Abstract
Duplications have long been postulated to be an important mechanism by which genomes evolve. Interspecies genomic comparisons are one method by which the origin and molecular mechanism of duplications can be inferred. By comparative mapping in human, mouse, and rat, we previously found evidence for a recent chromosome-fission event that occurred in the mouse lineage. Cytogenetic mapping revealed that the genomic segments flanking the fission site appeared to be duplicated, with copies residing near the centromere of multiple mouse chromosomes. Here we report the mapping and sequencing of the regions of mouse chromosomes 5 and 6 involved in this chromosome-fission event as well as the results of comparative sequence analysis with the orthologous human and rat genomic regions. Our data indicate that the duplications associated with mouse chromosomes 5 and 6 are recent and that the resulting duplicated segments share significant sequence similarity with a series of regions near the centromeres of the mouse chromosomes previously identified by cytogenetic mapping. We also identified pericentromeric duplicated segments shared between mouse chromosomes 5 and 1. Finally, novel mouse satellite sequences as well as putative chimeric transcripts were found to be associated with the duplicated segments. Together, these findings demonstrate that pericentromeric duplications are not restricted to primates and may be a common mechanism for genome evolution in mammals.
[Supplemental material is available online at www.genome.org.]
The evolution of genomic sequence is the primary molecular basis for the diversity within and among species. The associated sequence changes can arise by a number of mechanisms, including single-nucleotide substitutions, insertions and deletions, duplications, and other chromosomal rearrangements. Each of these can ultimately contribute to the phenotypic differences encountered between species and individuals.
Duplications are thought to play a particularly important role in evolution. Specifically, gene, segmental, and whole-genome duplications can lead to the presence of new genes that attain novel functions. Once a gene is duplicated, one duplicate copy can establish a new function while the other retains its ancestral role, or the two genes can partition the ancestral function between them. The subsequent functional loss of one of the duplicates can then contribute to the evolution of the species via the change in gene content. Indeed, gene duplications and losses appear to occur at a rate similar to that of single-nucleotide changes (Lynch and Conery 2000), making them very common events in genome evolution. Whole-genome duplications are thought to be much less frequent, perhaps occurring only once or twice in the vertebrate lineage (Gu et al. 2002; McLysaght et al. 2002). Segmental duplications represent an intermediate type of duplication, in which a portion of a chromosome is duplicated. These can involve a portion of a gene, a non-genic region, or a segment encompassing multiple genes.
Sequence-based evidence for segmental duplications has been uncovered for a range of species, including yeast (Fischer et al. 2001; Piskur 2001), Arabidopsis (Vision et al. 2000), and human (International Human Genome Sequencing Consortium 2001; Venter et al. 2001). Analyses of the human genome sequence indicate that recent segmental duplications (defined as sequences that are less then 10% diverged and greater than 1 kb in length) have arisen over the past 40 million years and now constitute as much as 5% of the human genome (Bailey et al. 2001). Briefly, these duplications have been broadly classified as being either interchromosomal or intrachromosomal: Interchromosomal duplicated segments tend to be located in pericentromeric (i.e., pericentromeric-directed duplications; Guy et al. 2000) or subtelomeric regions, whereas intrachromosomal duplicated segments tend to reside in euchromatic regions (Bailey et al. 2001; Eichler 2001). Both types of segmental duplications are comprised of a mosaic of sequence modules derived from distinct chromosomal regions and duplication events (Eichler et al. 1996; Horvath et al. 2000a; Bailey et al. 2002). The juxtaposition of modules containing portions of different genes can lead to the generation of chimeric transcripts and possibly new genes (Bailey et al. 2002).
Intrachromosomal duplicated segments have been demonstrated to be the molecular basis for a number of human diseases (Stankiewicz and Lupski 2002). However, no clear connection to human disease or any function has been established for recent segmental duplications located in the pericentromeric or subtelomeric regions, in part because no non-primate organism has been identified with similar duplications. Thus, recent segmental duplications are highly relevant to the study of both human genome evolution and genetic disease and would greatly benefit from the availability of a model system to study the potential function and molecular mechanisms associated with their origin and propagation.
In the course of establishing the orthologous positions of two human chromosome 7q21 (HSA7q21) genes in the mouse genome, we previously localized the site of an evolutionary breakpoint to a ∼300-kb interval between CDK6 and C7orf6 (Thomas et al. 1999). Both cytogenetic and genetic mapping data indicated that mouse Cdk6 resides near the centromere on mouse chromosome 5 (MMU5) and that the mouse C7orf6 ortholog (Estm25) resides at a similar position on MMU6. Subsequent mapping of the orthologous region on rat chromosome 4 (RNO4) revealed a genomic organization similar to that in human (Summers et al. 2001). Thus, we concluded that a chromosome-fission event split this region onto two separate chromosomes and that this event occurred in the mouse lineage following the divergence of mouse and rat. Interestingly, the initial cytogenetic mapping of mouse Cdk6 to MMU5 also revealed the potential presence of duplicated segments containing this locus near the centromeres of MMU4, MMU6, MMU7, MMU8, MMU12, MMU13, and MMU15 (Thomas et al. 1999).
To better understand this evolutionary rearrangement and to further characterize the apparent duplicated segments, we mapped and sequenced the relevant genomic regions in mouse and rat. Based on detailed analyses of the resulting data, we were able to refine the location of the evolutionary breakpoint, to discover recent segmental duplicates on multiple mouse chromosomes, and to uncover evidence for the presence of chimeric transcripts and novel satellite sequences associated with the duplicated segments. These studies provide the first evidence for recent, pericentromeric segmental duplications in a non-primate species and establish the mouse genome as a model system for studying this mechanism of genome evolution.
RESULTS
Physical Mapping of MMU5 and MMU6
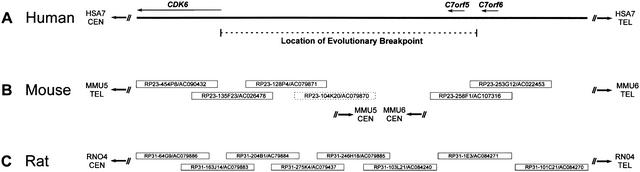
Previous genetic and cytogenetic mapping studies (Thomas et al. 1999) localized a human-mouse evolutionary breakpoint to an ∼300-kb interval on HSA7q21 flanked by the genes CDK6 and C7orf6(Fig. 1A). To characterize in detail the corresponding segments of the mouse genome, sequence-ready bacterial artificial chromosome (BAC) contigs were assembled that extended bi-directionally from Cdk6 on MMU5 and from Estm25 (the mouse ortholog of C7orf6) on MMU6 (Fig. 1B). One probe used for map assembly, which was derived from exon 3 of Cdk6, was found to localize to both the MMU5 and MMU6 contigs, potentially indicating the presence of a common duplicated segment.
