Abstract
In this paper we describe a new global alignment method called AVID. The method is designed to be fast, memory efficient, and practical for sequence alignments of large genomic regions up to megabases long. We present numerous applications of the method, ranging from the comparison of assemblies to alignment of large syntenic genomic regions and whole genome human/mouse alignments. We have also performed a quantitative comparison of AVID with other popular alignment tools. To this end, we have established a format for the representation of alignments and methods for their comparison. These formats and methods should be useful for future studies. The tools we have developed for the alignment comparisons, as well as the AVID program, are publicly available. See Web Site References section for AVID Web address and Web addresses for other programs discussed in this paper.
The comparison of biological sequences is one of the oldest problems in computational biology, and early work on the problem (Needleman and Wunsch 1970; Altschul et al. 1990) resulted in what were arguably the first highly successful and widely adopted applications of computer science to biology. It became apparent early on that alignment programs could be divided into two types:
Local alignment methods (e.g., BLASTZ; Schwartz et al. 2000) are designed to search for highly similar regions in two sequences, where the regions of similarity are not necessarily conserved in order and orientation. BLAST-like methods work by first finding very short common segments between the sequences, and then expanding out the matching regions as far as possible. Algorithms such as the Smith-Waterman dynamic programming (Smith and Waterman 1981) work by identifying the most likely significant matches according to an evolutionary model.
Global alignment algorithms (e.g., Needleman and Wunsch 1970) are suitable when an extra assumption holds, namely, that the highly similar regions in the sequences appear in the same order and orientation. These methods attempt to find the “global map” between the sequences, in the process rejecting alignments that overlap or cross over.
Local alignment algorithms are generally very useful in finding similarity between regions that may be related but are inverted or rearranged with respect to each other. There has also been evidence that transcription factor binding sites are prone to reordering and so are more suited to detection by local alignment methods. A problem with local alignment algorithms is that, because of the weaker assumptions in place, there is less power in finding weakly conserved regions; furthermore, identified conserved regions may not be true homologs (i.e., related via a common ancestor).
Global alignment algorithms have been found to be useful in many situations because biological sequences from related organisms tend to satisfy the order assumption, assuming that the regions being examined are sufficiently small. For example, on average, the human and mouse genome appear to have order and orientation preserved for regions up to 8 Mb in length (Mural et al. 2002). On the other hand, sophisticated scoring functions and global alignment models lead to slow algorithms that are also very memory intensive. Thus, until relatively recently, global alignment algorithms have generally only been applied to short sequences.
The AVID alignment method is our attempt to address existing shortcomings of global alignment programs. As we will show, AVID is sensitive in finding homologous regions, but is also specific and avoids the false-positive problem of local alignment programs. At the same time, it is fast and highly reliable. It has been used to align thousands of submitted sequence pairs from biological researchers worldwide, and it has also been used as a key component in an alignment of the entire human and mouse genomes (O. Couronne et al. 2003).
In order to assess the performance of AVID, we compared it to both local and global alignment programs. Despite the emergence of many alignment programs in recent years (Wiehe et al. 2000; Chain 2001; Miller 2001; Dubchak and Pachter 2002), there has not been a detailed performance comparison as has been done in the gene-finding field (Burset and Guigo 1996). This is partially due to the difficulty in assessing the “correctness” of an alignment. We have devised various ways of doing so and we describe the results following. Finally, we describe various applications of AVID (e.g., the alignment of assemblies), which have been prohibitive with previous computationally expensive approaches.
Algorithm
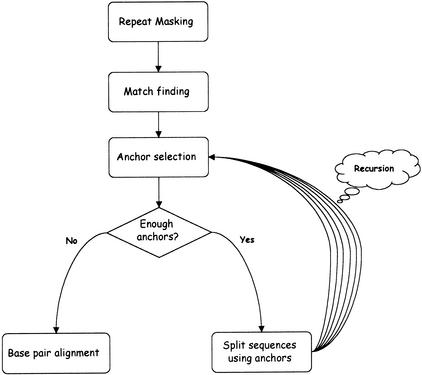
The AVID method is summarized in Figure 1. The input to the program consists of two genomic sequences; the output is a global alignment with additional information (e.g., an overall score). The details of the components are described as follows.
