Abstract
New molecular insight reveals novel points of attack for targeted cancer therapy. The recent advances in cancer genomics and novel insight into the complex biology of cancer make the promise of personalized, targeted cancer medicine closer than ever. The massive parallel sequencing endeavours performed by The Cancer Genome Atlas, the International Cancer Genome Consortium and by numerous individual investigators have provided a comprehensive genomic characterization of a wide range of cancers. The joint efforts enabled by the improved sequencing technology have demonstrated that individual cancers comprise mutational repertoires with only a few frequently recurrent driver genes. Thus, the identification of new drug targets and novel drugs have accelerated and renewed the hopes of personalized cancer therapy achieving clinical reality for a wider range of cancers. Together with cost-effective sequencing technology to perform comprehensive mutational profiling of each individual cancer, this provides the basis for a personalized cancer medicine revolution within the next few years. The aim of this MiniReview is to provide an overview of the history and evolution of targeted cancer therapy, exemplified by molecularly targeted drugs successfully implemented in the clinic. Furthermore, we aim to highlight novel molecular targets for therapeutic intervention, as well as the main present challenges including inter- and intratumor heterogeneity and cellular plasticity in addition to the importance of the tumor micro-environment. Many cancer patients already receive some form of tailored therapy, and recent evidence suggests that novel and highly innovative, targeted approaches are on their way into the clinic.
There is an old adage that cancer is a hundred diseases masquerading into one. In support of this, Hanahan and Weinberg have defined several hallmarks of cancer, common to most, if not all, cancers [1]. Application of improved DNA sequencing technologies developed during the Human Genome Project (HGP) has confirmed and extended this adage, revealing the fact that even within a single cancer group or subgroup, each cancer has a unique genetic make-up. New technology, availability and lowered sequencing costs, allow wider application and provide cancer researchers and clinicians with a comprehensive compendium of the genetic alterations present within an individual tumor sample. Indeed, recent findings from The Cancer Genome Atlas (TCGA) and the International Cancer Genome Consortium (ICGC) have further verified that although each cancer seems to be unique in its repertoire of genetic mutations, a range of signaling pathways are frequently affected within particular cancer types [2]. The present challenge is to filter the genetic alterations ‘driving’ tumor progression from the ‘passengers’, also referred to as ‘noise’ present due to extensive genomic instability. Ongoing efforts to detect alterations driving tumor progression can be identified at accelerated speed both within and across cancer subtypes. Integrated multi-disciplinary efforts combining insight into the underlying molecular processes, not only within a particular form of cancer, but also across different kind of cancers, is required to meet the expectations of development of tailored cancer treatment. Hopes are high that along with an improved molecular characterization, accelerated development of molecularly targeted drugs will provide the tools required to enable oncologists to tailor cancer treatment to the individual cancer patient based on tumor characteristics.
Oncology is a major field of focus for pharmaceutical and biotechnology companies. This interest stems from the unmet need for improved treatments of multiple types of cancer, as well as from the substantial market success of targeted cancer therapies launched in the past decade [3]. Targeted cancer therapies are drugs that are designed to address the genetic alterations required for cancer growth and progression and thereby prevent the survival, growth and spread of cancer cells. From a scientific point of view, specific alterations distinguishing cancer cells from normal cells may be referred to as molecular targets, and therapies that interfere with them are called targeted drugs or targeted therapies. Unlike conventional cancer therapies, targeted cancer therapies are designed to address specific molecular alterations harbored within a particular cancer. Targeted cancer therapies that have been approved for use in specific cancers include drugs that promote cancer cell death by interfering with cellular survival signaling, as well as specific targets responsible for maintaining supportive tumor micro-environment. The latter may be referred to as re-education of the micro-environment and can comprise attempts to prevent formation of tumor vasculature and stimulation of the immune system to attack and destroy cancer cells. Unlike traditional cytotoxic chemotherapies that have frequently entered the clinic without exact knowledge of their mechanism of action and have been discovered and developed on the basis of their ability to interfere with cell division, molecularly designed drugs, on the other hand, have been developed based on a particular known molecular target. Thus, a primary goal of molecularly targeted cancer therapies is to fight cancer with more precision than traditional chemotherapy regimens and hopefully with the additional benefit of less adverse side effects for the patient in treatment. It should be noted, however, that side effects of some molecularly targeted US Food and Drug Administration (FDA) approved drugs are not insignificant, and neither are the treatment costs, highlighting the importance of identifying reliable biomarkers to predict which patients will benefit from a particular treatment. Of note, most targeted therapy approaches are currently being tested within standard-of-care treatment protocols and will thus primarily function as a supplement to available treatment regimens. In addition, by combining molecularly targeted therapies with current standard-of-care, it is suggested that the efficacy of available treatment regimens will improve and may further reduce the risk of relapse and metastatic disease. Thus, targeted cancer therapy is suggested to be important in the management of cancers that are currently considered incurable, in a strategy where the aim is to keep the cancer under control and increase progression-free survival for prolonged periods of time, or even for the life-time of the patient.
Parameters such as histological grade and gene expression profiles may provide moderate statistical correlates to outcome, but do not define biological targets for therapeutic intervention. To make an analogy to clothes manufacturing; everybody spots the difference between having a full tailor-made suit or dress versus simply cutting a few centimetres of the length of some readymade garment to make it fit your length [4]. In parallel, targeted or tailor-made therapy should not be confused with implementing simple prognostic or predictive factors. These parameters, in general, do not define direct biological targets, but rather biological parameters revealing a variable statistical correlation to outcome [4]. Taking the analogy of clothes manufacturing, the definition of tailored therapy should implement targeted therapies based on identification of individual therapeutic targets such as HER2 [5] in the tumour tissue, providing a target exclusive to this tumor for therapeutic attack [6]. Thus, targeted therapy requires exact knowledge of a particular cancer and further empirical knowledge of predictive biomarkers.
In theory, a similar discussion should be applied not only to ‘targeted’ therapy, but also to anticancer strategies in general, including options like cytotoxic therapy. Some cytotoxic anticancer agents could be considered ‘targeted therapy’. This is perhaps best exemplified by the class of drugs named anthracyclines (or anthracycline antibiotics), which are amongst the most effective anticancer treatments ever developed. Anthracyclines are currently used in the treatment of a wide range of both solid and haematopoietic cancers. Similar to several other frequently used chemothera peutic agents; anthracyclines exert their cytotoxic effects by several mechanisms, one of which is inhibition of the topoisomerase II enzyme, thus blocking DNA transcription and replication. A parameter like topoisomerase II may be considered a therapeutic target per se. The fact that this enzyme is a direct target of anthracyclines, and amplification of its gene has been related to improved sensitivity to anthracycline therapy, suggests anthracycline-based chemotherapy to be a candidate tailored therapy for topoisomerase-II-amplified tumors [4]. It is, however, important to notice that topoisomerase II over-expression is not mandatory for anthracycline response, and evidence regarding its predictive role remains conflicting [7]. Thus, in this context, the use of antharcyclines in cancer therapy is not considered ‘targeted cancer therapy’.