Figure 1.
Physical mapping of the human, mouse, and rat genomic regions flanking an evolutionary breakpoint. The depicted segment of HSA7q21 (A) and the orthologous region of RNO4 (C) are contiguous in each species; however, the orthologous segment in the mouse genome is split between two regions (B), with portions located at the proximal (i.e., centromeric) ends of MMU5 and MMU6. The indicated location of the evolutionary breakpoint on HSA7q21 is based on genetic and cytogenetic mapping studies (Thomas et al. 1999). The positions of the three genes (CDK6, C7orf5, and C7orf6) on HSA7q21 are based on finished genomic sequence (GenBank nos. AC000065, AC004128, AC004011, AC002454, and AC000119). Also shown are the minimal tiling paths of BACs selected for sequencing the orthologous regions on MMU5, MMU6, and RNO4 (Thomas et al. 1999; Summers et al. 2001), with the clone name/GenBank accession number indicated for each. Note that the mouse BACs depicted here are the most proximal clones in larger (20-BAC) tiling paths assembled for both MMU5 and MMU6 (data not shown). One clone (RP23–104K20) was subsequently found not to map to MMU5 (indicated by a dashed box). The orientation of the depicted sequences and clones relative to each telomere (TEL) and centromere (CEN) is indicated. Note that the human sequence as well as the mouse and rat clones are not drawn to scale.
Given the potential difficulty of accurately mapping duplicated segments, multiple analyses of the BAC-derived sequences from MMU5 and MMU6 were performed to verify their position in the mouse genome. First, the high-quality sequence generated from each mouse BAC (see Methods; also note the GenBank record information provided in Fig. 1) was aligned with sequences derived from the putative neighboring clones to confirm the overlaps predicted by probe-content and restriction enzyme digest-based fingerprint maps. The sequence overlap between all clones (with the exception of RP23–104K20; see Fig. 1B) was consistent with the physical mapping results. RP23–104K20 was therefore excluded from the MMU5 sequence and subjected to independent analysis (see below). Sequences from each authenticated clone-tiling path were then assembled together to facilitate studies of gene content and orthology. The resulting fully ordered and oriented sequence contigs totaled 463,396 bp (Fig. 2A) and 290,131 bp (Fig. 2B) for MMU5 and MMU6, respectively.
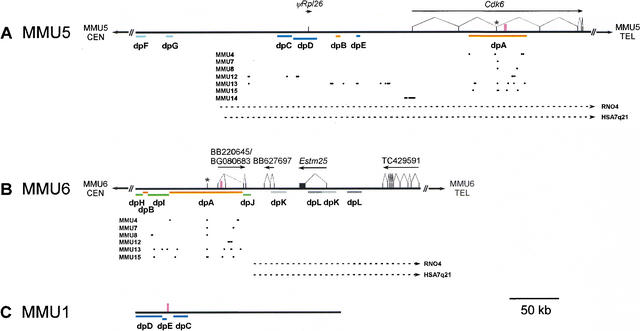
Figure 2.
Annotated features of the MMU5, MMU6, and MMU1 sequences. The generated sequences from MMU5 (A), MMU6 (B), and MMU1 (C) were compiled and annotated as described (see Methods), with the corresponding annotated sequence files available at www.nisc.nih.gov/data. The positions and intron-exon structures of the indicated genes were determined (arrows indicate the direction of transcription and black rectangles represent exons). Also indicated are the positions of satellite sequences (tall pink boxes) and the location of Cdk6 exon 3 (designated by *). The various colored lines labeled dpA through dpL depict the relative positions of duplicated segments (see Table 1). The positions of aligned, near-identical BAC-end sequences are represented by dots, with each sorted based on the mouse chromosome from which the originating BAC was mapped. The portions of the MMU5 and MMU6 sequences orthologous to RNO4 and HSA7q21 are denoted by the dashed arrows. The orientation of the MMU5 and MMU6 sequences relative to each telomere (TEL) and centromere (CEN) is indicated; note that the orientation of the MMU1 sequence relative to the centromere and telomere could not be determined. Additional details about the various annotated features are provided in the text.
To infer gene content, each assembled sequence was analyzed for the presence of matches to mouse transcripts and for concordance with established gene-based and comparative maps of the region. As expected, Cdk6 was identified in the MMU5 sequence and determined to have the same general intron-exon structure as human CDK6 (Fig. 2A). With the exception of a putative retrotransposon-derived Rpl26 pseudogene (ϕRpl26), no other genes were identified in the MMU5 sequence. Analysis of the MMU6 sequence also revealed the one expected gene, Estm25 (Fig. 2B). Like the human C7orf5 and C7orf6 genes (Thomas et al. 1999), Estm25 is predicted to contain a single large coding exon (4740 bp). In addition, matches to spliced ESTs indicated the presence of a 5′ untranslated Estm25 exon. In contrast to the single-copy nature of Estm25 in mouse, the orthologous segment of HSA7q21 contains two genes (C7orf5 and C7orf6) that are closely related to Estm25. Comparison of the predicted proteins encoded by these genes further supports the hypothesis that Estm25 is the ortholog of C7orf6 (Thomas et al. 1999).
Distal to Estm25 on MMU6 is a gene whose structure (Fig. 2B) could be readily established by comparing the generated genomic sequence with the TIGR gene consensus sequence TC429591 (www.tigr.org/tdb/tgi) and a series of ESTs. A human ortholog of this gene was detected at the predicted location distal to C7orf6 on HSA7q21. Proximal to Estm25 are two potential genes (Fig. 2B). Both match mouse ESTs and appear to contain three exons, but each is also associated with a notably short ORF (<100 amino acids) and, therefore, might not be translated. Unlike Estm25 and TC429591, no evidence (e.g., matching ESTs) could be found for the presence of a human ortholog for either of these two genes. Based on the available data, it is thus not clear whether these represent bona fide genes or reflect spurious transcripts. However, these two potential mouse genes are of interest due to their location relative to the other sequence features described below.
