods), in such a way that genomically proximate reads tended to be nearby in the new read order. Then for many computations which (a priori) required all reads to be in memory, we modified the algorithm so that 10% of the reads were brought into memory at a time, along with all other reads aligning to them (perhaps only another 10%, by virtue of the reordering). This change reduced memory usage, and in addition increased the locality of memory accesses, thereby speeding up their execution, while at the same time allowing the assembly modules to access all reads aligning to a given read.
In combination, these memory reductions allowed us to assemble the 41 million mouse reads using only 28 GB of memory. Vis-a-vis memory usage as reported in the original Arachne paper, this represents a fourfold reduction.
All in all, it was possible to assemble the mouse genome in 15 days using a single computer (Compaq ES40, see Methods), whose total cost including memory and disk storage was ∼$300,000.
DISCUSSION
Our mouse assembly is evidence that high-quality draft assembly of complex and repetitive genomes is possible from whole-genome shotgun sequence alone, which in combination with mapping data comprises a fast and cost-effective starting point for genome-wide sequencing projects. For perspective, we note in Table 1 the other eukaryotic genomes having published assemblies either from whole-genome shotgun reads or a hybrid including both whole-genome shotgun and clone-based sequence. The table should be interpreted cautiously in view of the diverse character of the sequencing data itself.
Table 1.
N50 Supercontig Sizes for Whole Genome Shotgun/Hybrid Assemblies of Eukaryotic Genomes
| Species | Genome (Gb) | Sequencing type | Sequence coverage | Clone coverage | Number of haplotypes | N50 supercontig (Mb) |
| D. melanogaster | 0.12 | hybrid | 16.6X | 65.5X* | 1 | 14.5* |
| F. rubripes | 0.37 | whole genome | 5.6X | 13.0X | 4 | 0.04* |
| O. sativa | 0.43 | whole genome | 4.2X | 7.7X | 1 | 0.01 |
| M. musculus | 2.5 | whole genome | 6.5X | 47.2X | 1 | 16.9 |
| H. sapiens | 2.9 | hybrid | 8.3X | 40.0X | >12 | 4.0* |
For assemblies of whole genome shotgun (WGS) data, and hybrid assemblies of WGS data together with clone-based sequence, some finished, we show the N50 supercontig size (exclusive of gaps), together with various parameters of the data set which might influence the N50 value. Asterisked entries were approximated from other data given in the relevant publication. (N50 values provide a standard measure of assembly connectivity, reflecting the nature of the bulk of the assembly rather than the cutoff which defines the smallest reportable assembly unit.) References: Myers et al. 2000, Aparicio et al. 2002, Yu et al. 2002, Venter et al. 2001.
A comparable mouse assembly has been produced from the same public data set using the Phusion assembler, and evaluated using comparable criteria: N50 contig, N50 supercontig, global accuracy (Mullikin and Ning 2003). Algorithmically, Phusion and Arachne share many features, but Phusion groups reads early in the assembly process, permitting parallel computations for each group. Phusion and Arachne both break contigs based on read pair inconsistencies, but Arachne is able to break contigs based on long-range inconsistencies, only visible at the supercontig level.
There are several reasons why the mouse genome, although large and repetitive, may have been relatively easy to assemble. First, we assembled sequence from an inbred organism: Assembly of highly polymorphic data is harder. Second, although the mouse genome has repeats, it may lack some more difficult repeat structures. For example, evidence suggests that it may have fewer recent segmental duplications than found in the human genome. Third, our data included end reads from three long-insert libraries (one fosmid, two BAC; Osoegawa et al. 2000; Zhao et al. 2001). Good coverage from long-insert clones is likely prerequisite to long-range assembly connectivity. For example, we found that if the BAC ends are excised from the full mouse assembly, and the supercontigs are then broken at gaps spanned by at most one link (as in method B1, above), then the N50 supercontig value drops from 17 Mb to 15 Mb. If one also deletes the fosmid ends (which are from 40-kb inserts), then the N50 value drops to 2 Mb. Moreover, an assembly from scratch could suffer much worse degradation if it lacked these long-insert reads. Fourth, we had high-quality maps at our disposal (genetic, Dietrich et al. 1996; physical, Gregory et al. 2002; radiation hybrid, Hudson et al. 2001) which anchored our assembly to the genome and provided feedback on the assembly process.
