Abstract
We have developed a unique comprehensive mouse radiation hybrid (RH) map of nearly 23,000 markers integrating data from three international genome centers and over 400 independent laboratories. We have cross-referenced this map to the 0.5-cM resolution recombination-based Jackson Laboratory (TJL) backcross panel map, building a complete set of RH framework chromosome maps based on a high density of known-ordered anchor markers. We have systematically typed markers to improve coverage and resolve discrepancies, and have reanalyzed data sets as needed. The cross-linking of the RH and recombination maps has resulted in a highly accurate genome-wide map with consistent marker order. We have compared these linked framework maps to the Ensembl mouse genome sequence assembly, and show that they are a useful medium resolution tool for both validating sequence assembly and elucidating chromosome biology.
[Supplemental material is available online at www.genome.org.]
The field of biomedical research has recently focused intense interest on obtaining the complete sequence of the human genome and those of model organisms. The availability of a complete and accurate genome sequence will engender a quantum leap forward in our ability to efficiently analyze underlying biology for both simple and complex biologic processes. Major genome sequencing efforts have recently produced preliminary assemblies of mammalian genome sequences, either based on hierarchical shotgun approaches (Venter et al. 2001) or whole-genome contig mapping (Gregory et al. 2002) and sequencing (Lander et al. 2001). Human sequence assemblies were announced in 2001 (Lander et al. 2001; Venter et al. 2001), followed closely by mouse genome assemblies from both private (http://www.celera.com) and public (http://www.ncbi.nlm.nih.gov/genome/seq/MmHome.html, http://www.ensembl.org/Mus_musculus/, http://genome.ucsc.edu/) efforts. Comparisons between the resulting independent genome sequences have revealed major differences (Hogenesch et al. 2001; Li et al. 2002) that raise the question of what is the correct sequence order and how will we know when we have it?
Prior to the availability of the sequence assemblies, other means of determining genome marker order have been of great informative value. The first maps were built by analyzing segregation of traits in genetic crosses combined with cytologic studies. High-quality recombination maps can reveal proven marker order up to the resolution limit of detecting a recombination event between markers under study. Because recombination is a function of the intact living genome, recombination-based maps are not subject to the problems of cloning and assembly. Thus, recombination map marker order may be used as a gold standard to measure the accuracy of emerging genome assemblies.
To maximize the usefulness of any measure of sequence accuracy, the standard of comparison itself must be as accurate as possible. Although many recombination-based maps have been generated and used for evaluating newer technologies, most of these maps have themselves been flawed either by experimental limitations as with the human genetic maps, or by error levels inherent in high throughput data gathering, or by the unavailability of the underlying reagents for further refinement of mapping data. When comparisons are made between such recombination maps and any new methodology, it has been difficult to determine how much of the discrepancy was due to problems in the older map or in the newer technology.
For these reasons we have undertaken to produce a highly accurate mouse genome map that can be used to assess the emerging sequence assemblies. We have chosen to use TJL interspecific backcross panels as the source of recombination mapping because these panels are readily available for experimental verification, are already densely typed for markers of all kinds, and provide sufficient resolution to support quality assessment and refinement of newer mapping technologies. In this article we report the completion of a comprehensive mouse radiation hybrid map that has been refined by correlation with this high-quality recombination map, and the results of our preliminary comparison of this framework map to the Ensembl v3 mouse genome sequence assembly.
In recent years new tools have been developed to define the mammalian/mouse genome in increasing detail. The use of interspecific backcross mapping panels (Copeland et al. 1993; European Backcross Collaborative Group 1994; Rowe et al. 1994; Rhodes et al. 1998) has produced genome wide recombination maps. Whole genome radiation hybrid panels, developed initially to assist map building in organisms where there is poor access to high-quality recombination studies (human: Gyapay et al. 1996; Schuler et al. 1996; Stewart et al. 1997; Nagaraja et al. 1998; other animals: cf. Steen et al. 1999; Gellin et al. 2000; Mellersh et al. 2000; Sun et al. 2001), offer increased resolving power, have a more random chromosome breakage distribution, and provide technical advantages to recombination mapping. The mouse T31 RH panel (McCarthy et al. 1997) has allowed the results of RH mapping analysis to be compared directly to the detailed mouse recombination map (Rowe et al. 2000).
Maps of the mouse genome based on radiation hybrid technology have been previously published. Genome Centers at The Whitehead Institute (Cambridge, MA) and The Medical Research Council (Harwell, UK) have jointly published a radiation hybrid map (Hudson et al. 2001, http://www-genome.wi.mit.edu/mouse_rh/index.html, http://websql.har.mrc.ac.uk/mps/maps/0/LOD_7/graphic.html) with 2280 simple sequence length polymorphism (SSLP) anchor markers and 11,109 expressed sequence tag (EST) loci, using RHMAPPER to assemble the data. A third Genome Center at Genoscope (Evry, France) has published an independent RH-based map (Avner et al. 2001, http://www.genoscope.cns.fr/externe/English/Projets/Projet_ZZZ/rhmap.html) from their own data including 1911 SSLP and 5904 EST markers using two different algorithms for map assembly: a multipoint maximum likelihood analysis and a traveling-salesman problem approach. Each Genome Center used a different methodology for data collection: Both PCR cycling conditions and product detection methods were unique to each project. The Whitehead/MRC included only their own data to build the radiation hybrid map, while the Genoscope map used 1066 combined data sets for MIT SSLP markers typed in common between the two Genome Centers as well as their own data for additional SSLPs and their own EST marker set. Both groups used simplifying assumptions to omit lower quality data sets from the map construction. SSLPs used by Genoscope to build the map were chosen to be concordant with the Whitehead (WI-MIT) genetic map. The completed Whitehead/MRC RH map was compared in global ways to the Mouse Genome Database (MGD) composite map position assignments, yielding estimates of map discordance of a few percent.
We have taken a different approach to build on the mouse genome map. We integrated all available data from the T31 mouse RH panel from the three international Genome Centers and from all other published and publicly accessible mapping projects into a single comprehensive RH database, and built the resulting RH map leveraging from the existing high quality mouse recombination maps. We observed that different projects independently mapped some of the same markers, and these duplicate data sets are not fully concordant. We showed that variations in PCR protocols can affect the set of positive cell lines detected for the same marker/primer pair, and that even with duplicate data for each marker, there is an overall error rate of 1%–2% per locus (Rowe et al. 2000). Observations like these led us to develop improved ways to analyze RH data. To eliminate the assumptions used in other software, we constructed our RH map based on maximum LOD/minimum break criteria using Map Manager QT software (Manly and Olson 1999) modified to include RH data analysis algorithms.
In confirming map order, we used recombination maps based on The Jackson Laboratory interspecific backcross panels (Rowe et al. 1994) of 188 N2 animals (http://www.jax.org/resources/documents/cmdata/bkmap/) chosen both for their excellent data coverage and their availability for additional marker map validation. Our map comparisons have allowed us to detect and correct errors of omission or interpretation in both the recombination and the RH maps.
The completed framework map underpins a higher level of accuracy in the comprehensive RH map. With the framework map support, the complete RH map may be used to evaluate the accuracy of the sequence assemblies. Where discordances are apparent, the underlying RH data and sequence data resources can be scrutinized to reveal and resolve order problems. We have made some preliminary comparisons of this map to the Ensembl mouse sequence assembly, and show that the improved genome maps will be of value in vetting the mouse sequence assemblies.
