Abstract
Discovery of the cellular and molecular mechanisms of induced pluripotency has been hampered by its low efficiency and slow kinetics. Here, we report an experimental system with multi-color time-lapse microscopy that permits direct observation of pluripotency induction at single cell resolution, with temporal intervals as short as five minutes. Using granulocyte-monocyte progenitors as source cells, we visualized nascent pluripotent cells emerge from a hematopoietic state. We engineered a suite of image processing and analysis software to annotate the behaviors of the reprogramming cells, which revealed the highly dynamic cell-cell interactions associated with early reprogramming. We observed frequent cell migration, which can lead to sister colonies, satellite colonies and colonies of mixed genetic makeup. In addition, we discovered a previously unknown morphologically distinct 2-cell intermediate of reprogramming, which occurs prior to other reprogramming landmarks. By directly visualizing the reprogramming process with E-cadherin inhibition, we demonstrate the requirement of E-cadherin for proper cellular interactions from an early stage of reprogramming, including the 2-cell intermediate. The detailed cell-cell interactions revealed by this imaging platform shed light on previously unappreciated early reprogramming dynamics. This experimental system could serve as a powerful tool to dissect the complex mechanisms of early reprogramming by focusing on the relevant but rare cells with superb temporal and spatial resolution.
INTRODUCTION
The discovery of induced pluripotent stem cells (iPSCs) 1–4 has made a profound conceptual impact on many biomedical disciplines, putting forward the hope to derive patient-specific therapeutic cell types and to study human diseases that had been previously difficult to model. Indeed, many somatic cell types can be reprogrammed to pluripotency by forced expression of defined factors such as Oct4, Sox2, Klf4 and c-Myc. However, this process generally takes a long time and the cells undergoing successful reprogramming are rare 5–6. Due to this inherent slow kinetics and inefficiency, mechanistic studies have relied on analyzing the bulk cell cultures, among which the relevant cells are the minority. Furthermore, even with the help of pluripotency reporters such as Oct-4-GFP or Nanog-GFP 7–8, assessment of reprogramming can only be made with cells that are already in the late reprogramming phase. Very little is known about the early cellular and molecular events preceding activation of the endogenous pluripotency circuitry 9.
Direct observation using time-lapse microscopy has been an important approach to define the molecular and cellular events during early reprogramming 10–13. However, the utility of these approaches has been compromised by one or more of the following technical limitations. 1) The imaging intervals are long (such as 6 hours or longer) and faithful single cell tracking can be difficult 11–12. Cellular events that occur faster than this temporal resolution are not detectible. 2) Constitutive iPS factor expression precludes accurate determination of the onset for factor action, which creates difficulty in identifying the initial founder cells of reprogrammed colonies 10–11, 13. 3) The slow kinetics of the reprogramming process (1–2 weeks) either requires medium change during imaging 10 or constrains imaging to a limited time window (e.g. from day 4.5 13). Medium change can disturb imaging continuity 10 as well as the local extracellular environment, and may physically dislodge cells and generate secondary colonies. 4) Monitoring pluripotency by colony morphology or post-reprogramming staining 11–12 does not allow real time visualization of the milestones of reprogramming.
Here, we describe an experimental method which overcomes many of the limitations and provides fine temporal and graphical details of early reprogramming. Hematopoietic cells have been often used as source cells to model and reveal reprogramming mechanisms 7–8. In particular, we used a well-characterized hematopoietic progenitor population, the granulocyte-monocyte progenitors (GMPs), as source cells for reprogramming since they have been meticulously defined both phenotypically and functionally 14, can be prospectively isolated to high purity, and support reprogramming with high efficiency 7. We found that highly dynamic cell migration occurs leading to the formation of sister colonies, satellite colonies, or mixed colonies containing cells of different genetic makeup. We report a 2-cell intermediate stage that is characterized by a unique dumbbell-like morphology, and induction of intracellular E-cadherin at this early stage. In addition, we demonstrate the feasibility of our system to accommodate additional reporters for interrogation of specific genetic effects during reprogramming by capturing perturbed reprogramming when E-cadherin is knocked down with shRNAs. Overall, this system provides a powerful tool to dissect the molecular and cellular mechanisms involved in factor-based pluripotency induction.
MATERIAL AND METHODS
Mouse Maintenance and Blastocyst Injection
The Oct-4-GFP:Rosa26-rtTA mice were derived by crossing Oct4-GFP mice with Rosa26-rtTA mice as described 7. The tet-O-Cre mice and E-cadherinfl/fl mice were purchased from the Jackson Laboratory and further crossed with the Rosa26-rtTA mice to achieve doxycycline inducible inactivation of E-cadherin. All mouse work was approved by the Institutional Animal Care and Use Committee of Yale University.
GMP-derived, doxycycline (dox) independent, GFP+ cells with typical ESC morphology were used for karyotype analysis by standard methods using Wright’s stain with 20 spreads counted per iPSC clone. For blastocyst injection, iPSCs were passaged in 2i medium 15–16 supplemented with leukemia inhibitory factor (LIF) 2–3 times prior to injection. C57Bl/6Tyrc-2J blastocysts were microinjected with 12–15 iPSCs per blast via standard methods 17. Injected embryos were transferred to uteri of pseudopregnant CD-1 females as described 17. Chimeras were measured by % agouti and black on the albino background provided by the host blastocyst. Male chimeras were mated to C57Bl/6Tyrc-2J females and germline transmission was assessed by observation of agouti coat color.
Cell Culture, Lentivirus Production and Transduction
For reprogramming source cells, MEFs were prepared from day 13.5 embryos of the Oct-4 GFP mice and reprogrammed as described 2. Wild type C57Bl6 mESCs were purchased from ATCC and transduced with retrovirus vectors expressing GFP or dsRed as described previously18.
For feeder cells, MEFs were derived from CF1 mice or purchased from Millipore. P3 MEFs were inactivated by 2 hour treatment of mitomycin C. Inactivated MEFs were plated on gelatinized dishes and used within 2 days. Lentivirus production was performed as described by the RNAi Consortium at the Broad Institute (http://www.broadinstitute.org/rnai/public/static/protocols/TRC_Protocols_Section_II_Viral_Production_100809.pdf). Lentiviral constructs for shRNAs against E-Cadherin are generous gifts from Dr. N. Geijsen 19. Hematopoietic progenitor cells were transduced in X VIVO15 medium supplemented with 100 ng/ml mSCF, 50 ng/ml mIL3, 50 ng/ml Flt3L and 50 ng/ml mTPO (all from PeproTech) supplemented with 4 μg/ml polybrene overnight, washed in PBS and plated on inactivated MEF feeders at a density of 20,000–50,000 cells/35mm dish in complete mESC culture medium (DMEM with 15% Hyclone FBS, Non-essential amino-acids, Pen/Strep, L-glutamine, β-mercaptoethanol and 1000U/ml LIF (Chemicon). Dox was added at 2 μg/ml to induce factor expression.
FACS Purification of Granulocyte-Monocyte Progenitors (GMPs)
GMPs were stained and FACS sorted on a BD Aria instrument as described previously 14,18. Briefly, mouse bone marrow cells were stained with antibody cocktail containing hematopoietic lineages antibodies, c-Kit APC, Sca PE or pacific blue, CD34 FITC and CD16/32 PEcy7 (all from BD Biosciences or eBiosciences). GMPs are defined as lineage negative, c-Kit positive, Sca negative, CD34 and CD16/32 double positive.
Immuno-fluorescent Staining and Confocal Analysis
The iPSC induction procedure is described as above. Starting from 24 hours after dox addition, cultures in 35mm dishes were processed for immuno-staining every 12 hours by fixation with 3.7% formaldehyde for 15 minutes, and permeabilization with 0.1% Triton-X-100 (Sigma-Aldrich) for 15 minutes. Fixed cells were maintained in PBS at 4°C before further processing. After all time points were collected, the cultures were stained at the same time with rabbit anti-E-Cadherin antibody (1:200, Cell Signaling). Bound antibody was detected using Alexa-555 or Alexa-488 labeled donkey anti-rabbit secondary antibody (Invitrogen). Fluorescent images were obtained on a Leica SP5 confocal microscope (Leica microsystem, Wetzlar, Germany).
Image Acquisition and Analysis
For all imaging studies, mitomycin C inactivated MEFs were plated on gelatinized dishes with enhanced optical properties (ibidi) on the day of GMP cell sorting. GMPs following overnight viral transduction were washed and plated on the MEF layer and placed in a VivaView Olympus incubator microscope with motorized stage. Images were acquired by the Metamorph software at defined time intervals with signals being recorded in the DIC, GFP and/or RFP channels with 10x or 20x magnification continuously for 3–5 days.