Finally, the position of each sequenced MMU5 and MMU6 clone in an independently derived, BAC-based physical map of the entire mouse genome (Gregory et al. 2002) was examined. Strikingly, both groups of clones resided at the proximal end of the most proximal BAC contig on the expected mouse chromosome. Further analysis to directly or indirectly link these clones to the centromere, based on the presence of mouse major or minor centromere-specific satellites either in the MMU5 or MMU6 sequences or BACend sequences from neighboring clones, was uninformative (details are available as supplementary material at www.genome.org). Thus, while the sequenced clones described above could not be definitively linked to centromeric regions within the whole-genome BAC map (Gregory et al. 2002), the predominance of the available mapping data indicates that these clones contain DNA from the pericentromeric regions of MMU5 and MMU6.
Refined Mapping of the Evolutionary Breakpoint
Establishing the genomic position and gene content of the generated MMU5 and MMU6 sequences allowed for the more precise mapping of the evolutionary breakpoint depicted in Figure 1. Towards that end, the MMU5 and MMU6 sequences were aligned to the orthologous HSA7q21 sequence. The resulting alignments, which revealed a series of orthologous genomic segments in the same relative order and orientation in both species, allowed the evolutionary breakpoint to be localized to an ∼50-kb interval between CDK6 and C7orf5 on HSA7q21, and 192 kb proximal to Cdk6 (Fig. 2A) and 46 kb proximal to Estm25 (Fig. 2B) on MMU5 and MMU6, respectively. Comparison to the RNO4 sequence (Fig. 1C) yielded similar results. Similarity searches of the remainder of the mouse sequence that failed to align with HSA7q21 and RNO4 did not reveal any evidence of orthology to other regions of the human genome. Interestingly, a portion of the proximal MMU6 sequence did show similarity to a small region of HSA7q21 and RNO4 that contains CDK6 exon 3.
Characterization of the Duplicated Segments on MMU5 and MMU6
To characterize duplicated segments on MMU5 and MMU6, a series of sequence comparisons were performed. The first duplicated segment identified (referred to as dpA in Fig. 2A,B and Table 1) is 96.3% identical, includes Cdk6 exon 3, and is present on both MMU5 and MMU6. There is no evidence for such a duplication in the orthologous rat sequence, which contains a single segment that is 84.3% identical to the MMU5 dpA sequence (Table 1). In addition, a smaller (5-kb) duplicated segment (called dpB) was found on MMU5 and MMU6 (Fig. 2A,B, Table 1). Given that dpA is present in a single-copy fashion in the orthologous region of HSA7q21 and RNO4 and that it contains a coding exon of the highly conserved Cdk6 gene, the most parsimonious ancestral location for dpA is likely MMU5. No such inference can be made for dpB, which does not align with the orthologous human or rat sequence.
Table 1.
Summary of Duplicated Segments
| Duplicated segmenta | Size of each duplicated segment (kb)b | Percent identityc |
| dpA (MMU5/MMU6) | 58.4/74.5 | 96.3% |
| dpA (MMU5/MMU8) | 58.4/ND | 96.1% |
| dpA (MMU6/MMU8) | 74.5/ND | 95.6% |
| dpA (MMU5/RNO4) | 58.4/ND | 84.3% |
| dpB (MMU5/MMU6) | 5.0/5.2 | 93.3% |
| dpC (MMU5/MMU1) | 15.1/14.0 | 95.2% |
| dpC (MMU5/RNO4) | 15.1/ND | 84.8% |
| dpD (MMU5/MMU1) | 24.2/26.8 | 94.8% |
| dpD (MMU5/RNO4) | 24.2/ND | 85.6% |
| dpE (MMU5/MMU1) | 4.4/4.4 | 93.8% |
| dpE (MMU5/RNO4) | 4.4/ND | 85.1% |
| dpF (MMU5/MMU7) | 10.3/ND | 96.1% |
| dpG (MMU5/MMU7) | 7.0/ND | 93.5% |
| dpH (MMU6/MMU8) | 8.3/ND | 96.4% |
| dpI (MMU6/MMU8) | 22.1/ND | 96.8% |
| dpJ (MMU6/MMU8) | 9.1/ND | 90.2% |
| dpK (MMU6/MMU6) | 16.0/14.6 | 97.3% |
| dpL (MMU6/MMU6) | 14.6/14.4 | 96.3% |
Each duplicated segment (dpA through dpL) is listed in a fashion that allows pair-wise comparisons between copies on the indicated mouse or rat chromosomes (in parentheses). Note that the location and sequence of each indicated duplicated segment are available in the annotated sequence files at www.nisc.nih.gov/data.
The size of each duplicated segment on the two chromosomes listed in the far-left column is provided. ND indicates that the size could not be accurately determined due to the lack of continuity of the available genomic sequence. Note that the genomic interval encompassing any two duplicated segments often varies due to both large and small insertions and deletions.
The percent identity between the sequences in gap-free alignments of the two duplicated segments listed in the far-left column is provided in each case.
Based on its location, dpA was the likely duplicated sequence responsible for the previously observed multi-chromosome cytogenetic mapping of a Cdk6-containing BAC (Thomas et al. 1999). To test this hypothesis, the dpA sequences were compared to all available mouse BAC-end sequences, and the resulting matches were investigated to determine the map position of the originating clone in the whole-genome BAC map (Gregory et al. 2002). Remarkably, nearly all of the matches were found to be derived from BACs mapping to the most proximal contig on the six chromosomes previously identified by cytogenetic mapping (MMU4, MMU7, MMU8, MMU12, MMU13, and MMU15; see Fig. 2A,B, with additional details available as supplementary material at www.genome.org). These results further demonstrate the presence of dpA at the proximal portion of multiple mouse chromosomes.
Comparative sequence analysis of the MMU7 and MMU8 regions (identified by cytogenetic studies as harboring duplicated segments) was facilitated by extensive data generated by mapping and sequencing of the syntenic regions of HSA19 (Dehal et al. 2001; Kim et al. 2001). In particular, these regions were scrutinized for the presence of dpA (details are available as supplementary material at www.genome.org). No dpA-containing alignments were detected with the available MMU7 sequence; however, two other likely duplicated segments shared with the proximal portion of the MMU5 sequence (designated dpF and dpG; see Fig. 2A, Table 1) were identified on MMU7. In the case of MMU8, dpA-containing alignments were identified. The MMU8 copy of dpA is 96.1% and 95.6% identical to copies on MMU5 and MMU6, respectively (Table 1). Three additional duplicated segments (designated dpH, dpI, and dpJ) are shared only between MMU6 and MMU8 (Fig. 2B, Table 1). These three duplicated segments are located within the proximal portion of MMU6 and either flank or reside between dpA and dpB. Thus, the comparisons to MMU7 and MMU8 clearly indicate a complex organization of genomic duplications residing near the centromeres on MMU5, MMU6, and elsewhere in the mouse genome.