Figure 1.
The AVID algorithm structure.
Repeat Masking (Optional)
The input sequences can be processed with the RepeatMasker program (http://ftp.genome.washington.edu/RM/RepeatMasker.html, Smit and Green), but, unlike standard alignment programs, both the masked and unmasked sequences are used in the alignment process. Matches are divided into two groups: those overlapping repeats that we call repeat matches, and those not overlapping repeats that we call clean matches (it is important to note that the term “match” here refers to a maximal match that is not necessarily unique). Clean and repeat matches are used in different ways by the program and we discuss this in more detail as follows.
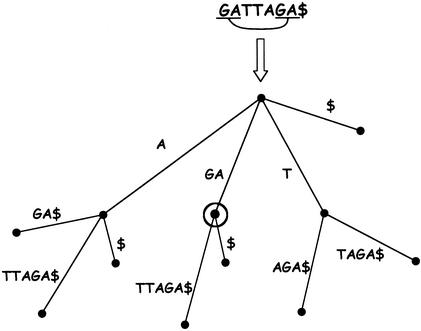
Finding Matches Using Suffix Trees
A maximal repeated substring in a string is a subsequence that has the property that every subsequence that contains it is not repeated in the string (Gusfield 1997). The problem of finding all maximal repeated substrings of a single string has a straightforward solution using suffix trees (Fig. 2). Maximal matches between two sequences are a pair of matching subsequences (one from each sequence) whose flanking bases are mismatches. In AVID, the problem of finding all maximal matches between two sequences is transformed to the problem of finding maximal repeated substrings in one string. This is done by concatenating the two sequences and placing the character N between them. A maximal repeat in this string that crosses the boundary between the two sequences represents a maximal match between the two sequences.
Figure 2.
Finding maximal matches using a suffix tree: The suffixes of the word at the root are represented by the characters along the paths from the root to the leaves. Branchings in the tree correspond to locations where different suffixes shared the same prefix, and therefore are matches. Every internal node in the tree is therefore a match (with the matching sequence corresponding to the path characters along the path from the root). Maximal matches can be efficiently detected by considering some additional criteria.
Anchor Selection
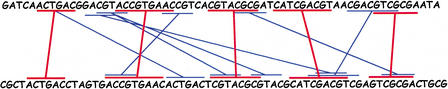
Once the match finding has been completed, AVID begins the recursive process of anchoring and aligning the sequences. An anchor set is a collection of nonoverlapping, noncrossing matches (e.g., the red matches shown in Fig. 3).
Figure 3.
Selecting anchors from the set of matches. Every maximal match is shown in blue. A set of good anchors is shown in red.
First, the entire match set is reduced to eliminate “noisy” matches from those being considered for anchors. Our current heuristic is to remove matches that are less than half the length of the longest match from initial consideration. The shorter matches will be reconsidered for anchoring in later rounds. The matches are then ordered, with clean matches appearing first (sorted by length), followed by repeat matches. Repeat matches will not be considered for anchoring until there are no more clean matches.
The anchors are selected using a variant of the Smith-Waterman algorithm. The gap score used is zero, and the mismatch score is—Infinity. The score assigned to a match is based on its length and the alignment score of the regions flanking the match (10 bp on each side). Anchors are also required to be nonoverlapping (hence a minor modification of the Smith-Waterman algorithm). This process of anchor selection is similar to the idea first adopted in the GLASS algorithm (Pachter 1999; Batzoglou et al. 2000)
There is no guarantee that the matches in the anchor set produced by this procedure are biologically significant. For regions that are too long to align by the Needleman-Wunsch algorithm, there is no choice but to use the anchors. For shorter regions, the use of anchors can speed up the alignment procedure, but may result in a lower quality alignment than that which would be arrived at using the Needleman-Wunsch algorithm. Therefore, the anchors should only be used if we are confident that they are correct. When AVID aligns regions short enough to perform an optimal alignment, it uses anchors only if the total length of the anchor set is >50% of the length of the sequence; otherwise the regions are aligned using the Needleman-Wunsch algorithm using standard parameters.
