Abstract
Cripto, the founding member of the EGF-CFC genes, plays an essential role in embryo development and is involved in cancer progression. Cripto is a GPI-anchored protein that can interact with various components of multiple signaling pathways, such as TGF-β, Wnt and MAPK, driving different processes, among them epithelial-mesenchymal transition, cell proliferation, and stem cell renewal. Cripto protein can also be cleaved and released outside the cell in a soluble and still active form. Cripto is not significantly expressed in adult somatic tissues and its re-expression has been observed associated to pathological conditions, mainly cancer. Accordingly, CRIPTO has been detected at very low levels in the plasma of healthy volunteers, whereas its levels are significantly higher in patients with breast, colon or glioblastoma tumors. These data suggest that CRIPTO levels in human plasma or serum may have clinical significance. However, very little is known about the variability of serum levels of CRIPTO at a population level and the genetic contribution underlying this variability remains unknown. Here, we report the first genome-wide association study of CRIPTO serum levels in isolated populations (n = 1,054) from Cilento area in South Italy. The most associated SNPs (p-value<5*10-8) were all located on chromosome 3p22.1-3p21.3, in the CRIPTO gene region. Overall six CRIPTO associated loci were replicated in an independent sample (n = 535). Pathway analysis identified a main network including two other genes, besides CRIPTO, in the associated regions, involved in cell movement and proliferation. The replicated loci explain more than 87% of the CRIPTO variance, with 85% explained by the most associated SNP. Moreover, the functional analysis of the main associated locus identified a causal variant in the 5’UTR of CRIPTO gene which is able to strongly modulate CRIPTO expression through an AP-1-mediate transcriptional regulation.
Author Summary
Cripto gene has a fundamental role in embryo development and is also involved in cancer. The protein is bound to the cell membrane through an anchor, that can be cleaved, causing the secretion of the protein, in a still active form. In the adult, CRIPTO is detected at very low levels in normal tissues and in the blood, while its increase in both tissues and blood is associated to pathological conditions, mainly cancer. As other GPI linked proteins such as the carcinoembryonic antigen (CEA), one of the most used tumor markers, CRIPTO is able to reach the bloodstream. Therefore, CRIPTO represents a new promising biomarker and potential therapeutic target, and blood CRIPTO levels might be associated to clinical features. Here we examined the variability of blood CRIPTO levels at a population level (population isolates from the Cilento region in South Italy) and we investigated the genetic architecture underlying this variability. We reported the association of common genetic variants with the levels of CRIPTO protein in the blood and we identified a main locus on chromosome 3 and additional five associated loci. Moreover, through functional analyses, we were able to uncover the mechanism responsible for the variation in CRIPTO levels, which is a regulation mediated by the transcriptional factor AP-1.
Introduction
Cripto, also known as Teratocarcinoma-derived growth factor 1 (TDGF1), is the original member of the Epidermal Growth Factor-Cripto/Fibroblast growth factor Receptor Ligand 1/Cryptic (EGF-CFC) family of vertebrate proteins involved in embryo development [1–3]. Cripto has been isolated in human and mouse [3] and is a GPI-anchored membrane protein [4] that can function in both membrane anchored and soluble form [5,6]. It is involved in multiple signaling pathways, such as TGF-β, Wnt and MAPK/ERK pathways, it regulates essential steps in early embryogenesis and it is also involved in processes such as cell migration, epithelial-mesenchymal transition (EMT), stem cell maintenance, all processes which are implicated in cancerogenesis [7–14]. Cripto has also a role in angiogenesis, being able to enhance the proliferation, migration and invasion of human umbilical endothelial cells, to stimulate their differentiation into vascular-like structures in Matrigel and is also able to induce tumor neovascularization in vivo [13]. Cripto is expressed at very low levels in different adult tissue types and organs, among them a higher expression is detected in colon, skeletal muscle, heart, cortex of adrenal gland and cerebellum (http://www.biogps.org/ [15], http://www.proteinatlas.org/ [16]). Pathological re-expression is seen in a number of solid cancers. Most studies focused on the role of Cripto in breast and colorectal cancer [17–20], in inflammatory conditions and also in a macaque model of neuroAIDS [21]. Numerous studies have demonstrated correlation between high expression levels of CRIPTO and malignant transformation, tumor invasiveness, metastatic spreading and hence poor prognosis [17,22–27]. In vitro and in vivo functional studies confirm a strong involvement of Cripto in cancer development and indicate that its effect on tumorigenesis might strictly depend on the cellular context in which it acts [28–33].
Moreover, many data indicate Cripto as a promising target for cancer therapy. Adkins et coll. demonstrated that the block of Cripto signaling with an anti-CFC domain antibody determined a strong inhibition of tumor cell growth in vivo [34]. Ever since, different approaches based on the use of oligonucleotides, vaccines or antibodies have been successfully applied to target Cripto by inhibiting its activity and/or expression in tumors and in neurodegenerative diseases [35]. Cripto inhibition by different approaches always resulted in inhibition of cancer cell proliferation in vitro and of tumor growth in vivo [36].
CRIPTO has been also detected at very low levels in the plasma of healthy controls, whereas significantly higher concentrations have been found in patients with breast, colon or cerebral tumors [19,37]. In both studies, tumor tissues and patient-matched blood samples have been analyzed, showing that CRIPTO high levels in the plasma correspond to re-expression of CRIPTO in tumor tissues [19]. These data suggest that CRIPTO, as the carcinoembryonic antigen (CEA) (another GPI anchored protein and a widely used tumor marker), is able to reach the bloodstream, being potentially released by tumor cells through GPI anchor cleavage. All together these data indicate that CRIPTO represents both a promising biomarker and a valid target for therapeutic intervention in cancer and that blood CRIPTO levels in humans may have clinical significance.
The two studies on the measure of circulating CRIPTO in the plasma published so far were both conducted on a very small group of individuals: the study of Bianco and coworkers analysed 21 healthy donors, 33 patients with colon carcinoma and 75 patients with breast carcinoma or benign breast lesions while the study of Pilgaard and coworkers included 28 Glioblastoma Multiforme (GBM) patients, 4 low-grade glioma patients and 8 healthy controls. In the first case no statistically significant correlations were observed between CRIPTO plasma concentration and various clinicopathologic variables, including tumor size, lymph node involvement and proliferative index and the degree of positivity for CRIPTO in tumor sections [19]. In the second case higher levels of the protein correlated with a shorter overall survival [37].
In the present study, CRIPTO serum levels were measured in a population-based sample from three isolated villages of the Cilento area, South Italy and a very high heritability (>80%) was estimated underscoring the importance of its genetic determinants. We present the first genome-wide association study (GWAS) for CRIPTO protein levels aiming at identifying genetic variants associated with the levels of circulating protein in the serum and we report the functional characterization of the main associated locus.
Results
Characteristics of the study participants are presented in Table 1. No difference was observed in the CRIPTO serum levels between men and women (p-value = 0.77 in the discovery; p-value = 0.65 in the replication). The median level of the protein was higher in the replication sample compared to the discovery sample (p-value<0.0001), because the proportion of individuals with CRIPTO serum levels below the threshold of assay quantification (< 1 pg/ml) is higher in the discovery (38%) than in the replication sample (27%) (see Methods). Significant heritability estimates were computed in both discovery (0.84, SE = 0.04) and replication (0.79, SE = 0.06) population samples (see Methods).
Table 1. Characteristics of the study subjects.
