Abstract
Multiplexed miRNA fluorescence in situ hybridization (miRNA FISH) is an advanced method for visualizing differentially expressed miRNAs, together with other reference RNAs, in archival tissues. Some miRNAs are excellent disease biomarkers due to their abundance and cell-type specificity. However, these short RNA molecules are difficult to visualize due to loss by diffusion, probe mishybridization, and signal detection and signal amplification issues. Here, we describe a reliable and adjustable method for visualizing and normalizing miRNA signals in formalin-fixed paraffin-embedded (FFPE) tissue sections.
Keywords: miRNA, Fluorescence in situ hybridization, Formalin-fixed and paraffin-embedded tissues, Molecular diagnostics, Multiplexing, Signal amplification methods
1 Introduction
RNA fluorescence in situ hybridization (RNA FISH) is a technique for visualizing RNA molecules in fresh-frozen and archival tissues. The technique is based on sequence complementarity between target transcripts and oligonucleotide probes that can be fluorescently labeled through direct or indirect approaches. Due to major advances in next-generation RNA sequencing, oligonucleotide synthesis, and fluorescence microscopy, RNA FISH is poised to become a major diagnostic and research technique.
miRNAs are short, noncoding RNAs (20–23 nt in length) that are useful disease biomarkers due to their abundance and cell-type specificity. miRNA biomarker studies typically involve extracting RNA from complex clinical samples containing multiple cell types and expression profiling using array-, PCR-, or sequencing-based methods. Despite providing valuable quantitative information on miRNA content, these methods are not suitable for assessing miRNA distribution within complex tissues unless labor-intensive laser microdissection technology is used.
To preserve visuospatial information in complex samples, we recently developed multiplexed miRNA FISH for use on FFPE tissues [1]. In our proof-of-concept study, we identified differentially expressed miRNAs in archived skin tumors of similar histologic appearance using barcoded small RNA sequencing and linear discriminant analysis [1, 2]. We subsequently used these reference miRNA profiles to develop miRNA FISH for FFPE tissues, identifying and successfully resolving three major issues involving miRNA fixation, probe specificity, and signal detection and amplification.
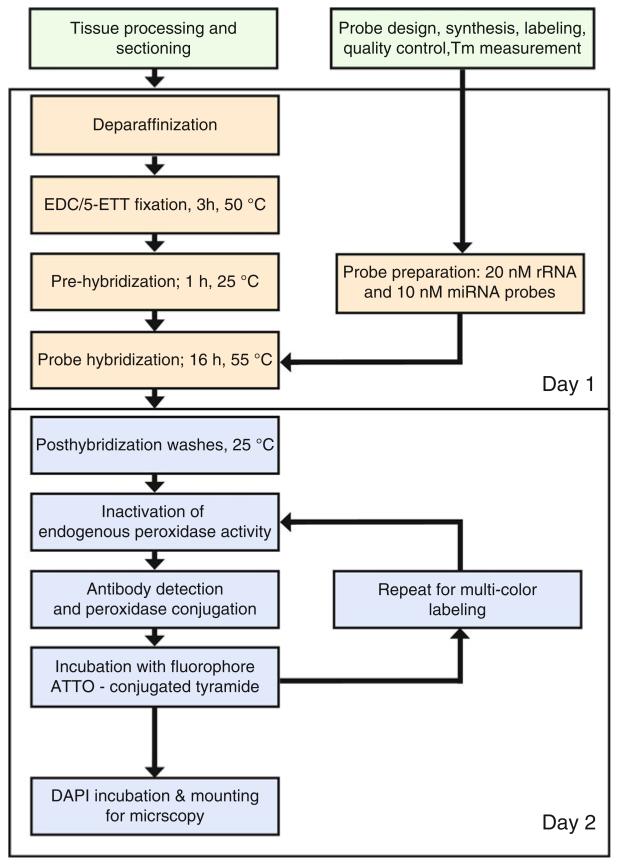
Here, we present our resulting miRNA FISH protocol (Fig. 1). The protocol is divided into two major parts: (1) oligonucleotide probe design and preparation for direct or indirect fluorescent detection (Subheadings 3.1–3.9) and (2) manual miRNA FISH for FFPE tissues (Subheadings 3.10–3.14). This protocol is designed to be flexible so that an individual researcher can easily modify the protocol to detect other miRNAs or RNAs of interest.
Fig. 1.
Protocol overview
2 Materials
Prepare all solutions using Millipore water (18.2 MΩ, 0.2 μm filtered) and analytical grade reagents. Prepare and store all reagents at room temperature unless indicated otherwise. Follow all waste disposal regulations when disposing of waste materials. Substitute reagents and/or equipment according to availability or preference. Materials are listed at first use only.
2.1 rRNA Probe Synthesis
3400 DNA synthesizer (Applied Biosystems).
Empty synthesis columns (Glen Research).
3′-Amino-modifier C7 CPG 500 (Glen Research).
DCI activator configured for ABI, 0.25 M (Proligo Reagent).
Cap A configured for ABI (Proligo Reagent).
Cap B configured for ABI (Proligo Reagent).
Oxidizer 0.02 M configured for ABI (Proligo Reagent).
DMT-dA (bz), DMT-dC (bz), DMT-dG (ib), DMT-dT phosphoramidites configured for ABI (Proligo Reagent).