Recursion
Once the anchors have been selected, they will form part of the final global alignment. We think of them as having been set. If n anchors have been set, there will be n + 1 regions between these anchors that remain to be aligned. By filtering the current list of matches, we produce n + 1 lists of matches, the ith list being the list of all maximal matches between the ith interanchor pair. This is done by checking for each match whether that match (or any sufficiently long part of that match) lies entirely between two sets of anchors. Once the maximal matches have been obtained, the smaller interanchor regions are realigned using the anchor selection step described earlier.
The recursion terminates when there are either no remaining bases to be aligned, or there are no significant matches in the remaining sequences. If the sequences are short (⪝4 kb each), they are aligned using the Needleman-Wunsch algorithm. For long sequences, we conclude that the lack of anchors indicates no significant alignment between them, and because a Needleman-Wunsch type algorithm is meaningless, we return a trivial alignment, where both sequences are completely gapped.
Draft
AVID has the ability to order and orient draft sequence by using comparisons to a finished sequence. If one of the sequences is submitted in draft format (in multiple FASTA format), then the contigs are first aligned separately to the finished genome. Alignments are performed in both orientations, and the resulting scores are used to determine the correct orientation. The matching locations of the 5′ ends of the contigs are then used to sort them, and consecutive contigs are then aligned to each other to determine the amount of overlap. The overlap information is then used to generate a “merged” draft sequence, which is then realigned to the finished sequence to produce a “finished-draft” alignment.
Testing
Despite a number of software packages for alignments of large genomic regions in recent years, there has been a lack of quantitative comparison between methods. One of the problems in comparing alignment methods is that local and global alignment methods have fundamentally different outputs, and comparison is complicated by the myriad of alignment formats and standards for output that are being used. We have developed convenient and general alignment formats, and tools for converting output of popular programs to these formats, in order to test sensitivity and specificity of the different aligners.
Sensitivity
The comparison of the sensitivity of different alignment programs is complicated by the fact that different programs are based on different sets of parameters, whose setting can greatly influence the amount of coverage. In order to fairly compare aligners, we adopted a technique first suggested by Jim Kent (Waterston et al. 2002), which is to filter the alignments after they have been generated to retain only those portions of the alignments that score above a certain threshold according to a suitable scoring matrix. After filtering, we computed two relevant statistics: the overall amount of coverage and the coverage of certain select features such as coding sequences and untranslated transcribed regions (UTRs).
Specificity
It is difficult to construct accurate tests for specificity of alignment programs because it is often hard to ascertain whether a reported alignment is biologically significant, or the result of random matching between the sequences. In the mouse analysis paper (Waterston et al. 2002), a “reverse” test suggested by Arian Smit has been used, in which the mouse genome was reversed (not reverse complemented) and aligned in order to measure the amount of alignment due to random matches. Here we have instead measured the average amount of overprediction resulting from alignments that are not order and orientation preserving. In other words, we have measured the amount of alignment when the alignments are required to be order and orientation preserving, versus the amount when they are not. This test was performed for BLASTZ thanks to its “chaining option”, which makes the measurement easy. It is not necessarily the case that all extra alignments are false positives, but, in the short regions we have analyzed, we have checked that order and orientation is preserved and thus extra alignments can be assumed to be mostly incorrect.
Testing Sets
We compiled a number of testing sets in order to analyze the performance of the programs:
Finished sequences from the cat (14), chicken (11), chimp (6), cow (16), dog (10), pig (27), and rat (33) for a total of 117 sequences with an average length of ∼170,000 bp. The corresponding human sequences were obtained together with RefSeq annotations.