RESULTS
The Comprehensive T31 RH Map
Our complete mouse genome RH map includes to date 3956 SSLP anchor markers, 18,284 EST loci, 115 other sequence tagged site (STS) loci, and 470 named genes (http://www.jax.org/resources/documents/cmdata/rhmap). The autosomal portion of this map contains no gaps in significant linkage at P = 0.001 that cannot be closed by elimination of one or a few poorly matching data sets. The X Chromosome (Chr) has two gaps in linkage support (data not shown, available from http://www.jax.org/resources/documents/cmdata/rhmap/rh.html). At the distal end there is a gap where the pseudoautosomal region fails to link with significance to the rest of the X Chromosome. The pseudoautosomal region is represented by the last three crossovers at the distal end of the recombination map, and has no cross-links to the distal RH map (see Fig. 1 and detailed maps in online supplementary material and poster included with this issue). More centrally there are gaps in the complete RH map database around DXMit149, just distal to the Xist locus. This is a region that contains many gaps in the Ensembl v3 sequence assembly (http://www.ensembl.org/Mus_musculus/), and some segments of the sequence between the gaps are rearranged with respect to the best fit RH and recombination map order. It is likely that there are sequences here that are not readily clonable or contain extensive repeats that make the assembly problematic in this region. The Y Chromosome RH map contains only four EST markers. These markers link to each other with LODs greater than 8.
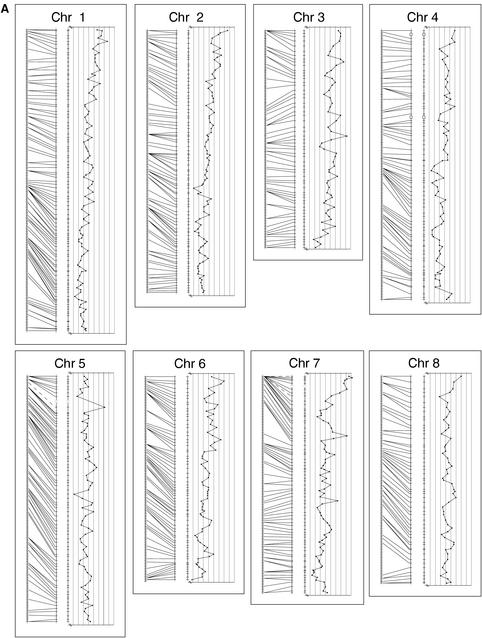
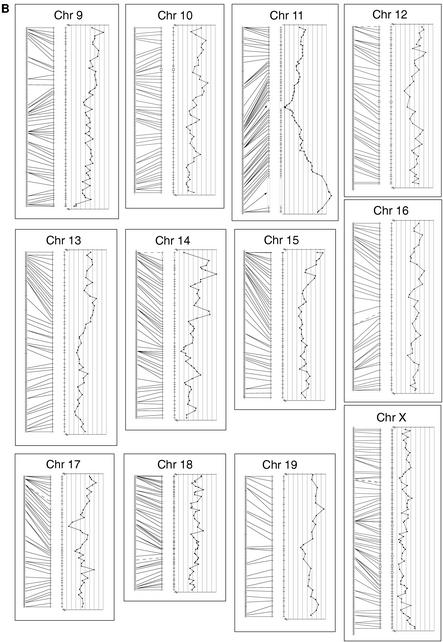
Figure 1.
The mouse recombination map linked to the T31 radiation hybrid map: alignment of mapped framework markers in both TJL interspecific backcross maps and T31radiation hybrid maps. All maps are drawn to a uniform cM scale, and the RH maps are set to equal the length of the corresponding recombination map. Thus, the cR scale for each chromosome may differ. The recombination map from the combined TJL BSB/BSS mapping panels is represented on the left. Each tick mark on the recombination map represents a single crossover event; one crossover/188 animals equals 0.56 cM between crossovers. The RH map in the center of each chromosome panel shows the framework markers spaced by the cR distance between each pair of adjacent framework markers based on the data for those two markers only. Missing scores for all markers are inferred if flanking data are concordant. Lines join the two maps where the same framework marker is mapped in both systems. These lines are dashed when the marker is mapped only in the 94 animals of the BSS cross due to failure of the Mus spretus allele to amplify from heterozygotes. Open boxes over the chromosome line indicate intervals whose LOD fails to meet the LOD > 6 criterion for significant linkage (see text for discussion). To the right of each chromosome framework map is graphed the RH retention frequency for each framework marker against its cR position in the map. Note that all retention graphs show the 15%–55% retention range, with the lower retention to the left, except the Chr 11 graph, which shows the range 0%–100% retention frequency.
Criteria for Framework Marker Assignment
To optimize the use of the recombination data in confirming the final mouse RH map, we selected anchor markers from our comprehensive RH map at a spacing that would give good statistical support of linkage from the RH data and mapped these loci onto TJL (C57BL/6J × M. spretus) × C57BL/6J and (C57BL/6JEi × SPRET/Ei) × SPRET/Ei interspecific backcross panels (hereafter, TJL BSB/BSS) (Fig. 1). This set of markers provides a confirmed framework order on which to base the order of other linked RH data sets. We note that our use of the term “framework marker” is distinct from the statistical construct definition used in RH mapping software like RHMAPPER (Stein et al. 1995). For our purposes, RH framework markers are those that are appropriately spaced to provide good genome coverage and whose local order is both well supported with interlocus LODs > 6 (see below) and confirmed by backcross mapping.
We note in this regard that any inversion differences between the Mus spretus genome and that of C57BL/6J will appear in the interspecific backcross data as regions of nonrecombination, because inversion heterozygotes show recombination suppression. One such inversion has been identified on proximal Chr 17 (Hammer et al. 1989) and includes the framework markers D17Mit246, D17Mit112, D17Mit156, and D17Mit113 that fail to recombine in TJL BSB/BSS. It is possible that an inversion between the parental strains in the proximal part of Chr 7 may explain the large number of loci that cosegregate in the backcrosses but that are resolved in the RH panel covering the proximal 13% of the RH framework map (see Fig. 2). Thus, any gene order determined from these crosses represents the regions of the genome where the two parental genomes are colinear, and the RH data (based on strain 129/SvEv) may be used to determine order for markers that cosegregate in the backcrosses.
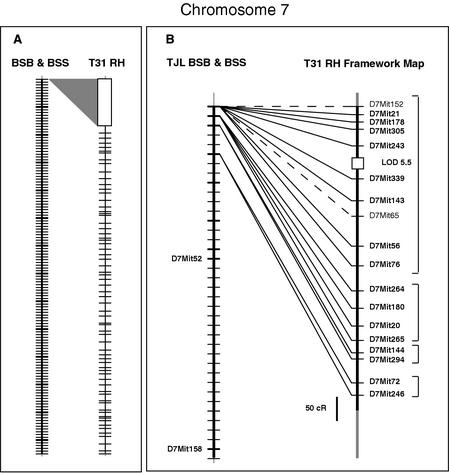
Figure 2.
Chromosome 7 framework map showing suppression of recombination near the centromere. All maps are drawn with the centromere at the top. (A) Shows on the left the entire Chr 7 recombination map from TJL BSB/BSS backcrosses with crosshatches for each crossover and heavy crosshatches every 10 crossovers. A gray triangle shows the proportion of the T31 RH framework map that is nonrecombinant at the proximal end of the backcross map. (B) Shows greater detail of this proximal region of the backcross and RH maps. Heavy crosshatches on TJL BSB/BSS chromosome figure indicate positions of framework markers. Brackets on the right indicate the groups of markers on the RH map that fail to recombine in the backcross. Locus symbols in plain text indicate markers that are mapped only in the 94 animals of the BSS cross due to failure of the Mus spretus allele to amplify in heterozygotes. An interval of LOD less than 6 is indicated with an open box over the RH chromosome line. Note that all locus symbols should be printed in italics, but are shown here in plain text for readability.