In some cases, when stitching was not necessary, Metamorph was used to annotate the images and generate movies. In most other cases, TIF images from the VivaView microscope were analyzed with the assistance of a custom suite of programs coded in Matlab, which has user friendly graphical interfaces (Fig. S3). Several steps were taken to process the images. First, to correct for vignetting, background illumination images for each scanning position and each channel were calculated by taking the median values of 60 to 80 time points (user-definable) at each pixel. Vignetting was corrected by dividing raw TIF images with the background illumination images. Second, images with adjacent positions were stitched together (up to 10 by 10). Stitching overlaps between neighboring images were calculated by minimizing the signal difference of the overlapping areas from the DIC channel of neighboring background-corrected images. The same stitching parameters were then applied to all light channels of the corresponding scanning positions. In the stitched images, overlap areas were represented by the mean of signals from neighboring images. Third, images from different light channels were mixed to create pseudocolor images. User is able to control the background and saturation of each channel, and the fraction by which each channel will be displayed in red, green and blue channels of the final pseudocolor image. To ensure accuracy of tracing, all iPSC lineages were inspected manually and annotated using a custom program coded in Matlab (Fig. S3B). For all movies, the first time points were stamped as +1 than real time such that they start as 00:30 min, 00:15 min and 00:05 for images acquired at 30 min, 15 min and 5 min intervals. For still images, the first time points were stamped as 00:00.
RESULTS
Granulocyte-Monocyte Progenitors (GMPs) as a Suitable Cell Source for Time-Lapse Imaging
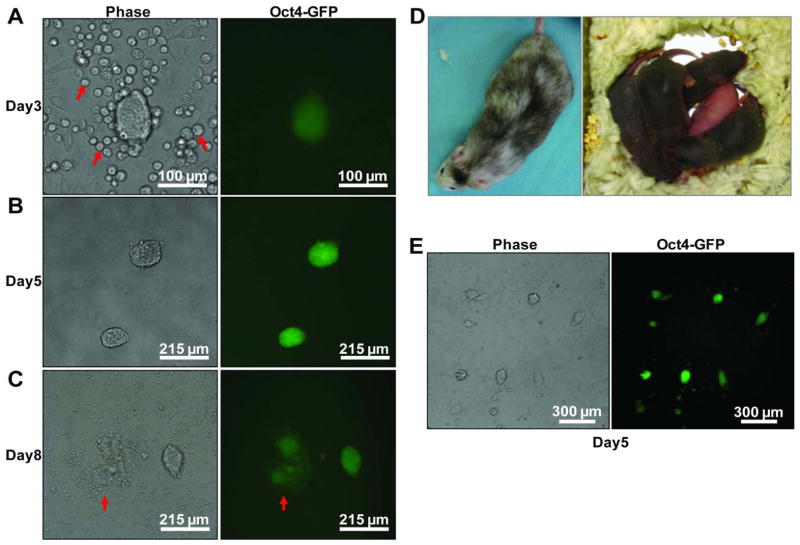
We sought to establish a reprogramming system that is fast, efficient, inducible and contains a marker for the activation of endogenous pluripotency circuitry 20. Importantly, the cells undergoing reprogramming should be clearly distinguishable from other cells present in the culture. We focused our attention on granulocyte-monocyte progenitors (GMPs) since they are well defined, can be prospectively isolated to high purity 14, and support reprogramming with sufficiently high efficiency 7 (5–10% apparent efficiency in our hands). We purified GMPs from mice genetically modified to express an Oct-4-GFP reporter as well as a universally expressed (Rosa26) reverse transactivator rtTA 7 (Fig S1). Following cell sorting, GMPs were infected with a polycistronic lentivirus encoding doxycycycline (dox) inducible expression of the transcription factors Oct-4, Sox2, Klf4 and c-Myc (OSKM) 21, plated on feeder cells, and induced to initiate reprogramming by adding dox. Oct-4-GFP+ colonies with embryonic stem cell (ESC)-like morphology appeared within the first 3 days following dox addition (Fig 1A). During this time, the number of cells present in the culture remains limited (<100,000 hematopoietic cells per 35mm dish containing 3.5 ml of medium) and medium change was not necessary. By 5 days, many of the colonies had grown to large sizes such that passaging was usually required (Fig 1B). When left without passaging (but with medium change) until day 8, many colonies displayed differentiation associated changes (Fig 1C). On day 5 post dox addition, primary GMPs routinely give rise to > 5 colonies for every 100 initial cells. The colonies could become dox-independent and be propagated with murine ESC culture conditions. The cells within the colonies have normal karyotypes (data not shown), support high degree chimeric mouse formation and are capable of germline transmission (Fig 1D), demonstrating their pluripotency.
Figure 1. GMPs are a suitable source cell type for imaging pluripotency induction.
GMPs from adult bone marrow of mice harboring Rosa26-rtTA and Oct4-GFP were transduced with a polycistronic lentivirus expressing Oct-4, Sox2, Klf4 and c-Myc (OSKM) and plated on inactivated feeder cells in mESC culture conditions supplemented with of 2 μg/ml dox. Representative images of Oct-4 GFP+ colonies after 3, 5 and 8 days are shown. A) Cells that were not reprogrammed (red arrows) show typical non-adherent, round morphology. B) By day 5, large, tightly compact colonies were present. C) Without passaging, spontaneous differentiation occurs in some colonies by day 8, as indicated by the loss of Oct-4 GFP expression at the periphery of the colony (red arrows). D) Dox-independent Oct-4 GFP+ single colonies were picked and injected into B6 albino blastocysts. High degree chimeras were observed as indicated by their mosaic coat color. Upon breeding with albino females, the chimeric males fathered completely agouti pups indicating germline transmission. E) A representative low power view of a reprogramming culture 5 days after dox addition. Note the lack of partially GFP+ colonies.
In addition to fast kinetics and high efficiency, GMP-based reprogramming displays unique imaging-friendly qualities. First, the developing Oct4-GFP+ cell clusters do not seem to contain Oct-4-GFP negative cells (Fig 1A, 1B, 1E). Partially GFP+ cell clusters are prominent when fibroblasts are used as source cells and are thought to represent various reprogramming intermediates or partially reprogrammed cells 10, 13. We examined thousands of GMP-derived colonies and have never encountered any colony that had a mixture of Oct-4-GFP positive and negative cells, as long as the cells were not differentiated due to overgrowth (Fig 1C, red arrows). Fig. 1E shows a typical field on a reprogramming culture dish. All colonies with ESC morphology are GFP positive and vice versa. This is true even for very small, nascent colonies (Fig 1A). Another imaging-friendly feature of GMP is that the cells that are not being reprogrammed retain their round morphology (Fig 1A, red arrows). Thus, the morphological changes associated with entry into early reprogramming are visually distinct.
Together, these key features indicate that GMP-based reprogramming system is well suited for imaging experiments. We thus performed all subsequent time-lapse experiments using this system.
High Temporal Resolution Microscopy Enables Tracing of Reprogrammed Colonies to Single Founder Cells
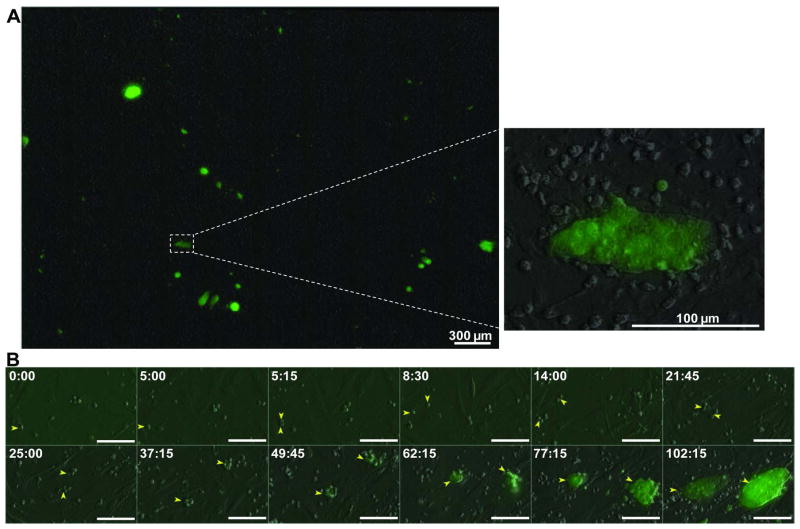
We used an incubator-enclosed microscope with a motorized stage, which permits continuous scanning and photographing of partially overlapping visual fields. These fields were stitched together in post experiment analysis, synthesizing a large area sufficient for capturing low-frequency reprogramming events (Fig S2 and Fig 2A). In addition, the imaging system records signals from differential interference contrast (DIC), GFP and additional channels when necessary. Through a 10X or 20X objective lens, the resulting images are of high optical resolution and provide fine cellular details (Fig 2A). Time-lapse microscopy for this entire synthetic field continues for 3–5 days at 5–30 minute intervals (Movie S1). We initiate image acquisition approximately one hour after adding dox (time required to stabilize and equilibrate the imaging system). Given that the polycistronic iPS vector needs time to produce RNA and protein after adding dox, this experimental scheme ensures that we capture the very beginning of the reprogramming process.