| Cohort | Discovery | Replication | |
|---|---|---|---|
| N° of Individuals | 1054 | 535 | |
| Women % | 56.5 | 54.2 | |
| Age (mean ± SD) | 52.4 ± 19.8 | 52.9 ± 19.5 | |
| Genealogical Kinship (mean ± SD) | 0.0032±0.0156 | 0.0083±0.0214 | |
| CRIPTO (pg/ml) | |||
| All | median | 27.5 | 184.4 |
| 95% CI | 21.3–34.3 | 133.2–211.7 | |
| Range | 0–1514.3 | 0–1253.2 | |
| Men | median | 25.9 | 168.4 |
| 95% CI | 17.6–42.7 | 71.6–217.2 | |
| Range | 0–1514.3 | 0–1253.2 | |
| Women | median | 29.0 | 188.9 |
| 95% CI | 19.6–35.6 | 117.7–221.4 | |
| Range | 0–1428.9 | 0–1174.2 | |
Loci associated with CRIPTO serum levels—discovery and replication
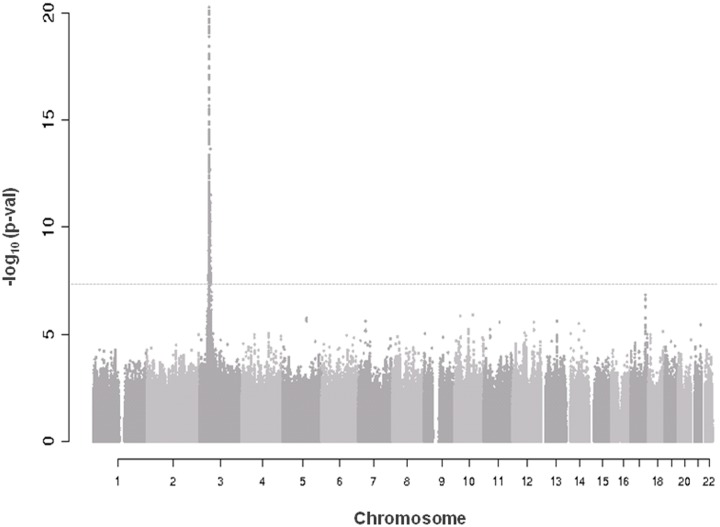
A quantile-quantile plot for the 6,222,455 investigated autosomal SNPs in discovery GWAS revealed many more SNPs with low observed p-values than expected (S1 Fig.). 455 SNPs associated with CRIPTO serum concentration at p-value<5*10–8 in the discovery stage were located on chromosome 3 in a region spanning 6.4 Mb (3p22.11–3p21.31), with the most associated SNP (rs3806702, p-value = 1.03*10–159) located in the CRIPTO gene region. (Figs. 1 and S2). The C allele of this SNP was associated with higher levels of CRIPTO (CC = 673.8±29.0pg/ml; CT = 310.5±6.9pg/ml; TT = 46.7±4.4pg/ml) (Table 2). When running a conditional GWAS adjusting for the most associated SNP on chromosome 3, no loci remain associated at the genome-wide significance level, but 700 SNPs were associated with p-value<1*10–4 on the genome (S1 Table). Of those, linkage disequilibrium (LD)-based independent variants were defined if the pair-wise LD (r2) was less than 0.01 and if they were separated by at least 1 Mb. In this way, 95 LD-based independent loci were identified that included SNPs associated with CRIPTO serum concentration at a p-value<1*10–4. For each of the independent loci, the SNP with the lowest p-value was carried forward to replication. Criteria for replication were defined as a p-value<0.05 in the independent replication sample, the effect in the same direction between discovery and replication, and a p-value in the meta-analysis of discovery and replication samples lower than that obtained in the discovery sample (see Methods). Table 2 shows the summary results for the main association and the additional 5 replicated loci for CRIPTO serum levels. Regional association plots provide a detailed overview of those loci (S2 Fig.). The proportion of serum CRIPTO variance explained by the replicated loci in the discovery sample was 87.0%, with 84.9% explained by rs3806702 alone and 2.1% explained by the remaining 5 associated SNPs. Similar results were obtained in the replication sample, with 87.1% of CRIPTO variance explained by the 6 associated variants.
Fig 1. Manhattan plot of genome-wide association results in discovery analysis.
Truncated Manhattan Plot showing -log10(p-values) for all SNPs of the CRIPTO discovery GWAS ordered by their chromosomal position. The dashed horizontal line represents the threshold for genome-wide significance (p-value<5*10-8). SNPs associated at the genome-wide significance level are all located on chromosome 3 (lowest p-value = 1.03*10–159).
Table 2. Association results of replicated SNPs.
| Discovery analysis | Replication analysis | Meta-analysis | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chr | SNP | Closest gene | Position (Mb) | Rsq | effect allele | effect allele frequency | Effect | s.e. of Effect | p-value | effect allele frequency | Effect | s.e. of Effect | p-value | Effect | s.e. of Effect | p-value |
| 3 | rs3806702 | CRIPTO | 46.62 | 1 | C | 0.23 | 1.325 | 0.049 | 1.03E-159 | 0.33 | 1.174 | 0.062 | 6.30E-79 | 1.267 | 0.039 | 8.64E-236 |
| 2 | rs6739316 | TNS1 | 218.77 | 0.65 | G | 0.76 | -0.108 | 0.027 | 9.03E-05 | 0.81 | -0.116 | 0.052 | 2.52E-02 | -0.109 | 0.025 | 8.80E-06 |
| 9 | rs17087824 | NTRK2 | 87.50 | 0.89 | T | 0.94 | -0.180 | 0.042 | 2.27E-05 | 0.89 | -0.119 | 0.054 | 2.69E-02 | -0.157 | 0.034 | 3.58E-06 |
| 14 | rs74062852 | SLC8A3 | 70.68 | 0.88 | C | 0.88 | 0.139 | 0.032 | 1.56E-05 | 0.85 | 0.136 | 0.048 | 4.72E-03 | 0.138 | 0.027 | 3.61E-07 |
| 15 | rs7168855 | MYO5A | 52.72 | 0.53 | T | 0.89 | -0.163 | 0.041 | 7.49E-05 | 0.94 | -0.275 | 0.101 | 6.55E-03 | -0.179 | 0.039 | 3.73E-06 |
| 17 | rs6503271 | GAS7 | 9.83 | 0.42 | G | 0.66 | 0.141 | 0.031 | 4.63E-06 | 0.66 | 0.115 | 0.052 | 2.88E-02 | 0.135 | 0.027 | 6.06E-07 |
Abbreviations: Chr-Chromosome, MAF-Minor Allele Frequency, s.e.-standard error, Rsq-imputation quality estimate
In the first row, the association result of the rs3806702 in the initial GWAS is shown. In the rows (2–6), the association results of the 5 replicated SNPs in the conditional GWAS adjusted for the rs3806702 genotype are reported.
Biological pathway analysis
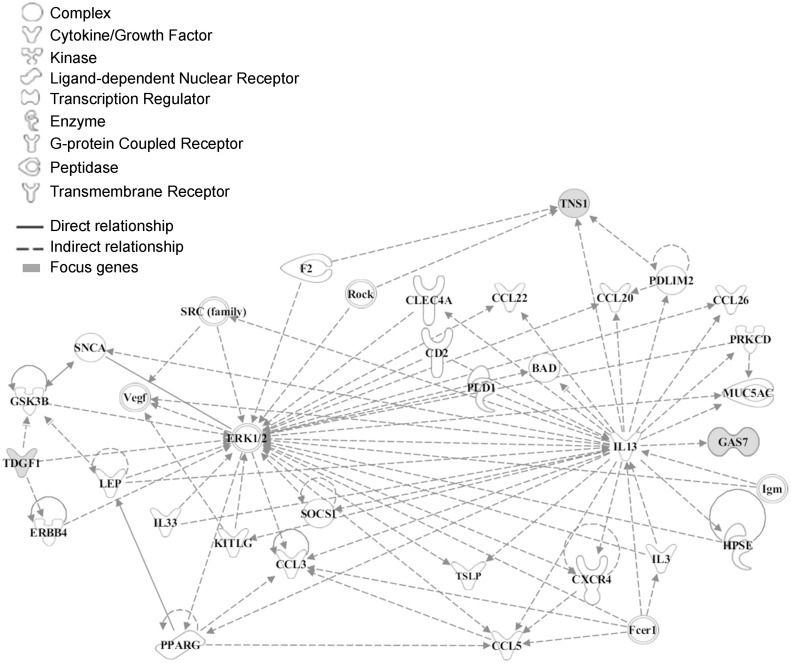
To explore the functional relationship between CRIPTO and the associated genes we used the Ingenuity Pathway Analysis software (IPA). CRIPTO gene and genes closest to the replicated SNPs were included in the analysis (see Methods). Those 10 seed genes were reported in the S2 Table. The IPA analysis identified a significant (p-value = 1*10–6) network of 35 molecules, 3 of which corresponded to CRIPTO associated loci (CRIPTO itself, GAS-7 and TNS-1) (Fig. 2). Function categories assigned to the whole gene network by IPA were: “cell to cell signaling and interaction, hematological system development and function, and immune cell trafficking”. In depth, function annotation sub-categories significantly enriched in genes involved in the network and including CRIPTO, were: cell migration, proliferation of tumor cells, cell differentiation, and blood vessel development (S3 Table).
Fig 2. Functional analysis of the associated genes.
The network was algorithmically constructed by Ingenuity Pathway Analysis (IPA) software on the basis of the functional and biological connectivity of genes. The network highlights the interconnections of 3 loci (marked in grey) identified from serum CRIPTO levels GWAS with a p-value = 1*10–6. CRIPTO gene is reported as TDGF1. Lines between genes represent known interactions and the nodes are displayed using various shapes that represent the functional class of the gene product (legend).