Framework Map Features
About one in four of the D-Mit- markers in the comprehensive map of the T31 RH data were used to make the final framework map. The framework map (Fig. 1, and in more detail in supplementary material online) contains 1161 D-Mit- markers linking the backcross and RH maps together at an average of one locus per 1.17 centimorgan (cM) genome-wide. TJL BSB/BSS combined recombination map has a total genome length of 1355.5 cM. The total centiRays3000 (cR) in the T31 RH framework map is 39,410. This number would be higher if a higher density of markers were used to build the map (Rowe et al. 2000). The correlation of cR to cM in this framework map is an average of 29.1 cR per cM. The 1161 RH data sets we used to make our framework map included 545 data sets from Whitehead Institute, 362 from Genoscope, 134 mapped in our own laboratory, and 119 from other contributing laboratories worldwide.
Two major statistical discontinuities in the framework map occur on Chr 11. These are caused by extreme marker retention frequencies in two regions of the chromosome (Fig. 3A). On central Chr 11, five consecutive MIT SSLP markers show a retention frequency of less than 12%, resulting in LODs of linkage that are below the LOD > 6.0 cutoff for significance. We placed these loci in an order that meets our criteria for minimum breaks through the region, and then compared the result with the Ensembl sequence assembly (Fig. 3B). The RH data accurately predict the sequence marker order, but cannot be used to calculate LODs of linkage or cR interval sizes due to the low retention frequencies. The retention frequency reaches a minimum of 0% for the D11Mit60 locus, and the markers on either side show increasing retention at increasing distance from the lowest retention marker until the LODs reach significance.
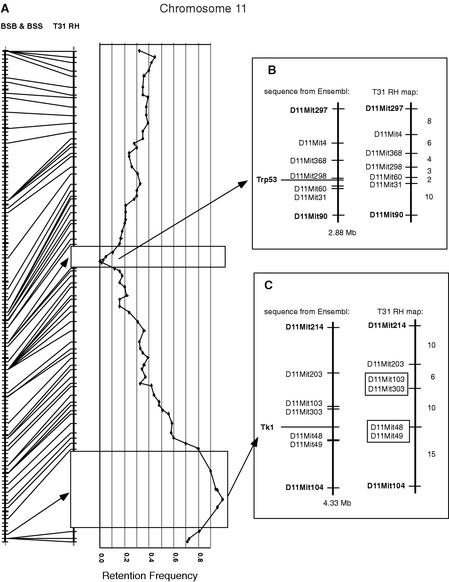
Figure 3.
Chromosome 11 framework map (A) showing two gaps in significant linkage due to extreme retention frequencies of markers. TJL BSB/BSS combined backcross map is shown on the left, with crosshatches at each crossover (0.56 cM) and heavier crosshatches every 10 crossovers. Lines join this map to the framework map of Chromosome 11 in the T31 RH data. RH markers are placed at their cR positions. Where the LOD of linkage drops below 6 (minimum significant LOD), an open rectangle is drawn across the RH chromosome line, with the flanking markers at their calculated cR distance apart. Within these low LOD intervals the marker spacing is drawn proportional to the minimum number of obligate segment breaks in each interval. Graphed on the right of (A) is the retention frequency for each framework marker, with x-axis intervals of 10%. The lowest retention frequency is zero and the highest is 100%. The presumed selected markers are assigned a retention rate of Trp53 = 0% and Tk1 = 100%. (B,C) The low-LOD intervals compared to sequence from Ensembl v3. Sequence maps are drawn to Mb scale. RH maps are set at the same length as the sequence map and RH distances are set proportional to the minimum number of obligate segment breaks in each interval. Numbers to the right of the RH map bars are number of breaks in the interval. Bold locus symbols indicate markers that link with significant LOD to the rest of the chromosome RH data. Plain text locus symbols indicate markers that have interlocus LODs less than 6. Presumed selected marker is shown in bold out to the left of each sequence.
The reciprocal case occurs on the distal end of the Chr 11 RH map (Fig. 3C): Here five loci show retention frequencies over 80% and intermarker LODs less than 6. Similarly, the RH marker order derived using the minimum break criterion is fully concordant with the order given by the Ensembl sequence assembly. In this region the highest retention frequency for an MIT SSLP marker is 96%, shown by D11Mit49 and D11Mit48 on the distal side of the Tk1 locus used to select the hybrid cells, while D11Mit303 and D11Mit103 on the proximal side are retained at 94%. Although we did not map the Tk1 locus itself, we can assume that its retention frequency is 100%.
Comparing the Different T31 RH Maps
The Whitehead Institute Genome Center RH map (Hudson et al. 2001) includes 734 of the markers in our framework map. In the Release 10 version of the Whitehead map there are 45 genomic regions that contain two to four misordered markers by comparison to our curated framework marker TJL BSB/BSS recombination data. D13Mit66 and D14Mit64 are both placed at significant distance from our framework map positions. There are additional differences between our comprehensive RH map positions for markers compared to the Whitehead RH map, and some examples of these differences are discussed below.
Five hundred twenty-three markers in our framework map are also included in the Genoscope RH map build (Avner et al. 2001). There are 12 genomic regions where the Genoscope map is discordant with TJL BSB/BSS map order. This low number of order conflicts may be in part due to the lower density of markers in common making detection of conflicts less sensitive. The Genoscope map building methodology included multiple pruning steps aimed at improving accuracy of the resulting maps, and these may also improve the agreement with our framework map.
Comparing the Framework Map to the Sequence Assembly
Although a detailed comparison of the complete framework-supported comprehensive RH map to the emerging sequence assemblies will be the subject of future studies, we have undertaken a comparison of the 1161 marker framework map to the Ensembl v3 sequence assembly to assess the potential value of this work. We found the agreement in locus order to be very good overall, which we took to indicate that both the sequence and our framework map are likely to have good accuracy.
One hundred ninety-nine of the 1161 framework markers were not annotated in the Ensembl database but could be identified in the sequence by BLAST analysis. An additional 108 of the framework markers were only annotated by their Whitehead assay names (example: D1Mit10 found as A117). Eleven of the framework markers could not be found in the sequence assembly by BLAST, but a sequence gap was found in a location that coincided with the expected location of the missing marker sequence. Seven of the framework markers were not detectable by BLAST with no obvious sequence gap in the expected region.
Six framework markers were anomalously assembled in the sequence. D2Mit200 is in a distinctly distant location on Chr 2 next to a sequence gap. D3Mit54 is placed on distal Chr 4 in the sequence with a concomitant gap in its expected location on Chr 3. Similarly, D3Mit139 is placed on Chr 2 with a gap in its expected location on Chr 3, and D12Mit140 is annotated on Chr 14 with a gap in its expected location on Chr 12. D7Mit178 is assembled to distal Chr 7 instead of proximally and the complete sequence for this marker is not in the assembly, and D14Mit151 is out of correct order and surrounded by sequence gaps.
Local marker order conflicts between the framework mapping and the sequence assembly were notably rare. D4Mit146 and D4Mit43 are in opposite orders in the two maps, as are D6Mit148 and D6Mit104, D6Mit340 and D6Mit372, DXMit86 and DXMit193. In all of these cases the two markers cosegregate in TJL BSB/BSS, and data for one to three RH cell lines determines the RH-based order. It is possible that either RH data errors or some sequencing anomaly is causing the reversal of relative marker order.