Figure 2. Tracing reprogramming to single founder cells.
A) Representative scanned field showing overlay of DIC and GFP signals from stitching 10 x10 adjacent imaging frames at the end of image acquisition for 5 days. Areas of interest can be viewed with sufficient detail to allow cell tracing (enlarged inset on right). B) A reprogramming lineage imaged at 15 min intervals was traced to a founder cell, indicated by the yellow arrow at time 0:00 (~1 hour after plating on MEF and adding dox). The founder cell produced two Oct-4 GFP+ colonies similar in size, which we term sister colonies (two yellow arrows). Bar = 100 μm. Time stamp in hour:minute format.
For image processing, analysis and annotation, we created a suite of user-friendly computational tools (Fig S3). During image analysis, we tracked cell movement from Oct-4-GFP+ colonies back in time to a single cell. This is made possible by the ability of the imaging system to capture cell migratory and divisional events at high frequency (every 5–15 minutes), generating reliable accounts for the origin of every emerging cell in view. Many of the reprogramming lineages allowed us to trace back to a single cell at the beginning of image acquisition (Fig. 2B and Movie S2) when factor induction in these hematopoietic cells had occurred for just one hour. We define these single cells as “founder” cells. It is important to note that our definition of the founder cells is much more stringent than traceable single progenitors (or ancestors) used in previous methods. For example, a previous study tracked back to single cell progenitors in the middle of reprogramming 13, after 4.5 days of factor transduction. In such cases, it was difficult to determine the identity of these cells as factor expression had been on for an extended time. Cellular events that happened before this traceable progenitor stage would have been missed.
Given the fast reprogramming kinetics and high efficiency, lack of Oct-4 GFP negative cells in the cell clusters, timed factor expression and unique cellular morphology, our experimental system is a significant improvement over previous methodologies to characterize the early reprogramming process.
Dynamic Cellular and Clonal Behaviors During Early Reprogramming
Time-lapse observation shows that the cells, either singularly or as cell clusters, display active migratory behavior during the entire imaged process (Movie S1, S2). For example, single cells appeared to roll on top of the feeder cells, which themselves underwent constant morphologic remodeling. The migratory motions appeared to be random in terms of direction, duration and distance, but were widespread and often brought distant cells together. Such migratory behaviors predict that two remote cells and/or colonies could come together resulting in clonal mixing, which can be affected by the local density of migratory cells.
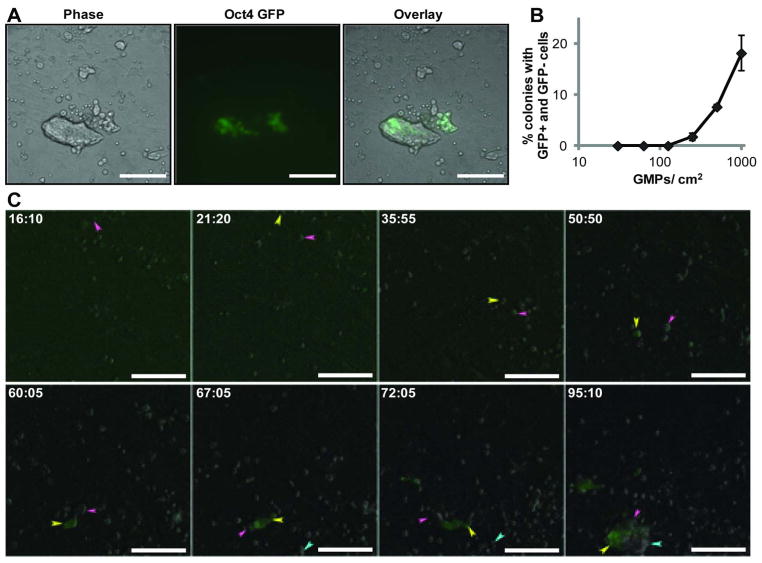
Tracking lineage pedigrees revealed that many of the colonies formed are indeed not clonal, but rather contained cells of different reprogramming origins, including from different founder cells (Fig. 3 and Movie S3). To overcome any potential ambiguity associated with tracing single cells, we plated factor transduced GMPs from Rosa26-rtTA mice with or without the Oct-4 GFP reporter. As discussed earlier, all reprogrammed colonies from the Oct-4 GFP reporter-containing GMPs are GFP+. Thus, such a co-plating strategy would produce colonies containing both GFP+ and GFP- cells if clonal mixing does occur (Fig. S4). Indeed, we observed numerous colonies containing various amounts of GFP- cells (Fig. 3A). We estimated the extent of mixing by co-plating factor transduced GMPs at different densities (Fig. 3B). Mixing was not detectable when GMPs were plated at 130 or fewer per cm2 growth area. Of note, this is a vast underestimation of mixing behavior, since GFP+ vs. GFP+ and GFP- vs. GFP- mixing could not be detected by this method, which also would miss mixed colonies that contained few cells of a mixing component (i.e. a largely GFP+ colony with few GFP- cells is likely not to be scored as mixed).
Figure 3. Cell migration during early reprogramming generates colonies of mixed clonal origin.
A) GMPs from Rosa26-rtTA mice with or without the Oct-4 GFP reporter were induced to reprogram on the same culture dish. Colonies containing GFP+ (originated from the Oct-4 GFP reporter GMPs) and GFP− (originated from the non reporter GMPs) cells were observed. B) The occurrence of mixed colonies containing both GFP+ and GFP− cells when transduced GMPs were plated at different densities. C) Time-lapse observation capturing the formation of a mixed colony. The arrows track the movement of different cells during the process. The mixed colony (containing both GFP+ and GFP- cells) was generated by cell migration and is indicated by three arrows of different colors at the end of image acquisition. Time stamp in hour:minute format. All bars = 100 μm.
To demonstrate that the mixed colonies are formed by cell migration, we performed time-lapse imaging with co-plated GMPs (Fig. 3C and Movie S3). We tracked a mixed colony at the end of image acquisition back in time and found that the mixing components entered the imaging field from distinct locations at different times (colored arrows follow the traceable events). Migration occurred with the single cells and continued as the cells developed into small cell clusters (Fig. 3C and Movie S3). Once within close proximity, the cells established cell-cell contacts and the entire cell aggregate continued migrating while also becoming more compact. As a result, a single colony containing both GFP+ and GFP- cells with normal ESC morphology formed. Since the founder cells are genetically distinguishable by the presence of GFP upon emergence of pluripotency, this observation demonstrates that mixed colonies could occur during pluripotency induction and the morphologic clonal appearance is not always equivalent to true cellular clonality.
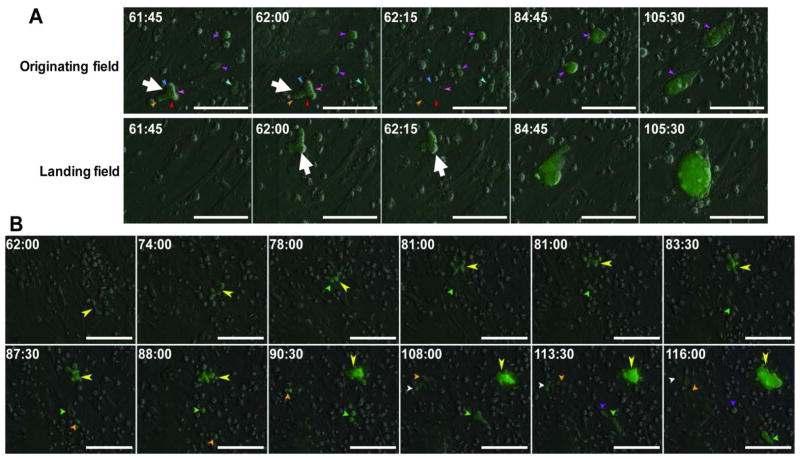
In addition to clonal mixing, cellular migration also led to clonal dispersal. Developing small cell clusters are highly dynamic structures, with parts of the cell cluster transiently extending well beyond the focus plane (Fig. 3C and Movie S3). Others could break off, land in remote sites and continue to develop into normal looking colonies (Fig. 4A and Movie S4). This kind of migration would be less dependent on cell plating density, and more on random differences in the local cell micro-environment and/or sheer force.
Figure 4. Cell migration during early reprogramming can lead to clonal dispersal.
A) Time-lapse observation of a colony breaking off from the originating field of view (top row), landing at another site and continuing to develop into a normal colony (bottom row). The colony at the time of breaking away and landing is indicated by the large white arrow. The imaging system captured this event during its scan at 62 hours from the originating field to the landing field following a path as illustrated in Fig S2. B) Time lapse images of single cells (small arrows) breaking off from a developing Oct-4 GFP+ colony (large yellow arrow). These single cells established smaller colonies in the vicinity of the main colony. We termed the smaller colonies “satellite” colonies. Time stamp in hour:minute format. All bars = 100 μm.