Another possible duplication in the MMU5 sequence was identified based on similar, but not identical, overlap with BAC RP23–104K20, which was subsequently localized to mouse chromosome 1A2 by cytogenetic mapping (data not shown) and to the proximal-most MMU1 contig in the whole-genome BAC map (Gregory et al. 2002). Analysis of the MMU1 and MMU5 sequence comparison identified three duplicated segments (designated dpC, dpD, and dpE; see Fig. 2A,C, Table 1). Interestingly, the MMU1 and MMU5 copies of these duplicated segments seem to have undergone rearrangements relative to one another. The difference in the relative positions of dpC, dpD, and dpE on MMU1 and MMU5 is consistent with a model whereby dpC was transposed to its current position on MMU1. Based on the physical arrangements of these sequences on HSA7q21 and RNO4, which are both in the same order and orientation as on MMU5 (Fig. 2A), it seems most likely that these segments originated on MMU5 and then underwent rearrangements after the duplication event(s) that put them on MMU1.
To identify any other potential duplicated segments in the MMU5 and MMU6 sequences in common with other chromosomes, each was compared to all available BAC-end sequences (Fig. 2A,B). In the case of the MMU5 sequence, a large segment starting just proximal to dpC and ending distal to dpE matched multiple BAC-end sequences mapping to MMU12 and the proximal-most MMU13 contig. A cluster of BAC-end sequences mapping to MMU14 also matched a small segment near Cdk6 exon 1. In the case of the MMU6 sequence, matches were found between dpI and BAC-end sequences mapping to proximal MMU13 and proximal MMU15 contigs.
Finally, the MMU5 and MMU6 sequences were searched for local intrachromosomal duplications. There was no indication of an intrachromosomal duplication in the MMU5 sequence. However, in the MMU6 sequence, two duplicated segments (dpK and dpL) were found, with the first pair flanking the Estm25 coding exon (Fig. 2B, Table 1). There were no dpK- or dpL-related duplicated segments in the orthologous positions on HSA7q21, but there was evidence for similar duplications in the orthologous rat sequence. However, given the high degree of sequence similarity between the mouse duplicated segments and the presence of five copies of genes similar to Estm25 in the orthologous rat sequence (J.W. Thomas and E.D. Green, unpubl.), it seems likely that the duplicated rat sequences were derived from an independent event(s).
Detection of Novel Mouse Satellite Sequences
In the human genome, duplicated segments of recent origin are sometimes associated with short, interspersed repetitive sequences that reside at the boundaries of duplication integration sites (Eichler et al. 1999). To search for the presence of similar repeats in the mouse genome, the unmasked sequences flanking all the duplicated segments on MMU1, MMU5, and MMU6 (Table 1) were carefully scrutinized. No repetitive motifs were identified in the flanking regions on MMU5 and MMU6; however, a novel 27-bp satellite sequence was found to reside between dpE and dpC on MMU1 (Figs. 2C, 3A). Interestingly, by inspecting pair-wise and self-self alignments of the MMU5 and MMU6 sequence, we were able to detect a second novel satellite sequence within (as opposed to flanking) dpA (Figs. 2A,B, 3B). This 36-bp satellite was not detected in the orthologous human or rat sequence; therefore, it appears to be of a recent, mouse lineage-specific origin.
Figure 3.
Consensus sequences of novel mouse satellites. (A) Consensus sequence of the 27-bp satellite derived by MEME analysis of sequence flanking dpE on MMU1. The consensus represents 83 monomers, each 27 bp in length and occurring in a tandem, head-to-tail fashion within BAC RP23–104K20. The information content (in bits) is included to indicate the degree of conservation at each position in the consensus. The base with the greatest probability of occurrence at each position is shown in the first line of the multilevel consensus sequence; alternative bases are indicated on the second line only if they occur with a probability greater than 0.2. (B) Consensus sequence of the 36-bp satellite derived by MEME analysis of sequence within dpA. The consensus represents 106 monomers, each 36 bp in length, derived from several genomic locations: 62 monomers from MMU5, 40 from MMU6, and four from MMU8. Monomers of this 36-bp satellite are arranged in a tandem, head-to-tail orientation.
Identification of Chimeric Transcripts Emanating From the Duplicated Segments
Duplicated genomic segments containing part of a gene have been shown to give rise to novel chimeric transcripts as a result of the juxtaposition of duplicon sequences with new flanking sequences (Bailey et al. 2002). Annotation of the MMU6 sequence reported here indicated the presence of two putative genes proximal to Estm25 that were associated with matching ESTs but not significant ORFs. The positions of these putative MMU6 genes relative to the duplicated segments (Fig. 2B) suggested that they were likely producing chimeric transcripts. In the case of the proximal putative gene, both alternative first exons reside within dpA, with the second two exons positioned within dpJ. Since dpA likely originated on MMU5 and dpJ is not present on MMU5, the transcripts emanating from this putative gene likely reflect a chimeric product brought about by the novel duplication-induced configuration of that region of MMU6. Similarly, the first exon of the other putative gene resides in dpK, with the second two exons positioned within a single-copy portion of MMU6. Interestingly, within the two copies of dpK on MMU6, the positions of the first exon of this putative gene and the 5′ exon of Estm25 overlap. Thus, it is possible that the endogenous Estm25 promoter was duplicated and now drives the expression of a chimeric transcript in the second copy of dpK.
DISCUSSION
Elucidating and classifying the different mechanisms by which genomes change over time is important for understanding how biological diversity is achieved. Duplications—in particular, recent, pericentromeric segmental duplications—represent one such mechanism, and have been extensively studied in primates (Eichler et al. 1996; Jackson et al. 1999; Horvath et al. 2000b; Bailey et al. 2002) and hypothesized to play an important role in primate diversity. Using comparative mapping and sequencing to study specific genomic regions in human, mouse, and rat, we report the first evidence for recent, pericentromeric duplications in the laboratory mouse, thereby demonstrating a broader role for this mechanism of genomic rearrangement in mammalian evolution.