LNA-A (bz), LNA-mC (bz), LNA-G (dmf), LNA-T phosphoramidites configured for ABI (Exiqon).
Acetonitrile anhydrous, DNA synthesis grade ≥99.9 % (Fisher).
Methylene chloride, Optima (Fisher).
TCA deblock (Sigma-Aldrich).
2.2 miRNA Probe Design and Synthesis
Protected biotinLC serinol phosphoramidite (Glen Research).
6-Fluorescein serinol phosphoramidite (Glen Research).
Spacer phosphoramidite 18 (Glen Research).
3′-6-Fluorescein serinol CPG (Glen Research).
3′-Protected biotinLC serinol CPG (Glen Research).
2.3 rRNA and miRNA Probe Deprotection
Diethylamine puriss. p.a. ≥99.5 % (Sigma-Aldrich).
Diethylamine/acetonitrile anhydrous (10 % v/v) solution, 100 ml.
Piperidine puriss (Sigma-Aldrich).
N, N-Dimethylformamide (DMF, Sigma).
Piperidine/DMF (20 % v/v) solution, 100 ml.
Screw cap micro tubes, 2 ml (Sarstedt).
Ammonium hydroxide (28–30 %) solution (EMD).
Eppendorf shaking thermomixer.
1-Butanol (Fisher).
Polypropylene tubes, 13 ml (Sarstedt).
Sorvall RC 5C Plus (floor model) centrifuge with SS-34 rotor and adapters (Thermo Scientific).
Eppendorf vacufuge concentrator.
3 M NaCl solution.
Absolute ethanol (Pharmco-Aaper).
Sorvall Legend Micro 21R (benchtop) centrifuge (Thermo Scientific).
2.4 rRNA and miRNA Probe Quality Control
SmartSpec Plus UV spectrophotometer (Bio-Rad).
Polyacrylamide gel electrophoresis apparatus (203 × 203 mm, Whatman) with a 20-well comb and 0.4 mm spacers.
Ammonium persulfate (10 % w/v) solution: In a 15 ml Falcon tube, dissolve 1 g ammonium persulfate (National Diagnostics) in 8 ml water. Make up to 10 ml with water (see Note 1).
Denaturing polyacrylamide gel (20 %) solution, 30 ml: 3 ml UreaGel buffer (National Diagnostics), 3 ml UreaGel diluent (National Diagnostics), 24 ml UreaGel concentrate (National Diagnostics), 240 μl ammonium persulfate (10 % w/v) solution, and 12 μl N, N, N ′, N ′-Tetramethylethylenediamine (TEMED, National Diagnostics). Mix solution and pour gel immediately.
10× TBE running buffer, 7 l: In a large plastic (carboy) bottle combine 377.3 g Tris base (Fisher), 192.6 g boric acid (Fisher), and 26.1 g ethylenediaminetetraacetic acid disodium salt dihydrate (Na2EDTA · 2H2O, Sigma). Make up to 7 l with water and mix well. Dilute to 1× TBE as required.
8 M urea solution.
Bromophenol blue sodium salt (Sigma).
Loading dye solution: In a 100 ml glass bottle, dissolve 10 mg bromophenol blue sodium salt in 100 ml 8 M urea solution.
Saran wrap.
Fluorescence-indicator-coated silica gel plate, 20 × 20 cm, F254s (Whatman).
2.5 rRNA Probe Direct Fluorescent Labeling
100 mM Na2CO3 solution.
100 mM NaHCO3 solution.
100 mM sodium carbonate buffer, pH 9.0: In a 100 ml glass bottle, combine 11 ml 100 mM Na2CO3 solution and 89 ml 100 mM NaHCO3 solution.
DMF, anhydrous (Solulink).
ATTO-647N NHS ester (10 mM) solution: In a 1.5 ml Eppendorf tube, dissolve 1 mg ATTO-647N NHS ester (ATTO TEC, GmbH) in 119 μl anhydrous DMF.
2.6 Fluorophore-Labeled rRNA Probe Purification
Polyacrylamide gel electrophoresis apparatus (395 × 225 mm) with an 8-well comb and 2 mm spacers.
Denaturing polyacrylamide gel (18 %) solution, 200 ml: In a 500 ml glass beaker, combine 20 ml UreaGel buffer (National Diagnostics), 36 ml UreaGel diluent (National Diagnostics), 144 ml UreaGel concentrate (National Diagnostics), 1.6 ml ammonium persulfate (10 % w/v) solution, and 0.08 ml TEMED (National Diagnostics). Mix solution and pour gel immediately.
Disposable scalpels.
Eppendorf tubes, 2 ml.
Labquake rotisserie rotator (Thermo Scientific).
2.7 Fluorophore-Labeled rRNA Probe Concentration Determination
Cary 100 Series UV-Vis spectrophotometer (Agilent Technologies).
Hellma quartz cuvettes, 10 mm light path, 1 ml (Hellma Analytics).
ATTO-Tyramide Conjugation for miRNA Signal-Amplification-Based Fluorescent Detection
ATTO-488 NHS ester (ATTO TEC, GmbH).