There are several cases of significant inversions or translocations of sequence in the assembly, problems that are revealed by the framework map order confirmed by TJL backcross data. The proximal ends of Chr 5, 7, 14, 17, and 19 all have several framework markers misordered consistent with several megabases of sequence being inverted from the centromeric end to a gap or several gaps in the sequence. Examples of these inversions are discussed in detail in the Discussion.
Browsing through the Ensembl sequence we noted 50 annotated MIT SSLP markers that are not mapped in TJL BSB/BSS or in the T31 RH panel that are placed in the sequence on unexpected chromosomes. Seventeen more MIT SSLP marker sequences that are assembled to unexpected chromosomes are confirmed by T31 RH data, and in some cases by interspecific backcross mapping data that agree with the sequence assembly placement. Thirteen MIT SSLP markers annotated on unexpected chromosomes conflict with mapping data that confirm them in the location expected from the original WI-MIT genetic mapping. D10Mit216 is annotated on Chr 7 in Ensembl. Data from Whitehead mapped this marker to Chr 10 with a maximum LOD of 11. Noting several nonlinking positive scores in this vector, we reassayed this marker and found that there were two mouse band sizes produced, one that mapped to the Chr 10 position and another that mapped to Chr 7. The sequence annotation correlates with the Chr 7 band map position. D8Mit351 is annotated on Chr 7 in Ensembl, but is mapped to Chr 10 by RH data from Genoscope.
DISCUSSION
Although we recognize the importance of independently validating each new technology, we also value the insights available from detailed intermethodology comparison. Previously reported mouse RH mapping analyses have been made mostly independently of the existing recombination maps, comparing the two systems to estimate frequencies of nonconcordance after the maps were built or limiting analysis to concordant markers (Van Etten et al. 1999; Avner et al. 2001; Hudson et al. 2001).
The genetic maps previously used for intermethodology comparison have themselves contained a significant error rate. In screening markers for inclusion in our framework map we identified 82 individual markers confirmed by our backcross analysis to be out of order on their respective chromosomes in the WI-MIT genetic map, while the proximal ends of Chrs 1 and 14 both contain multiple misordered markers.
Forty-eight of the 3956 unique MIT SSLP markers that are mapped in the T31 RH panel map to a different chromosome from that reported in the original WI-MIT mapping experiments (Dietrich et al. 1996). In several cases multiple sources have mapped the same marker to the same unexpected location in the RH panel. In some cases the new position is confirmed in the backcross panel maps. Four MIT SSLP markers have already been renamed to reflect the corrected genomic location for the marker. D3Mit217 is confirmed on Chr 1 and is renamed D1Mit1000, D19Mit72 is also confirmed on Chr 1 and is renamed as D1Mit1001, D8Mit46 is on Chr 9 as D9Mit1000, and D8Mit112 is also on Chr 9 as D9Mit1001.
Some of the mismapped MIT SSLP markers have been used as RHMAPPER framework markers in the Whitehead T31 RH map (Hudson et al. 2001), thus causing several linked markers to appear to be poorly linked to surrounding data. A case in point is D10Mit278, which in fact maps to Chr 15 in the RH data. The highest LOD to any marker on Chr 10 is 3.9, while this locus has LODs up to 24.4 on Chr 15. Because D10Mit278 was placed as a framework marker on Chr 10 in the Whitehead RH map, nine ESTs that map to Chr 15 have also been placed on Chr 10, including BB317391 (LOD 26 to Chr 15) and BB315977 (LOD 19 to Chr 15). The LOD between this group of misplaced markers and the nearest Chr 10 marker in the Whitehead Map is 1.4, well below significant linkage. Similarly, D9Mit170 maps to Chr 6, but, used as a framework marker in the Whitehead RH analysis, it brings AA987181 (LOD 9.7 to Chr 6) and BB137539 (LOD 22 to Chr 6) with it to the proximal end of Chr 9. Note that AA987181 is an EST for Hoxa9, which has been mapped to Chr 6 by other methods, and is also mapped to the same Chr 6 position in this T31 RH panel by another laboratory. These are examples of problems that can arise using RH map-building software that requires framework designation for mapping.
Mismapping of framework markers has been cited (Gregory et al. 2002) for causing discrepancy between other T31 RH maps and the optimum contig assembly underlying the mouse sequence assembly. We compared our T31 RH map to the Whitehead T31 RH map in the 80–90 Mb region of Chr 2 that Gregory et al. found to disagree with the contig data. Our comprehensive T31 RH map order agrees well with the contig and sequence order, and has several local differences in order with the Whitehead T31 RH map.
In our study, by examining in detail the underlying data for both the recombination map and the RH map and using each to improve the other, we have achieved a higher level of confidence in the overall maps, both in analysis methodology and in the final improved data. By bringing the two map sources into full concordance we are able to leverage the quality of the mouse genome map.
We note that many previous publications of correlations between RH maps and recombination maps have shown multiple order incongruencies (examples: Schalkwyk et al. 1998; Elliott et al. 1999; Hopitzan et al. 2000; Arkell et al. 2001). We believe that there are several explanations, in addition to the improved accuracy of the framework-supported data, for the contrasting complete congruence of our whole-genome framework map. First, many comparisons have used a composite genetic map, either from MGD compilation at http://www.informatics.jax.org or from the Chromosome Committees http://www.informatics.jax.org/ccr. Although composite maps are useful for obtaining a comprehensive overview of all markers mapped to particular chromosomes and for identifying potential candidate loci mapping in a region of interest, their construction from multiple mapping sources using multiple methodologies precludes the high resolution accuracy needed to support a detailed comparison between RH and recombination mapping. Second, many recombination maps have been based on data from a few animals or on high throughput data that contain undetected errors. By careful checking of TJL BSB/BSS scores for all 188 progeny, we were able to achieve a higher level of accuracy in the recombination data. Third, by using a subset of RH data that link in the LOD 6–15 range, we are using the resolving power of the T31 panel to its highest advantage. Many previous comparisons have included data at a density that is beyond saturating the T31 map, resulting in poor support for one local marker order over alternative orders.
Gaps in the Framework Map
Because we limited our framework map to the available SSLP marker set, some of the gaps remaining in the RH framework map may be due to extended genomic regions that do not contain mapped microsatellite sequences. The single gap in significant LOD on the Chr 10 framework map (see Fig. 1 and the online supplementary material and poster included with this issue) between D10Mit45 and D10Mit53 represents over 3 Mb of sequence according to the Ensembl sequence assembly. Ensembl shows no microsatellite loci in this region. The complete RH map contains 16 EST loci mapped in this interval and the lowest intermarker LOD is 13.3, so the overall map is well supported by the RH data in the absence of microsatellite markers. The Ensembl sequence in this interval includes 4 RIKEN cDNA markers, three known genes (Rev3l, Lama4, Hdac2) and the EST marker AW552119 (the Fyn oncogene) that is also mapped to this region in the RH map. Including AW552119 as a framework marker would close the gap in significant linkage in the RH data.