Since duplicated segments in the pericentromeric region have thus far only been reported in primates, we evaluated the mouse duplications in terms of their similarities and differences to those observed in human. We found that the hallmark features of pericentromeric duplicated segments in the human genome were also present in the mouse. Specifically, duplicated segments were found to be mosaics of sequence modules from different chromosomal regions (Eichler et al. 1996; Horvath et al. 2000a; Bailey et al. 2002). For example, the duplicated segment on MMU6 consists of modules that are shared with MMUS and MMU8, while others are exclusive to MMU6 and MMU8. In addition, the lack of sequence alignment between the proximal portion of the MMU5 sequence (outside of the duplicated sequence modules) and either HSA7q21 or RNO4 also suggests that the entire region might be part of a larger duplicon originating from elsewhere in the genome. The mosaic structure of the duplicated region on MMU6 appears to give rise to chimeric transcripts, as indicated by the presence of novel mouse ESTs. While no function has been assigned to these transcripts nor is it known whether they encode a gene, mosaic transcripts have been shown to be derived from the novel juxtaposition of gene segments in the human genome (Bailey et al. 2002). Rearrangements involving the duplicated segments were also noted in the mouse, again similar to that found with human duplications (Horvath et al. 2000a). Finally, both sequence and physical mapping analyses reported here support the occurrence of duplicated segments in the pericentromeric regions of eight of the nineteen mouse autosomes, suggesting that they are a common feature of pericentromeric regions in the mouse and human genomes.
In contrast to the above similarities, some possible differences were also noted between pericentromeric duplications in mouse and human. Though the single-nucleotide divergence was in the same general range for both species, insertions and deletions within the duplicated modules appear to be more common in the mouse (Bailey et al. 2001). It is also possible that our specific approach and thresholds for aligning sequences led to some of these differences; however, we believe this is unlikely since interspecies comparison of orthologous mouse and rat sequences showed a substantial amount of insertions and deletions within the duplicated segments. A second possible difference is the absence of common, short repeated sequences flanking the duplicated segments, as were encountered with human pericentromeric duplications and suggested to be involved in the duplication events (Eichler et al. 1999). We searched for such sequences both within and flanking the duplicated segments on MMU1, MMU5, and MMU6, but found no common sequence motifs. However, a novel 27-bp satellite repeat was found between two of the duplicated segments on MMU1. In addition, a novel 36-bp satellite repeat was found at the same position within the dpA copies on MMU5, MMU6, and MMU8. Whether or not these newly identified mouse satellite sequences are common to regions harboring duplicated segments can be established once the mouse genome sequence is finished.
The process of segmental duplication in the human genome appears to have been ongoing for the last ∼40 million years (International Human Genome Sequencing Consortium 2001). From the small number of duplicated segments characterized here, a similar estimate cannot be made for the mouse genome. However, based on the degree of sequence divergence between the mouse duplicated segments (2.7%–9.8%), the duplication events seem to have occurred very recently and most likely at different times. Comparisons to the orthologous rat sequences also provide a means for establishing the origin and timing of the duplications. In particular, since the degree of sequence divergence between the mouse and rat sequences are two to four times higher than between the mouse duplicons and assuming a divergence time of ∼14 million years for mouse and rat (Jacobs and Pilbeam 1980), then the mouse duplication events likely occurred in the last 3–7 million years. This calculation only reflects a general time estimate and does not account for sequence homogenization that may have occurred since the originating duplication event. In addition to helping to establish the timing of the mouse duplication events, comparisons with the rat sequence can be used to infer other details about the duplications. For example, in the case of dpC, dpD, and dpE (present on both MMU1 and MMU5), the ancestral location can be inferred to be MMU5, since these sequences are within the orthologous RNO4 segment. Thus, future inferences regarding the origin of the mouse duplications will be greatly aided by comparisons to the rat genome sequence.
Multiple evolutionary breakpoints between the human and mouse genomes have been examined at the sequence level (Lund et al. 2000; Dehal et al. 2001; Puttagunta et al. 2000). In many cases, these breakpoint sequences were found to be enriched for simple-sequence repeats and L1 repetitive elements or associated with clustered gene families, gene family expansions, and more limited gene duplications. Together, these findings suggest that duplications or duplicated sequences are often involved in chromosomal rearrangements. The results presented here are consistent with these previous data, in that large duplicated blocks of sequence were identified on both chromosomes MMU5 and MMU6 involved in the chromosome-fission event. However, the duplicated segments on MMU5 and MMU6 are distinct from those identified previously at evolutionary breakpoints because of their pericentromeric location and their association with the genesis of two new centromeres.
Based on the information reported here and in previous studies in primates (Eichler et al. 1996, 1997; Regnier et al. 1997), we would propose the following model for the evolution of the pericentromeric regions of MMU5 and MMU6. First, a chromosome-fission event occurred between Cdk6 and Estm25 on the ancestral MMU5/MMU6 (precursor) chromosome, which was likely similar to the present-day RNO4 (Walentinsson et al. 2001). The subsequent repair involved addition of centromeric sequences to each side of the double-stranded break, stabilizing the chromosome-fission products and creating two new centromeres and chromosomes. The newly formed proximal regions of MMU5 and MMU6 became ‘activated’ for exchanging duplicated segments with other existing pericentromeric regions. Specifically, dpA spread from MMU5 to multiple other chromosomes; similarly, dpC, dpD, and dpE on MMU5 duplicated to occupy positions on MMU1. Other sequences were likely accepted onto MMU5, including the most proximal segment (of at least ∼90 kb) that encompasses dpF and dpG. In general, this model is consistent with the two-step process previously proposed for human pericentromeric duplications, as reviewed by Horvath et al. (2001). However, the case reported here involved a chromosome-fission event and the creation of new centromeres. Thus, it is possible that the mechanism of pericentromeric duplication in the mouse is intimately related to the derivation of new centromeres. Further characterization of the origin and timing of these and other pericentromeric duplications in the mouse genome should shed light on this issue.
In conclusion, we have uncovered convincing evidence for the occurrence of pericentromeric duplications in the laboratory mouse associated with a chromosome-fission event involving the present-day MMU5 and MMU6. These duplications appear to be recent in origin, perhaps occurring 3–7 million years ago, and suggest that pericentromeric duplications are an important facet of genome evolution in non-primate species. Thus, these studies provide the first indication that pericentromeric duplications represent a common mechanism of genome evolution and might play an important role in creating the observed diversity among mammals.
METHODS
Mapping and Analysis of Mouse BACs
The RPCI-23 mouse BAC library (Osoegawa et al. 2000; see www.chori.org/bacpac) was screened by hybridization using ‘overgo’ probes (Vollrath 1999; Thomas et al. 2000, 2002) orthologous to human chromosome 7 in the region flanking the previously described evolutionary rearrangement (Thomas et al. 1999). Additional BAC insert-end sequences were used for designing new overgo probes for contig expansion. Sequence-ready BAC contigs were assembled based on probe-content data (Thomas et al. 2000) and restriction enzyme digest-based fingerprint analysis (Marra et al. 1997), and a minimal set of overlapping clones were selected for sequencing.