ATTO-488 stock solution A: ATTO-488 NHS ester (10 mg/ml): In a 1.5 ml Eppendorf tube, dissolve 5 mg ATTO-488 NHS ester in 500 μl anhydrous DMF (Solulink).
ATTO-532 NHS ester (ATTO TEC, GmbH).
ATTO-532 stock solution A: ATTO-532 NHS ester (10 mg/ml): In a 1.5 ml Eppendorf tube, dissolve 5 mg ATTO-532 NHS ester in 500 μl anhydrous DMF (Solulink).
Tyramine-HCl (Sigma).
Triethylamine (TEA, Fisher).
Tyramine-HCl stock solution B: Tyramine-HCl (10 mg/ml): In a 1.5 ml Eppendorf tube, dissolve 10 mg tyramine-HCl in 1 ml anhydrous DMF (Solulink). Add 10 μl TEA.
Ethanolamine (Sigma).
Ethanolamine/anhydrous DMF (1 % v/v) stock solution C, 200 μl.
Dimethyl sulfoxide (DMSO, Sigma).
2.8 Melting Temperature Curve Analysis of Oligonucleotide Duplexes
Hellmanex cuvette cleaning concentrate (Fisher).
Hellmanex cuvette cleaning solution: In a 500 ml squeeze wash bottle, add 2.5 ml Hellmanex cuvette cleaning concentrate to 497.5 ml water.
Vakuwash cuvette washer (Aldrich).
Formamide, deionized (Chemicon).
1 M Na2HPO4 solution.
1 M NaH2PO4 solution.
Dry bath incubator heat block (Fisher).
Mineral oil (Sigma-Aldrich).
Space saver vacuum dessicator (Bel-Art Products).
2.9 FFPE Tissue Sectioning
Automated rotary microtome Leica RM2255 (Leica Biosystems).
Water bath for paraffin sections Leica HI1210 (Leica Biosystems).
Colorfrost plus microscopic slides (Fisher).
Fine-tipped watercolor brush.
InSlide out oven with stainless steel slide racks and aluminum trays (Boekel Scientific).
2.10 Deparaffinization and EDC/5-ETT Fixation
Glass Coplin jars.
Histo-Clear II (National Diagnostics).
1 M HCl solution (see Note 2).
1-Methylimidazole (Sigma, see Note 3).
0.1 M 1-Methylimidazole buffer, pH 8.0: In a 1 l glass bottle, combine 7.93 ml 1-methylimidazole, 100 ml 3 M NaCl solution, and 800 ml water. Adjust pH to 8.0 by adding about 13 ml 1 M HCl. Make up to 1 l with water.
50× Denhardt’s solution (AppliChem, see Note 4).
10 M NaOH solution: In a 100 ml glass bottle, weigh 40 g NaOH and add 70 ml water. Transfer solution to a 100 ml graduated cylinder. Make volume up to 100 ml with water. Store solution in an airtight plastic bottle.
5-Ethylthio-1H-tetrazole (5-ETT, American International Chemical, Inc.).
N-(3-Dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC-HCl, Sigma).
EDC/5-ETT fixation solution: In a 15 ml Falcon tube, add 97 μl 10 M NaOH to 9.5 ml 0.1 M 1-methylimidazole buffer, pH 8.0. Vortex. Add 130 mg 5-ETT. Vortex vigorously until dissolved. Measure pH and adjust with 10 M NaOH or 1 M HCl, as necessary, to ensure that the pH is 8.0. Immediately prior to use, add 0.4 ml 50× Denhardt’s solution, and dissolve 192 mg EDC-HCl in the freshly prepared mixture (see Note 5).
1 M Tris buffer (pH 7.4): In a 1 l glass bottle, combine 820 ml 1 M Tris–HCl buffer and 180 ml 1 M Tris base solution.
1× TBS buffer: In a 1 l glass bottle, combine 970 ml water, 33.3 ml 3 M NaCl, and 10 ml 1 M Tris buffer (pH 7.4).
2.11 Hybridization
Formamide, ≥99.0 % (GC, Sigma).
Baker’s yeast tRNA (20 mg/ml) stock solution (Sigma, see Note 6).
Salmon sperm DNA (AppliChem).
1 M Tris buffer (pH 8.5): In a 1 l glass bottle, combine 266 ml 1 M Tris–HCl solution and 734 ml 1 M Tris base solution.
Tween-20 (Sigma).
3[(3-Cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS, Sigma).
Hybridization buffer: In a 100 ml glass bottle, combine 25 ml formamide (Sigma), 16.7 ml 3 M NaCl solution, 3.75 ml 1 M Tris buffer (pH 8.5), 1 ml 50× Denhardt’s solution, 0.625 ml Baker’s yeast tRNA stock solution, 25 mg salmon sperm DNA, 77 mg CHAPS, 0.25 ml Tween-20, and 2.675 ml water (see Note 7).
Siliconized Eppendorf tubes.
Wash buffer 1: In a 1 l glass bottle, combine 500 ml formamide, 83.3 ml 3 M NaCl solution, 75 ml 1 M Tris buffer (pH 8.5), and 1 ml Tween-20. Make up to 1 l with water.
Wash buffer 2: In a 1 l glass bottle, combine 16.7 ml 3 M NaCl solution, 75 ml 1 M Tris buffer (pH 8.5), and 1 ml Tween-20. Make up to 1 l with water.