Some of the gaps remaining in the RH framework map are likely to be due to major differences in apparent retention frequency between nearby markers. An example of this type of gap can be found at the proximal end of the Chr 14 map. D14Mit48 has an apparent retention frequency of 20%, while the next framework marker, D14Mit98 (which cosegregates with D14Mit48 in TJL BSB/BSS crosses for a genetic distance of less than 0.56 ± 0.56 cM) has a retention frequency of 47%. Other markers on proximal Chr 14 show similarly higher retention frequencies. Under all our standard PCR conditions, amplifications of the hamster and mouse progenitor DNAs for the RH panel with D14Mit48 primers yielded only a faint primer dimer. We suspect that the assay for this marker is problematic, possibly due to the string of twelve consecutive cytidine residues flanking the simple sequence repeat, and that with a more reliable assay the retention frequency would likely be higher, improving the LOD of linkage. The low retention frequency in the D14Mit48 data make the minimum obligate break placement at the proximal end of the map, because the data create many new segment breaks in any interval.
Because D14Mit48 and D14Mit98 cosegregate in the backcross data, it is possible that D14Mit48 may, in fact, map more distally. In this region of the Ensembl mouse sequence assembly, the proximal part of Chr 14 is inverted compared to the known genetic map order from TJL BSB/BSS crosses, probably from the centromere to a sequence contig gap at 14.6 Mb. Reversing the order of this section of the Chr 14 sequence would bring the sequence assembly into agreement with the recombination and RH data, and would suggest that the D14Mit48 sequence may lie just distal to D14Mit98 and D14Mit220, and proximal to D14Mit49. This exercise in comparison of the different independent map data sources is a good example of the power these different methodologies can contribute to the accurate assembly of the overall mouse genome.
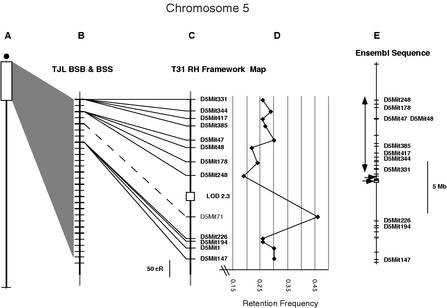
There is a similar inversion between the Ensembl sequence assembly and the recombination map of proximal Chr 5 (Fig. 4). As with Chr 14, a significant gap in the sequence appears to delineate the “inversion breakpoint.” Two crossovers in the BSS data (raw data available online at http://www.jax.org/resources/documents/cmdata/bkmap/) clearly place the proximal MIT SSLP data into the order proximal −[D5Mit331, D5Mit344, D5Mit385, D5Mit417] − [D5Mit47, D5Mit48, D5Mit178, D5Mit248] − D5Mit71 − distal. The RH data order agrees with the backcross data, but the unusually high retention frequency for D5Mit71 disrupts the linkage continuity near this locus. The D5Mit71 marker is not annotated in the Ensembl sequence. BLAST analysis of the genome assembly with the primer sequences gives several moderate-identity matches, the nearest of which is to a Chr 5 position somewhat distal to its genetic map position, in a region where other annotated markers appear in an order that is congruent between the sequence assembly and the maps. We speculate that the D5Mit71 sequence that is mapped in the backcrosses and RH panel may be contained in the missing segment of proximal Chr 5 sequence, and that the high RH retention frequency may reflect some amplification from the related sequences found by BLAST analysis.
Figure 4.
Proximal Chromosome 5 compared to the Ensembl v3 sequence assembly. (A) Shows the portion of the Chr 5 map detailed in the rest of the figure. (B) Shows TJL BSB/BSS backcross recombination map, with crosshatches for each crossover and heavier crosshatches for each position that contains a framework marker. (C) The T31 RH framework map for this proximal region. A 50-cR scale bar is included to the left of the chromosome line. Note that there are four markers that cosegregate at the top of the recombination map and another four that map two crossovers distally. D5Mit71, shown in plain text with a dashed line joining the maps, could only be mapped in the BSS cross, and its BSB position is estimated from data based on reading copy number of C57BL/6J alleles. The backcross mapping places this marker clearly proximal to D5Mit226 and D5Mit194. (D) The retention frequency graph for this part of Chr 5 shows the unusually high retention of the D5Mit71 marker that causes it to link poorly to the surrounding data. (E) The Ensembl v3 sequence assembly for this region. The sequence is shown to Mb scale (a 5-Mb scale bar is included to the right of the figure), with crosshatches for each MIT SSLP marker annotated in the sequence. Right-pointing arrows indicate gaps in the contig. The vertical double-headed arrow shows the region that is in inverted order relative to the recombination/RH map.
Selection Affects Marker Retention Variation
The retention frequencies in the vicinity of two loci on Chr 11 demonstrate the effect of selection on the RH map (Fig. 3). The Tk1 locus on the distal end of Chr 11 was used as a selectable marker in the creation of the T31 radiation hybrid cell lines. Thus, all hybrid cell lines should contain at least a single fragment of mouse DNA from distal Chr 11 including this mouse gene. Markers spanning 1.81 Mb of sequence (Ensembl) around the Tk1 locus have very high retention frequencies that preclude LOD and cR calculation by the standard algorithms. Conversely, on the central part of Chr 11 the retention frequencies become too low to calculate LOD or cR. The retention frequency of D11Mit60 is zero, even though two independent 129-strain control DNA templates both yield a very strong mouse positive band. We agree with Behboudi et al. (2002) that it is likely that expression from the Trp53 gene that maps between D11Mit298 (retention frequency 3%) and D11Mit60 is causing the cell lines that carry this mouse gene to fail to grow. These two reciprocal types of selection on the RH cell lines are clear extreme examples of the potential affect of selection on retention frequency that may render RH data analysis problematic. Such selection violates two of the underlying assumptions used in some RH mapping algorithms, that nearby loci will have similar retention patterns, and that there is no selection affecting retention. Despite the complications arising from selection, however, we note that in both of these regions we were able to use a simple minimum break criterion to build an accurate locus order (Fig. 3B, C), and that the relative intermarker distances based on break count are in good agreement with the sequence spacing.
Many chromosomes show a higher retention frequency near the centromere, and sometimes a slight increase of retention at the telomere. Some selection in favor of centromere and telomere sequences is likely to result from the ability of a chromosome fragment to form a stable minichromosome in the recipient hamster cell. For many of the mouse chromosome fragments the retention frequency tends to decline with distance from the centromere (examples: Chrs 1, 2, 3, and 6). The X Chromosome has a lower retention frequency over its entire length that can be entirely accounted for by the hemizygosity of the male mouse donor to the hybrid cells. This reduced retention rate makes the ordering of X Chromosome loci based on the RH data less certain than in higher retention genomic regions.
Distribution of Recombination
Comparing the spacing of the framework markers between the recombination and RH maps (Fig. 1, and in more detail in online supplementary material and poster included with this issue) reveals regions that have high or low recombination per cR. High resolution recombination hotspots and cold spots are well documented in previously reported data (e.g., Wahls 1998; Isobe et al. 2002). This study reveals the distribution of recombination on a whole-chromosome and genome-wide scale. On most chromosomes the frequency of recombination near the centromere is reduced and there is a high frequency of recombination per cR in the adjacent part of the chromosome (exceptions: Chrs 2, 4, 10, 16, and X). In an extreme example of this pattern the proximal 13% of the Chr 7 RH map fails to recombine in the backcrosses (Fig. 2). The recombination hotspot distal to this region of recombination suppression contains 20 crossovers (11 in BSB, 9 in BSS) between the position of D7Mit52 and the position of D7Mit158, while these two adjacent framework markers are only 52.7 cR apart in the RH map (expect 1.8 crossovers). The Ensembl sequence contains several significant gaps in this region, and places D7Mit52 about 6 Mb proximal to D7Mit158.