Clonal dispersal, by which one founder cell gives rise to one or more colonies, could also result from single cell migration. One founder cell could produce one or more distinctive colonies similar in size, which we term “sister” colonies (Fig. 2B and Movie S2). Alternatively, single cells can break away from developing large cell clusters, and establish separate colonies (Fig. 4B and Movie S5). Because the resultant new colonies are smaller in size than the main colony, we term them “satellite colonies”. This observation may explain the previously reported satellite colonies arising in fibroblast-based reprogramming cultures without obvious origin 7, 12.
As the migratory behavior is widespread, continuous and apparently random, colony mixing and dispersal co-exist for many colonies, involving multiple founder cells (Fig. 3C and Movie S3) and/or multiple alternating rounds of mixing and dispersal (Movie S4), yielding colonies of complex clonal origins. Importantly, the migratory behaviors are not specific for hematopoietic-cell-originated pluripotency, as we observed similar mixing and dispersal with other types of pluripotent cells (Movies S6–8) including iPS cells derived from fibroblasts and embryonic stem cells themselves.
Taken together, these data demonstrate that dynamic cell migration accompanies the fate transition into pluripotency and affects the clonality of the colonies formed. Reprogramming efficiency may not be accurately described by the number of morphologically distinct colonies at the end of a reprogramming experiment. More stringent approaches involving the separation of founder cells may be required 7–8. Importantly, assessment of iPS cell clonality should be more vigorously performed.
Cell-Cell Interactions During Early Reprogramming Yield a Novel Morphologic Landmark
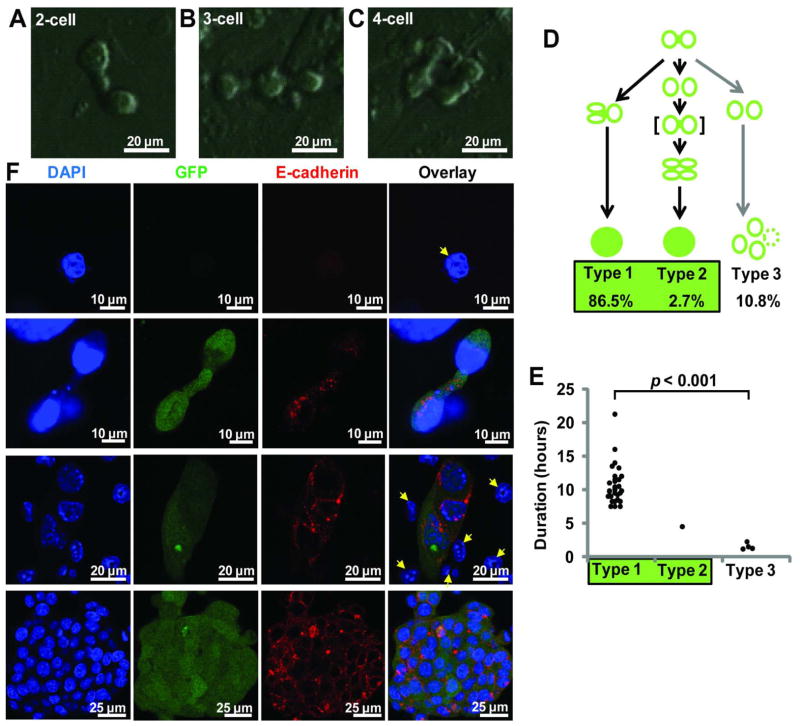
When back-tracking reprogramming from Oct-4-GFP+ colonies to founder cells, we identified a novel 2-cell intermediate stage characterized by a unique “dumbbell” morphology, in which two cells appeared to be connected by an extended thin cellular process (Fig. 5A). Noticeably, the dumbbell intermediates occurred before the Oct4-GFP signal was detectable by time-lapse microscopy. Such 2-cell dumbbells were formed as a result of cell division from a single parental cell. The two daughter cells did not fully segregate after division, but appeared “sticky” and become connected by a bridge-like structure, resulting in a dumbbell-shaped intermediate (Fig. 5A, could be seen in Movie S4). Fig. 4B also shows a dumbbell at time point 62:00 (arrow, from Movie S5). Often without clear segregation, these two cells within a dumbbell continued with their next cell division(s) and became a partially connected multi-cell structure containing 3–4 cells (Fig 5B–C). We traced 37 dumbbell structures across 6 independent reprogramming experiments and compared their duration and fate outcome (Fig. 5D–E). 33 (all Type 1 and 2) of them reprogrammed successfully and 4 (all Type 3) failed. There is a highly significant difference in the duration of the dumbbells: all dumbbells that lasted until the next cell division (10.5 ± 2.8 hours, Type 1, n = 32) led to a colony or migrated to become part of another colony. In contrast, the dumbbells that failed to reprogram existed only briefly (1.5 ± 0.5 hours, Type 3, n = 4), quickly broke apart and adopt a morphology similar to that of hematopoietic cells. We did observe one pair of cells (Type 2, n = 1) that dissociated transiently, but then came together again before proceeding with reprogramming (Figure 5D, the dumbbell in brackets and Movie S2). In this case, the pair of cells remained in contact with each other for at least 4.5 hours, although the connecting process between the two cells was not as obvious due to the highly dynamic cell behavior. As such, all traceable colonies originated from a dumbbell structure and no colony was seen to have bypassed this stage. This unique structure could also be detected by conventional methods such as immuno-fluorescence and confocal microscopy (Fig. 5F, second row). Since the brief cell-pairing is minority (10.8%) and much shorter-lived (1.5 ± 0.5 hours as compared to 10.5 ± 2.8 hours) (Fig 5D–E), the great majority of the dumbbell intermediates captured in these assays could be regarded as an early landmark of cells undergoing reprogramming.
Figure 5. A reprogramming intermediate and the initiation of E-cadherin expression.
A–C) Typical reprogramming intermediates detected by time-lapse imaging. Prior to Oct-4 GFP becoming detectible by time-lapse imaging, the reprogramming cells appeared to be connected by bridge-like structures. A) shows a 2-cell dumbbell intermediate. A dumbbell can extend further when one or both cells in the structure divide, yielding B) 3 or C) 4 inter-connected cells. D) Scheme illustrating the three observed outcomes when the cell pair in a dumbbell developed further. The duration of a cell pair is defined as from the time of mitosis that created the two cells, to the time of next mitosis (Type 1) or when the cells broke apart (Type 2 and Type 3). Shown is a summary of 37 traced cell pairs across 6 independent experiments. The majority of cell pairs in a dumbbell proceeded to successful reprogramming (all Type 1 and Type 2, 89.2%). In Type 2, the broken cell pair re-established cell-cell interaction, indicated with “[ ]”, before proceeding to reprogramming. All reprogrammed colonies could be traced to a dumbbell. E) There is a highly significant difference in the duration of cell pairing. The lasting cell pairs (10.5 ± 2.8 hours) led to reprogramming and the brief pairing (1.5 ± 0.5 hours) did not. F) Reprogramming cultures at day 0–5 after dox addition were stained with DAPI, anti-GFP and anti-E-cadherin antibodies and examined by confocal microscopy. E-cadherin was not expressed in GMPs and non-reprogrammed cells (yellow arrows). A two-cell dumbbell-shaped intermediate detected by confocal microscopy is shown in the second row. At the 2-cell stage, E-cadherin is mostly intra-cellular and peri-nuclear. Later, its distribution is along the cell periphery (middle and bottom rows).
Induction of E-cadherin is associated with the pluripotent state22–24, but the exact timing and cellular stage when E-cadherin is induced remains unclear. With the hypothesis that E-cadherin might be responsible for the cell-cell interaction of the dumbbell stage, we explored the induction of E-cadherin at the 2-cell dumbbell stage and subsequent cell clusters using immuno-fluorescence and confocal microscopy. GMPs and the non reprogramming cells did not express E-cadherin at a level above the background fluorescence (Fig. 5F top row, yellow arrows of 3rd row and S5A). E-cadherin protein became detectible at the 2-cell dumbbell stage, but its distribution is mostly intra-cellular and peri-nuclear (Fig. 5F, second row). Not until the stage of compacting Oct-4-GFP+ cell cluster formation did E-cadherin start to distribute along the cellular periphery, a distribution pattern that was maintained afterwards (Fig. 5F bottom two rows). Interestingly, Oct-4-GFP signal could be detected by immuno-fluorescence even at the 2-cell dumbbell stage (Fig. 5F second row), possibly due to the higher detection sensitivity of low-level GFP by immuno-staining as compared to direct fluorescence. These observations suggest that E-cadherin induction occurs very early and coincides with the dumbbell appearance during GMP initiated reprogramming. The distribution pattern of E-cadherin changes to the cell periphery as reprogramming progresses.