The BAC-based physical map of the whole mouse genome (Gregory et al. 2002), consisting of clones from the RPCI-23 and RPCI-24 C57BL/6J libraries, was downloaded from the Washington University Genome Sequencing Center Web site (genome.wustl.edu) on February 2, 2002. This map contained 305,916 clones and 314 contigs. Each contig was numbered to reflect its relative position on the chromosome, such that the most proximal contig on each chromosome was designated as 1 (i.e., 5001 was the most proximal contig on MMU5).
To identify regions of the mouse genome containing duplicated segments similar to those reported here (see Table 1), a total of 454,634 repeat-masked, mouse BAC-end sequences (from the RPCI-23 and RPCI-24 libraries) were downloaded from the TIGR Web site (ftp.tigr.org) and compared to the generated mouse genomic sequences (see below) using MegaBLAST (Zhang et al. 2000). BAC-end sequences that aligned to the genomic sequence with an expectation value (E value) of less than 1e-33 were scrutinized further. Those deemed to reflect unmasked common repetitive elements (based on alignments to multiple mouse and rat genomic sequences in the htgs division of GenBank) were eliminated. For the remaining sequences, the positions of the originating clones in the mouse whole-genome BAC map were established.
To identify the relative positions of centromeres in the mouse BAC map, the mouse major (GenBank nos. AJ296902, AJ296890, AJ296871, AJ296867, AJ296864, AJ296860, and X03556) and minor (GenBank nos. X14462-X14470, Z22152-Z22170, and M62681) centromeric satellite sequences were compared to the gss division of GenBank using MegaBLAST (E value cutoff of <1e-90). Aligning RPCI-23 and RPCI-24 BAC-end sequences were then extracted from the MegaBLAST output, and the positions of the originating clones within the mouse BAC map were established.
BAC Sequencing
BACs were sequenced using a standard shotgun-sequencing strategy (Wilson and Mardis 1997; Green 2001). In brief, purified BAC DNA (genome.wustl.edu/tools/protocols) was kinetically sheared with a Hydroshear instrument (GeneMachines), and the resulting fragments were end repaired with T4 DNA polymerase and Klenow. BstXI/EcoRI linkers (Invitrogen) were ligated to the end-repaired fragments, and the ligated DNA was then size selected (to 1.5–3.0 kb) by agarose gel electrophoresis and subcloned into the plasmid pOTWI3. Sequence reads were generated from both insert ends of randomly selected subclones using BigDye dye-terminator chemistry and model 3700 automated DNA sequencing instruments (Applied Biosystems). Following the generation of an estimated ∼10-fold sequence redundancy (based on the measured insert size of each starting BAC), sequences were assembled and edited using the Phred/Phrap/Consed suite of programs (Ewing and Green 1998; Ewing et al. 1998; Gordon et al. 1998; see www.phrap.org). Manual inspection of the assembled sequences allowed obvious errors, artifacts, and misassemblies to be corrected. In addition, low-quality sequence data were trimmed from contig ends. Finally, contigs were ordered and oriented based on read-pair associations of gap-spanning subclones, sequence overlaps between neighboring clones, and (in rare cases) PCR amplification of genomic segments between adjacent contigs. Additional details and protocols are available on request.
Sequence Assimilation and Analysis
The mouse genomic sequences generated from a given set of overlapping BACs were assembled into a single sequence file with the minimum number of gaps using Sequin (www.ncbi.nlm.nih.gov/Sequin/index.html). The resulting MMU5 sequence consists of 23 ordered and oriented contigs that total 463,396 bp, the MMU6 sequence consists of 10 ordered and oriented contigs that total 290,131 bp, and the MMU1 sequence consists of 11 ordered and oriented contigs that total 210,516 bp. The annotation of each mouse sequence included establishing the positions of common repetitive elements with RepeatMasker (A.F.A. Smit and P. Green, unpubl.; see repeatmasker.genome.washington.edu), each overgo probe sequence used for BAC-contig construction, and the originating clone for each aligned BAC-end sequence. Gene structures were inferred by comparison of ESTs and available mRNA sequences to the genomic sequence with the program Spidey (www.ncbi.nlm.nih.gov/IEB/Research/Ostell/Spidey). In the case of Estm25, the position of the gene in the genomic sequence was determined based on both EST alignments and the identification of a predicted ORF with ORF Finder (www.ncbi.nlm.nih.gov/gorf/gorf.html). For the identification, characterization, and annotation of satellite sequences, versions 3.0 and 2.2 of MEME (Bailey and Elkan 1994; see meme.sdsc.edu/meme/website) were used. Other sequence features, such as the location of duplicated segments and predicted orthology, were annotated based on sequence alignments. The assembled and annotated sequences are available at www.nisc.nih.gov/data.
PipMaker (Schwartz et al. 2000; see bio.cse.psu.edu) was used for genomic sequence alignments. To identify all potential alignments, self-self and pair-wise comparisons between sequences were performed using the ‘all-matches’ option. The resulting dot-plots and percent-identity-plots (PIPs) as well as the alignments generated with the ‘single-coverage’ option were evaluated to detect the presence of any inter- or intrachromosomal duplicated sequences. At the same time, the alignment data were examined for any evidence of inversions or transpositions between aligning sequences. In the case of aligned, ordered and oriented sequence that did not show any gross rearrangements, the ‘single-strand’ and ‘chaining’ options were used to generate the definitive alignment for percent-identity calculations. In the case of aligned, ordered and oriented sequence that showed evidence of a gross rearrangement, the ‘single-coverage’ option was used to generate the definitive alignment. The latter routine was also used for situations where one of the query sequences contained unordered sequence contigs. In all instances, the resulting alignments were manually inspected and edited to remove spurious matches and to ensure that each base in a sequence was present at most once in the final alignment. The percent identity of each aligned region was then calculated based on gap-free alignments. Regions sharing sequence identity of greater than 90% over at least 3 kb of gap-free alignments were considered to be duplicated segments.
WEB SITE REFERENCES
www.nisc.nih.gov/data; supplementary data, including annotated sequence, for the studies reported here.
www.chori.org/bacpac; BACPAC Resources at Children's Hospital Oakland Research Institute.
genome.wustl.edu; Washington University Genome Sequencing Center.
genome.wustl.edu/tools/protocols; laboratory protocols available from the Washington University Genome Sequencing Center.
www.tigr.org/tdb/tgi; The Institute for Genome Research (TIGR) Gene Index.
ftp.tigr.org; The Institute for Genome Research (TIGR) ftp site.
repeatmasker.genome.washington.edu; Repeatmasker program.
www.phrap.org; Phred/Phrap/Consed suite of programs.
bio.cse.psu.edu; PipMaker program.
www.ncbi.nlm.nih.gov/IEB/Research/Ostell/Spidey; Spidey program.
www.ncbi.nlm.nih.gov/Sequin/index.html; Sequin program.
meme.sdsc.edu/meme/website; MEME program.
www.ncbi.nlm.nih.gov/gorf/gorf.html; ORF Finder program.