The vast majority of current targeted therapies are either monoclonal antibodies or small-molecule drugs. Most monoclonal antibodies cannot penetrate the cell's plasma membrane and are directed against extracellular targets, like the ligand-binding sites of receptors or the ligand itself. Small-molecule drugs are typically able to diffuse into cells and thus have the advantageous ability to act on targets localized inside the cell.
For the purpose of therapy, the monoclonal antibodies were primarily designed to target the ligand-binding domain at a membrane-bound receptor molecule, thus acting as an antagonist, preventing the ligand/receptor interaction, as well as the activation of its downstream signalling cascade. There are currently 14 antibodies approved by the FDA for use in clinical oncology, eight of which are indicated for solid tumours, and six for haematopoietic cancers [8]. As the first therapeutic antibody, Rituximab (Rituxan®/Mabthera®; Novartis: Basel, Switzerland) targeting the B-lymphocyte antigen, CD20, was approved by FDA in 1997 for the use in patients suffering from non-Hodgkin's lymphoma and chronic lymphocytic leukaemia (CLL). Antibodies approved for treatment of haematological cancers and B-cell-associated targets (CD20, CD52) act mainly by inducing apoptosis in the target cell upon binding [9],[10]. More recent antibody-based therapeutic approaches propose the use of monoclonal antibodies as promising vehicles for the targeted delivery of potent chemotherapeutic agents as well as powerful tools to manipulate anticancer immune responses.
Small molecules are, as the name implies, defined and restricted by their size. By definition, small molecules cannot exceed a mass of 900 Dalton. However, a maximum size of 500 Dalton is recommended for small-molecule drug development, due to the improved ability of smaller molecules to penetrate the plasma membrane. From a pharmacological point of view, the term ‘small molecule’ is often restricted to a compound that acts as effector by binding to a specific macromolecule and altering its activity and function. A macromolecule may be a specific protein or nucleic acid. Small-molecule inhibitors (SMIs) can be designed to inhibit a specific function of a multi-functional protein or impede protein–protein interactions. As will be discussed later in this work, the development of small-molecule inhibitors for clinical oncology was pioneered by the first attempts to develop antihormonal drugs for breast cancer.
The generic name of a targeted agent provides clues to the type of agent and its cellular target. Briefly, monoclonal antibodies (mAbs) are denoted with the suffix ‘-mab’, while small-molecule inhibitors end with ‘-ib’. In addition, the specification of the mAbs is included in the name, designated ‘-ximab’ for chimeric human-mouse antibodies or ‘-zumab’ for humanized mAbs and ‘-mumab’ for fully human antibodies. Furthermore, depending on whether the target is circulatory or a tumor target, the substems ‘-ci’ or ‘-tu’ are utilized for mAbs, whereas for small-molecule inhibitors, the stem ‘-tin’ is used for tyrosine kinase inhibitors, and ‘-zom’ is used for proteasome inhibitors. Examples of the naming of targeted therapies are hence cetuximab, where the tyrosine kinase is the target, and the mAb is a human–mouse chimera. Imatinib Mesylate (Gleevec®; Novartis) is an example of a small-molecule inhibitor in which a tyrosine kinase is the target. In this MiniReview, generic names for each drug are given, with brand name and pharmaceutical company in brackets. A complete overview of targeted drugs in clinical trials can be found at: http://www.clinicaltrials.com.
Targeted Therapy in Clinical Practice Directed Against Cancer Cells
Antihormonal therapy
The first established tailored therapy implemented in oncological practice is the antihormonal therapy in breast cancer. Antihormonal therapy is still one of the most widely used targeted therapies, as two of three of breast cancers in post-menopausal women are oestrogen receptor (ER) positive. Antihormonal therapy works by depriving tumour cells of ligand ER activation either by use of anti-oestrogens or through suppression of oestrogen synthesis by aromatase inhibitors, such as anastrozole (Arimidex®; AstraZeneca: Wilmington, DE, USA) [4]. The widely used Tamoxifen (discovered by AstraZeneca) belongs to the collection of drugs termed selective estrogen receptor modulators (SERMs), which is a group of small-molecule inhibitors with strong binding affinity for the oestrogen receptor, thus acting as oestrogen competitors at the binding site at the oestrogen receptor antagonizing the effect of oestrogen. The development of antihormonal therapy for breast cancer has been reviewed extensively elsewhere [11].
Tyrosine kinase inhibitors
Tyrosine kinases (TKs) are a group of enzymes responsible for the activation of multiple downstream proteins in cellular signal transduction cascades. Activation is presented by protein phosphorylation. Tyrosine kinase inhibitors (TKIs) are designed to inhibit the catalytic activity occurring during phosphorylation of the TKs. Depending on the TK of interest and the site of target, TKIs may be designed to compete with binding affinity of the co-enzyme energy carrier molecule ATP or the enzyme substrate (or both), or by allosteric inhibition resulting in unfavourable conformational changes, resulting in blocked or reduced enzymatic activity.
TKs are frequently deregulated in cancer, and due to their importance in maintaining survival signaling and proliferation of cancer cells, they represent attractive targets for cancer therapy. However, protein phosphorylations by TKs are important regulators of cellular communication within all cells, cellular proliferation and maintenance of both normal and stem cell populations. Thus, targeting TKs by TKIs may also affect non-canceros cell populations, which in turn may result in adverse side effects. Although TKIs overall appear to be a well-tolerated drug class, most TKIs are associated with short-term haematological side effects like anaemia, thrombopenia and neutropenia. The most common short-term extra-haematologic adverse effects are oedema, nausea, hypothyroidism, vomiting and diarrhoea. Regarding possible long-term effects, use of TKI has been associated with cardiotoxicity [12].