Inhibition of E-Cadherin Disrupts Cell-Cell Interactions of Early Reprogramming Intermediates and Prevents the Progression of Reprogramming
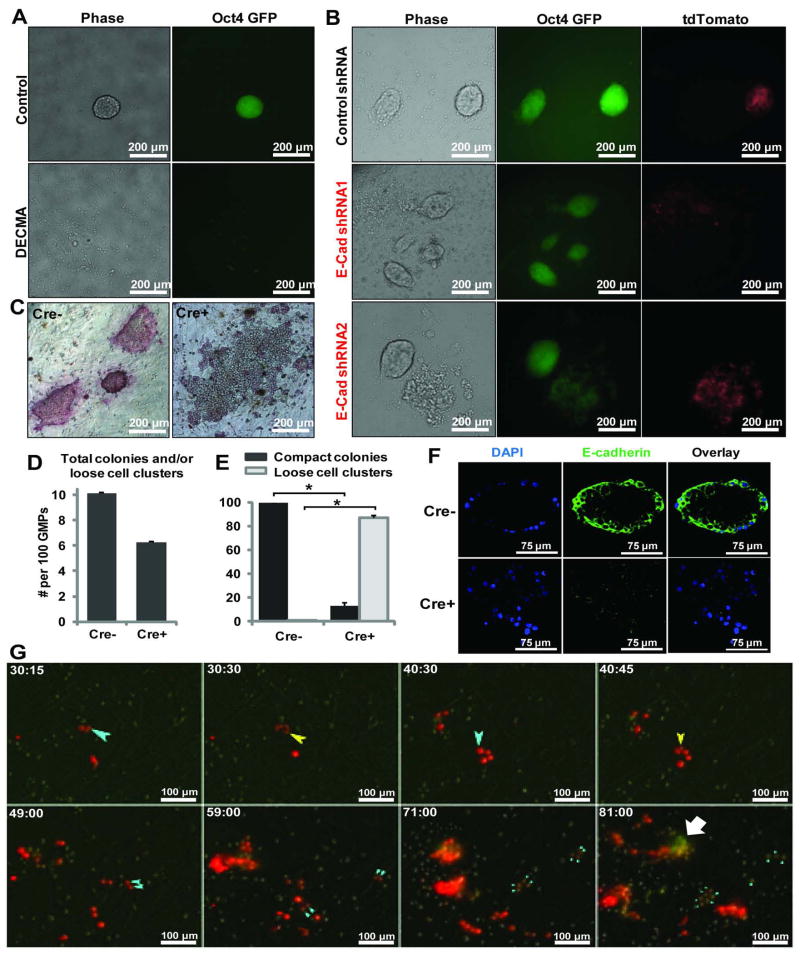
Since the 2-cell dumbbell intermediate was seen in all lineages that successfully led to Oct-4 GFP+ colonies, and E-cadherin expression could be observed from this stage onward, we sought to determine the requirement for E-cadherin in forming this intermediate stage. Although E-cadherin is required during fibroblast-based reprogramming 22–25, a similar role in hematopoietic cell initiated reprogramming has not be reported 26. We first examined the function of E-cadherin during GMP-based reprogramming using an E-cadherin blocking antibody 22, 27. Similar to MEF-based reprogramming, this antibody abolished the formation of Oct-4 GFP+ colonies with tight compact ESC morphology (Fig 6A). Upon closer examination, some scattered weak GFP+ cells could be detected. In addition, we also used two validated shRNAs against E-cadherin 19 expressed from viral constructs that also contain tdTomato, such that the transduced cells are marked by red fluorescence. Normal compact colonies developed with the control tdTomato virus (GFP+ and tdTomato+) (Fig. 6B top panel). However, only disseminated colonies (weakly GFP+ and tdTomato+) were seen when shRNAs against E-cadherin were expressed (Fig. 6B middle and bottom panels). The existence of tdTomato negative, Oct-4 GFP+ colonies of normal morphology in all of the cultures suggests that the dysmorphic colony appearance is due to E-cadherin knockdown (Fig 6B). The scattered weakly GFP positive cells could not be propagated under mESC conditions while the control cells could (data not shown).
Figure 6. E-cadherin is required for the 2-cell dumbbell intermediate formation and reprogramming from hematopoietic cells.
A) An E-cadherin blocking antibody (DECMA) was added to the GMP-initiated reprogramming culture. While colonies of normal morphology were observed in control cultures, the DECMA treated cultures were devoid of compact colonies. Some loosely scattered, weakly GFP+ cells were detected. B) GMPs were transduced with a control lenti-virus or two independent shRNAs against E-cadherin, along with the OSKM lentivirus. tdTomato fluorescence marks transduced cells. While normal colonies formed with the control virus (top row), viruses of E-cadherin shRNAs led to dysmorphic colonies consisting of loosely scattered cells. C) GMPs from the Rosa26-rtTA:tetO-Cre+:E-cadherinfl/fl mice or the tetO-Cre- littermate control were reprogrammed and stained for alkaline phosphatase activity on Day6 post Dox addition. D) The total colonies in Cre- cultures and the total cell clusters (loose or compact) from Cre+ cultures per 100 transduced GMPs. E) The percentage of compact colonies in Cre- (intact E-cadherin) and Cre+ (inactivated E-cadherin) cultures. n = 2 each, *: p < 0.01. F) Immuno-fluoresence staining and confocal microscopy with the E-cadherinfl/fl cells following reprogramming culture. E-cadherin protein was absent in the Cre+ cells, which form loose cell clusters similar to those seen with shRNA knockdown. G) Time-lapse microscopy was performed by recording DIC, GFP and RFP images at 15 min intervals. GMPs were transduced with the same E-cadherin shRNAs as in B. The light blue arrow tracks the same cell lineage over time, with the yellow arrows indicate the first and second cell divisions of that lineage. For ease of viewing, only one of the daughter lineages of these two divisions is denoted by arrows, with sequentially smaller blue arrows indicating successive cell divisions. The dumbbell-shaped intermediate indicating cell-cell interaction and subsequent cell compaction is not seen. The white block arrow indicates a cluster of Oct-4-GFP+ tdTomatolow cells that arose possibly by silencing the shRNA construct.
To further confirm the role of E-cadherin, we performed reprogramming using GMPs from mice with a conditional E-cadherin allele 28. For this purpose, E-cadherinfl/fl mice were crossed with mice containing the Rosa26-rtTA7 and tetO-Cre 29 alleles, such that E-cadherin could be inactivated specifically upon the addition of Dox (Fig S5B), which also initiates the reprogramming factor expression. While GMPs from tetO-Cre- mice gave rise to colonies of normal iPSC morphology that also stained positive for alkaline phosphatase activity, GMPs from the tetO-Cre+ littermates produced only loosely scattered cell clusters (Fig 6C–F). Inactivation of E-cadherin did not overtly inhibit the number of total cell clusters (Fig 6D) if morphological abnormality is not considered, even though the lack of clearly defined morphologic boundaries for loose cell clusters may result in an underestimation of their numbers. However, when compactness of the cell clusters was assessed, the Cre+ cultures (hence the E-cadherin gene could be inactivated) were dominated by loose cell clusters (Figure 6E) while all colonies in the Cre- cultures were compact. The few compact colonies present in the Cre+ cultures could be due to incomplete Cre-mediated gene inactivation 18, although it is also possible that some cell lineages could better sustain the loss of E-cadherin. The loose cell clusters in Cre+ cultures lacked E-cadherin protein expression as detected by immuno-fluorescence staining and confocal microscopy (Fig 6F), which also showed little alkaline phosphatase activity (Fig 6C). These data confirm that, as for MEFs, E-cadherin is required for GMP-initiated reprogramming. Inhibition of E-cadherin results in loosely scattered cell clusters that lack the phenotypes of pluripotent cells.
We then performed time-lapse imaging to examine whether the dumbbell intermediate was perturbed by E-cadherin knockdown, and if so, how reprogramming was affected. We recorded signals from DIC, GFP and RFP channels at 15 minutes intervals. In the presence of E-cadherin shRNAs, as indicated by tdTomato, the 2-cell dumbbell intermediate was never observed (Fig. 6G, Movie S9). The cells continued to divide and some of the daughter cells even stayed in close proximity for an extended time. However, a cell-cell interaction mediated by a connecting process was never observed. While cell-cell interaction leading up to compaction is clearly visible beginning at 40.2 ± 4.7 hours post dox addition in un-perturbed reprogramming (from n = 25 reprogramming events across 4 independent experiments, see Fig. 2B for one example), the cells with E-cadherin shRNAs showed no sign of cell-cell interaction even at 81 hours post dox addition (Fig. 6G). Oct-4 GFP expression was observed, but the intensity was much reduced (Fig. 6G, small light blue arrows). Of note, some of the cells appeared to have lost their red fluorescence, possibly due to silencing of the viral shRNA construct. These cells appear more normal in morphology and brighter for GFP (Fig. 6G, white block arrow). These observations revealed the perturbed early cell-cell interaction and subsequent formation of loose cell clusters.