1× TBS-T buffer: In a 1 l glass bottle, combine 999 ml 1× TBS and 1 ml Tween-20.
2.12 Antibody Detection and Signal Amplification
H2O2 solution, 30 % (Fisher, see Note 8).
Goat serum (Sigma, see Note 9).
Antibody-blocking solution: In a 15 ml Falcon tube, combine 0.5 ml goat serum and 9.5 ml 1× TBS-T buffer.
Anti-fluorescein-HRP antibody (Perkin Elmer).
Anti-fluorescein-HRP (1:100) solution: In a 1.5 ml siliconized Eppendorf tube, combine 950 μl antibody-blocking solution with 10 μl anti-fluorescein-HRP antibody and 40 μl 50× Denhardt’s solution (see Note 10).
Streptavidin-HRP antibody (Perkin Elmer).
Streptavidin-HRP (1:500) solution: In a 1.5 ml siliconized Eppendorf tube, combine 958 μl antibody-blocking solution with 2 μl streptavidin-HRP antibody and 40 μl 50× Denhardt’s solution (see Note 11).
4-Bromophenylboronic acid (Aldrich).
Bromophenylboronic acid solution: In a 1.5 ml Eppendorf tube, weigh 20 mg 4-bromophenylboronic acid and dissolve in 400 μl DMSO (see Note 12).
Pre-amplification buffer (pH 8.0): In a 100 ml glass beaker, combine 2.34 ml 1 M Tris base, 2.66 ml 1 M Tris–HCl, 66.6 ml 3 M NaCl solution, 28 ml water, and 400 μl 4-bromophenylboronic acid.
Amplification buffer: In a 15 ml Falcon tube, combine 10 ml pre-amplification buffer (pH 8.0) with 5.1 μl 30 % H2O2 (see Note 13).
Amplification solution: In a 15 ml Falcon tube, combine 4.9 ml amplification buffer with 0.1 ml either ATTO-488 or −532 tyramide as required (see Note 14).
2.13 Slide Mounting and Microscopy
4′,6-Diamidino-2-phenylindole dihydrochloride (DAPI, Sigma).
DAPI (5 mg/ml) stock solution.
DAPI working solution: In a 15 ml Falcon tube, combine 10 μl DAPI (5 mg/ml) stock solution in 10 ml 1× TBS-T buffer.
0.2 M Tris buffer (pH 6.8): In a 100 ml glass beaker, combine 1 ml 1 M Tris base, 19 ml 1 M Tris–HCl, and 80 ml water. Mix and transfer to a glass bottle.
Mounting solution: In a 50 ml glass bottle, slowly add 2.4 g MOWIOL (Polysciences Inc.) to 6.0 g glycerol (Fisher) over the course of a few hours. Mix solution using a stir bar and magnetic stirrer. Add 6 ml water and incubate overnight at 25 °C. Add 12 ml 0.2 M Tris buffer (pH 6.8) and heat for 10 min at 50 °C with occasional mixing (see Note 15).
Glass cover slips, 24 × 50 mm (Thermo Scientific).
VS 110 fluorescence virtual slide scanning system (Olympus).
3 Methods
Carry out all procedures at room temperature unless otherwise indicated.
3.1 rRNA Probe Synthesis
Synthesize LNA-spiked oligonucleotide probes targeting human or mouse ribosomal 28S RNA (i.e., rRNA probes, see below) at a 1.0 μmol scale on a 3400 DNA synthesizer using 3′-amino-modifier C7 CPG (500 Å) solid glass support and DNA and LNA phosphoramidites (see Note 16).
Human and mouse 28S rRNA probe set (see Note 17)
LNA1: 5′-CTttTCtGggGTcTGaT-(X-CPG) (TM 83.5 °C).
LNA2: 5′-CAGcGCcATcCAtTTtCAG-(X-CPG) (TM 78.5 °C).
LNA3: 5′-CATCTcTcAGGAcCgAcT-(X-CPG) (TM 77.3 °C).
LNA4: 5′-GGTgCctCtCGtACTgA-(X-CPG) (TM 78.2 °C).
(Upper case: DNA nucleotides; lower case: LNA modification; X-CPG: 3′-amino-modifier C7 CPG 500.)
Alternately, order these probes following the exact sequence from a commercial vendor.
3.2 miRNA Probe Design and Synthesis
Design antisense LNA-modified probes targeting miRNAs of interest using mature miRNA sequences from miRBase (www.miRBase.org). To minimize rRNA cross-hybridization, avoid probe sequences with greater than six consecutive nucleotide matches to rRNA and shorten probes to 14–15 nt in length as required. Assess hairpin formation and self-dimerization using MFold (http://mfold.rna.albany.edu). Insert LNA modifications in regions of no secondary structure or self-hybridization (see Note 18).
Synthesize LNA-modified miRNA probes on a 1.0 μmol scale as described for rRNA probes above. To enable miRNA detection through signal amplification, incorporate hapten (e.g., biotin or fluorescein) and spacer (aka linker) phosphoramidites at the 3′ end of the desired probe sequence.