Targeting the Bcr-Abl fusion protein
A unique example of a target expressed solely by cancer cells was provided in 1960 by the discovery of the Philadelphia chromosome in chronic myeloid leukaemia (CML). This was the very first specific chromosomal alteration described in cancer [13]. Later, it was discovered that most cases of CML are caused by the expression of the fusion protein Bcr-Abl, encoded by the fusion gene named BCR-ABL [14]. This fusion gene is formed when pieces of chromosome 9 (BCR) and chromosome 22 (ABL) break off and trade places. The resulting fused chromosome is referred to as the Philadelphia chromosome. The TK protein normally produced by the ABL gene is a signaling molecule that plays an important role in controlling cell proliferation and predominantly interacts tightly with other signaling molecules in order to be activated. However, Abl signaling is always active when expressed in the form of the fusion protein Bcr-Abl. Thus, the constitutive activity of Bcr-Abl promotes continuous proliferation of CML cells, and CML may thus be considered an oncogene addictive cancer. As the protein Bcr-Abl is expressed solely by cancer cells, it represents an ideal target for TKIs. Imatinib Mesylate (Gleevec®; Novartis) was originally designed for inhibition of PDGFR but was later identified as an efficient inhibitor of the Abl kinase domain [15]. The introduction of a molecularly designed drug that efficiently targeted Bcr-Abl represented a shift in paradigm in the field of clinical oncology. As for several other targeted drugs, Imatinib was initially launched for a narrow indication, but its indications were later expanded to include other malignancies. While Imatinib has been shown to significantly limit tumour growth in CML, resistant clones, most often caused by point mutations in the kinase domain of Bcr-Abl, still represent a therapeutic challenge [16]. In treatment of CML, Imatinib was initially replaced by more potent second-generation drugs, Dasatinib (Sprycel®; Bristol-Myerz Squibb; New York City, NY, USA) and Nilotinib (Tasigna®; Novartis), that have proven effective for patients who have relapsed or are refractory to treatment with Imatinib. However, patients carrying the mutation in the Abl kinase domain T315I are resistant to both Dasatinib and Nilotinib. A third-generation drug, Ponatinib (Iclusig®; ARIAD Pharmaceuticals: Cambridge, MA, USA), was shown to overcome the resistance due to the T315I mutation in CML patients and had an accelerated FDA approval. However, severe side effects observed in clinical trials led to the withdrawal of Ponatinib from the US market. The history of Ponatinib highlights the importance of implementing clinical evaluation and reliable biomarkers to predict which patients have an elevated risk of adverse side effects.
Targeting the epidermal growth factor receptor (EGFR)
The receptor tyrosine kinase (RTK) epidermal growth factor receptor (EGFR) is known to be deregulated and represents a key player in oncogenesis in several solid cancers including non-small-cell lung carcinoma (NSCLC), head and neck cancer, colon cancer and glioblastoma (GMB). EGFR is aberrantly activated by various mechanisms including amplifications and mutations, resulting in over-expression, decreased internalization and constitutive activation [17]. Although not unique to cancer cells, EGFR has long been an attractive molecular target for cancer therapy. Attempts to target EGFR have been explored by both monoclonal antibodies, as well as small-molecule inhibitors. Unfortunately, the clinical efficacy of EGFR-targeted therapy for some cancer types like GBM, both in the case of antibody therapy and small-molecule inhibitors, has shown variable levels of success and has not met its expectations. While Cetuximab (Erbitux®; Bristol Myers Squibb) as an adjuvant therapy in head and neck cancer has significantly improved both progression-free survival and overall survival, the majority of clinical trials implementing anti-EGFR treatment in other solid cancers have failed to induce prolonged survival [18,19]. In many cases, a temporary response to EGFR inhibitory treatment may be observed. However, rapid tumor re-growth and resistance to therapy are frequently seen [20]. Several explanations have been postulated to explain the lack of response and resistance to EGFR-targeted therapy, including mutations in the binding sites of the targeted drug, as well as secondary effector mutations affecting downstream signaling molecules in the EGFR-activated pathway. Secondary mutations in EGFR (T790M) or up-regulation of the MET kinase are found in over 50% of resistant tumors [21]. Moreover, in some cancers with de-regulated EGFR, the fraction of cells harboring alterations in the EGFR regulations varies considerably, and this heterogeneity may further explain the modest efficacy of anti-EGFR treatment. Importantly, for EGFR-targeted therapy, EGFR itself may be misleading as a biomarker for stratification to EGFR-targeted therapy, as it often fails to predict therapeutic response [18,22]. An excellent example is provided in the case of metastatic colon cancers, where the downstream effectors of EGFR, such as the mutation status of KRAS, have been implemented as a predictive biomarker for selection of patients to EGFR-targeted therapies [23]. Panitumumab (Vectibix®; Amgen: Thousand Oaks, CA, USA) was approved by the FDA in 2006 and was the first antibody directed against EGFR to demonstrate the use of KRAS as a predictive biomarker. Later, in 2009, Cetuximab was approved by the FDA for the use in KRAS wild-type, EGFR-expressing, metastatic colorectal cancer in combination with Irinotecan (topoisomerase I inhibitor)-based chemotherapy regimens [24]. In addition to KRAS mutations, which accounts for 35–40% of resistant cases, also NRAS mutations, B-RAF mutations [25], mutations in PI3KCA [26] and inactivation of PTEN [27] have been shown to contribute to anti-EGFR resistance in several cancers. Implementation of these and other predictive biomarkers in clinical trials validating anti-EGFR treatment may significantly improve the clinical trial design. Interestingly, the presence of the RTK AXL has been shown to limit the response of EGFR-targeted inhibitors in NSCLC [28] and ErbB2/Her-2 inhibitors in triple-negative breast cancer cells [29], thus identifying AXL as a promising therapeutic target for overcoming resistance to TKIs targeting members of the EGFR family of TKs. The identification of potential biomarkers and resistance mechanisms may further aid in the development of new potential targets for therapy, as seen in the case of NSCLC, where AXL were found to be over-expressed in the therapy-resistant clones. Since this breakthrough discovery, AXL was further suggested as an applicable target [28], and the first-in-class AXL inhibitor BGB324 is now entering clinical trials [30].
Targeting HER-2
In the case of solid cancers, the first molecularly targeted antibodies directed at epitopes of the cancer cells themselves were anti-EGFR (ErbB1) and anti-HER2 (ErbB2), transmembrane RTKs. Contrasting the partially unmet potential of EGFR-targeted therapies, HER-2 targeted therapies have revolutionized the treatment of the 15–30% of breast cancers harboring amplification of the ERBB2 gene or over-expression of the HER-2 protein [31]. In the era prior to HER-2-targeted therapies, ERBB2-amplified breast cancers were associated with a particularly poor prognosis. Trastuzumab (Herceptin®; Genentech: San Fransisco, CA, USA), a humanized mAb interfering with the HER2/Neu receptor, was approved by the FDA in 1998 and has since then had a major impact on the treatment of HER2-positive breast cancers as an adjuvant therapy. Early stages of HER-2-positive breast cancers (stage 0–3) respond well to Trastuzumab, and importantly, 50% of these cases do not develop recurrent tumors. Furthermore, in a phase-III study of HER2-positive stage 4 (metastatic) breast cancers, Trastuzumab increased the overall survival (OS) from 20 to 25 months [32]. However, in the case of metastatic breast cancer, innate resistance or acquired resistance, which does eventually occur in virtually all cases, points at a major challenge common to all kinds of oncology treatment. Recently, however, a differently modified version of trastuzumab, referred to as trastuzumab emtansine (T-DM1, Kadkyla®; Genentech), has been approved as targeted therapy for HER2-positive metastatic breast cancers [33]. T-DM1 qualifies as an antibody-drug conjugate (ADC) as it is a covalently modified cytotoxic microtubule antagonist and thus represents a form of targeted chemotherapy. Recent and ongoing phase III clinical trials including T-DM1 present significantly prolonged PFS and OS [34]. Importantly, these results show for the first time that an antibody conjugate alone can be more effective than standard chemotherapy with Doxetaxel plus Trastusumab as a first-line treatment for metastatic HER-2-positive breast cancer [34]. In addition to a higher tumor response rate, there was significantly better disease control as well as significantly fewer adverse side effects. Based on the recent success accomplished with T-DM1, multiple ADCs are currently in clinical development.