Taken together, these experiments demonstrate that E-cadherin is required from the earliest stages of the reprogramming process. The earliest identifiable feature associated with productive reprogramming is the appearance of a unique dumbbell-shaped intermediate which subsequently grows in size as the cells become more compact. Disruption of E-cadherin prevents the formation of this intermediate and normal iPS colonies. Without E-cadherin, the pluripotency network may initiate, as suggested by the appearance of weakly GFP+ cells, but it cannot sustain and mature without proper E-cadherin function due to defective cell-cell interactions (Fig. 7).
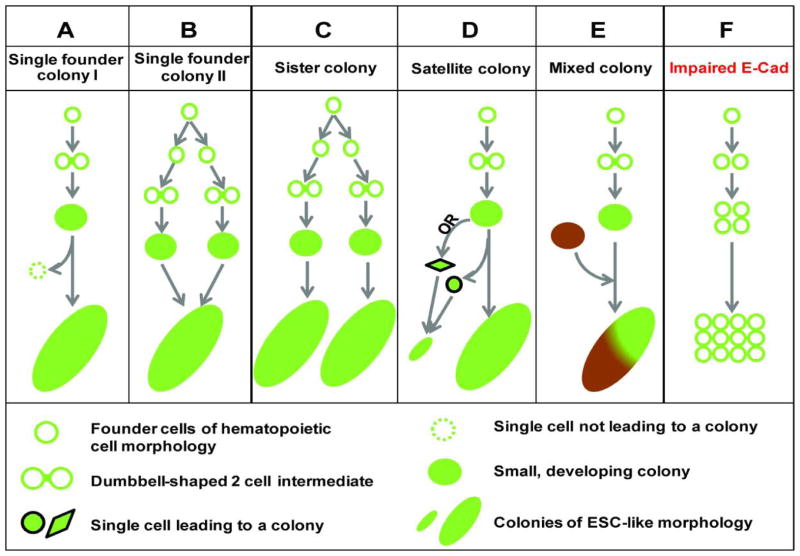
Figure 7. Schematic illustration of common modes of cell interaction and migration during early reprogramming.
A–D) One founder cell could give rise to one or more colonies and E) one colony could contain cells from one or more founder cells. The occurrence of a 2-cell intermediate of a dumbbell morphology is related to the proper induction of E-cadherin, disruption of which F) inhibits reprogramming.
DISCUSSION
We describe a novel experimental system with which we directly visualized the earliest phase of pluripotency induction by Yamanaka factors. In addition to high-temporal resolution up to every 5 minutes, the system overcomes multiple limitations of previously described methods enabling us to confidently trace reprogramming events back to the founder cells in their native somatic state. This system allows study of reprogramming at a stage much earlier than that accessible by conventional methods. It is seen that dynamic migration and cell-cell interactions are integral to pluripotency emergence. These processes not only are crucial for reprogramming to proceed, but also could compromise the interpretation of conventional reprogramming experiments. A simple founder-colony relationship is inaccurate due to this complex and dynamic process (Fig. 7).
We demonstrate that the dynamic cellular migration associated with pluripotency induction can lead to mixed colonies containing derivatives from multiple founder cells. Clonal mixing (Fig. 7E) challenges the assumption that all cells within the same morphologic boundary share the same genetic make-up. Clonality is a key assumption when interpreting data on pluripotency of iPSC colonies, their molecular similarity and their safety in potential clinical uses. Our data caution against the practice to pick and assume clonality of iPSC colonies only based on their final morphological boundary without rigorously ascertaining their clonality.
Clonal mixing has been suggested by Warlich et al. using fibroblast as source cells, in which colony mixing was observed 13. However, it is important to point out that since Warlich et al. imaged MEF reprogramming from day 4.5 post viral transduction, it is not known whether the mixed colonies were coming from different founder cells, or from different lineages downstream of the same founder cell. We affirmed the existence of mixed colonies with the enhanced resolution of our imaging system, which was further validated by co-plating experiments using genetically distinct cells. The mixing behaviors of early reprogramming across different cell types, the existence of satellite colonies in fibroblast-initiated reprogramming12 and the conserved requirement for E-cadherin argue against the dynamic behaviors being the peculiarity of GMP-initiated reprogramming. We demonstrate highly analogous migration behavior also for cells with fully established pluripotency including MEF-derived iPSCs and ESCs, suggesting that these dynamic behaviors are associated with the pluripotent state, rather than being unique to a particular source somatic cell state.
We uncovered a novel reprogramming intermediate with unique morphology. At least in this GMP-initiated reprogramming system, a dumbbell-shaped 2-cell intermediate that lasts for about 10 hours appears to be a faithful marker for productive reprogramming. Furthermore, we demonstrate the involvement of E-cadherin in forming this intermediate. E-cadherin is an important mediator for the mesenchymal-to-epithelial transition following Yamanaka factor expression 23, 25–26. As E-cadherin is expressed by pluripotent cells and can replace Oct-4 in the original Yamanaka cocktail 24, it is conceivable that the signaling transduced by E-cadherin could feed into the core pluripotency network 9, 20 and provide a stabilizing effect. Alternatively, this mechanism may provide a crucial survival signal for cells in pluripotency but be dispensable otherwise. In support of this notion, it was reported that disruption of E-cadherin-mediated cell-cell adhesion activates the Rho/ROCK/MLC2 signaling cascade specifically in an epiblast-equivalent pluripotent cell state 30. Our data is consistent with a cell-autonomous role of E-cadherin in allowing the fate transition into pluripotency to progress further. When E-cadherin function is perturbed, proper cell-cell interactions reflecting cell fate transition into early pluripotency fail leading to impaired reprogramming. Our work provides the first direct visualization of this disrupted process and confirms the role of E-cadherin in the very early stages of pluripotency induction. Clearly, this system can be used to address multiple key questions regarding early reprogramming. For instance, when coupled with specific molecular perturbations, molecular machineries that underlie early reprogramming can be probed. Additional markers of cellular states can be incorporated to generate richer real-time information. We also anticipate that new cellular features that inform stochastic and deterministic behaviors 6, 31 may be uncovered.
Supplementary Material
Signals from DIC and GFP were recorded, stitched and overlayed. There were no GFP+ cells during the early part of the image series. GFP+ cells/colonies start to be visible around 60 hours. Image quality is reduced to show the overview of the entire synthetic field. Time stamps are in hours:minutes format. Bar = 100 μm.
High resolution images were acquired by time-lapse imaging, processed and annotated. Images in the beginning show cells of hematopoietic morphology, which lack detectible Oct-4 GFP expression. Arrows follow the movement of cell(s) that developed into GFP+ colonies at the end of image acquisition. The founder cell is followed by the yellow arrow, which split and established two similar sized colonies at the end of image acquisition. This is a typical example of sister colony formation, with some minor satellite colonies seen as well (not marked). Overlayed images of DIC and GFP channels are shown. Time stamps are in hours:minutes format. Bar = 100 μm.
GMPs from Rosa26-rtTA mice with or without the Oct-4 GFP reporter were induced to reprogram on the same culture dish. Time-lapse observation captured the formation of one mixed colony. The arrows track the movement of different cells as they continued the reprogramming process. They entered the field of view from distinct locations at different times. The migration brought them into close proximity of each other. The small cell clusters originated from these cells continued to grow and compact without distinguishing the cells of different origins. The resulted colony contained both GFP+ and GFP- cells, originating from cells indicated by the pink, yellow and blue arrows. Also present in view are colonies consisting of only GFP+ cells and of only GFP- cells (not marked). Time stamps are in hours:minutes format. Bar = 100 μm.
The founder cell divided on the 2nd frame of acquisition (00:30) and its two immediate daughters are followed by the yellow and blue arrows. All cells in view marked by arrows are descendents of the same founder. Addition of a new colored arrow indicates the birth of a visually distinct new daughter cell. Note the complex splitting, migrating and mixing behavior. Some arrows disappeared in the movie, which indicate either apoptotic appearance of the cell or loss of Oct4-GFP, or becoming out of view. Note the appearance of several dumbbell intermediates. Around 62 hours, two of the larger colonies disappeared suddenly from view. Images from another field (Right) show one of these colonies landed a remote site and continued its development afterwards. The identity of the break-away small cell cluster was informed by its shape and the timing when it disappeared from view in one field and appeared in another field. Overlayed images of DIC and GFP channels are shown. Time stamps are in hours:minutes format. Bar = 100 μm.
Large yellow arrow follows the main reprogramming colony. Various colored smaller arrows point to the break-away cells, which continued to grow and form smaller “satellite” colonies. Overlayed images of DIC and GFP channels are shown. Time stamps are in hours:minutes format. Bar = 100 μm.