Functional element analysis in associated loci
To identify functional elements in the associated loci, ENCODE data related to chromatin modifications and hypersensitivity DNAse elements were analyzed in 3 cell lines (NTERA-2, HepG2, H1-hESC) selected as expressing CRIPTO mRNA. Among the replicated SNPs and variants in LD with them (r2>0.5), 3 variants (rs3791936 in the intronic region of TNS1, rs112481213 in the 5’UTR of CRIPTO, and rs61117007 in the intronic region of NTRK2) were located in promoter or enhancer histone marks and 1 of those (rs112481213) was also located in DNA hypersensitivity elements in the selected cell lines, suggesting a potential functional role of those associated variants.
Functional characterization of the associated rs112481213 variant
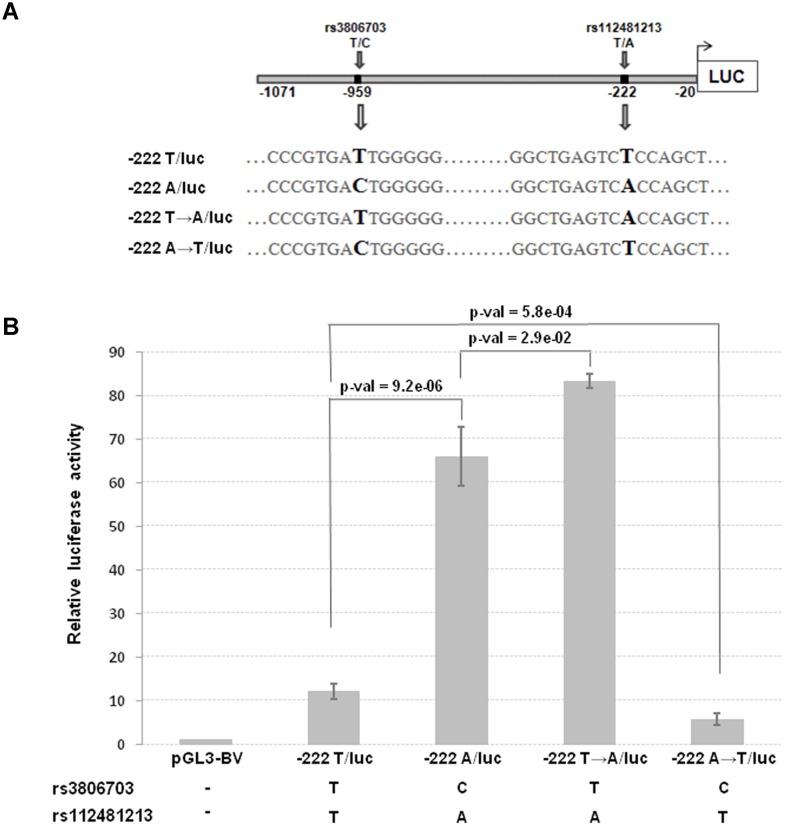
The top 8 associated SNPs in the CRIPTO gene region were all included in a unique LD block (S3A–S3B Fig.). Among them the rs112481213 variant (LD with rs3806702, r2 = 0.99, p-value of association = 1.53*10–158), reported to be in a regulative region from ENCODE data (see the above paragraph), was also predicted to create an AP-1 binding site by bioinformatics analysis with MatInspector program and TRANSFAC database (S3C Fig.). To explore the possibility that this SNP, located in the 5’UTR of CRIPTO gene, might influence CRIPTO transcription we tested the SNP allele effect on the transcriptional activity in the NTERA2 teratocarcinoma cell line, expressing high levels of CRIPTO [38]. Two constructs, both containing a 1,051 bp region upstream the CRIPTO ATG start codon (including almost all the 5’UTR and also 342 bp upstream the transcription start site), but differing for the rs112481213 allele (-222A/luc and-222T/luc, respectively) were transfected in NTERA2 cells. Overall, 15 SNPs were included in that region. As one of those SNPs, the rs3806703 variant was in linkage disequilibrium with rs112481213 (r2 = 0.72), two additional constructs (-222T→A/luc, -222A→T/luc) were also produced by site-directed mutagenesis to discriminate the effect of each of the two variants on the transcription efficiency (Fig. 3A). The construct containing the rs112481213 A allele (-222A/luc) produced an about 5-fold increase (p-value = 9.2*10–6) of the luciferase activity compared to the construct containing the T allele (-222T/luc) indicating that rs112481213 SNP affects the transcription (Fig. 3B). Also, the luciferase activity was reported to high levels in the site-mutated-222T→A/luc construct demonstrating that the main effect on the transcription can be attributed to the rs112481213 variant. Interestingly, the activity produced by the-222T→A/luc is significantly higher than that observed for the-222A/luc (p-value = 2.9*10–2) and inverse results were obtained for-222A→T/luc and-222T/luc (p-value = 5.8*10–4), indicating that rs3806703 might also have an effect, although modest, on the transcription. A cooperative role of rs112481213 and rs3806703 was also statistically supported by an interaction model tested for the two variants (βrs112481213 = -1.18, SErs112481213 = 0.04, βrs3806703 = 0.27, SErs3806703 = 0.06, βinter = -0.15, SEinter = 0.03, p-valueinter = 7.08*10–8).
Fig 3. Transcriptional activity and schematic representation of reporter constructs.
(A) The two constructs (222A/luc and-222T/luc) contain a 1,051 bp region including the 5’UTR and 342 bp upstream the transcription start site of CRIPTO and differ for the rs112481213 allele. The-222A→T/luc and 222T→A/luc constructs derive from a site-directed mutagenesis at the rs112481213 SNP position. (B) NTERA2 cell line was transfected with firefly luciferase reporter constructs carrying 1,051bp of the CRIPTO gene with either the A allele or the T allele at rs112481213. The rs112481213 A allele produced a 5-fold increase in luciferase activity compared to the T allele. Site-mutated constructs at the rs112481213 allowed to discriminate the effect of rs112481213 and rs3806703 on the transcription. Data are presented as fold-induction compared with promoter-less vector (pGL3-BV). Data are shown as mean±SD from five experiments.
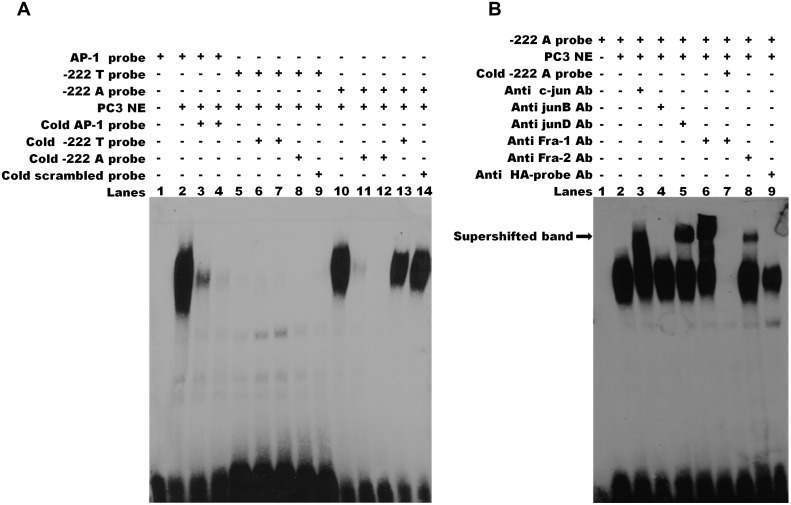
Therefore, the promoter activity data demonstrated that rs112481213 is a functional regulatory element of CRIPTO transcription which effect might be modulated by rs3806703. We next verified that the mechanism through which rs112481213 variant might influence CRIPTO transcription is the creation of an AP-1 consensus binding site, as predicted by bioinformatics analysis. Indeed, Electrophoretic Mobility Shift Assay (EMSA) using an AP-1 containing PC3 nuclear extract suggested that the A allele-containing oligonucleotide probe might robustly bind AP-1 complex while an oligonucleotide probe containing the T-allele weakly bound the complex (Fig. 4A). The AP-1 binding was confirmed by addition of specific antibodies for components of AP-1 complex, revealing a visibly supershifted band, representing a DNA-protein-antibody complex (Fig. 4B). All together, these data demonstrate the specificity of the observed DNA-protein interaction as well as a differential interaction of the AP-1 complex with the rs112481213 alleles.
Fig 4. The rs112481213 variant alter the AP-1 binding site.