Targeted Therapy Directed at the Micro-environment
During the past few years, it has become increasingly evident to acknowledge the importance of the stromal contribution to cancer initiation and progression. The complex intercommunication between tumor and micro-environment is obvious in all stages of cancer and may represent a unique niche in which novel molecular markers and targets for therapy have yet to be identified. Several studies have revealed that the micro-environment is capable of normalizing tumor cells, suggesting that re-education of the stromal compartment, rather than targeted ablation per se, may be an effective strategy for treating cancer [35]. Furthermore, including direct intracellular changes, the initiation and progression of a tumor are dependent on intercellular contact, both with neighbouring cancer cells and normal cells composing the micro-environment. Components of tumor stroma include cancer-associated fibroblasts, various cells of the immune system and vasculature. It has been postulated that mechanisms of therapy resistance are conferred primarily by alterations in the tumor micro-environment [36]. The most prominent component of the tumor stroma is perhaps the vasculature. In 2004, Bevacizumab (Avastin®; Genentech/Roche), the only FDA-approved therapeutic antibody directed against the secreted vascular endothelial growth factor (VEGF), was approved by the FDA for use in several different cancers. VEGF is considered as a key pro-angiogenic factor, required for the sprouting of blood vessels, a physiological process termed angiogenesis. Angiogenesis is a hallmark of cancer [1], frequently associated with a poor prognosis, and thus, targeting the formation of tumor vasculature represents an attractive area for targeted cancer therapy. Unfortunately, in clinical trials, the treatment efficacy of Bevacizumab has been rather disappointing. However, as seen in the case of metastatic colorectal cancer, Bevacizumab provided a 2.5-month survival advantage when used in combination with the chemotherapy regimen FOLFOX4 (Oxaliplatin (Elxatin®; Sanofi-Aventis: Bridgewater, NJ, USA) plus Leucovorin plus 5-fluorouracil) [37]. Bevacizumab has further been implemented in the treatment of recurrent GBM, a highly angiogenic grade-IV brain tumour, where it has been shown to reduce oedema and relieve intracranial pressure. However, it has not been associated with an increase in OS. Two recent reports in NEJM, the AVAglia report [38] and RTOG 0825 [39] trials, address the clinical benefit of adding Bevacizumab to the best standard treatment for newly diagnosed GBM. The trials both show prolongation of PFS, but did, however, not translate into an increase in OS [38]. Although numerous attempts have been made to identify reliable predictive biomarkers for Bevacizumab, so far no reliable and reproducible biomarker have been identified [40] (fig.1).
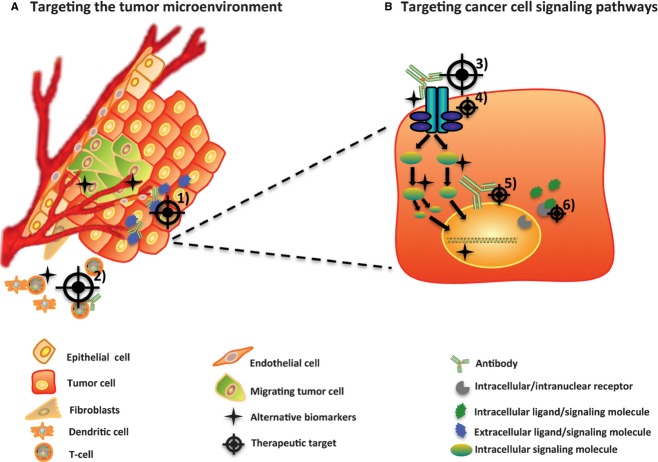
Fig. 1.
Targeted cancer therapies directed at targets within the cancer cells or micro-environmental targets. Targeted therapy is currently focused on (A) tumor micro-environmental targets such as (1) angiogenic blood vessels (e.g. Bevacuzimab) and (2) re-education of the immune system (e.g. Ipilimumab), or (B) targets on the cancer cells themselves. Targeting cancer cell signaling pathways include (3) targeting extracellular domains of transmembrane receptors (e.g. Cetuximab and Trastuzumab) or (4) intracellular targets, such as TK domains and downstream signaling (e.g. Imatinib). Furthermore, intracellular targets also comprise nuclear signaling (5 and 6; e.g. SERMs). As highlighted in this figure, alternative biomarkers may either be expressed by cells composing tumor micro-environment or by the tumor cells, and importantly, cancer biomarkers may differ from the target itself.
As for the tumor micro-environment at large, the process of vascularization is complex, and a panel of selected drugs may be needed in order to efficiently inhibit formation of new blood vessels supplying nutrients and oxygen to the growing tumors. Alternative therapeutic approaches for targeting tumor vascularization includes blocking of the pro-angiogenic receptors including VEGFRs and PDGFR (Sorafinib Tosylate (Nexavar®; Bayer and Onyx Pharmaceuticals: San Fransisco, CA, USA), Sunitinib Malate (Sutent®; Pfizer: New York City, NY, USA) – both of which are approved by the FDA for various cancers in specific therapeutic windows. As in the case of EGFR, the process of vascularization is not unique for tumorigenesis, and the risk of severe adverse events including serious soft-tissue and vascular toxicities may present [41]. However, while the anti-angiogenic strategies have not fulfilled their expectations, preclinical and initial clinical evidence reveals that normalization of the abnormal tumor vasculature is emerging as a complementary therapeutic paradigm, as it may recreate a normal blood flow and thus increase the efficacy of other radiotherapy and systemically delivered anticancer treatments [42]. Tumor micro-environment is a field of intense study, and the importance of inflammation in tumor initiation and progression has been highlighted in recent studies. In the cases of chronic inflammatory diseases, multiple targeted agents inhibiting pro-inflammatory cytokines are currently approved for clinical administration. Importantly, numerous pro-inflammatory cytokines have also been verified as potent pro-angiogenic factors, suggesting promising therapeutic interventions by implementing targeted anti-inflammatory agents in existing cancer treatment regimens [43,44]. Currently, several drugs inhibiting integrin activation are implemented in clinical trials and may in addition improve anti-angiogenic, as well as anti-invasion and antimigratory therapy [45]. As previously mentioned, activation of the AXL kinase has shown to be a predictive biomarker for anti-EGFR therapy resistance [46]. AXL is associated with poor prognosis for many kinds of cancers, including breast, NSCLC and GBMs, mediating the event of epithelial to mesenchymal transition, stemness, as well as survival, cell proliferation and chemo resistance. Interestingly, recent studies have provided evidence indicating that the ligand for AXL, growth arrest-specific gene 6 (GAS6), is mainly produced by bone-marrow-derived stem cells (BDSC) [47]. This paracrine axis between AXL-GAS6 highlights the importance of intercellular communication and regulated connection between cancer cells and the surrounding micro-environment. Furthermore, AXL-targeted drugs have been shown to improve the anti-EGFR drug sensitization and may thus represent another promising candidate for personalized medicine [47].