Examples of successfully validated miRNA probes
miR-205: 5′-GGTGGAAtgaAgga-(L)3-F-L-F-L-F-L-F-L-F-(F-CPG) (TM 61.6 °C).
miR-375: 5′-AGCCGaaCGaAcaaA-(L)3-B-L-B-L-B-L-B-L-B-(B-CPG) (TM 62.1 °C).
(Upper case: DNA nucleotides; lower case: LNA modification; L: spacer phosphoramidite 18, F: 6-fluorescein serinol; F-CPG: 3′-6-fluorescein serinol CPG, B: 3′-protected biotinLC serinol CPG.)
Alternately, order these probes following the exact sequence from a commercial vendor.
3.3 rRNA and miRNA Probe Deprotection
Deprotect rRNA probes with piperidine washes (see Note 19). Wash synthesis columns three times with 3 ml diethylamine/acetonitrile anhydrous (10 % v/v) solution for 3 min (see Note 20) and rinse once with 10 ml acetonitrile anhydrous. Wash columns twice with 3 ml piperidine/DMF (20 % v/v) solution for 5 min, and rinse twice with 2 ml DMF, and twice with 3 ml acetonitrile anhydrous. Gently blow air through column for 10 s to dry.
Remove CPGs from their corresponding synthesis column. Using 2 ml screw cap tubes, combine each CPG with 1.7 ml ammonium hydroxide (28–30 %) solution. Incubate tubes in an Eppendorf shaking thermomixer set at 650 rpm for 16 h at 55 °C. Cool tubes on ice for 5 min. Using 13 ml polypropylene tubes, combine each CPG mixture with 11 ml 1-butanol, vortexing thoroughly. Collect oligonucleotide pellets by centrifugation in a floor model centrifuge at 20,000 × g (10,500 rpm) for 30 min at 4 °C. Dry pellets in an Eppendorf vacufuge concentrator set to aqueous mode at 45 °C. Resuspend pellets in 400 μl water and transfer to 2 ml Eppendorf tubes.
Precipitate resuspended rRNA probes. Add 50 μl 3 M NaCl and 1,350 μl chilled absolute ethanol. Vortex and store for 1 h on ice (see Note 21). Collect pellets in a benchtop centrifuge at 16,200 × g (13,000 rpm) for 20 min at 4 °C. Dry pellets in an Eppendorf vacufuge concentrator set to aqueous mode at 25 °C. Resuspend pellets in 400 μl water.
Deprotect miRNA probes using different solutions from above. Wash synthesis columns three times with 3 ml diethylamine/acetonitrile anhydrous (10 % v/v) solution for 3 min and rinse each column with 10 ml acetonitrile anhydrous. Gently blow air through column for 10 s to dry. Combine CPG and ammonium hydroxide (28–30 %) solution, and incubate tubes in an Eppendorf shaking thermomixer set at 650 rpm for 16 h at 55 °C. Cool tubes on ice for 5 min, and combine each CPG mixture with 11 ml 1-butanol in 13-ml polypropylene tubes, vortexing thoroughly. Collect pellets by centrifugation and dry pellets as described above. Resuspend pellets in 400 μl water. Transfer to 1.5 ml Eppendorf tubes. Proceed to the quality control step below (see Note 22).
3.4 rRNA and miRNA Probe Quality Control
Assess rRNA and miRNA probe quality through spectrophotometry and polyacrylamide gel electrophoresis.
Determine probe concentrations using a SmartSpec Plus UV spectrophotometer.
Prepare a 20 % denaturing polyacrylamide gel. Run in 1× TBE running buffer. Pre-run the gel at 20 W for 30 min. Combine 0.4 OD260 units oligonucleotide probe with 15 μl loading dye solution. Incubate samples at 95 °C for 3 min. Load each well with up to 35 μl sample. Run gel at 10 W for 3–4 h until the dye reaches the bottom of the gel. Remove glass plates and cover gel with Saran wrap. Place gel on a fluorescence-indicator-coated silica gel plate and UV-shadow to detect starting material and product.
3.5 rRNA Probe Direct Fluorescent Labeling
Aliquot 50 nmol (approx. 14 OD260 units) each unlabeled rRNA probe into separate 1.5 ml Eppendorf tubes. Dry aliquots in an Eppendorf vacufuge concentrator. Dissolve pellets in 50 μl 100 mM sodium carbonate buffer, pH 9.0 to obtain an rRNA probe concentration of 1 mM.
For direct fluorescent labeling of rRNA probes, combine 50 μl 1 mM rRNA probe solution and 50 μl 10 mM ATTO-647N NHS ester solution (see Note 23). Mix well, vortexing as needed to dissolve visible precipitates. Incubate tubes in an Eppendorf shaking thermomixer overnight at 37 °C. Precipitate probe by adding 11 μl 3 M NaCl solution and 333 μl chilled absolute ethanol (see Note 21). Vortex and store for 1 h on ice or overnight at −20 °C. Centrifuge in a benchtop centrifuge at 16,200 × g (13,000 rpm) for 20 min at 4 °C. Remove supernatant. Dissolve pellet in 70 μl water. Add 140 μl loading dye solution.