The smaller chromosomes tend to show recombination suppression near the centromere, a central region of high recombination, and a distal region of average cM per cR. Examples of this type of crossover distribution can be seen on Chrs 13, 14, 15, 17, 18, and 19. The larger chromosomes tend to have a region of high recombination frequency near the distal end. The larger mouse chromosomes also show several regions of recombination clusters throughout the central part of the chromosome. These patterns of recombination may reflect the effect of high crossover interference between multiple crossovers on the larger mouse chromosomes (Broman et al. 2001). It should be noted that TJL BSB/BSS backcross panels used in this study are based on female recombination only, as male (C57BL/6J × SPRET/Ei) F1 animals are sterile. The distribution of recombination in the genome seen by comparing TJL BSB/BSS recombination map to the RH map agrees well with previous studies (e.g., Lawrie et al. 1995) that examined chiasmata distribution using cytologic methods. This similarity suggests that at least on a gross scale cR distance correlates well with physical distance. Because calculated cR distances are dependent on data density, it is difficult to use RH data to assess the possibility of local differences in radiation sensitivity in the genome.
At this moment in the history of the mouse genome project, with the arrival of the genome-wide contig and sequence assemblies, it is tempting to conclude that the genetic maps are outdated. We take a different view, however. We have used an improved recombination map, with lower resolution but higher order confidence, to discover and repair flaws in the RH map. With an improved RH map, regions of difference between the maps and the sequence indicate foci for further study and development of sequence coverage and/or assembly methods. In addition, these mapping panel resources provide efficient cross checking for any sequence under study to independently confirm or reject a genomic placement based on the sequence assemblies, as well as allowing placement of markers that have as yet either not proven clonable or which are problematic to the sequence assembly algorithms. Finally, by comparing the recombination maps in detail to the final sequence assembly, we can begin to assign Mb units to recombination distribution, opening a new avenue for the study of the biology of recombination.
METHODS
Radiation Hybrid Typing
MIT SSLP loci were screened using 50 ng A23 Hamster DNA, 50 ng 129/J Mouse DNA, and a mixture of 25 ng A23 Hamster DNA and 25 ng 129/J Mouse DNA (Invitrogen Corporation). Reactions were 22 μL total of: 50 mM KCl, 10 mM Tris HCl pH 8.3, 1.5 mM MgCl2, 0.01% or 0.001% gelatin, 200 μM each dNTP (Amersham Biosciences AB), 0.12 μM forward primer, 0.12 μM reverse primer (Invitrogen Corporation and Integrated DNA Technologies), 0.02 units/μL AmpliTaq DNA Polymerase (Applera Corporation). PCR conditions: 94°C 3 m, 38 cycles of (94°C 30 sec, 55°C 35 sec, 72°C 30 sec), 72°C 7 m, 4° hold. PCR products were separated on 2%–4% MetaPhor agarose, SeaKem GTE agarose or NuSieve GTE agarose gels with 1× SYBR Green I Nucleic Acid Stain (Cambrex Corporation).
All 100 of the cell line DNAs of the T31 Mouse Radiation Hybrid Panel (Invitrogen Corporation) were typed in duplicate with a second set of independent A23 Hamster and 129/J Mouse controls. Most of the loci were mapped using the PCR and gel conditions used for the screenings. Some markers with weaker bands were typed using 0.24 μM each primer rather than the standard 0.12 μM. Some markers were typed using 50° or 52°C annealing temperature. A few markers were retyped using a hot start protocol with AmpliTaq Gold Polymerase (Applera Corporation) or HotStarTaq DNA Polymerase (Qiagen) per manufacturer's instructions. All RH mapping data and relevant protocol notes for each marker are available at http://www.jax.org/resources/documents/cmdata/rhmap/rh.html.
Backcross Typing
The Jackson Laboratory BSB panel includes 94 N2 animals from the cross (C57BL/6J × Mus spretus) F1 × C57BL/6J. The Jackson Laboratory BSS panel includes 94 N2 animals from the reciprocal cross (C57BL/6JEi × SPRET/Ei) F1 × SPRET/Ei (Rowe et al. 1994). MIT SSLP loci were screened using 12.5 ng C57BL/6J DNA, 12.5 ng SPRET/Ei DNA, and a mixture of 6.25 ng C57BL/6J DNA and 6.25 ng SPRET/Ei DNA. Reactions were 12 μL total, otherwise the same as for RH typing. Where the segregating band sizes were very close, markers were screened using the addition of 0.5 μCi (32P dCTP (Amersham Biosciences AB) per 10 μL reaction. Radioactive PCR conditions: 94°C 3 m, 25 cycles of (94°C 15 sec, 55°C 2 m, 72°C 2 m), 72°C 7 m, 4° hold. PCR products were separated on 7%–12% Long Ranger acrylamide gels (Cambrex Corporation) or 7%–9% Acrylamide/bis-Acrylamide gels (Sigma-Aldrich Corporation). All backcross mapping data and relevant protocol notes for each marker are available at http://www.jax.org/resources/documents/cmdata/bkmap/index.html.
Data Analysis
Data were stored and analyzed using the Map Manager QT program (Manly and Olson 1999) that includes algorithms for radiation hybrid mapping. Duplicate assays of new radiation hybrid data were merged into a single consensus data set for each locus. Any discordance between duplicate assays was resolved by repeating the PCR in duplicate for those samples. Markers were initially placed at the position of highest LOD of linkage and then adjusted to minimize the number of breaks needed to explain the locus order. Where necessary, linking positive scores was given priority over linking negative scores. New breaks in otherwise continuous segments were reexamined for possible error. TJL backcross data were ordered on the map by minimizing crossovers. Any assays yielding apparent single-locus double crossovers were retested. There are no single-locus double crossovers in the final backcross framework data set and no missing crossover typings.
Constructing the Comprehensive T31 RH Map
Using Map Manager QT software, we assembled T31 data as they became available from laboratories around the world. Early MIT SSLP data were compared to the WI-MIT genetic map to assist in placing data that were too sparse to support linkage from the RH data alone. As more data became available, the maps were continually refined, increasingly based on RH linkage as determined from maximum LOD and minimum break criteria. We began framework marker mapping before the large data sets from the Genome Centers were released, and continued this process one chromosome at a time. As framework markers were placed on TJL BSB/BSS, the T31 chromosomal maps were reanalyzed in light of new marker order information.
Building the Framework Maps
DNA microsatellite SSLP (D-Mit-) markers were selected from the RH data for use as framework loci with a first criterion that the RH mapping data for the marker fit well with surrounding data. Where more than one data set was available for a marker, we chose to use the single data set that fit the surrounding data with the fewest required breaks. To include as much of the chromosome length within the framework as possible, we included as a framework marker the most proximally and the most distally mapping SSLP marker on each chromosome. In regions where the RH data lacked good continuity, we chose additional markers to add to the RH map to try to fill the gaps. If there was a poorly matching data set that appeared to map in a gap, we first repeated the RH mapping experiment for the marker to see if we could obtain better matching data for the marker.
Some poorly fitting RH data were improved by minor technical alteration in the assay protocol. When our standard PCR protocol produced weak mouse-specific bands, we tested using a reduced annealing temperature, increased primer concentration, or both. We were able to improve the fit of D17Mit175 data, for example, by reducing the annealing temperature to 50 degrees. D17Mit16 data were improved by using a 45-degree annealing temperature. D18Mit141, D18Mit57, and D6Mit196 data were improved by using 52-degree annealing. D18Mit50 data fit better when we used double primer concentration and 52-degree annealing temperature. If the first screen of a marker showed a complex pattern of bands in the hamster control that might make scoring the mouse band difficult, we tested with a hotstart protocol, with or without the reduced annealing temperature. Examples of assays improved by hotstart were D4Mit9, D7Mit52, D8Mit58, D16Mit26, and D18Mit67. A combination of hotstart and reduced annealing temperature was used to retype D16Mit6 and DXMit149. Usually one of these PCR protocol changes improved the readability of the assays, and consequently the match of the data to surrounding data in the map. Details of all specific protocols used are available from our Web site.