Therapies Activating Immune Response
The theory of immune surveillance was first conceptualized in the early 1900s by Paul Erlich. He suggested that cancer cells frequently arise in the body, but are recognized as foreign and efficiently eliminated by the immune system. Some 50 years later, Burnet and Thomas suggested that the cell-mediated branch of the immune system had evolved to patrol the body and eliminate cancer cells. According to these concepts, cancers arise only if the cancer cells are able to escape immune surveillance, either by reducing their expression of tumor antigen or by impairment in the immune response to these cells, or both. However, despite the theory proposed by Burnet and Thomas concerning immune surveillance, the significance of the immune system in patrolling the body and preventing cancer initiation and progression remained controversial, until the important work by Schreiber and colleagues in 2011 [48], demonstrating that the immune system can, and often does prevent tumors from developing, and thus that the immune system plays an important role in protecting the organism against the development and progression of cancer [49]. This work established that tumour cells are indeed recognized and destroyed through immune surveillance. However, some tumor cells may evolve to avoid recognition by the immune system, through a process referred to as immune editing [48]. Clinically, it is known that tumors can remain dormant and asymptomatic in patients for years or even decades before they become clinically apparent and that patients appearing to be in full remission still may have tumor cells circulating in their blood, which may cause cancer relapse. Thus, a promising therapeutic strategy like re-activating the immune system to target the cancer cells may synergize with standard-of-care therapy to keep tumors in dormancy for prolonged periods of time to prevent relapse and metastatic spread of tumors [35]. The very idea behind cancer immunotherapy is to induce a targeted immune response against cancer cells [50]. Evidence is now accumulating on the issue of various effects of immune cells infiltrating cancers, and infiltration of T cells is for many cancers the most prominent prognostic factor, and thus, the mechanisms underlying differences in immune status amongst cancer patients are currently being investigated [51]. The first approved immune checkpoint blockade therapy was the CTLA4 blocking antibody Ipilimumab (Yervoy®; Bristol-Myers Squibb). The FDA approval of Ipilimumab in 2011 represents a breakthrough in the field of immunotherapy, as well as a new therapeutic paradigm for cancer. Actually, the study by Hodi and coworkers was the first report in 10 years to show improved survival in patients with malignant melanoma [52]. Elevated expression of the cytotoxic T-lymphocyte-associated antigen 4 (CTLA4) in cancer cells suppresses the immune system by down-regulating the amplitude of T-cell activation [52]. By targeting the immunological synapse immune checkpoint CTLA4, Ipilimumab allows for a re-activation of the patient's own immune system to fight cancer cells. Although Ipilimumab induces long-lasting clinical responses in about 10% of melanoma patients, it is associated with severe side effects, again highlighting the need for reliable predictive biomarkers.
New Development of Targeted Cancer Therapy
From a researcher point of view, the century of 2000 was initiated with the access to the unique data set produced in the HGP, allowing complete insight into the sequence of every human gene, acknowledging, for the first time, the complete genetic blueprint of the human body. This project has had great implications in multiple aspects of research, ranging from evolution to molecular medicine. In 2003, the project was completed, and the enormous data set obtained from HCP is available online and represents a unique tool for characterization of the genomic changes occurring in the initiation and progression of cancer. In the wake of the HGP, efforts of the Cancer Genome Association (TCGA; http://cancergenome.nih.gov/) and ICGG (http://icgc.org/) provide complete characterization of cancer genomes. TCGA is a consortium initiated in 2005 aiming to describe and catalogue genetic changes in a panel of different cancers by combining high throughput techniques and bioinformatic tools. TCGA is supervised by a cooperation of the National Cancer Institute and the National Human Genome Research Institute, funded by the US government. In 2006, TCGA initiated a 3-year pilot project with the aim of characterizing three types of cancers, namely GBM, NSCLC and ovarian cancer, by the use of high throughput techniques including gene expression profiling, copy number variation profiling, small nucleotide polymorphisms (SNP) genotyping, genome-wide DNA methylation profiling, microRNA profiling and exon sequencing. The consortium has now revealed detailed genomic signatures of numerous cancer types [2]. Clearly, various research groups had already established many of the genomic alterations identified by the TCGA. However, as well as providing statistically relevant information of established genetic alterations in cancers, the TCGA share detailed information of additional genetic changes that can be used by cancer researchers throughout the world. Complementing the data sets obtained on a panel of different cancer types, the TCGA has published a unique data set in 2013, referred to as the pan-cancer initiative analysis project where they compare the first 12 tumor types that were profiled. Primary cancers included in the pan-cancer initiative were GBM, acute myeloid leukaemia (AML), head and neck cancer, squamous cell cancer, as well as breast-, lung-, kidney-, ovarian-, bladder-, colon-, rectal-, cervical- and endometrial adenocarcinomas. Undoubtedly, these findings will be of great importance in the search for finding common genetic signatures across cancer types [2]. Hopes are high that the massive parallel sequencing efforts may provide clues on how to overcome resistance to targeted therapies across different cancer types. If, or when, deep sequencing technologies are implemented in the clinic, it may be possible to predict whether a particular cancer will respond to a given treatment and thus allow the oncologist to tailor-make the treatment to the individual cancer patient. A key point is that exomic sequencing needs to be conducted together with analysis of gene expression and transcriptomic alterations by, for example, RNA sequencing, to detect whether the genomic alterations predicted by DNA sequencing are actually expressed. Furthermore, as cancer cells frequently exhibit genomic instability and epigenetic changes such as aberrant methylation and altered transcription factor binding, mapping of epigenetic alterations and analysis of gross chromosomal rearrangements should also be included in a complete cancer genomics analysis. Of particular importance is the establishment of comprehensive bio-banks containing not only tumor tissue, but in addition, information regarding how each particular tumor responds to defined treatment regimens, whether the tumor gave rise to metastases and information regarding time to progression and survival. By comparing biological information with clinical outcome, we enable identification of common denominators responsible for specific malignant phenotypes. The clinically important issue of drug resistance may be best studied in clinically relevant tumor models and further in prospectively designed clinical trials. With regard to the clinical challenge of metastasis, tumor bio-banks should further be expanded to include analyses of not only primary tumors but also metastatic/recurrent lesions or circulating tumor cells (CTC) and disseminated tumor cells (DTC). The focus of CTC, DTC or metastatic lesions is particularly important as we know that it is the metastatic spread of a small population of tumor cells that eventually may compromise a normal tissue or organ and are responsible for most cancer-related deaths.