(A) Electrophoretic mobility shift assay (EMSA) for AP-1 binding in nuclear extracts (NEs) of PC-3 cell line. The binding activities of NEs (lanes 2–14) were analyzed by EMSA using biotin-labeled AP-1 probe (lanes 2–4), -222 T probe (lanes 5–9) and-222 A probe (lanes 10–14). Competition was performed with 50 and 200 fold molar excess of the cold probe (lanes 3–4, 6–7, 11–12). Cross competition was performed using the cold probe carrying the alternative allele (lane 8 and 13) and the cold scrambled probe (lane 9 and lane 14) at 200 fold molar excess. No binding was detectable for-222 T probe (lanes 5–9). The DNA-protein complex of-222 A probe (lane 10) can be eliminated by 50 or 200 fold molar excess of cold-222 A probe (lanes 11, 12) but not by 200 fold molar excess of cold-222 T probe and cold scrambled probe. (B) Specificity of AP-1 binding activity. Supershift assay was carried out for-222 A probe using specific antibodies for the AP-1 components c-Jun (lane 3), JunB (lane 4), JunD (lane 5), Fra-1 (lane 6) and Fra-2 (lane 8). A supershifted DNA-protein-antibody complex (indicated by the arrow) is observed (lanes 3, 5, 6, 8). Lane 7 shows that the specific complexes were eliminated by addition of 200 fold molar excess of cold-222 A probe. No supershifted complex was observed adding HA-probe antibody used as negative control (lane 9). The results are representative of three independent determinations.
Discussion
Cripto is a typical example of an oncodevelopmental gene having key functions in early embryogenesis, and being re-expressed in the adult during tumorigenesis. Cripto is a GPI-anchored membrane protein, that can also be cleaved and released in the medium and is able to induce cellular proliferation, EMT, migration, and invasion, as well as to stimulate tumor angiogenesis both in vitro and in vivo [36]. Cripto promotes oncogenesis via modulation of TGF-β ligand signaling and through mechanisms that are independent of TGF-β ligands and their signaling receptors [39]. The effect of TGF-β ligands and Cripto on tumorigenesis is also dependent on the cellular context [33,39]. Interestingly, Cripto protein is an obligatory co-receptor for some TGF-β family members such as Nodal, enabling them to bind to Activin receptorial complexes and activate Smad cascade and is also able to antagonize the signaling of other members of the TGF-β family, (i.e., Activins and TGF-β), inhibiting their antioncogenic effect [34,35,40]. Moreover, Cripto acts via separate, non-overlapping mechanisms to enhance the canonical Wnt/β-catenin signaling pathway by binding to low-density lipoprotein receptor-related protein (LRP) 5 and LRP6 [41] and to activate ras/raf/MAPK and PI3K/Akt pathways via c-Src [28]. More recently, novel Cripto-interacting proteins, also involved in cancer, have been identified including the chaperonin glucose regulated protein-78 (Grp78) and Notch1 [39].
In humans, CRIPTO is expressed at very low levels in both normal tissues and plasma, while its expression was found increased in patients with cancer (also in both tumor tissues and plasma), suggesting that CRIPTO blood levels might have a great clinical relevance [19,37]. However in these two articles only a small number of healthy volunteers was analysed and no data on a general population sample were reported.
Our study is the first GWAS of circulating CRIPTO levels. It was undertaken in 1,589 individuals from three population isolates of the Cilento area, South Italy and represents the largest survey of CRIPTO measurement in a population-based sample. In our study, the variability of the circulating protein, according to the heritability estimation, was found to be highly determined by genetic factors. The GWAS identified the strongest association on chromosome 3 at rs3806702 located in the CRIPTO gene region. GWAS conditional to that variant showed that this locus represents the main genetic contribution to the modulation of CRIPTO in the serum. Indeed, 85% of the inherited component of circulating CRIPTO levels is explained by the rs3806702 variant. In accordance with this finding, all individuals with CRIPTO protein levels below the detection threshold (34% of the entire sample) were found homozygous for the T allele of the rs3806702 variant. The lower levels of CRIPTO in the discovery sample compared to the replication sample can also be explained by the difference in allele frequency (for T allele 0.77 and 0.67 respectively).
A LD block included rs3806702 as well as the top 7 associated SNPs in the CRIPTO gene region. Some of those were reported by bioinformatics analysis as potential candidates affecting transcription factor binding sites. Among those variants, rs112481213, located in the 5’UTR of CRIPTO gene, was identified by functional experiments as a causal SNP for CRIPTO transcriptional regulation. Regulation of Cripto expression during embryogenesis and tumorigenesis was still incompletely defined. So far different binding sites have been found in the promoter region of the Cripto gene: for Smad-proteins [42], the T-cell factor/lymphoid enhancer factor (Tcf/Lef) [43], the Hypoxia-Inducible Factor 1 (HIF-1) [44], the Nkx2–5 early cardiac transcription factor [45], and the orphan nuclear Liver Receptor Homolog-1 (LRH-1) [46]. HIF-1 and Nkx2–5 are able to transcriptionally activate Cripto during cardiac differentiation, HIF-1 also activates CRIPTO expression in human embryonal carcinoma cells, following hypoxic conditions [44,46]. Conversely, CRIPTO is directly repressed by the orphan nuclear receptor germ cell nuclear factor (GCNF) which binds to the promoter during retinoic acid-induced differentiation of human embryonic carcinoma cells and by the miR-15a/16 cluster which bind to the 3’UTR of CRIPTO mRNA [47,48]. However, this is the first time that an AP-1 transcriptional activation of CRIPTO has been described. We have indeed demonstrated that CRIPTO expression is regulated by AP-1 transcription factor and that this regulation depends on rs112481213 genotype. Transcriptional activity data also suggest that in addition to rs112481213, rs3806703, another SNP present in that region, may have a role in modulating the CRIPTO protein levels, possibly through the involvement of GATA binding transcriptional factors. In support to this hypothesis, a statistical interaction between rs112481213 and rs3806703 was also found. Due to the complexity of Cripto gene regulation and its dependency on the specific biological context, additional regulatory mechanisms might occur in the case of cancer-related cell dysfunction.
Further, five additional loci were associated to CRIPTO serum levels at p-values<1*10–4 independently from the main signal. Although these associations did not reach genome-wide significance in the discovery, likely because of lack of power of our study, they were replicated in an independent sample and might represent good candidate loci as modulators of the circulating CRIPTO. Moreover, three of the replicated variants were located in regions involved in regulative processes associated to chromatin accessibility.
A single network of 35 molecules including CRIPTO and two other associated loci, the growth-arrest-specific gene7 (GAS-7) and the Tensin1 (TNS-1), was identified by IPA analysis. The network included 30 genes implicated in cell migration, 18 genes involved in tumor cell proliferation, 25 genes in cell differentiation and, interestingly, 17 genes implicated in blood vessel development.
The analysis of CRIPTO associated loci showed that these are mainly linked to the MAPK/ERK signaling pathway with ERK1/2 as one of the principal players of the network. Aberrant regulation of MAPK cascades is known to strongly contribute to cancer and other human diseases.
Like CRIPTO, TNS-1 and GAS-7, the other two genes in the associated regions present in the network, are both involved in breast and colon cancer [49–52]. TNS-1 binds to actin filaments [53] and serves as a link between signal transduction pathways and the actin cytoskeleton by forming a structural platform that regulates the assembly of focal adhesion components, phosphoproteins, and signaling molecules for processes such as cell migration [54]. TNS-1 is expressed in normal tissues [55] while its expression is greatly reduced in human breast, prostate, head and neck squamous cell carcinomas, and melanoma suggesting a role as tumor suppressor [56] as well as in the maintenance of cell polarization, and the suppression of invasion that are involved in metastasis [57]. The TNS-1 phosphotyrosine binding (PTB) 1 domain binds the cytoplasmic tail of beta-integrin, presumed to be the basis for focal adhesion localization. Interestingly, overexpression of Cripto in vitro and in vivo has been associated with increased expression of fibronectin and various integrins and with increased activation of focal adhesion kinase [58].
Similarly to TNS-1, GAS-7 binds actin and participates in cytoskeleton dynamics, executing different functions in different cellular processes, such as vesicle trafficking, cell migration and morphological differentiation [59,60]. GAS-7 hypermethylation has been found in breast and colon cancers whereas increased expression has been detected in medulloblastoma [61]. GAS-7 expression is regulated by ERK signaling pathway [62], in which Cripto is also involved.
In the same CRIPTO associated locus on chromosome 17, besides GAS-7, is also located the G-protein-coupled receptor (GLP-2R) gene [63–65]. Interestingly, GLP-2R activation also induces ERK1/2 MAP kinase activation and is able to both stimulate the expression of the immediate early genes c-Fos, c-Jun, JunB and Egr-1 and to activate AP1-driven gene transcription in a PKA-dependent manner [66,67].
Two other genes included in the associated loci, Myosin VA (MYOVA) gene on chromosome 15 and Neurotrophin tyrosine kinase receptor 2 (NTRK2) on chromosome 9, are both overexpressed in cancer. In particular, MYOVA is highly expressed in a number of highly metastatic cancer cell lines and metastatic colorectal cancer tissues and is able to interfere with metastatic capabilities by influencing cell migration. MYOVA expression is upregulated by the transcription factor Snail, one of the molecular switches for the EMT program involved in cancer metastasis [68]. NTRK2 has been found frequently overexpressed in human cancers, including pancreatic and prostate carcinoma, Wilms’ tumor and neuroblastomas, particularly those with aggressive behavior and poor prognosis. As Cripto, NTRK2 activates both phosphatidylinositol-3-kinase (PI3K) and MAPK/ERK signaling.