Pluripotent cells were prepared by reprogramming MEFs from the Oct-4 GFP mice. The pluripotent state is similarly indicated by the expression of Oct-4 GFP. Single cells were plated on feeder cells following trypsinization. Movie S6: Clonal mixing is seen with MEF-derived iPSCs. Two Oct4-GFP+ cells migrated toward each other and merged to form one colony. Movie S7: Colony dispersal is seen with MEF-derived iPSCs. Note the dispersing behavior of the colony growing on the bottom half of the imaging field. A non-dispersing colony is also in view. Movie S8: Clonal mixing is seen with ESCs. ESCs from wild type C57Bl6 mice were transduced with a retrovirus that expresses either GFP or dsRed. Fluorescently marked cultures were trypsinized to single cells and co-plated at 1:1 ratio on feeder cells. Cell migration yielded colonies that contain both GFP+ and dsRed+ cells. The migratory behaviors from these pluripotent cells are highly similar to those seen with GMP-derived pluripotent cells. Time stamps are in days:hours:minutes format. Bars = 50 μm.
GMPs were transduced with OSKM together with a lentivirus expressing E-cadherin shRNA plus tdTomato. Red fluorescence indicates shRNA expressing cells. At the beginning, the large blue arrow points to a single cell that gave rise to all the later cells marked by arrows. The two yellow flashes indicate two times of cell division in this traced lineage. Out of the 4 daughter cells from these 2 divisions, only one of the lineages is marked out for easier view. All the cells from the later divisions are marked by blue arrows. With each cell division, the arrow becomes one size smaller. Note the absence of the 2-cell dumbbell intermediate as well as compaction among the cells. Weak GFP+ (seen as a yellowish hue), loosely scattered cells were present at the end of image acquisition. Overlayed images of DIC, GFP and tdTomato channels are shown. Time stamps are in hours:minutes format. Bar = 100 μm.
Figure S1. Schematic illustration of the key genetic players used for imaging the emergence of Oct-4 GFP+ cells. A) Compound mice harboring Rosa26-rtTA and Oct-4 GFP alleles were used as donors for all experiments. rtTA is expressed but inactive in the absence of dox. Oct-4 locus is inactive. B) GMPs were introduced with the reprogramming factors by a poly-cistronic lentivirus. Reprogramming factors are not expressed in the absence of dox. Oct-4 locus is inactive. C) Adding dox activates rtTA and induces factor expression. Oct-4 locus undergoes epigenetic changes, preparing for potential activation. D) In rare cells, Oct-4 locus is activated and GFP protein is made. Microscopy detects the GFP+ cells at various reprogramming stages.
Figure S2. Schematic illustration of the scanning and stitching strategy. Camera photographs consecutive frames in a scanning motion with the dashed arrows showing its scanning path. It records signals from DIC and GFP (also RFP channels when necessary) at regular time intervals of 5–30 minutes. Image acquisition initiates about one hour after adding dox and continues for 3–5 days, when GFP+ cells emerge and develop into ESC-like colonies. During post experiment image analysis, the neighboring frames were stitched together to synthesize a large field of view. This large field is necessary for capturing low frequency events and tracking the complete trajectory of a reprogramming lineage given the active cell migration. An area containing a reprogramming event could be zoomed in and examined in close detail. Shown is a hypothetical 10×10 synthetic field.
Figure S3. The interface of the custom software used for image processing, analysis and annotation. This software is coded in Matlab with graphical interface. A) is for image processing and B) for image annotation.
Figure S4. Schematic illustration of the formation of mixed colony containing both GFP+ and GFP- cells. This involves using GMPs from two genetically distinct mice, one that contains the Oct-4 GFP reporter and the other that does not. Both types of GMPs contain the same rtTA transgene at the Rosa26 locus to allow dox-inducible factor expression. Since all colonies from the Oct-4 GFP reporter containing mice are entirely GFP+, and all colonies from the non reporter containing GMPs are GFP-, three types of colonies are expected: all GFP+, all GFP- and a mix of GFP+ and GFP- if clonal mixing occurs during reprogramming.
Figure S5. A) Background fluorescence with immuno-fluorescence staining and detected by confocal microscopy. Staining was performed by omitting the primary E-cadherin antibody and processed in parallel. Similar background level of fluorescence is seen with GMPs and non reprogrammed cells as shown in Fig. 5F top row and Fig. 6F (Cre+), demonstrating a lack of E-cadherin expression in those cells. B) A conditional allele allows for dox inducible inactivation of the E-cadherin gene. PCR detection of the recombined/inactivated E-cadherin allele (Δ) in Cre+ cells when Dox was added.
Acknowledgments
We would like to thank Dr. Haifan Lin for insightful discussion. This study was supported by NIH grants DK082982 (to SG), DK086267 and DK072442 (to DK and Yale Center of Excellence in Molecular Hematology) and Connecticut Innovations (to JL and SG).
Footnotes
Author Contributions:
Cynthia M. Megyola: Collection and/or assembly of data, data analysis and interpretation, manuscript writing
Yuan Gao: Collection and/or assembly of data, data analysis and interpretation
Alexandra M. Teixeira: Provision of study material or patients, collection and/or assembly of data
Jijun Cheng: Collection and/or assembly of data
Kartoosh Heydari: Collection and/or assembly of data
Ee-chun Cheng: Provision of study material or patients
Timothy Nottoli: Collection and/or assembly of data, data analysis and interpretation
Diane S. Krause: Provision of study material or patients, manuscript writing
Jun Lu: Conception and design, financial support, data analysis and interpretation, manuscript writing
Shangqin Guo: Conception and design, financial support, collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript
References
- 1.Wernig M, Meissner A, Foreman R, et al. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 3.Park IH, Zhao R, West JA, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 4.Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 5.Hochedlinger K, Plath K. Epigenetic reprogramming and induced pluripotency. Development. 2009;136:509–523. doi: 10.1242/dev.020867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanna JH, Saha K, Jaenisch R. Pluripotency and cellular reprogramming: facts, hypotheses, unresolved issues. Cell. 2010;143:508–525. doi: 10.1016/j.cell.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eminli S, Foudi A, Stadtfeld M, et al. Differentiation stage determines potential of hematopoietic cells for reprogramming into induced pluripotent stem cells. Nat Genet. 2009;41:968–976. doi: 10.1038/ng.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanna J, Saha K, Pando B, et al. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 2009;462:595–601. doi: 10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jaenisch R, Young R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell. 2008;132:567–582. doi: 10.1016/j.cell.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Araki R, Jincho Y, Hoki Y, et al. Conversion of ancestral fibroblasts to induced pluripotent stem cells. Stem Cells. 2010;28:213–220. doi: 10.1002/stem.282. [DOI] [PubMed] [Google Scholar]
- 11.Chan EM, Ratanasirintrawoot S, Park IH, et al. Live cell imaging distinguishes bona fide human iPS cells from partially reprogrammed cells. Nat Biotechnol. 2009;27:1033–1037. doi: 10.1038/nbt.1580. [DOI] [PubMed] [Google Scholar]
- 12.Smith ZD, Nachman I, Regev A, et al. Dynamic single-cell imaging of direct reprogramming reveals an early specifying event. Nat Biotechnol. 2010;28:521–526. doi: 10.1038/nbt.1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Warlich E, Kuehle J, Cantz T, et al. Lentiviral vector design and imaging approaches to visualize the early stages of cellular reprogramming. Mol Ther. 2011;19:782–789. doi: 10.1038/mt.2010.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Akashi K, Traver D, Miyamoto T, et al. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404:193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- 15.Ying QL, Wray J, Nichols J, et al. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nichols J, Silva J, Roode M, et al. Suppression of Erk signalling promotes ground state pluripotency in the mouse embryo. Development. 2009;136:3215–3222. doi: 10.1242/dev.038893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nagy A, Gertsenstein M, Vintersten K, et al. Manipulating the Mouse Embryo. (3) 2003 [Google Scholar]
- 18.Guo S, Lu J, Schlanger R, et al. MicroRNA miR-125a controls hematopoietic stem cell number. Proc Natl Acad Sci U S A. 2010;107:14229–14234. doi: 10.1073/pnas.0913574107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chou YF, Chen HH, Eijpe M, et al. The growth factor environment defines distinct pluripotent ground states in novel blastocyst-derived stem cells. Cell. 2008;135:449–461. doi: 10.1016/j.cell.2008.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marson A, Levine SS, Cole MF, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carey BW, Markoulaki S, Hanna J, et al. Reprogramming of murine and human somatic cells using a single polycistronic vector. Proc Natl Acad Sci U S A. 2009;106:157–162. doi: 10.1073/pnas.0811426106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen T, Yuan D, Wei B, et al. E-cadherin-mediated cell-cell contact is critical for induced pluripotent stem cell generation. Stem Cells. 2010;28:1315–1325. doi: 10.1002/stem.456. [DOI] [PubMed] [Google Scholar]
- 23.Li R, Liang J, Ni S, et al. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010;7:51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 24.Redmer T, Diecke S, Grigoryan T, et al. E-cadherin is crucial for embryonic stem cell pluripotency and can replace OCT4 during somatic cell reprogramming. EMBO Rep. 2011;12:720–726. doi: 10.1038/embor.2011.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Samavarchi-Tehrani P, Golipour A, David L, et al. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 2010;7:64–77. doi: 10.1016/j.stem.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 26.Polo JM, Hochedlinger K. When fibroblasts MET iPSCs. Cell Stem Cell. 2010;7:5–6. doi: 10.1016/j.stem.2010.05.018. [DOI] [PubMed] [Google Scholar]
- 27.Li D, Zhou J, Wang L, et al. Integrated biochemical and mechanical signals regulate multifaceted human embryonic stem cell functions. J Cell Biol. 2010;191:631–644. doi: 10.1083/jcb.201006094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boussadia O, Kutsch S, Hierholzer A, et al. E-cadherin is a survival factor for the lactating mouse mammary gland. Mech Dev. 2002;115:53–62. doi: 10.1016/s0925-4773(02)00090-4. [DOI] [PubMed] [Google Scholar]
- 29.Perl AK, Wert SE, Nagy A, et al. Early restriction of peripheral and proximal cell lineages during formation of the lung. Proc Natl Acad Sci U S A. 2002;99:10482–10487. doi: 10.1073/pnas.152238499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohgushi M, Matsumura M, Eiraku M, et al. Molecular pathway and cell state responsible for dissociation-induced apoptosis in human pluripotent stem cells. Cell Stem Cell. 2010;7:225–239. doi: 10.1016/j.stem.2010.06.018. [DOI] [PubMed] [Google Scholar]
- 31.Stadtfeld M, Hochedlinger K. Induced pluripotency: history, mechanisms, and applications. Genes Dev. 2010;24:2239–2263. doi: 10.1101/gad.1963910. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Signals from DIC and GFP were recorded, stitched and overlayed. There were no GFP+ cells during the early part of the image series. GFP+ cells/colonies start to be visible around 60 hours. Image quality is reduced to show the overview of the entire synthetic field. Time stamps are in hours:minutes format. Bar = 100 μm.