Recent advances in sequencing technology have overcome the limitations of cost and scale, and thousands of somatic mutations can now be identified in a single cancer sample even when several mutational processes are operative [2]. Massive parallel sequencing combined with various bioinformatic algorithms can aid in further exploration of the landscape of mutational signatures and possibly also to further distinguish the ‘driver’ mutations from the ‘passenger’ mutations [2]. This can be established by means of molecular sequencing of DNA from tumor biopsy specimens, determining which genetic or epigenetic alterations have developed at the time of clinical appearance or the time of evidence of drug resistance. Furthermore, multiple fusion genes that may represent new therapeutic targets have been discovered in various cancers as a consequence of the major advances and availability of high throughput techniques such as whole-genome sequencing. However, interpretation and implementation of the findings from the sequencing consortia will require comprehensive multi-disciplinary efforts for translation into the clinic.
Present Challenges for Translating Promising Targets to Successful Cancer Treatment
Comprehensive characterization of multiple tumor specimens obtained from the same patient illustrates that the remarkable intratumor heterogeneity might exist between distinct geographical regions within the same tumor, as well as between the primary tumor and subsequent local or distant recurrence in the same patient. Indeed, inter- and intratumor heterogeneity poses a significant challenge to personalized cancer medicine [53]. As previously discussed, intracellular crosstalk between signaling pathways as seen with the RTKs AXL and EGFR, dynamic signaling interactions between tumor cells and stromal cells represent sophisticated mechanisms of drug resistance [54]. Another mechanism of resistance may be seen in the situation of intercellular crosstalk and paracrine signaling. Furthermore, as seen in the situation of EGFRvIII, a tumor-specific EGFR mutant has been shown to increase the tumorigenecity of its neighbouring tumor cells by stimulating the secretion of the pro-tumorigenic interleukins IL-6 and LIF [55]. To overcome the issue of resistance, the challenge is to design an optimal combination of targeted drugs. Moreover, defining optimal therapeutic windows may be critical. It has been hypothesized that various and specific niches within the tumor, as well as in the surrounding tissue, contribute to maintain and enhance the plasticity of cancer cells. In this context, it should be noted that the efficacy of a particular targeted drug in the clinical setting is not solely dependent on the specific genetic make-up of the tumor cell population, but also dependent on micro-environmental ability to sustain the different tumor cell populations. This heterogeneity may also be subject to selection and evolution of the developing tumor upon treatment.
The key for unravelling the code for cancer development may not be achieved solely by detailed sequencing of the cancer genome. Pinpointing genetic alterations and ‘driver’ mutations may aid in defining the cancer cells, but in order to truly understand the initiation and development of cancer, as well as mechanisms involved in therapy resistance, we need to focus on the interactions between the cancer cells and the tumor micro-environment. To unravel the functional consequences of intercellular communication and specific mutational signatures on cellular signaling pathways in cancer, it is crucial to develop model systems that truly recapitulate the disease. Accurately, modelling tumor complexity and heterogeneity poses a significant challenge. Moreover, further development of reproducible, yet complex and representative tumor models both in vitro and in vivo is urgently needed.
Relevant Biomarkers in Targeted Cancer Therapy
Due to the significant cost and possible side effects of targeted drugs, a new drug should ideally not be introduced to the market without a reliable biomarker to predict which patients will benefit from the treatment and which patients are at particularly high risk of experiencing serious side effects of the drug. The challenge may be to determine the right combination of drugs or lines of therapy for different subtypes of cancer. In this respect, and as previously mentioned, the data showing that patients with colorectal cancer with KRAS mutations do not benefit from treatment with Cetuximab, are setting the stage for increased use of molecular diagnostics that could guide oncologists to select the most effective treatment options for their patients.
Importantly, the currently used models of clinical trials are in most cases not suitable for evaluation of therapy tailored to treat a single cancer patient or smaller groups of cancer patients harbouring cancers with particular genetic alterations in common. Optimal trial design is a pre-requisite for efficient translation of novel drugs and represents a major challenge to medical oncology, clinical trials and modern health care in general. The success of combining a targeted drug with chemotherapy has led to the hypothesis that efficacy could be enhanced by adding several targeted agents [56], particularly with regard to fight the mechanisms of treatment resistance. Implementation of relevant biomarkers may assist in overcoming the challenges involved in optimal design of clinical trials, optimization of drug combinations, timing and dose regimens, and may thus be an important step towards the goal of implementation of novel, targeted treatment regimens in the clinic.
Conclusions and Future Perspectives
‘Molecularly targeted cancer therapies have a short but rich history, an exciting present and a promising future’ [8]. Despite the availability of improved drugs and new therapeutic strategies, including targeted cancer therapies, cancer is still one of the leading causes of death worldwide. However, due to novel technology and improved knowledge of the genetic and epigenetic make-up of a particular cancer, cancer treatment may change radically over the next few years. Never the less, the most prominent clinical challenges in cancer treatment, namely metastatic spread and development of therapy-resistant clones, are responsible for most cancer-related deaths, and substantial multi-disciplinary efforts are required to overcome these hurdles.
Looking forward, perhaps the greatest promise comes from the notion that re-educating a dysfunctional tumor micro-environment could yield striking results in cancer control and remission, as evidenced by the accumulating success stories in the cancer immunotherapy field. Moreover, the discovery and implementation of relevant biomarkers may assist substantially in the development of targeted cancer therapy. Although more research is warranted, the results recently achieved by targeted cancer therapy efforts suggest that tailored therapy for most, if not all, cancer patients may become a realistic approach in the near future.
Acknowledgments
We apologize in advance to those authors whose work could not be cited due to reference limitations. We thank our colleagues for interesting and inspiring discussions. Our sincere thanks to the cancer patients who voluntarily enrol in clinical trials and participate in cancer research projects. Without your valuable participation, the recent progress in clinical cancer research and improvement in cancer treatment could never have been realized.