In summary, our data showed that CRIPTO protein is measurable in the serum of the majority of individuals in a population-based sample. Further, we identified the largest genetic contribution to the CRIPTO variability and demonstrated that a functional variant located in the 5’UTR of CRIPTO gene is able to modulate CRIPTO expression through an AP-1-mediated transcriptional regulation. We also provided support for additional associated loci that will need to be confirmed in larger samples. Nevertheless, many of those associations converge in cancer phenotypes, mainly in cell movement and proliferation functions. As any association has been detected at these CRIPTO associated loci in large-scale cancer GWAS, further studies looking at CRIPTO variability in serum together with genotyping of the functional variant in specific cohorts of patients, focusing the analysis on specific cancer phenotypes, as metastasis formation, aggressiveness, prognosis, would be useful to better investigate possible associations between variants modulating CRIPTO protein levels and cancer features.
Methods
Population samples and CRIPTO measurement
The discovery sample includes 1,054 individuals recruited through a population-based sampling strategy in two small isolated villages of the Cilento region, South Italy (Gioi and Cardile) [69]. In silico replication was performed in additional 535 subjects from another village (Campora) of the same region [70]. The study design was approved by the ethics committee of Azienda Sanitaria Locale Napoli 1. The study was conducted according to the criteria set by the declaration of Helsinki and each subject signed an informed consent before participating to the study.
Blood samples were collected in the morning after the participants had been fasting for at least 12 h. Aliquots of serum were immediately prepared and stored at -80°C, and were subsequently used for the assessment of CRIPTO levels. CRIPTO (pg/ml) was measured using an enzyme-linked immunosorbent assay, according to the manufacturer’s instructions (DRG Instruments GmbH, Germany). An intra-assay coefficient of variation of the CRIPTO measure of 7.74% was obtained from 10 times measurements of 10 serum samples. Individuals with CRIPTO levels below the detection threshold were included in the study and a value of 0pg/ml was assigned to them.
Mann-Whitney U test was used to compare median CRIPTO serum levels among the samples.
A normal quantile transformation was applied to the trait and the transformed trait was used in all statistical analyses. The heritability of CRIPTO serum levels was estimated by SOLAR software [71] using extended genealogies of discovery and replication population samples and adjusting the phenotype for gender and age.
GWAS and replication study
Genotyping was performed with 370K and Omniexpress Illumina chips, phasing and imputation were conducted separately by platform with the MaCH [72] and minimac (http://genome.sph.umich.edu/wiki/Minimac) software respectively, using 1000G v3 data as reference. SNP allele frequencies in Cilento samples versus 1000 Genomes reference allele frequencies for all genotyped SNPs were reported in the S4 Fig. Quality control filters applied before imputation were call rate >95% for SNPs and samples and minor allele frequency (MAF) >0.01. GWAS was carried out through a mixed model linear regression where the variance/covariance matrix is the genomic kinship to account for relatedness between individuals. Age and gender were used as covariates and an additive genetic model was considered. The analysis was performed with GenABEL package [73] for genotyped SNPs and ProbABEL [74] for imputed data. SNPs with imputation quality (Rsq in MACH) <0.4 or MAF <0.05 were excluded.
Conditional analysis was carried out in the discovery and replication samples adding the additive effect of rs3806702 as covariate in the association model.
To select linkage disequilibrium (LD)-based independent association signals among the CRIPTO associated SNPs, we conducted the clumping procedure implemented in PLINK [75] and picked the index SNPs with the most significant association p-value from each clumped association region based on the GWAS. The 1000G v3 genotypes were used as reference panel; the physical threshold for clumping was 1 Mb, and the r2 threshold for clumping was 0.01.
To assess evidence for replication, test-statistics of discovery and in silico replication samples were meta-analysed using a fixed effect model weighted by inverse variance, using Metal [76]. SNPs were considered replicated if the SNP p-value was <0.05 in the replication sample alone, the effect was in the same direction between discovery and replication, and the p-value in the meta-analysis was lower than in the discovery sample.
The percentage of the variance of the CRIPTO levels explained by the replicated SNPs was calculated both in the discovery and replication samples. Three linear mixed effects models were fitted, in which the CRIPTO was regressed, respectively, on: 1) gender and age (basic model); 2) gender, age, additive effect of a single SNP (single SNP model); 3) gender, age, additive effect of each of the replicated SNPs (multiple SNP model). The variance explained by each SNP was calculated as the difference between the variance explained by the single SNP model and that explained by the basic model. Similarly, the variance explained by the replicated SNPs all together was estimated as the difference between the variance explained by the multiple SNP model and that explained by the basic model. The lmekin function (R package), which uses the genomic kinship matrix to correct for relatedness between individuals, was applied.
To test for interaction between rs112481213 and rs3806703, a linear mixed effect model, (including gender, age, additive effect of the two SNPs and the interaction between the two SNPs effects) implemented in the lmekin function (R package) was used. The variance inflation factor (VIF) was checked to be below ten to exclude collinearity problems [77].
Pathway analysis
For each replicated SNP, seed genes were selected as located within a region of 100 kb upstream and 100 kb downstream the region delimited by SNPs in LD (r2>0.5) with it. For the main associated locus on chromosome 3, CRIPTO gene was included as seed. Overall, 12seed genes were analyzed with Ingenuity Pathway Analysis software (IPA, Ingenuity Systems, www.ingenuity.com) to explore the functional relationship between the proteins encoded by those genes. IPA tests a set of genes for enrichment in defined canonical pathways or functions and generates de novo networks of interacting genes or gene products. IPA computes a p-value, based on a Fisher’s exact test, that represents the likelihood of the core genes in a network and biological function being found together due to random chance. Direct and indirect interactions, a high confidence (experimentally observed or highly predicted) and a maximum size of 35 genes/proteins per network were used as parameters in the analysis.
DNA functional element analysis
The associated loci were investigated for presence of chromatin histone marks and hypersensitive DNAse elements using data from ENCODE included in Haploreg software (http://www.broadinstitute.org/mammals/haploreg/) [78]. The replicated SNPs and variants in LD with them (r2>0.5) were analyzed. Cell lines where the CRIPTO mRNA was reported to be expressed in ENCODE database (www.genome.gov/encode/) were selected for the analysis.
Constructs and transcription assays
A 1051-bp fragment upstream the ATG of the CRIPTO gene (position-1071/-20, RefSeq Gene NG_017049.1) was amplified by PCR from genomic DNA of individuals homozygous for either the A allele or the T allele of the rs112481213 at position-222 using as primers 5’-CGACGCGTCAAGCGGCACATCAGAGTC-3’ and 5’-GAAGATCTGAAAAGAGGCGTTAGCATCG-3’. The PCR products were digested with MluI/BglII and directionally cloned into the MluI and BglII sites of the luciferase reporter pGL3-basic vector (Promega, Madison, WI) to obtain-222A/luc and-222T/luc reporter constructs. The integrity of constructs was confirmed by DNA sequencing. Site-directed mutagenesis of both-222A/luc and-222T/luc constructs was performed using GeneArt Site-Directed Mutagenesis System (Invitrogen) according to the manufacturer’s protocol using as primers 5’-GAATCCCCGGAAAGGCTGAGTCACCAGCTCAAGGTCAAAACGTCC-3’ and 5’-GGACGTTTTGACCTTGAGCTGGTGACTCAGCCTTTCCGGGGATTC-3’ to perform the mutagenesis of-222T/luc (-222T→A/luc) and 5’-GAATCCCCGGAAAGGCTGAGTCTCCAGCTCAAGGTCAAAACGTCC-3’ and 5’-GGACGTTTTGACCTTGAGCTGGAGACTCAGCCTTTCCGGGGATTC-3’ for-222A/luc (-222A→T/luc). The constructs were then sequenced to confirm the sequence changes.
NTERA2 cells, cultured in Dulbecco’s modified Eagle’s medium F-12 (Gibco-Invitrogen) supplemented with 10% FBS at 37°C and 5% CO2, were transiently transfected using JetPRIME transfection reagent (PolyPlus Transfection) following the manufacturer’s protocol. Briefly, 0.5 μg of either pGL3-Basic vector, -222T/luc, -222A/luc, -222T→A/luc, -222A→T/luc together with 10 ng of Renilla luciferase reporter plasmid (Promega) were cotransfected. Luciferase activity was assayed at 48h using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s protocol. Measurement of the firefly luciferase activity was normalized relative to the activity of the Renilla luciferase. Each construct was tested in triplicate in at least 3 independent experiments.