3.6 Fluorophore-Labeled rRNA Probe Purification
Prepare an 18 % denaturing polyacrylamide gel and 1× TBE running buffer. Pre-run gel at 50 W for 30 min. Incubate samples for 3 min at 95 °C. Load 30 μl probe/loading dye solution into each of the seven wells and 30 μl unlabeled probe/loading dye solution (use approx. 7–8 nmol unlabeled probe) in the remaining well as a reference. Run the gel at 30 W for 7 h (see Note 24). Disassemble apparatus, removing both glass plates, and cover gel with Saran wrap. Place gel on a fluorescence-indicator-coated silica gel plate and UV-shadow to detect the starting material and product (see Note 25). Circle fluorophore-labeled probes with a marker. Use a clean scalpel to cut out products and place in a 2 ml Eppendorf tube. Weigh each gel piece-containing tube, subtracting the weight of each empty tube. Based on these weights, elute rRNA probes by adding two volumes of (about 1.5 ml) 0.3 M NaCl solution to each tube and rotating on a rotisserie rotator overnight. Transfer supernatants to 13 ml polypropylene centrifuge tubes. Add 3 volumes of chilled absolute ethanol (see Note 21). Vortex and incubate for at least 1 h on ice or overnight at −20 °C. Collect pellets in a floor model centrifuge at 20,000 × g (10,500 rpm) for 30 min at 4 °C. Dissolve each pellet in 100 μl water.
3.7 Fluorophore-Labeled rRNA Probe Concentration Determination
Measure fluorophore-labeled rRNA probe concentration in a Cary Series 100 UV-Vis Spectrophotometer (see Note 26). Load sample into a Hellma quartz cuvette. Measure absorbance spectrum (from 200 to 800 nm) of ATTO-647N-labeled probe aliquot. Determine corrected concentration of fluorophore-labeled probe using the following equation: A260, corrected = A260, observed − (CF260, ATTO-647N × A644); CF260 is the correction factor for A260 readings (see Note 27), and A644 is the absorbance reading at the absorption maximum (λmax) of ATTO-647N.
3.8 ATTO-Tyramide Conjugation for miRNA Signal-Amplification-Based Fluorescent Detection
Mix 500 μl ATTO-488 or ATTO-532 NHS ester stock solution A with 84 or 76 μl tyramine-HCl stock solution B, respectively. Incubate in the dark for 2 h. Add 5.9 or 5.3 μl ethanolamine stock solution C, respectively. Stir for 5 min. Make stock solutions up to 5 ml with DMSO. Aliquot ATTO-tyramide solution, termed stock solution D, into light-protected Eppendorf tubes (see Note 28).
3.9 Melting Temperature Curve Analysis of Oligonucleotide Duplexes
Wash cuvettes with Hellmanex cleaning solution (see Note 29). Invert cuvettes on top of a Vakuwash cuvette washer. Pour water in funnel, directing jet to the base of the cuvette. Drain under vacuum. Pour 20 ml 70 % ethanol in funnel and drain as above. Air-dry cuvettes.
Prepare 300 μl oligonucleotide (probe-target) mixture with an A260 of ~0.5 in phosphate buffer (see Note 30). In a 1.5 ml siliconized Eppendorf tube, combine 150 μl formamide, 100 μl 3 M NaCl solution, 10 μl 1 M Na2 HPO4, 5 μl 1 M NaH2PO4, 2 μM (final concentration) target oligonucleotide, and 2 μM (final concentration) complementary oligoribonucleotide probe. Make up to 300 μl with water.
Slowly anneal the probe-target mixture in a heatblock or PCR machine using the following conditions: 5 min at 95 °C, 5 min at 80 °C, gradual decrease to 50 °C over 3 h, gradual decrease to 25 °C over 1 h, and hold at 25 °C overnight (see Note 31). Transfer mixture to a clean cuvette. Carefully overlay 500 μl mineral oil. Fill the reference cuvette with 1 ml sample buffer without probe and do not cover with mineral oil. Degas samples twice for 10 min in a vacuum dessicator. Cap all cuvettes except for the reference cuvette. Measure absorbance at 280 nm (see Note 32) from 20 °C up to 90 °C and from 90 °C down to 20 °C with ramping speed of 0.50 °C/min. Determine melting temperature in a Cary 100 UV-Vis spectrophotometer.
3.10 FFPE Tissue Sectioning
The following manual miRNA protocol is a modified version (see Note 33) of an established method [1, 3]. First, prepare or obtain FFPE tissue blocks (see Note 34). Trim paraffin blocks to an optimal cutting surface using a rotary microtome. Cut 5 μm sections and place paraffin ribbon in ice-cold water bath. Using a glass slide, transfer one section to a 42 °C water bath, allow to expand, and position on a glass slide with a wet brush (see Note 35). Dry sections at room temperature for 1 h until the water trapped between the tissue and slide has evaporated. Dry sections in a laboratory oven at 40 °C for 1 h. Bake sections in a hybridization oven at 56 °C for 1 h prior to use (see Note 36).
3.11 Deparaffinization and EDC/5-ETT Fixation
In a glass Coplin jar, using 50 ml volumes, deparaffinize sections through serial immersion in Histo-Clear II (National Diagnostics) twice for 5 min, 100 % ethanol for 2 min, 95 % ethanol twice for 1 min, 70 % ethanol for 1 min, and 50 % ethanol for 1 min. Rinse slides several times in cold tap water and proceed immediately to the next step.