Disclosure Statement
The authors declare no conflict of interest.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–21. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aggarwal S. Targeted cancer therapies. Nat Rev Drug Discov. 2010;9:427–8. doi: 10.1038/nrd3186. [DOI] [PubMed] [Google Scholar]
- 4.Lonning PE. Tailored targeted therapy for all: a realistic and worthwhile objective? Breast Cancer Res. 2009;11(Suppl 3):S7. doi: 10.1186/bcr2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–82. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 6.Romond EH, Perez EA, Bryant J, Suman VJ, Geyer CE, Jr, Davidson NE, et al. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N Engl J Med. 2005;353:1673–84. doi: 10.1056/NEJMoa052122. [DOI] [PubMed] [Google Scholar]
- 7.Tubbs R, Barlow WE, Budd GT, Swain E, Porter P, Gown A, et al. Outcome of patients with early-stage breast cancer treated with doxorubicin-based adjuvant chemotherapy as a function of HER2 and TOP2A status. J Clin Oncol. 2009;27:3881–6. doi: 10.1200/JCO.2008.20.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sliwkowski MX, Mellman I. Antibody therapeutics in cancer. Science. 2013;341:1192–8. doi: 10.1126/science.1241145. [DOI] [PubMed] [Google Scholar]
- 9.Mathas S, Rickers A, Bommert K, Dorken B, Mapara MY. Anti-CD20- and B-cell receptor-mediated apoptosis: evidence for shared intracellular signaling pathways. Cancer Res. 2000;60:7170–6. [PubMed] [Google Scholar]
- 10.McLendon R, Friedman A Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sainsbury R. The development of endocrine therapy for women with breast cancer. Cancer Treat Rev. 2012;39:507–17. doi: 10.1016/j.ctrv.2012.07.006. [DOI] [PubMed] [Google Scholar]
- 12.Rottlaender D, Motloch LJ, Reda S, Larbig R, Hoppe UC. Cardiac arrest due to long QT syndrome associated with excessive consumption of energy drinks. Int J Cardiol. 2011;158:e51–2. doi: 10.1016/j.ijcard.2011.10.017. [DOI] [PubMed] [Google Scholar]
- 13.Nowell PC, Hungerford DA. Chromosome studies on normal and leukemic human leukocytes. J Natl Cancer Inst. 1960;25:85–109. [PubMed] [Google Scholar]
- 14.Kurzrock R, Gutterman JU, Talpaz M. The molecular genetics of Philadelphia chromosome-positive leukemias. N Engl J Med. 1988;319:990–8. doi: 10.1056/NEJM198810133191506. [DOI] [PubMed] [Google Scholar]
- 15.Schindler T, Bornmann W, Pellicena P, Miller WT, Clarkson B, Kuriyan J. Structural mechanism for STI-571 inhibition of abelson tyrosine kinase. Science. 2000;289:1938–42. doi: 10.1126/science.289.5486.1938. [DOI] [PubMed] [Google Scholar]
- 16.Bixby D, Talpaz M. Seeking the causes and solutions to imatinib-resistance in chronic myeloid leukaemia. Leukemia. 2011;25:7–22. doi: 10.1038/leu.2010.238. [DOI] [PubMed] [Google Scholar]
- 17.Jorissen RN, Walker F, Pouliot N, Garrett TP, Ward CW, Burgess AW. Epidermal growth factor receptor: mechanisms of activation and signalling. Exp Cell Res. 2003;284:31–53. doi: 10.1016/s0014-4827(02)00098-8. [DOI] [PubMed] [Google Scholar]
- 18.Cunningham D, Humblet Y, Siena S, Khayat D, Bleiberg H, Santoro A, et al. Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N Engl J Med. 2004;351:337–45. doi: 10.1056/NEJMoa033025. [DOI] [PubMed] [Google Scholar]
- 19.Jonker DJ, O'Callaghan CJ, Karapetis CS, Zalcberg JR, Tu D, Au HJ, et al. Cetuximab for the treatment of colorectal cancer. N Engl J Med. 2007;357:2040–8. doi: 10.1056/NEJMoa071834. [DOI] [PubMed] [Google Scholar]
- 20.Benvenuti S, Sartore-Bianchi A, Di Nicolantonio F, Zanon C, Moroni M, Veronese S, et al. Oncogenic activation of the RAS/RAF signaling pathway impairs the response of metastatic colorectal cancers to anti-epidermal growth factor receptor antibody therapies. Cancer Res. 2007;67:2643–8. doi: 10.1158/0008-5472.CAN-06-4158. [DOI] [PubMed] [Google Scholar]
- 21.Wheeler DL, Huang S, Kruser TJ, Nechrebecki MM, Armstrong EA, Benavente S, et al. Mechanisms of acquired resistance to cetuximab: role of HER (ErbB) family members. Oncogene. 2008;27:3944–56. doi: 10.1038/onc.2008.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chung KY, Shia J, Kemeny NE, Shah M, Schwartz GK, Tse A, et al. Cetuximab shows activity in colorectal cancer patients with tumors that do not express the epidermal growth factor receptor by immunohistochemistry. J Clin Oncol. 2005;23:1803–10. doi: 10.1200/JCO.2005.08.037. [DOI] [PubMed] [Google Scholar]
- 23.Messersmith WA, Ahnen DJ. Targeting EGFR in colorectal cancer. N Engl J Med. 2008;359:1834–6. doi: 10.1056/NEJMe0806778. [DOI] [PubMed] [Google Scholar]
- 24.Van CE, Kohne CH, Hitre E, Zaluski J, Chang CC, Makhson A, et al. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med. 2009;360:1408–17. doi: 10.1056/NEJMoa0805019. [DOI] [PubMed] [Google Scholar]
- 25.Di Nicolantonio F, Martini M, Molinari F, Sartore-Bianchi A, Arena S, Saletti P, et al. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol. 2008;26:5705–12. doi: 10.1200/JCO.2008.18.0786. [DOI] [PubMed] [Google Scholar]
- 26.Perrone F, Lampis A, Orsenigo M, Di Bartolomeo M, Gevorgyan A, Losa M, et al. PI3KCA/PTEN deregulation contributes to impaired responses to cetuximab in metastatic colorectal cancer patients. Ann Oncol. 2009;20:84–90. doi: 10.1093/annonc/mdn541. [DOI] [PubMed] [Google Scholar]