Electrophoretic mobility shift and antibody supershift assays
All reactions included double-stranded, biotin-labeled oligonucleotide probe at 40fmol concentration. EMSAs were performed by using the LightShift Chemiluminescent EMSA kit (Pierce Biotechnology) according to the manufacturer’s protocol. PC3 cell line was used because of high expression of AP-1 components [79]. Nuclear extracts were prepared using Subcellular Protein Fractionation Kit for Cultured Cells (Pierce Biotechnology) according to the manufacturer’s protocol. Nuclear extracts (5μg) were incubated at room temperature either with the biotin-labeled probe alone or with the biotin-labeled probe and 50 or 200-fold molar excess unlabeled competitor probe for 20 min, before loading on a 4% nondenaturing acrylamide gel and subjected to autoradiography. The following double-stranded biotin-labeled oligonucleotides were used as probe: AP-1 control 5’-biotin-CGCTTGATGACTCAGCCGGAA-3’; -222T probe 5’-biotin-GAAAGGCTGAGTCTCCAGCTC-3’; -222A probe 5’-biotin-GAAAGGCTGAGTCACCAGCTC-3’. Also, a scrambled oligonucleotide (5’-GAAAGGCTTGACGACCAGCTC-3’) was used for competition at 200-fold molar excess.
Supershift assays were performed identically except for the addition of 3μg of antibody for 3h in ice before the addition of-222A probe. Antibodies used were anti-c-Jun, anti-JunB, anti-JunD, anti-Fra1 and anti-Fra2 [79] from Santa Cruz Biotechnology. HA-probe antibody against the influenza hemagglutinin (HA) protein (Santa Cruz Biotechnology), was used as negative control.
Supporting Information
For each variant tested, the observed -log10(p-value) (y-axis) is plotted against the expected -log10(p-value) under the null hypothesis (red line).
(DOCX)
Regional association plots (panels A-F) show -log10(p-values) for all SNPs ordered by their chromosomal position within all regions of the replicated loci. For the rs3806702 the p-value of the discovery GWAS is reported, for the other SNPs p-values of the discovery conditional GWAS are reported. Each SNP is colored according to its correlation with the replicated SNP within the region as specified in the color scheme. Correlation structures correspond to hg19/1000 Genomes EUR Mar 2012. Plots were generated with LocusZoom [80]. The CRIPTO gene is reported as TDGF1.
(DOCX)
(A) The genomic localization of the top 8 associated SNPs (red) respect to the TDGF1 gene is shown using the GRCh38 primary assembly as reference sequence. The transcript (NM_003212.03) and the coding (CCDS2742.1) sequence are also shown. The functional SNP rs112481213 is underlined and reported in italic. The black rectangle indicates the region of 1.052 bp, upstream the ATG start codon, cloned in the luciferase reporter pGL3-basic vector. The rs3806703 SNP (orange and with a dashed line) is present in the cloned region but is not included in the top 8 associated SNPs (LD < 0.8 for rs112481213). (B) LD between the top 8 associated SNPs is reported. The analysis is carried out on the 1000 genome data (European panel) using the Haploview software. The distance in kb from the ATG start codon is shown on the left column. (C) MatInspector analysis. The results are reported only for the SNPs for which: 1) SNP alleles differently affect the TF biding site. In the schema, “Change” of the allele is-1 if the Alt allele disrupts the TF binding site and is 1 if the Alt allele creates a new TF binding site; 2) The value of the matrix similarity is higher than 0.9; 3) The position of the allele in the sequence logo shows an information content > 0.2 (y axis of the sequence logo). The black star represents the position of the SNP in the TF consesus binding site.
(DOCX)
Allele frequencies of all genotyped SNPs in the discovery or replication samples (y axis) are plotted against allele frequencies in 1000 Genomes European panel reference (x axis). RAF = Reference Allele Frequency.
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We thank the populations of Cilento for their participation in the study. We thank Prof. Georg Hess and Dr. Matthias Herkert from DRG Instruments GmbH for providing material to measure CRIPTO protein in the serum. We thank Dr. Genesia Manganelli for helpful suggestions and Dr. Pasquale Verde for critical discussions. We thank IAC-CNR for the use of the cluster Lilligridbio. This study is dedicated to the memory of Dr. Graziella Persico who discovered Cripto gene and designed and set up a genetic study in Cilento populations.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by grants from: the Interomics Flagship Project, http://www.interomics.eu/; the Assessorato Ricerca Regione Campania, http://www.regione.campania.it/; the Fondazione con il SUD (2011-PDR-13) http://www.fondazioneconilsud.it/; the Istituto Banco di Napoli – Fondazione http://www.ibnaf.it/ to MC. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Shen MM, Schier AF (2000) The EGF-CFC gene family in vertebrate development. Trends Genet 16: 303–309. [DOI] [PubMed] [Google Scholar]
- 2. Bianco C, Normanno N, Salomon DS, Ciardiello F (2004) Role of the cripto (EGF-CFC) family in embryogenesis and cancer. Growth Factors 22: 133–139. [DOI] [PubMed] [Google Scholar]
- 3. Minchiotti G, Parisi S, Liguori GL, D’Andrea D, Persico MG (2002) Role of the EGF-CFC gene cripto in cell differentiation and embryo development. Gene 287: 33–37. [DOI] [PubMed] [Google Scholar]
- 4. Minchiotti G, Parisi S, Liguori G, Signore M, Lania G, et al. (2000) Membrane-anchorage of Cripto protein by glycosylphosphatidylinositol and its distribution during early mouse development. Mech Dev 90: 133–142. [DOI] [PubMed] [Google Scholar]
- 5. Brandt R, Normanno N, Gullick WJ, Lin JH, Harkins R, et al. (1994) Identification and biological characterization of an epidermal growth factor-related protein: cripto-1. J Biol Chem 269: 17320–17328. [PubMed] [Google Scholar]
- 6. Yan YT, Liu JJ, Luo Y, E C, Haltiwanger RS, et al. (2002) Dual roles of Cripto as a ligand and coreceptor in the nodal signaling pathway. Mol Cell Biol 22: 4439–4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ding J, Yang L, Yan YT, Chen A, Desai N, et al. (1998) Cripto is required for correct orientation of the anterior-posterior axis in the mouse embryo. Nature 395: 702–707. [DOI] [PubMed] [Google Scholar]
- 8. Xu C, Liguori G, Persico MG, Adamson ED (1999) Abrogation of the Cripto gene in mouse leads to failure of postgastrulation morphogenesis and lack of differentiation of cardiomyocytes. Development 126: 483–494. [DOI] [PubMed] [Google Scholar]
- 9. Liguori GL, Echevarria D, Improta R, Signore M, Adamson E, et al. (2003) Anterior neural plate regionalization in cripto null mutant mouse embryos in the absence of node and primitive streak. Dev Biol 264: 537–549. [DOI] [PubMed] [Google Scholar]
- 10. Liguori GL, Echevarria D, Bonilla S, D’Andrea D, Liguoro A, et al. (2009) Characterization of the functional properties of the neuroectoderm in mouse Cripto(-/-) embryos showing severe gastrulation defects. Int J Dev Biol 53: 549–557. 10.1387/ijdb.082650gl [DOI] [PubMed] [Google Scholar]
- 11. Wechselberger C, Ebert AD, Bianco C, Khan NI, Sun Y, et al. (2001) Cripto-1 enhances migration and branching morphogenesis of mouse mammary epithelial cells. Exp Cell Res 266: 95–105. [DOI] [PubMed] [Google Scholar]
- 12. Watanabe K, Bianco C, Strizzi L, Hamada S, Mancino M, et al. (2007) Growth factor induction of Cripto-1 shedding by glycosylphosphatidylinositol-phospholipase D and enhancement of endothelial cell migration. J Biol Chem 282: 31643–31655. [DOI] [PubMed] [Google Scholar]
- 13. Bianco C, Strizzi L, Ebert A, Chang C, Rehman A, et al. (2005) Role of human cripto-1 in tumor angiogenesis. J Natl Cancer Inst 97: 132–141. [DOI] [PubMed] [Google Scholar]
- 14. Miharada K, Karlsson G, Rehn M, Rorby E, Siva K, et al. (2011) Cripto regulates hematopoietic stem cells as a hypoxic-niche-related factor through cell surface receptor GRP78. Cell Stem Cell 9: 330–344. 10.1016/j.stem.2011.07.016 [DOI] [PubMed] [Google Scholar]
- 15. Wu C, Orozco C, Boyer J, Leglise M, Goodale J, et al. (2009) BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biology 10 10.1186/gb-2009-10-12-115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Uhlen M, Bjorling E, Agaton C, Szigyarto CA, Amini B, et al. (2005) A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol Cell Proteomics 4: 1920–1932. [DOI] [PubMed] [Google Scholar]