Fix miRNAs in FFPE tissue sections using EDC/5-ETT fixation solution. Place slides face up in a stainless steel slide rack in a metal tray (see Note 37). Add 500 μl EDC/5-ETT fixation solution to each slide, seal tray, and incubate in the hybridization oven above for 3 h at 50 °C. Wash samples twice with 50 ml 1× TBS for 3 min.
3.12 Hybridization
Place slides on a stainless steel slide rack. Pre-hybridize sections by adding 500 μl freshly prepared hybridization buffer to each slide. Fully cover tissue sections. Incubate slides in a sealed metal tray in a humidified chamber for 1 h at 25 °C. Decant hybridization buffer.
Select the desired rRNA and miRNA probes and preheat ovens to a hybridization temperature approximately 10 °C below the lowest TM of the hapten-conjugated miRNA probes. Prepare an rRNA and miRNA probe cocktail in hybridization buffer; rRNA and miRNA probes should be present at a final concentration of 20 nM and 10 nM, respectively (see Note 38). Add 100–200 μl probe hybridization solution to each slide covering the entire tissue. Incubate slides as above for 16 h at the hybridization temperature.
In glass Coplin jars, using 50 ml volumes, wash slides twice with wash buffer 1 for 10 min and once with wash buffer 2 for 5 min. Rinse in 1× TBS-T buffer for 3 min.
3.13 Antibody Detection and Signal Amplification
Block endogenous peroxidase activity by immersing slides in 50 ml 3 % H2O2 solution for 25 min (see Note 39). Wash slides three times in 50 ml 1× TBS-T buffer for 3 min. Place slides in a humidified slide rack.
Detect miRNAs of interest through consecutive addition of HRP-conjugated antibodies targeting known haptens in the respective miRNA probes and tyramide signal amplification. Add 500 μl antibody-blocking solution to each slide. Incubate slides for 1 h at 25 °C. Decant antibody-blocking solution. Dilute anti-fluorescein-HRP antibody (1:100) in antibody-blocking solution. Add 500 μl dilute antibody solution to each slide and incubate for 1 h at 25 °C. Wash slides twice in 50 ml 1× TBS-T for 10 min. Place slides in a humidified slide rack. Add 400 μl amplification solution containing tyramide-ATTO-532 (1:50 dilution) onto each slide. Incubate in the dark for 30 min.
Wash slides three times in 50 ml 1× TBS-T buffer for 3 min. Incubate slides in 50 ml 3 % H2O2 solution for 25 min (see Note 39). Wash slides three times in 50 ml 1× TBS-T buffer for 3 min. Place slides in a humidified slide rack. Dilute streptavidin-HRP antibody (1:500) in antibody-blocking solution. Add 500 μl dilute antibody solution to each slide and incubate for 1 h at 25 °C. Wash slides twice in 50 ml 1× TBS-T for 10 min. Place slides in a humidified slide rack. Add 400 μl amplification solution containing tyramide-ATTO-488 solution (1:50 dilution) onto each slide. Incubate in the dark for 30 min. Wash slides three times in 50 ml 1× TBS-T for 3 min.
3.14 Slide Mounting and Microscopy
Place slides in a humidified slide rack. Add 500 μl DAPI working solution to each slide and incubate for 10 min. Wash slides twice in 50 ml 1× TBS for 3 min. Place slides in a humidified slide rack. Place two drops of mounting solution on each tissue section. Carefully position a 24 × 50 mm glass cover slip over each tissue section, avoiding air bubbles. Air-dry for 10 min. Store samples in a dark slide rack. Perform microscopy, obtaining images and gaining fluorescence data for rRNA and miRNAs; normalize miRNA signals against rRNA signals (see Note 40). Examples of miRNA FISH images and normalization of fluorescence data are available in our recent manuscript on the same topic [1].
Acknowledgements
N.R. is supported through a K08 award (NS072235) from the National Institute of Neurological Disorders and Stroke. T.T. is an HHMI investigator and supported through R01 funding from NIH CA159227 and MH080442 and a grant by the Starr Cancer Consortium. The project described was supported through the Rockefeller University Bridges to Better Medicine Technology Innovation Fund. The project was also partially supported by Grant Award Number (UL1RR024143) from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH) and NIH Roadmap for Medical Research, and its contents are solely the responsibility of the authors and do not necessarily represent the official view of NCRR or NIH.
Footnotes
Store ammonium persulfate (10 % w/v) solution for 1–2 weeks at 4 °C.
Work in a fume hood. Concentrated HCl is caustic. In a 500 ml graduated cylinder, add 450 ml water. In a 50 ml graduated cylinder, measure 41.3 ml conc. HCl (37.3 %, 12.1 M, Fisher). Slowly add the smaller to the larger volume. Make up to 500 ml with water. Transfer solution to a glass bottle for storage at 25 °C.
1-Methylimidazole is a colorless liquid that should be replaced if discolored yellow. Handle liquid with a volumetric glass syringe. Store 1-methylimidazole under argon for a few weeks at 25 °C.
Store 50× Denhardt’s solution at −20 °C.
Prepare EDC/5-ETT solution immediately prior to use. Take the EDC-HCl bottle out of the freezer 1 h before weighing out sample to allow it to reach room temperature and avoid condensate reacting with the reagent. Store anhydrous EDC-HCl powder under argon at −20 °C.