- 27.Frattini M, Saletti P, Romagnani E, Martin V, Molinari F, Ghisletta M, et al. PTEN loss of expression predicts cetuximab efficacy in metastatic colorectal cancer patients. Br J Cancer. 2007;97:1139–45. doi: 10.1038/sj.bjc.6604009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Z, Lee JC, Lin L, Olivas V, Au V, LaFramboise T, et al. Activation of the AXL kinase causes resistance to EGFR-targeted therapy in lung cancer. Nat Genet. 2012;44:852–60. doi: 10.1038/ng.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meyer AS, Miller MA, Gertler FB, Lauffenburger DA. The receptor AXL diversifies EGFR signaling and limits the response to EGFR-targeted inhibitors in triple-negative breast cancer cells. Sci Signal. 2013;6:ra66. doi: 10.1126/scisignal.2004155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sheridan C. First Axl inhibitor enters clinical trials. Nat Biotechnol. 2013;31:775–6. doi: 10.1038/nbt0913-775a. [DOI] [PubMed] [Google Scholar]
- 31.Burstein HJ. The distinctive nature of HER2-positive breast cancers. N Engl J Med. 2005;353:1652–4. doi: 10.1056/NEJMp058197. [DOI] [PubMed] [Google Scholar]
- 32.Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344:783–92. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 33.LoRusso PM, Weiss D, Guardino E, Girish S, Sliwkowski MX. Trastuzumab emtansine: a unique antibody-drug conjugate in development for human epidermal growth factor receptor 2-positive cancer. Clin Cancer Res. 2011;17:6437–47. doi: 10.1158/1078-0432.CCR-11-0762. [DOI] [PubMed] [Google Scholar]
- 34.Verma S, Miles D, Gianni L, Krop IE, Welslau M, Baselga J, et al. Trastuzumab emtansine for HER2-positive advanced breast cancer. N Engl J Med. 2012;367:1783–91. doi: 10.1056/NEJMoa1209124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013;19:1423–37. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meads MB, Gatenby RA, Dalton WS. Environment-mediated drug resistance: a major contributor to minimal residual disease. Nat Rev Cancer. 2009;9:665–74. doi: 10.1038/nrc2714. [DOI] [PubMed] [Google Scholar]
- 37.Kerbel RS. Antiangiogenic therapy: a universal chemosensitization strategy for cancer? Science. 2006;312:1171–5. doi: 10.1126/science.1125950. [DOI] [PubMed] [Google Scholar]
- 38.Chinot OL, Wick W, Mason W, Henriksson R, Saran F, Nishikawa R, et al. Bevacizumab plus radiotherapy-temozolomide for newly diagnosed glioblastoma. N Engl J Med. 2014;370:709–22. doi: 10.1056/NEJMoa1308345. [DOI] [PubMed] [Google Scholar]
- 39.Gilbert MR, Dignam JJ, Armstrong TS, Wefel JS, Blumenthal DT, Vogelbaum MA, et al. A randomized trial of bevacizumab for newly diagnosed glioblastoma. N Engl J Med. 2014;370:699–708. doi: 10.1056/NEJMoa1308573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Junttila MR, de Sauvage FJ. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature. 2013;501:346–54. doi: 10.1038/nature12626. [DOI] [PubMed] [Google Scholar]
- 41.Higa GM, Abraham J. Biological mechanisms of bevacizumab-associated adverse events. Expert Rev Anticancer Ther. 2009;9:999–1007. doi: 10.1586/era.09.68. [DOI] [PubMed] [Google Scholar]
- 42.Wang J, Svendsen A, Kmiecik J, Immervoll H, Skaftnesmo KO, Planaguma J, et al. Targeting the NG2/CSPG4 proteoglycan retards tumour growth and angiogenesis in preclinical models of GBM and melanoma. PLoS ONE. 2011;6:e23062. doi: 10.1371/journal.pone.0023062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454:436–44. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 44.Knowles RG. Development of anti-inflammatory drugs – the research and development process. Basic Clin Pharmacol Toxicol. 2014;114:7–12. doi: 10.1111/bcpt.12130. [DOI] [PubMed] [Google Scholar]
- 45.Chamberlain MC, Cloughsey T, Reardon DA, Wen PY. A novel treatment for glioblastoma: integrin inhibition. Expert Rev Neurother. 2012;12:421–35. doi: 10.1586/ern.11.188. [DOI] [PubMed] [Google Scholar]
- 46.Zang ZJ, Cutcutache I, Poon SL, Zhang SL, McPherson JR, Tao J, et al. Exome sequencing of gastric adenocarcinoma identifies recurrent somatic mutations in cell adhesion and chromatin remodeling genes. Nat Genet. 2012;44:570–4. doi: 10.1038/ng.2246. [DOI] [PubMed] [Google Scholar]
- 47.Ben-Batalla I, Schultze A, Wroblewski M, Erdmann R, Heuser M, Waizenegger JS, et al. Axl, a prognostic and therapeutic target in acute myeloid leukemia mediates paracrine crosstalk of leukemia cells with bone marrow stroma. Blood. 2013;122:2443–52. doi: 10.1182/blood-2013-03-491431. [DOI] [PubMed] [Google Scholar]
- 48.Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity's roles in cancer suppression and promotion. Science. 2011;331:1565–70. doi: 10.1126/science.1203486. [DOI] [PubMed] [Google Scholar]
- 49.Shankaran V, Ikeda H, Bruce AT, White JM, Swanson PE, Old LJ, et al. IFNgamma and lymphocytes prevent primary tumour development and shape tumour immunogenicity. Nature. 2001;410:1107–11. doi: 10.1038/35074122. [DOI] [PubMed] [Google Scholar]
- 50.Banchereau J, Steinman RM. Dendritic cells and the control of immunity. Nature. 1998;392:245–52. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- 51.Fridman WH, Pages F, Sautes-Fridman C, Galon J. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer. 2012;12:298–306. doi: 10.1038/nrc3245. [DOI] [PubMed] [Google Scholar]
- 52.Hodi FS, O'Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363:711–23. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Burrell RA, McGranahan N, Bartek J, Swanton C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature. 2013;501:338–45. doi: 10.1038/nature12625. [DOI] [PubMed] [Google Scholar]
- 54.Rosell R, Bivona TG, Karachaliou N. Genetics and biomarkers in personalisation of lung cancer treatment. Lancet. 2013;382:720–31. doi: 10.1016/S0140-6736(13)61715-8. [DOI] [PubMed] [Google Scholar]
- 55.Inda MM, Bonavia R, Mukasa A, Narita Y, Sah DW, Vandenberg S, et al. Tumor heterogeneity is an active process maintained by a mutant EGFR-induced cytokine circuit in glioblastoma. Genes Dev. 2010;24:1731–45. doi: 10.1101/gad.1890510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sawyers CL. Cancer: mixing cocktails. Nature. 2007;449:993–6. doi: 10.1038/449993a. [DOI] [PubMed] [Google Scholar]