Original framework markers were chosen from the RH data with a goal of LOD 10–15 between adjacent markers. An intermarker LOD minimum was set at LOD 6, below which we considered the linkage to be unsupported by the RH data, and we treated such regions as gaps in the framework map. We chose to use the LOD > 6 criterion for significance of linkage because individual locus data sets find multiple LODs under 6 to spurious positions, in addition to much higher LODs to their correct chromosomal locations; see Rowe et al. 2000 for a more complete discussion.
Loci chosen as framework markers were screened for polymorphism between the parental strains (C57BL/6J and SPRET/Ei) for TJL BSB/BSS backcrosses. Where the marker density was high, we chose markers whose reported allele sizes (http://www-genome.wi.mit.edu/cgi-bin/mouse/index) had a >5% length difference to permit rapid typing on agarose gels. If one allele amplified poorly in the presence of the other (it is usually the Mus spretus allele that matches less well to primer sequences based on C57BL/6J sequence), we tested reducing the PCR annealing temperature to improve the relative band intensities. A few markers could be typed only in the BSS cross (94 segregants) due to failure of the Mus spretus allele to amplify in heterozygotes. The map positions for these markers are therefore of lower confidence.
Aligning the Maps
When the marker order determined from the backcross mapping was at variance with that from the RH data, we examined both data sources more closely. The data for informative backcross recombinants were reexamined, and the local RH data in the comprehensive database were rechecked for minimum break order. In some cases, one or a few poorly matching RH data sets for nearby markers had obscured a better local order that did match the order from the recombination data. In some cases, sections of the RH data had been inverted at “breakpoints” where the LODs were low in either order. In some cases, previous typing errors in the recombination data for key crossover animals had mislocated a marker. In some cases, a primer pair had been mislabeled by the manufacturer, and a newly synthesized primer pair mapped in congruence with the expected location. In all cases, by careful examination of the underlying data we were able to reconcile the two framework maps to a single best locus order.
WEB SITE REFERENCES
http://www-genome.wi.mit.edu/mouse_rh/index.html; WICGR mouse RH map home page.
http://www-genome.wi.mit.edu/cgi-bin/mouse/index; Whitehead Institute genetic and physical maps of the mouse genome.
http://websql.har.mrc.ac.uk/mps/maps/0/LOD_7/graphic.html; UK Mouse Genome Centre EST RH maps.
http://www.genoscope.cns.fr/externe/English/Projets/Projet_ZZZ/rhmap.html;Genoscope / EU mouse radiation hybrid mapping project.
http://www.informatics.jax.org; Mouse Genome Informatics at The Jackson Laboratory.
http://www.informatics.jax.org/ccr; Chromosome Committee reports.
http://www.jax.org/resources/documents/cmdata/; The Jackson Laboratory mapping panels.
http://www.jax.org/resources/documents/cmdata/bkmap/; The Jackson Laboratory backcross DNA panel mapping resource.
http://www.jax.org/resources/documents/cmdata/rhmap; The Jackson Laboratory T31 mouse radiation hybrid database.
http://www.jax.org/resources/documents/cmdata/rhmap/rh.html; The Jackson Laboratory T31 mouse radiation hybrid database, public RH map raw data by chromosome.
http://www.ensembl.org/Mus_musculus/; Ensembl mouse genome server.
http://www.ncbi.nlm.nih.gov/genome/seq/MmHome.html; NCBI mouse genome sequencing.
http://genome.ucsc.edu/; UCSC genome bioinformatics.
http://www.celera.com; Celera Corporation.
Acknowledgments
This work was supported by a grant HG00941 from NHGRI. The authors thank Kristy Grant for technical assistance, and Charles Donnelly and Marge May for writing the scripts that expedited handling the large RH data sets, and Jennifer Smith for the poster design. Scientific Resources at The Jackson Laboratory are supported in part by a Cancer Center Core Grant CA34194 from the National Cancer Institute. We also thank all the investigators who contributed data to both the recombination and RH maps.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
E-MAIL lbr@jax.org; FAX (207) 288-6072.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.858103. Article published online before print in December 2002.
REFERENCES
- 1.Arkell R.M., Cadman, M., Marsland, T., Southwell, A., Thaung, C., Davies, J.R., Clay, T., Beechy, C.V., Evans, E.P., Strivens, M.A., et al. 2001. Genetic, physical, and phenotypic characterization of the Del13Svea36H mouse. Mamm. Genome 12: 687-694. [DOI] [PubMed] [Google Scholar]
- 2.Avner P., Bruls, T., Poras, I., Eley, L., Gas, S., Ruiz, P., Wiles, M.V., Sousa-Nunes, R., Kettleborough, R., Rana, A., et al. 2001. A radiation hybrid transcript map of the mouse genome. Nat. Genet. 29: 194-200. [DOI] [PubMed] [Google Scholar]
- 3.Behboudi A., Roshani, L., Lundin, L., Stahl, F., Levan, K.K., and Levan, G. 2002. The functional significance of absence: The chromosomal segment harboring Tp53 is absent from the T55 rat radiation hybrid mapping panel. Genomics 79: 844-848. [DOI] [PubMed] [Google Scholar]
- 4.Broman K.W., Rowe, L.B., Churchill, G.A., and Paigen, K. 2001. Crossover interference in the mouse. Genetics 160: 1123-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Copeland N.G., Jenkins, N., Gilbert, D., Eppig, J.T., Maltais, L.J., Miller, J.C., Dietrich, W.F., Weaver, A., Lincoln, S.E., Steen, R.G., et al. 1993. A genetic linkage map of the mouse: Current applications and future prospects. Science 262: 57-66. [DOI] [PubMed] [Google Scholar]
- 6.Dietrich W.F., Miller, J., Steen, R., Merchant, M.A., Damron-Boles, D., Husain, Z., Dredge, R., Daly, M.J., Ingalls, K.A., O'Connor, T.J., et al. 1996. A comprehensive genetic map of the mouse genome. Nature 380: 149-152. [DOI] [PubMed] [Google Scholar]
- 7.Elliott R.W., Manly, K.F., and Hohman, C. 1999. A radiation hybrid map of mouse Chromosome 13. Genomics 57: 365-370. [DOI] [PubMed] [Google Scholar]
- 8.European Backcross Collaborative Group (EUCIB) 1994. Towards high resolution maps of the mouse and human genomes—A facility for ordering markers to 0.1 cM resolution. Hum. Mol. Genet. 3: 621-627. [PubMed] [Google Scholar]