Acknowledgments
We wish to acknowledge the dedicated work of the technical and computational staff of the NIH Intramural Sequencing Center (NISC) for their efforts in the NISC Comparative Sequencing Program. The work reported here was supported with funds provided by the National Human Genome Research Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
E-MAIL egreen@nhgri.nih.gov; FAX (301) 402-4735.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.791403. Article published online before print in December 2002.
REFERENCES
- 1.Bailey J.A., Yavor, A.M., Massa, H.F., Trask, B.J., and Eichler, E.E. 2001. Segmental duplications: Organization and impact within the current Human Genome Project assembly. Genome Res. 11: 1005-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bailey J.A., Yavor, A.M., Viggiano, L., Misceo, D., Horvath, J.E., Archidiacono, N., Schwartz, S., Rocchi, M., and Eichler, E.E. 2002. Human-specific duplication and mosaic transcripts: The recent paralogous structure of chromosome 22. Am. J. Hum. Genet. 70: 83-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bailey T. and Elkan, C., 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. In Conference on intelligent systems for molecular biology, pp. 28–36. AAAI Press, Menlo Park, CA. [PubMed]
- 4.Dehal P., Predki, P., Olsen, A.S., Kobayashi, A., Folta, P., Lucas, S., Land, M., Terry, A., Ecale Zhou, C.L., Rash, S., et al. 2001. Human chromosome 19 and related regions in mouse: Conservative and lineage-specific evolution. Science 293: 104-111. [DOI] [PubMed] [Google Scholar]
- 5.Eichler E.E. 2001. Recent duplication, domain accretion and the dynamic mutation of the human genome. Trends Genet. 17: 661-669. [DOI] [PubMed] [Google Scholar]
- 6.Eichler E.E., Lu, F., Shen, Y., Antonacci, R., Jurecic, V., Doggett, N.A., Moyzis, R.K., Baldini, A., Gibbs, R.A., and Nelson, D.L. 1996. Duplication of a gene-rich cluster between 16p11.1 and Xq28: A novel pericentromeric-directed mechanism for paralogous genome evolution. Hum. Mol. Genet. 5: 899-912. [DOI] [PubMed] [Google Scholar]
- 7.Eichler E.E., Budarf, M.L., Rocchi, M., Deaven, L.L., Doggett, N.A., Baldini, A., Nelson, D.L., and Mohrenweiser, H.W. 1997. Interchromosomal duplications of the adrenoleukodystrophy locus: A phenomenon of pericentromeric plasticity. Hum. Mol. Genet. 6: 991-1002. [DOI] [PubMed] [Google Scholar]
- 8.Eichler E.E., Archidiacono, N., and Rocchi, M. 1999. CAGGG repeats and the pericentromeric duplication of the hominoid genome. Genome Res. 9: 1048-1058. [DOI] [PubMed] [Google Scholar]
- 9.Ewing B. and Green, P. 1998. Base-calling of automated sequencer traces usingPhred. II. error probabilities. Genome Res. 8: 186-194. [PubMed] [Google Scholar]
- 10.Ewing B., Hillier, L., Wendl, M.C., and Green, P. 1998. Base-calling of automated sequencer traces using Phred. I. accuracy assessment. Genome Res. 8: 175-185. [DOI] [PubMed] [Google Scholar]
- 11.Fischer G., Neuveglise, C., Durrens, P., Gaillardin, C., and Dujon, B. 2001. Evolution of gene order in the genomes of two related yeast species. Genome Res. 11: 2009-2019. [DOI] [PubMed] [Google Scholar]
- 12.Gordon D., Abajian, C., and Green, P. 1998. Consed: A graphical tool for sequence finishing. Genome Res. 8: 195-202. [DOI] [PubMed] [Google Scholar]
- 13.Green E.D. 2001. Strategies for the systematic sequencing of complex genomes. Nat. Rev. Genet. 2: 573-583. [DOI] [PubMed] [Google Scholar]
- 14.Gregory S.G., Sekhon, M., Schein, J., Zhao, S., Osoegawa, K., Scott, C.E., Evans, R.S., Burridge, P.W., Cox, T.V., Fox, C.A., et al. 2002. A physical map of the mouse genome. Nature 418: 743-750. [DOI] [PubMed] [Google Scholar]
- 15.Gu X., Wang, Y., and Gu, J. 2002. Age distribution of human gene families shows significant roles of both large- and small-scale duplications in vertebrate evolution. Nat. Genet. 31: 205-209. [DOI] [PubMed] [Google Scholar]
- 16.Guy J., Spalluto, C., McMurray, A., Hearn, T., Crosier, M., Viggiano, L., Miolla, V., Archidiacono, N., Rocchi, M., Scott, C., et al. 2000. Genomic sequence and transcriptional profile of the boundary between pericentromeric satellites and genes on human chromosome arm 10q. Hum. Mol. Genet. 9: 2029-2042. [DOI] [PubMed] [Google Scholar]
- 17.Horvath J.E., Schwartz, S., and Eichler, E.E. 2000a. The mosaic structure of human pericentromeric DNA: A strategy for characterizing complex regions of the human genome. Genome Res. 10: 839-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horvath J.E., Viggiano, L., Loftus, B.J., Adams, M.D., Archidiacono, N., Rocchi, M., and Eichler, E.E. 2000b. Molecular structure and evolution of an α satellite/non-α satellite junction at 16p11. Hum. Mol. Genet. 9: 113-123. [DOI] [PubMed] [Google Scholar]
- 19.Horvath J.E., Bailey, J.A., Locke, D.P., and Eichler, E.E. 2001. Lessons from the human genome: Transitions between euchromatin and heterochromatin. Hum. Mol. Genet. 10: 2215-2223. [DOI] [PubMed] [Google Scholar]
- 20.International Human Genome Sequencing Consortium 2001. Initial sequencing and analysis of the human genome. Nature 409: 860-921. [DOI] [PubMed] [Google Scholar]
- 21.Jackson M.S., Rocchi, M., Thompson, G., Hearn, T., Crosier, M., Guy, J., Kirk, D., Mulligan, L., Ricco, A., Piccininni, S., et al. 1999. Sequences flanking the centromere of human chromosome 10 are a complex patchwork of arm-specific sequences, stable duplications and unstable sequences with homologies to telomeric and other centromeric locations. Hum. Mol. Genet. 8: 205-215. [DOI] [PubMed] [Google Scholar]