- 17. Normanno N, De Luca A, Bianco C, Maiello MR, Carriero MV, et al. (2004) Cripto-1 overexpression leads to enhanced invasiveness and resistance to anoikis in human MCF-7 breast cancer cells. J Cell Physiol 198: 31–39. [DOI] [PubMed] [Google Scholar]
- 18. Gong YP, Yarrow PM, Carmalt HL, Kwun SY, Kennedy CW, et al. (2007) Overexpression of Cripto and its prognostic significance in breast cancer: a study with long-term survival. Eur J Surg Oncol 33: 438–443. [DOI] [PubMed] [Google Scholar]
- 19. Bianco C, Strizzi L, Mancino M, Rehman A, Hamada S, et al. (2006) Identification of cripto-1 as a novel serologic marker for breast and colon cancer. Clin Cancer Res 12: 5158–5164. [DOI] [PubMed] [Google Scholar]
- 20. Jiang PC, Zhu L, Fan Y, Zhao HL (2013) Clinicopathological and biological significance of cripto overexpression in human colon cancer. World J Gastroenterol 19: 8630–8637. 10.3748/wjg.v19.i46.8630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Friess H, Yamanaka Y, Buchler M, Kobrin MS, Tahara E, et al. (1994) Cripto, a member of the epidermal growth factor family, is over-expressed in human pancreatic cancer and chronic pancreatitis. Int J Cancer 56: 668–674. [DOI] [PubMed] [Google Scholar]
- 22. Ertoy D, Ayhan A, Sarac E, Karaagaoglu E, Yasui W, et al. (2000) Clinicopathological implication of cripto expression in early stage invasive cervical carcinomas. Eur J Cancer 36: 1002–1007. [DOI] [PubMed] [Google Scholar]
- 23. De Luca A, Lamura L, Strizzi L, Roma C, D’Antonio A, et al. (2011) Expression and functional role of CRIPTO-1 in cutaneous melanoma. Br J Cancer 105: 1030–1038. 10.1038/bjc.2011.324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wu Z, Li G, Wu L, Weng D, Li X, et al. (2009) Cripto-1 overexpression is involved in the tumorigenesis of nasopharyngeal carcinoma. BMC Cancer 9: 315 10.1186/1471-2407-9-315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhong XY, Zhang LH, Jia SQ, Shi T, Niu ZJ, et al. (2008) Positive association of up-regulated Cripto-1 and down-regulated E-cadherin with tumour progression and poor prognosis in gastric cancer. Histopathology 52: 560–568. 10.1111/j.1365-2559.2008.02971.x [DOI] [PubMed] [Google Scholar]
- 26. Miyoshi N, Ishii H, Mimori K, Sekimoto M, Doki Y, et al. (2010) TDGF1 is a novel predictive marker for metachronous metastasis of colorectal cancer. Int J Oncol 36: 563–568. [DOI] [PubMed] [Google Scholar]
- 27. Tysnes BB, Satran HA, Mork SJ, Margaryan NV, Eide GE, et al. (2013) Age-Dependent Association between Protein Expression of the Embryonic Stem Cell Marker Cripto-1 and Survival of Glioblastoma Patients. Transl Oncol 6: 732–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bianco C, Strizzi L, Normanno N, Khan N, Salomon DS (2005) Cripto-1: an oncofetal gene with many faces. Curr Top Dev Biol 67: 85–133. [DOI] [PubMed] [Google Scholar]
- 29. Strizzi L, Bianco C, Normanno N, Seno M, Wechselberger C, et al. (2004) Epithelial mesenchymal transition is a characteristic of hyperplasias and tumors in mammary gland from MMTV-Cripto-1 transgenic mice. J Cell Physiol 201: 266–276. [DOI] [PubMed] [Google Scholar]
- 30. Strizzi L, Bianco C, Hirota M, Watanabe K, Mancino M, et al. (2007) Development of leiomyosarcoma of the uterus in MMTV-CR-1 transgenic mice. J Pathol 211: 36–44. [DOI] [PubMed] [Google Scholar]
- 31. Sun Y, Strizzi L, Raafat A, Hirota M, Bianco C, et al. (2005) Overexpression of human Cripto-1 in transgenic mice delays mammary gland development and differentiation and induces mammary tumorigenesis. Am J Pathol 167: 585–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wechselberger C, Strizzi L, Kenney N, Hirota M, Sun Y, et al. (2005) Human Cripto-1 overexpression in the mouse mammary gland results in the development of hyperplasia and adenocarcinoma. Oncogene 24: 4094–4105. [DOI] [PubMed] [Google Scholar]
- 33. Giorgio E, Liguoro A, D’Orsi L, Mancinelli S, Barbieri A, et al. (2014) Cripto haploinsufficiency affects in vivo colon tumor development. Int J Oncol 45: 31–40. 10.3892/ijo.2014.2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Adkins HB, Bianco C, Schiffer SG, Rayhorn P, Zafari M, et al. (2003) Antibody blockade of the Cripto CFC domain suppresses tumor cell growth in vivo. J Clin Invest 112: 575–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bianco C, Salomon DS (2010) Targeting the embryonic gene Cripto-1 in cancer and beyond. Expert Opin Ther Pat 20: 1739–1749. 10.1517/13543776.2010.530659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Klauzinska M, Castro NP, Rangel MC, Spike BT, Gray PC, et al. (2014) The multifaceted role of the embryonic gene Cripto-1 in cancer, stem cells and epithelial-mesenchymal transition. Semin Cancer Biol. 10.1016/j.semcancer.2014.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pilgaard L, Mortensen JH, Henriksen M, Olesen P, Sorensen P, et al. (2014) Cripto-1 Expression in Glioblastoma Multiforme. Brain Pathol. 10.1111/bpa.12241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ciccodicola A, Dono R, Obici S, Simeone A, Zollo M, et al. (1989) Molecular characterization of a gene of the ‘EGF family’ expressed in undifferentiated human NTERA2 teratocarcinoma cells. EMBO J 8: 1987–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gray PC, Vale W (2012) Cripto/GRP78 modulation of the TGF-beta pathway in development and oncogenesis. FEBS Lett 586: 1836–1845. 10.1016/j.febslet.2012.01.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gray PC, Shani G, Aung K, Kelber J, Vale W (2006) Cripto binds transforming growth factor beta (TGF-beta) and inhibits TGF-beta signaling. Mol Cell Biol 26: 9268–9278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nagaoka T, Karasawa H, Turbyville T, Rangel MC, Castro NP, et al. (2013) Cripto-1 enhances the canonical Wnt/beta-catenin signaling pathway by binding to LRP5 and LRP6 co-receptors. Cell Signal 25: 178–189. 10.1016/j.cellsig.2012.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mancino M, Strizzi L, Wechselberger C, Watanabe K, Gonzales M, et al. (2008) Regulation of human Cripto-1 gene expression by TGF-beta1 and BMP-4 in embryonal and colon cancer cells. J Cell Physiol 215: 192–203. [DOI] [PubMed] [Google Scholar]
- 43. Hamada S, Watanabe K, Hirota M, Bianco C, Strizzi L, et al. (2007) beta-Catenin/TCF/LEF regulate expression of the short form human Cripto-1. Biochem Biophys Res Commun 355: 240–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bianco C, Cotten C, Lonardo E, Strizzi L, Baraty C, et al. (2009) Cripto-1 is required for hypoxia to induce cardiac differentiation of mouse embryonic stem cells. Am J Pathol 175: 2146–2158. 10.2353/ajpath.2009.090218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Behrens AN, Ren Y, Ferdous A, Garry DJ, Martin CM (2012) Nkx2–5 Regulates Tdgf1 (Cripto) Early During Cardiac Development. J Clin Exp Cardiolog Suppl 11: 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bianco C, Castro NP, Baraty C, Rollman K, Held N, et al. (2013) Regulation of human Cripto-1 expression by nuclear receptors and DNA promoter methylation in human embryonal and breast cancer cells. J Cell Physiol 228: 1174–1188. 10.1002/jcp.24271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hentschke M, Kurth I, Borgmeyer U, Hubner CA (2006) Germ cell nuclear factor is a repressor of CRIPTO-1 and CRIPTO-3. J Biol Chem 281: 33497–33504. [DOI] [PubMed] [Google Scholar]
- 48. Chen F, Hou SK, Fan HJ, Liu YF (2014) MiR-15a-16 represses Cripto and inhibits NSCLC cell progression. Mol Cell Biochem 391: 11–19. 10.1007/s11010-014-1981-y [DOI] [PubMed] [Google Scholar]
- 49. Kotepui M, Thawornkuno C, Chavalitshewinkoon-Petmitr P, Punyarit P, Petmitr S (2012) Quantitative real-time RT-PCR of ITGA7, SVEP1, TNS1, LPHN3, SEMA3G, KLB and MMP13 mRNA expression in breast cancer. Asian Pac J Cancer Prev 13: 5879–5882. [DOI] [PubMed] [Google Scholar]