- 9.Gellin J., Brown, S., Graves, J.A.M., Rothschild, M., Schook, L., Womack, J., and Yerle, M. 2000. Comparative gene mapping workshop: Progress in agriculturally important animals. Mamm. Genome 11: 140-144. [DOI] [PubMed] [Google Scholar]
- 10.Gregory S.G., Sekhon, M., Schein, J., Zhao, S., Osoegawa, K., Scott, C.E., Evans, R.S., Burridge, P.W., Cox, T.V., Fox, C.A., et al. 2002. A physical map of the mouse genome. Nature 418: 743-750. [DOI] [PubMed] [Google Scholar]
- 11.Gyapay G., Schmitt, K., Fizames, C., Jones, H., Vega-Czarny, N., Spillett, D., Muselet, D., Prud'Homme, J.F., Dib, C., Auffray, C., et al. 1996. A radiation hybrid map of the human genome. Hum. Mol. Genet. 5: 339-346. [DOI] [PubMed] [Google Scholar]
- 12.Hammer M.F., Schimenti, J., and Silver, L.M. 1989. Evolution of mouse Chromosome 17 and the origin of inversions associated with t haplotypes. Proc. Natl. Acad. Sci. 86: 3261-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hogenesch J.B., Ching, K.A., Batalov, S., Su, A.I., Walker, J.R., Zhou, Y., Kay, S.A., Schultz, P.G., and Cooke, M.P. 2001. A comparison of the Celera and Ensembl predicted gene sets reveals little overlap in novel genes. Cell 106: 413-415. [DOI] [PubMed] [Google Scholar]
- 14.Hopitzan A., Himmelbauer, H., Spevak, W., and Castanon, M. J. 2000. The mouse Psma1 gene coding for the α-type C2 proteasome subunit: Structural and functional analysis, mapping, and colocalization with Pde3b on mouse Chromosome 7. Genomics 66: 313-323. [DOI] [PubMed] [Google Scholar]
- 15.Hudson T.J., Church, D.M., Greenaway, S., Nguyen, H., Cook, A., Steen, R.G., Van Etten, W.J., Castle, A.B., Strivens, M.A., Trickett, P., et al. 2001. A radiation hybrid map of mouse genes. Nat. Genet. 29: 201-205. [DOI] [PubMed] [Google Scholar]
- 16.Isobe T., Yoshino, M., Mizuno, K., Lindahl, K., Koide, T., Gaudieri, S., Gojobori, T., and Shiroishi, T. 2002. Molecular characterization of the Pb recombination hotspot in the mouse major histocompatibility complex class II region. Genomics 80: 229-235. [DOI] [PubMed] [Google Scholar]
- 17.Lander E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860-921. [DOI] [PubMed] [Google Scholar]
- 18.Lawrie N.M., Tease, C., and Hultén, M.A. 1995. Chiasma frequency, distribution and interference maps of mouse autosomes. Chromosoma 104: 308-314. [DOI] [PubMed] [Google Scholar]
- 19.Li S., Liao, J., Cutler, G., Hoey, T., Hogenesch, J.B., Cooke, M.P., Schultz, P.G., and Ling, X.B. 2002. Comparative analysis of human genome assemblies reveals genome-level differences. Genomics 80: 138-139. [DOI] [PubMed] [Google Scholar]
- 20.Manly K.F. and Olson, J.M. 1999. Overview of QTL mapping software and introduction to Map Manager QT. Mamm. Genome 10: 327-334. [DOI] [PubMed] [Google Scholar]
- 21.McCarthy L.C., Terrett, J., Davis, M.E., Knights, C.J., Smith, A.L., Critcher, R., Schmitt, K., Hudson, J., Spurr, N.K., and Goodfellow, P.N. 1997. A first-generation whole genome-radiation hybrid map spanning the mouse genome. Genome Res. 7: 1153-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mellersh C.S., Hitte, C., Richman, M., Vignaux, F., Priat, C., Jouquand, S., Werner, P., André, C., DeRose, S., Patterson, D.F., et al. 2000. An integrated linkage-radiation hybrid map of the canine genome. Mamm. Genome 11: 120-130. [DOI] [PubMed] [Google Scholar]
- 23.Nagaraja R., MacMillan, S., Jones, C., Maisisi, M., Pengue, G., Porta, G., Miao, S., Casamassimi, A., D'Urso, M., Brownstein, B., et al. 1998. Integrated YAC/STS physical and genetic map of 22.5 Mb of Human Xq24-q26 at 56-kb inter-STS resolution. Genomics 52: 247-266. [DOI] [PubMed] [Google Scholar]
- 24.Rhodes M., Straw, R., Fernando, S., Evans, A., Lacey, T., Dearlove, A., Greystring, J., Walker, J., Watson, P., Weston, P., et al. 1998. A high-resolution microsatellite map of the mouse genome. Genome Res. 8: 531-542. [DOI] [PubMed] [Google Scholar]
- 25.Rowe L.B., Nadeau, J.H., Turner, R., Frankel, W.N., Letts, V.A., Eppig, J.T., Ko, M.S.H., Thurston, S.J., and Birkenmeier, E.H. 1994. Maps from two interspecific backcross DNA panels available as a community genetic mapping resource. Mamm. Genome 5: 253-274. [DOI] [PubMed] [Google Scholar]
- 26.Rowe L.B., Barter, M.E., and Eppig, J.T. 2000. Cross-referencing radiation hybrid data to the recombination map: Lessons from mouse Chromosome 18. Genomics 69: 27-36. [DOI] [PubMed] [Google Scholar]
- 27.Schalkwyk L.C., Weiher, M., Kirby, M., Cusack, B., Himmelbauer, H., and Lehrach, H. 1998. Refined radiation hybrid map of mouse Chromosome 17. Mamm. Genome 9: 807-811. [DOI] [PubMed] [Google Scholar]
- 28.Schuler G.D., Boguski, M.S., Stewart, E.A., Stein, L.D., Gyapay, G., Rice, K., White, R.E., Rodriguez-Tome, P., Aggarwal, A., Bajorek, E., et al. 1996. A gene map of the human genome. Science 274: 540-546. [PubMed] [Google Scholar]
- 29.Steen R.G., Kwitek-Black, A.E., Glenn, C., Gullings-Handley, J., Van Etten, W., Atkinson, O.S., Appel, D., Twigger, S., Muir, M., Mull, T., et al. 1999. A high-density integrated genetic linkage and radiation hybrid map of the laboratory rat. Genome Res. 9: AP1-AP8. [PubMed] [Google Scholar]
- 30.Stein L., Kruglyak, L., Slonim, D., and Lander, E., 1995. RHMAPPER, unpublished software. Whitehead Institute/MIT Center for Genome Research, Cambridge, MA.
- 31.Stewart E.A., McKusick, K.B., Aggarway, A., Bajorek, E., Brady, S., Chu, A., Fang, N., Hadley, D., Harros, M., Hussain, S., et al. 1997. An STS-based radiation hybrid map of the human genome. Genome Res. 7: 422-433. [DOI] [PubMed] [Google Scholar]
- 32.Sun S., Murphy, W.J., Menotti-Raymond, M., and O'Brien, S.J. 2001. Integration of the feline radiation hybrid and linkage maps. Mamm. Genome 12: 436-441. [DOI] [PubMed] [Google Scholar]
- 33.Van Etten W.J., Steen, R.G., Nguyen, H., Castle, A.B., Slonim, D.K., Ge, B., Nusbaum, C., Schuler, G.D., Lander, E.S., and Hudson, T.J. 1999. Radiation hybrid map of the mouse genome. Nat. Genet. 22: 384-387. [DOI] [PubMed] [Google Scholar]
- 34.Venter J.C., Adams, M.D., Myers, E.W., Li, P.W., Mural, R.J., Sutton, G.G., Smith, H.O., Yandell, M., Evans, C.A., and Holt, R.A. 2001. The sequence of the human genome. Science 291: 1304-1351. [DOI] [PubMed] [Google Scholar]
- 35.Wahls W.P. 1998. Meiotic recombination hotspots: Shaping the genome and insights into hypervariable minisatellite DNA change. Curr. Top. Dev. Biol. 37: 37-75. [DOI] [PMC free article] [PubMed] [Google Scholar]