Store Baker’s yeast tRNA stock solution in 0.625 ml aliquots at −20 °C.
Store hybridization buffer at −20 °C.
Make sure that commercial H2O2 does not exceed the expiration date. Store diluted solution at 4 °C for up to 1 week.
Store goat serum at −20 °C.
A 1:100 antibody dilution is recommended but may vary per batch.
A 1:500 antibody dilution is recommended but may vary per batch.
Bromophenylboronic acid solution can be stored for at least a year at 25 °C.
We prepare amplification buffer with H2O2 immediately prior to use.
Dilute ATTO-tyramides 1:50 in amplification buffer.
Store MOWIOL solution for several months at −20 °C.
For detailed instructions on automated single-stranded oligonucleotide synthesis using the ABI 3400 DNA synthesizer, refer to the web-accessible user guide [4].
Buffer composition and pH and probe concentrations for these TM measurements are provided in Subheadings 2.12 and 3.12, respectively.
We typically incorporate 4–6 LNAs in a 15-mer probe, avoiding positions that would stabilize interactions with rRNAs.
Use piperidine washes to remove FMOC protection group from 3′-amino-modifier C7.
In this step, cyanoethyl protecting groups are removed from the phosphate backbone.
Use 10 % additional volume ethanol to accommodate pipetting errors.
Store fluorescein hapten-containing oligonucleotides in light-protected Eppendorf tubes.
Use approximately tenfold excess ATTO dye NHS ester in the direct fluorescent labeling reaction.
Place a metal plate in front of the glass plate to ensure even distribution of heat. Run until the dye has migrated to the bottom of the gel (approx. 7 h). Running at 60 W will likely crack the gel.
Unlabeled probes migrate faster than labeled probes. Labeled probes are the dominant product.
For detailed instructions, refer to the Cary Spectrophotometers Users Guide [5].
CF260, ATTO-647N = 0.06. Correction factors (CFs) can typically be found in the manufacturer’s manuals and dye specifications. CFs can also be experimentally determined by measuring the absorbance spectrum of a particular dye (from 200 to 800 nm) and calculating the ratio A260,dye/Aλmax of the dye·
Stock solution D can be stored for at least 8 months at 4 °C or 1 year at −20 °C [6].
For accurate absorbance measurements, remove all liquids from the cuvettes, including mineral oil residue from previous use.
The pH of the phosphate buffer is approx. pH 7.0.
Slower annealing prevents hairpin formation.
Measure absorbance at 280 nm because formamide absorbs at 260 nm.
In this protocol, we omit proteinase K permeabilization, 4 % paraformaldehyde fixation, acetylation, and endogenous biotin-blocking steps.
There is no universal protocol for FFPE tissue preparation. Review each protocol and avoid steps (e.g., unnecessary contact with non-fixative solutions) that allow miRNA diffusion.
To prevent paraffin melting and miRNA diffusion, set the water bath at 42 °C and avoid lengthy (>10 s) immersion of tissue sections in water.
After drying, slides can be reliably stored for at least 1 year at 4 °C.
Ensure that slides are facing upwards for the remainder of the protocol.
Use directly labeled rRNA probes to normalize signals from hapten-conjugated miRNA probes. rRNA probes also enable assessment of RNA retention, integrity, and specificity for probe hybridization.
Probe validation should be performed on characterized tissues of known miRNA content.
Block endogenous peroxidase activity with 3 % H2O2 solution before adding antibody-HRP solution. Prepare 3 % H2O2 solution immediately prior to use; expired solutions can result in high background signals.
Please refer to the user guide or a knowledgeable resource for fluorescence microscopy. With our microscopy system, we captured images using 20 and 60× UPlanSApo objectives. Signal intensity histograms were obtained to delineate specific RNA from background signals. Following background removal, pixel intensities for miRNAs and rRNA were multiplied by the corresponding sum of pixels. Use these values to normalize miRNA against reference rRNA signals.
References
- 1.Renwick N, Cekan P, Masry PA, et al. Multicolor microRNA FISH effectively differentiates tumor types. J Clin Invest. 2013;123:2694–2702. doi: 10.1172/JCI68760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hafner M, Renwick N, Farazi TF, et al. Barcoded cDNA library preparation for small RNA profiling by next-generation sequencing. Methods. 2012;58:164–170. doi: 10.1016/j.ymeth.2012.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pena JT, Sohn-Lee C, Rouhanifard SH, et al. miRNA in situ hybridization in formaldehyde and EDC-fixed tissues. Nat Methods. 2009;6:139–141. doi: 10.1038/nmeth.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. http://www3.appliedbiosystems.com/cms/groups/mcb_support/documents/generaldocuments/cms_095581.pdf.
- 5. http://www.chem.agilent.com/Library/usermanuals/Public?1972_7000.pdf.
- 6.Hopman AH, Ramaekers FC, Speel EJ. Rapid synthesis of biotin-, digoxigenin-, trinitrophenyl-, and fluorochrome-labeled tyramides and their application for in situ hybridization using CARD amplification. J Histochem Cytochem. 1998;46:771–777. doi: 10.1177/002215549804600611. [DOI] [PubMed] [Google Scholar]