High resolution images were acquired by time-lapse imaging, processed and annotated. Images in the beginning show cells of hematopoietic morphology, which lack detectible Oct-4 GFP expression. Arrows follow the movement of cell(s) that developed into GFP+ colonies at the end of image acquisition. The founder cell is followed by the yellow arrow, which split and established two similar sized colonies at the end of image acquisition. This is a typical example of sister colony formation, with some minor satellite colonies seen as well (not marked). Overlayed images of DIC and GFP channels are shown. Time stamps are in hours:minutes format. Bar = 100 μm.
GMPs from Rosa26-rtTA mice with or without the Oct-4 GFP reporter were induced to reprogram on the same culture dish. Time-lapse observation captured the formation of one mixed colony. The arrows track the movement of different cells as they continued the reprogramming process. They entered the field of view from distinct locations at different times. The migration brought them into close proximity of each other. The small cell clusters originated from these cells continued to grow and compact without distinguishing the cells of different origins. The resulted colony contained both GFP+ and GFP- cells, originating from cells indicated by the pink, yellow and blue arrows. Also present in view are colonies consisting of only GFP+ cells and of only GFP- cells (not marked). Time stamps are in hours:minutes format. Bar = 100 μm.
The founder cell divided on the 2nd frame of acquisition (00:30) and its two immediate daughters are followed by the yellow and blue arrows. All cells in view marked by arrows are descendents of the same founder. Addition of a new colored arrow indicates the birth of a visually distinct new daughter cell. Note the complex splitting, migrating and mixing behavior. Some arrows disappeared in the movie, which indicate either apoptotic appearance of the cell or loss of Oct4-GFP, or becoming out of view. Note the appearance of several dumbbell intermediates. Around 62 hours, two of the larger colonies disappeared suddenly from view. Images from another field (Right) show one of these colonies landed a remote site and continued its development afterwards. The identity of the break-away small cell cluster was informed by its shape and the timing when it disappeared from view in one field and appeared in another field. Overlayed images of DIC and GFP channels are shown. Time stamps are in hours:minutes format. Bar = 100 μm.
Large yellow arrow follows the main reprogramming colony. Various colored smaller arrows point to the break-away cells, which continued to grow and form smaller “satellite” colonies. Overlayed images of DIC and GFP channels are shown. Time stamps are in hours:minutes format. Bar = 100 μm.
Pluripotent cells were prepared by reprogramming MEFs from the Oct-4 GFP mice. The pluripotent state is similarly indicated by the expression of Oct-4 GFP. Single cells were plated on feeder cells following trypsinization. Movie S6: Clonal mixing is seen with MEF-derived iPSCs. Two Oct4-GFP+ cells migrated toward each other and merged to form one colony. Movie S7: Colony dispersal is seen with MEF-derived iPSCs. Note the dispersing behavior of the colony growing on the bottom half of the imaging field. A non-dispersing colony is also in view. Movie S8: Clonal mixing is seen with ESCs. ESCs from wild type C57Bl6 mice were transduced with a retrovirus that expresses either GFP or dsRed. Fluorescently marked cultures were trypsinized to single cells and co-plated at 1:1 ratio on feeder cells. Cell migration yielded colonies that contain both GFP+ and dsRed+ cells. The migratory behaviors from these pluripotent cells are highly similar to those seen with GMP-derived pluripotent cells. Time stamps are in days:hours:minutes format. Bars = 50 μm.
GMPs were transduced with OSKM together with a lentivirus expressing E-cadherin shRNA plus tdTomato. Red fluorescence indicates shRNA expressing cells. At the beginning, the large blue arrow points to a single cell that gave rise to all the later cells marked by arrows. The two yellow flashes indicate two times of cell division in this traced lineage. Out of the 4 daughter cells from these 2 divisions, only one of the lineages is marked out for easier view. All the cells from the later divisions are marked by blue arrows. With each cell division, the arrow becomes one size smaller. Note the absence of the 2-cell dumbbell intermediate as well as compaction among the cells. Weak GFP+ (seen as a yellowish hue), loosely scattered cells were present at the end of image acquisition. Overlayed images of DIC, GFP and tdTomato channels are shown. Time stamps are in hours:minutes format. Bar = 100 μm.
Figure S1. Schematic illustration of the key genetic players used for imaging the emergence of Oct-4 GFP+ cells. A) Compound mice harboring Rosa26-rtTA and Oct-4 GFP alleles were used as donors for all experiments. rtTA is expressed but inactive in the absence of dox. Oct-4 locus is inactive. B) GMPs were introduced with the reprogramming factors by a poly-cistronic lentivirus. Reprogramming factors are not expressed in the absence of dox. Oct-4 locus is inactive. C) Adding dox activates rtTA and induces factor expression. Oct-4 locus undergoes epigenetic changes, preparing for potential activation. D) In rare cells, Oct-4 locus is activated and GFP protein is made. Microscopy detects the GFP+ cells at various reprogramming stages.
Figure S2. Schematic illustration of the scanning and stitching strategy. Camera photographs consecutive frames in a scanning motion with the dashed arrows showing its scanning path. It records signals from DIC and GFP (also RFP channels when necessary) at regular time intervals of 5–30 minutes. Image acquisition initiates about one hour after adding dox and continues for 3–5 days, when GFP+ cells emerge and develop into ESC-like colonies. During post experiment image analysis, the neighboring frames were stitched together to synthesize a large field of view. This large field is necessary for capturing low frequency events and tracking the complete trajectory of a reprogramming lineage given the active cell migration. An area containing a reprogramming event could be zoomed in and examined in close detail. Shown is a hypothetical 10×10 synthetic field.
Figure S3. The interface of the custom software used for image processing, analysis and annotation. This software is coded in Matlab with graphical interface. A) is for image processing and B) for image annotation.
Figure S4. Schematic illustration of the formation of mixed colony containing both GFP+ and GFP- cells. This involves using GMPs from two genetically distinct mice, one that contains the Oct-4 GFP reporter and the other that does not. Both types of GMPs contain the same rtTA transgene at the Rosa26 locus to allow dox-inducible factor expression. Since all colonies from the Oct-4 GFP reporter containing mice are entirely GFP+, and all colonies from the non reporter containing GMPs are GFP-, three types of colonies are expected: all GFP+, all GFP- and a mix of GFP+ and GFP- if clonal mixing occurs during reprogramming.
Figure S5. A) Background fluorescence with immuno-fluorescence staining and detected by confocal microscopy. Staining was performed by omitting the primary E-cadherin antibody and processed in parallel. Similar background level of fluorescence is seen with GMPs and non reprogrammed cells as shown in Fig. 5F top row and Fig. 6F (Cre+), demonstrating a lack of E-cadherin expression in those cells. B) A conditional allele allows for dox inducible inactivation of the E-cadherin gene. PCR detection of the recombined/inactivated E-cadherin allele (Δ) in Cre+ cells when Dox was added.