In addition, we should emphasize that assembly from whole-genome shotgun reads produces only draft sequence. Clone-based sequence still appears essential to produce a finished genome sequence.
Given these caveats, and given that some new genomes will require algorithmic innovation to assemble optimally, we release Arachne 2 as a publicly available system which can assemble genomes of mammalian size (see Methods). Now, given the public availability of both a mammalian whole-genome shotgun data set and a program capable of assembling them, it should be possible for researchers to undertake experiments involving assemblies of subsets of the data to shed light on fundamental questions regarding sequencing strategy. In particular, such experiments could tell us what would be lost if coverage were reduced or if a less diverse range of insert sizes were used. Answers to these questions will provide insight for sequencing the next mammalian genome.
METHODS
Supercontig Joining: How We Solved Problem (J)
Given a single link between two supercontigs, and given an offset for them (i.e., the difference between their starting positions), the amount which the link is stretched by is defined to be the absolute value of the difference between the given offset and the offset inferred from the link, divided by the standard deviation of the insert length for the link. For a given order of supercontigs linked off the end of a given supercontig, let the vector x denote the variable vector which contains the separations between the ends of successive supercontigs in the order. Because we assume that the supercontigs do not overlap, the entries of x are nonnegative. For an appropriate constant matrix A and vector b, the problem of minimizing link stretching may be recast as one of minimizing |Ax-b|, that is, of minimizing the maximum of the absolute values of the entries of the vector Ax-b. Our solution to this problem involved successive random perturbations, leading in the end to an approximate answer.
Negative Breaking Method B3
In principle, this should break a supercontig at places where only sequence holds it together. As implemented, the method breaks at points in a supercontig which satisfy all of the following criteria: (1) The point is not in the first or last 25 kb of the supercontig; (2) the point is not covered by the unsequenced part of a template (the part between the end reads); (3) the reads before the point do not link forward to another contig, which the reads after the point also link forward to; and (4) the reads before the point do not link backward to another contig which the reads after the point also link back to.
Positive Breaking: How We Defined Correlated Clusters of Links
A read pair link between two supercontigs defines an offset for their positions. This offset has a standard deviation associated with it (which is the standard deviation for the insert length corresponding to the read pair). Two such read pair links between the same two supercontigs define two offsets o1, o2, and two associated standard deviations, d1 and d2. If |o1−o2| < 2.5 * max(d1, d2), we say that the two links are correlated. This extends to define an equivalence relation on the set of all links between two given supercontigs. A correlated cluster is an equivalence class for this relation.
Computational Platform
Assemblies were performed on a Compaq Alpha ES40, equipped with four 833 MHz processors and 32 GB of memory (RAM). The mouse assembly (including the raw data and all intermediate files) occupies 215 GB of disk space. The Arachne source code is in C++, compiled under g++.
Reordering of Reads
Start with the first read in the inputted list of reads. Make this the first read in the new order. Append to the new order all reads aligning to the first read. Then find the first read in the new order which has a read aligning to it, not already in the new order, and append this aligning read to the new order. (If there is no such read, go back to the input list to get a read.) Repeat. Exception: If a read aligns to more than 100 reads, ignore the alignments of it to other reads.
Software Availability
Arachne 2 is available (2/2002) at http://www.genome.wi.mit.edu/wga. The license terms are described on that site.
Mouse Assembly, Auxiliary Files
Certain auxiliary input files used in the Arachne assembly of the public (MGSC) mouse data set are available at ftp://wolfram.wi.mit.edu/pub/mouse_contigs/Mar10_02/auxiliary_input_files.
Acknowledgments
We thank Jim Mullikin and Zemin Ning for sharing their PHUSION mouse assemblies and for valuable discussion.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
E-MAIL jaffe@genome.wi.mit.edu; FAX (617) 258-9108.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.828403.