- 50. Burghel GJ, Lin WY, Whitehouse H, Brock I, Hammond D, et al. (2013) Identification of candidate driver genes in common focal chromosomal aberrations of microsatellite stable colorectal cancer. PLoS One 8: e83859 10.1371/journal.pone.0083859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ashktorab H, Daremipouran M, Goel A, Varma S, Leavitt R, et al. (2014) DNA methylome profiling identifies novel methylated genes in African American patients with colorectal neoplasia. Epigenetics 9: 503–512. 10.4161/epi.27644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ronneberg JA, Fleischer T, Solvang HK, Nordgard SH, Edvardsen H, et al. (2011) Methylation profiling with a panel of cancer related genes: association with estrogen receptor, TP53 mutation status and expression subtypes in sporadic breast cancer. Mol Oncol 5: 61–76. 10.1016/j.molonc.2010.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lo SH, Janmey PA, Hartwig JH, Chen LB (1994) Interactions of tensin with actin and identification of its three distinct actin-binding domains. J Cell Biol 125: 1067–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chen H, Lo SH (2003) Regulation of tensin-promoted cell migration by its focal adhesion binding and Src homology domain 2. Biochem J 370: 1039–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chen H, Ishii A, Wong WK, Chen LB, Lo SH (2000) Molecular characterization of human tensin. Biochem J 351 Pt 2: 403–411. [PMC free article] [PubMed] [Google Scholar]
- 56. Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, et al. (2004) ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 6: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hall EH, Daugherty AE, Choi CK, Horwitz AF, Brautigan DL (2009) Tensin1 requires protein phosphatase-1alpha in addition to RhoGAP DLC-1 to control cell polarization, migration, and invasion. J Biol Chem 284: 34713–34722. 10.1074/jbc.M109.059592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Strizzi L, Postovit LM, Margaryan NV, Seftor EA, Abbott DE, et al. (2008) Emerging roles of nodal and Cripto-1: from embryogenesis to breast cancer progression. Breast Dis 29: 91–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. She BR, Liou GG, Lin-Chao S (2002) Association of the growth-arrest-specific protein Gas7 with F-actin induces reorganization of microfilaments and promotes membrane outgrowth. Exp Cell Res 273: 34–44. [DOI] [PubMed] [Google Scholar]
- 60. Gotoh A, Hidaka M, Hirose K, Uchida T (2013) Gas7b (growth arrest specific protein 7b) regulates neuronal cell morphology by enhancing microtubule and actin filament assembly. J Biol Chem 288: 34699–34706. 10.1074/jbc.M113.513119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ebinger M, Senf L, Wachowski O, Scheurlen W (2006) Expression of GAS7 in childhood CNS tumors. Pediatr Blood Cancer 46: 325–328. [DOI] [PubMed] [Google Scholar]
- 62. Chang Y, Ueng SW, Lin-Chao S, Chao CC (2008) Involvement of Gas7 along the ERK1/2 MAP kinase and SOX9 pathway in chondrogenesis of human marrow-derived mesenchymal stem cells. Osteoarthritis Cartilage 16: 1403–1412. 10.1016/j.joca.2008.03.018 [DOI] [PubMed] [Google Scholar]
- 63. Munroe DG, Gupta AK, Kooshesh F, Vyas TB, Rizkalla G, et al. (1999) Prototypic G protein-coupled receptor for the intestinotrophic factor glucagon-like peptide 2. Proc Natl Acad Sci U S A 96: 1569–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yusta B, Huang L, Munroe D, Wolff G, Fantaske R, et al. (2000) Enteroendocrine localization of GLP-2 receptor expression in humans and rodents. Gastroenterology 119: 744–755. [DOI] [PubMed] [Google Scholar]
- 65. Lovshin J, Estall J, Yusta B, Brown TJ, Drucker DJ (2001) Glucagon-like peptide (GLP)-2 action in the murine central nervous system is enhanced by elimination of GLP-1 receptor signaling. J Biol Chem 276: 21489–21499. [DOI] [PubMed] [Google Scholar]
- 66. Yusta B, Somwar R, Wang F, Munroe D, Grinstein S, et al. (1999) Identification of glucagon-like peptide-2 (GLP-2)-activated signaling pathways in baby hamster kidney fibroblasts expressing the rat GLP-2 receptor. J Biol Chem 274: 30459–30467. [DOI] [PubMed] [Google Scholar]
- 67. Velazquez E, Ruiz-Albusac JM, Blazquez E (2003) Glucagon-like peptide-2 stimulates the proliferation of cultured rat astrocytes. Eur J Biochem 270: 3001–3009. [DOI] [PubMed] [Google Scholar]
- 68. Lan L, Han H, Zuo H, Chen Z, Du Y, et al. (2010) Upregulation of myosin Va by Snail is involved in cancer cell migration and metastasis. Int J Cancer 126: 53–64. 10.1002/ijc.24641 [DOI] [PubMed] [Google Scholar]
- 69. Colonna V, Nutile T, Ferrucci RR, Fardella G, Aversano M, et al. (2009) Comparing population structure as inferred from genealogical versus genetic information. Eur J Hum Genet 17: 1635–1641. 10.1038/ejhg.2009.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Colonna V, Nutile T, Astore M, Guardiola O, Antoniol G, et al. (2007) Campora: a young genetic isolate in South Italy. Hum Hered 64: 123–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Almasy L, Blangero J (1998) Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet 62: 1198–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR (2010) MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol 34: 816–834. 10.1002/gepi.20533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Aulchenko YS, Ripke S, Isaacs A, van Duijn CM (2007) GenABEL: an R library for genome-wide association analysis. Bioinformatics 23: 1294–1296. [DOI] [PubMed] [Google Scholar]
- 74. Aulchenko YS, Struchalin MV, van Duijn CM (2010) ProbABEL package for genome-wide association analysis of imputed data. BMC Bioinformatics 11: 134 10.1186/1471-2105-11-134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Willer CJ, Li Y, Abecasis GR (2010) METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26: 2190–2191. 10.1093/bioinformatics/btq340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Neter J (1996) Applied linear statistical models. Chicago; London: Irwin. [Google Scholar]
- 78. Ward LD, Kellis M (2012) HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res 40: D930–934. 10.1093/nar/gkr917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Kajanne R, Miettinen P, Tenhunen M, Leppa S (2009) Transcription factor AP-1 promotes growth and radioresistance in prostate cancer cells. Int J Oncol 35: 1175–1182. [DOI] [PubMed] [Google Scholar]
- 80. Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, et al. (2010) LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 26: 2336–2337. 10.1093/bioinformatics/btq419 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
For each variant tested, the observed -log10(p-value) (y-axis) is plotted against the expected -log10(p-value) under the null hypothesis (red line).
(DOCX)
Regional association plots (panels A-F) show -log10(p-values) for all SNPs ordered by their chromosomal position within all regions of the replicated loci. For the rs3806702 the p-value of the discovery GWAS is reported, for the other SNPs p-values of the discovery conditional GWAS are reported. Each SNP is colored according to its correlation with the replicated SNP within the region as specified in the color scheme. Correlation structures correspond to hg19/1000 Genomes EUR Mar 2012. Plots were generated with LocusZoom [80]. The CRIPTO gene is reported as TDGF1.
(DOCX)
(A) The genomic localization of the top 8 associated SNPs (red) respect to the TDGF1 gene is shown using the GRCh38 primary assembly as reference sequence. The transcript (NM_003212.03) and the coding (CCDS2742.1) sequence are also shown. The functional SNP rs112481213 is underlined and reported in italic. The black rectangle indicates the region of 1.052 bp, upstream the ATG start codon, cloned in the luciferase reporter pGL3-basic vector. The rs3806703 SNP (orange and with a dashed line) is present in the cloned region but is not included in the top 8 associated SNPs (LD < 0.8 for rs112481213). (B) LD between the top 8 associated SNPs is reported. The analysis is carried out on the 1000 genome data (European panel) using the Haploview software. The distance in kb from the ATG start codon is shown on the left column. (C) MatInspector analysis. The results are reported only for the SNPs for which: 1) SNP alleles differently affect the TF biding site. In the schema, “Change” of the allele is-1 if the Alt allele disrupts the TF binding site and is 1 if the Alt allele creates a new TF binding site; 2) The value of the matrix similarity is higher than 0.9; 3) The position of the allele in the sequence logo shows an information content > 0.2 (y axis of the sequence logo). The black star represents the position of the SNP in the TF consesus binding site.
(DOCX)
Allele frequencies of all genotyped SNPs in the discovery or replication samples (y axis) are plotted against allele frequencies in 1000 Genomes European panel reference (x axis). RAF = Reference Allele Frequency.
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.