- 22.Jacobs L.L. and Pilbeam, D. 1980. Of mice and men: Fossil-based divergence dates and molecular “clocks.” J. Hum. Evol. 9: 551-555. [Google Scholar]
- 23.Kim J., Gordon, L., Dehal, P., Badri, H., Christensen, M., Grosa, M., Ha, C., Hammond, S., Vargas, M., Wehri, E., et al. 2001. Homology-driven assembly of a sequence-ready mouse BAC contig map spanning regions related to the 46-Mb gene-rich euchromatic segments of human chromosome 19. Genomics 74: 129-141. [DOI] [PubMed] [Google Scholar]
- 24.Lund J., Chen, F., Hua, A., Roe, B., Budarf, M., Emanuel, B.S., and Reeves, R.H. 2000. Comparative sequence analysis of 634 kb of the mouse chromosome 16 region of conserved synteny with the human velocardiofacial syndrome region on chromosome 22q11.2. Genomics 63: 374-383. [DOI] [PubMed] [Google Scholar]
- 25.Lynch M. and Conery, J.S. 2000. The evolutionary fate and consequence of duplicate genes. Science 10: 1151-1155. [DOI] [PubMed] [Google Scholar]
- 26.Marra M.A., Kucaba, T.A., Dietrich, N.L., Green, E.D., Brownstein, B., Wilson, R.K., McDonald, K.M., Hillier, L.W., McPherson, J.D., and Waterston, R.H. 1997. High throughput fingerprint analysis of large-insert clones. Genome Res. 7: 1072-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McLysaght A., Hokamp, K., and Wolfe, K.H. 2002. Extensive genomic duplication during early chordate evolution. Nat. Genet. 31: 200-204. [DOI] [PubMed] [Google Scholar]
- 28.Osoegawa K., Tateno, M., Woon, P.Y., Frengen, E., Mammoser, A.G., Catanese, J.J., Hayashizaki, Y., and de Jong, P.J. 2000. Bacterial artificial chromosome libraries for mouse sequencing and functional analysis. Genome Res. 10: 116-128. [PMC free article] [PubMed] [Google Scholar]
- 29.Piskur J. 2001. Origin of the duplicated regions in the yeast genome. Trends Genet. 17: 302-303. [DOI] [PubMed] [Google Scholar]
- 30.Puttagunta R., Gordon, L.A., Meyer, G.E., Kapfhamer, D., Lamerdin, J.E., Kantheti, P., Portman, K.M., Chung, W.K., Jenne, D.E., Olsen, A.S., et al. 2000. Comparative maps of human 19p13.3 and mouse chromosome 10 allow identification of sequences at evolutionary breakpoints. Genome Res. 10: 1369-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Regnier V., Meddeb, M., Lecointre, G., Richard, F., Duverger, A., Nguyen, V.C., Dutrillaux, B., Bernheim, A., and Danglot, G. 1997. Emergence and scattering of multiple neurofibromatosis (NF1)-related sequences during hominoid evolution suggest a process of pericentromeric interchromosomal transposition. Hum. Mol. Genet. 6: 9-16. [DOI] [PubMed] [Google Scholar]
- 32.Schwartz S., Zhang, Z., Frazer, K.A., Smit, A., Riemer, C., Bouck, J., Gibbs, R., Hardison, R., and Miller, W. 2000. PipMaker—A web server for aligning two genomic DNA sequences. Genome Res. 10: 577-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stankiewicz P. and Lupski, J.R. 2002. Genome architecture, rearrangements and genomic disorders. Trends Genet. 18: 74-82. [DOI] [PubMed] [Google Scholar]
- 34.Summers T.J., Thomas, J.W., Lee-Lin, S.-Q., Maduro, V.V.B., Idol, J.R., and Green, E.D. 2001. Comparative physical mapping of targeted regions of the rat genome. Mamm. Genome 12: 508-512. [DOI] [PubMed] [Google Scholar]
- 35.Thomas J.W., Lee-Lin, S.-Q., and Green, E.D. 1999. Human-mouse comparative mapping of the genomic region containing CDK6: Localization of an evolutionary breakpoint. Mamm. Genome 10: 764-767. [DOI] [PubMed] [Google Scholar]
- 36.Thomas J.W., Summers, T.J., Lee-Lin, S.-Q., Maduro, V.V.B., Idol, J.R., Mastrian, S.D., Ryan, J.F., Jamison, D.C., and Green, E.D. 2000. Comparative genome mapping in the sequence-based era: Early experience with human chromosome 7. Genome Res. 10: 624-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomas J.W., Prasad, A.B., Summers, T.J., Lee-Lin, S.-Q., Maduro, V.V.B., Idol, J.R., Ryan, J.F., Thomas, P.J., McDowell, J.C., and Green, E.D. 2002. Parallel construction of orthologous sequence-ready clone contig maps in multiple species. Genome Res. 12: 1277-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Venter J.C., Adams, M.D., Myers, E.W., Li, P.W., Mural, R.J., Sutton, G.G., Smith, H.O., Yandell, M., Evans, C.A., Holt, R.A., et al. 2001. The sequence of the human genome. Science 291: 1304-1351. [DOI] [PubMed] [Google Scholar]
- 39.Vision T.J., Brown, D.G., and Tanksley, S.D. 2000. The origins of genomic duplications in Arabidopsis. Science 290: 2114-2117. [DOI] [PubMed] [Google Scholar]
- 40.Vollrath D., et al. 1999. DNA markers for physical mapping. In Genome analysis: A laboratory manual (ed. B. Birren), pp. 187–215. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York.
- 41.Walentinsson A., Helou, K., and Levan, G. 2001. A dual-color FISH gene map of the proximal region of rat Chromosome 4 and comparative analysis in human and mouse. Mamm. Genome 12: 900-908. [DOI] [PubMed] [Google Scholar]
- 42.Wilson R.K., Mardis, E.R., et al. 1997. Shotgun sequencing. In Genome analysis: A laboratory manual (ed. B. Birren), pp. 397–454. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York.
- 43.Zhang Z., Schwartz, S., Wagner, L., and Miller, W. 2000. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 7: 203-214. [DOI] [PubMed] [Google Scholar]