REFERENCES
- 1.Aparicio S., Chapman, J., Stupka, E., Putnam, N., Chia, J., Dehal, P., Christoffels, A., Rash, S., Hoon, S., Smit, A., et al. 2002. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science 297: 1301-1310. [DOI] [PubMed] [Google Scholar]
- 2.Batzoglou S., Jaffe, D.B., Stanley, K., Butler, J., Gnerre, S., Mauceli, E., Berger, B., Mesirov, J.P., and Lander, E.S. 2002. Arachne: A whole-genome shotgun assembler. Genome Res. 12: 177-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dietrich W.F., Miller, J., Steen, R., Merchant, M.A., Damron-Boles, D., Husain, Z., Dredge, R., Daly, M.J., Ingalls, K.A., O'Connor, T.J., et al. 1996. A comprehensive genetic map of the mouse genome. Nature 380: 149-152. [DOI] [PubMed] [Google Scholar]
- 4.Edwards A., Voss, H., Rice, P., Civitello, A., Stegemann, J., Schwager, C., Zimmermann, J., Erfle, H., Caskey, C.T., and Ansorge, W. 1990. Automated DNA sequencing of the human HPRT locus. Genomics 6: 593-608. [DOI] [PubMed] [Google Scholar]
- 5.Fleischmann R.D., Adams, M.D., White, O., Clayton, R.A., Kirkness, E.F., Kerlavage, A.R., Bult, C.J., Tomb, J., Dougherty, B.A., and Merrick, J.M. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269: 496-512. [DOI] [PubMed] [Google Scholar]
- 6.Gregory S.G., et al. 2002. A physical map of the mouse genome. Nature 418: 743-750. [DOI] [PubMed] [Google Scholar]
- 7.Hudson T.J., Church, D.M., Greenaway, S., Nguyen, H., Cook, A., Steen, R.G., Van Etten, W.J., Castle, A.B., Strivens, M.A., Trickett, P., et al. 2001. A radiation hybrid map of mouse genes. Nat. Genet. 29: 201-205. [DOI] [PubMed] [Google Scholar]
- 8.Mouse Genome Sequencing Consortium. Initial sequencing and comparative analysis of the mouse genome. 2002. Nature (this issue). [DOI] [PubMed]
- 9.Mullikin, J.C. and Ning, Z. 2003. The Phusion assembler. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- 10.Mural R.J., Adams, M.D., Myers, E.W., Smith, H.O., Miklos, G.L., Wides, R., Halpern, A., Li, P.W., Sutton, G.G., Nadeau, J., et al. 2002. A comparison of whole-genome shotgun-derived mouse chromosome 16 and the human genome. Science 296: 1661-1671. [DOI] [PubMed] [Google Scholar]
- 11.Myers E.W., Sutton, G.G., Delcher, A.L., Dew, I.M., Fasulo, D.P., Flanigan, M.J., Kravitz, S.A., Mobarry, C.M., Reinert, K.H., Remington, K.A., et al. 2000. A whole-genome assembly of Drosophila. Science 287: 2196-2204. [DOI] [PubMed] [Google Scholar]
- 12.Osoegawa K., Tateno, M., Woon, P.Y., Frengen, E., Mammoser, A.G., Catanese, J.J., Hayashizaki, Y., and de Jong, P.J. 2000. Bacterial artificial chromosome libraries for mouse sequencing and functional analysis. Genome Res. 10: 116-128. [PMC free article] [PubMed] [Google Scholar]
- 13.Sanger F., Nicklen, S, and Coulsen, A.R. 1977. DNA sequencing with chain terminating inhibitors. Proc. Natl. Acad. Sci. 74: 5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanger F., Coulson, A.R., Hong, G.F., Hill, D.F., and Peterson, G.B. 1982. Nucleotide sequence of bacteriophage λ DNA. J. Mol. Biol. 162: 729-773. [DOI] [PubMed] [Google Scholar]
- 15.Venter J.C., Adams, M.D., Myers, E.W., Li, P.W., Mural, R.J., Sutton, G.G., Smith, H.O., Yandell, M., Evans, C.A., Holt, R.A., et al. 2001. The sequence of the human genome. Science 291: 1304-1351. [DOI] [PubMed] [Google Scholar]
- 16.Yu J., Hu, S., Wang, J., Wong, G.K., Li, S., Liu, B., Deng, Y., Dai, L., Zhou, Y., Zhang, X., et al. 2002. A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296: 79-92. [DOI] [PubMed] [Google Scholar]
- 17.Zhao S., Shatsman, S., Ayodeji, B., Geer, K., Tsegaye, G., Krol, M., Gebregeorgis, E., Shvartsbeyn, A., Russell, D., Overton, L., et al. 2001. Mouse BAC ends quality assessment and sequence analyses. Genome Res. 11: 1736-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]