The Celera and EDGP assemblies of the tip of the X chromosome in Drosophila (Benos et al. 2001).
Celera mouse chromosome 16 (Mural et al. 2002) for alignment to the public mouse genome.
The test sets are available for download at http://baboon.math.berkeley.edu/∼syntenic/avid/tests/.
Programs
We attempted to test as many programs as possible, and we report on comparisons of AVID, MUMmer (Delcher et al. 1999, 2002), BLASTZ (Schwartz et al. 2000), CHAOS (Brudno and Morgenstern 2002), and GLASS (Pachter 1999; Batzoglou et al. 2000). Other programs such as DBA (Jareborg et al. 1999), WABA (Kent and Zahler 2000), and DIALIGN (Morgenstern et al. 1998, 2002) were not tested for one or more of a number of reasons: problems obtaining or working with the code, difficulty in parsing the output for comparison, or slow running time for large sequences. The main problem with DIALIGN was the speed of the program—runs on a few hundred kilobases did not finish in hours. We did not succeed in merging the CHAOS and DIALIGN programs for a significant speedup (Morgenstern et al. 2002), but this is possible. All the programs were tested with their default parameters. It is possible that certain results could improve with different parameters, but such analysis for each program is beyond the scope of this paper. BLASTZ was run with and without the chaining option.
Running Times and Memory Usage
We measured the running time for each of the programs on a typical Linux-based PC (2-GHz processor). Repeat Masking times were not included. Memory usage was not reported because we found that memory usage varied substantially with the alignment structure of the sequences and not just their length (this is probably due to the heuristic nature of the alignment programs). We did find that AVID and BLASTZ require approximately 100 Mb to align bacterial artificial chromosome (BAC)-sized sequences.
Formats
A major problem in comparing aligners is that every alignment program has its own output format, and these vary greatly between local and global aligners, and multiple alignment programs. We have introduced a new alignment format we call AVX, which is a hybrid of the CLUSTALW and FASTA formats and allows for recording multiple local or global alignments. The exact specification is described in http://baboon.math.berkeley.edu/avid/.
RESULTS
Table 1 summarizes the results of the BAC testing. The table shows the amount of coding exon coverage in base pairs rather than percentages because of the difficulty in determining how much of the annotated coding regions were truly coverable by alignments. It is reassuring to note that most of the programs are good at identifying coding exons, and this holds true for all the organisms tested. It is interesting that the local and global aligners did not exhibit much difference in this measure, AVID and BLASTZ (with the chaining option) having very similar coverage. BLASTZ did have slightly higher coding exon coverage without chaining, but this is associated with a considerable increase in total coverage (∼10%), most of which cannot be homologous alignment. Nevertheless, the overall coverage of BLASTZ in chaining mode is very similar to the AVID coverage. We believe that the large discrepancy in coverage between the BLASTZ chaining and nonchaining modes is mostly due to false-positive alignments. The reason for this is that these finished sequences were selected for containing the same genes in the same order with no rearrangements between the sequences. It is possible that a part of the 10% is due to alignments of transcription factor binding sites that are not preserved in order, and to repetitive sequence.
Table 1.
Coverage Results for the Different Programs on Human Sequence Alignments With Cat, Chicken, Cow, Dog, Pig, and Rat
| Coverage (bp) | Refseq | UTR | Time (sec) | |
| Cat | ||||
| AVID | 1200310 | 21966 | 13742 | 78 |
| BLASTZ | 1270696 | 22383 | 13632 | 101 |
| BLASTZ (chaining) | 1170350 | 21993 | 13632 | 52 |
| CHAOS | 360436 | 19022 | 7604 | 128 |
| GLASS | 1179833 | 21942 | 13426 | 9215 |
| MUMmer | 1092441 | 20604 | 12205 | 111 |
| Chicken | ||||
| AVID | 77114 | 17886 | 1742 | 35 |
| BLASTZ | 56370 | 19204 | 1357 | 11 |
| BLASTZ (chaining) | 56370 | 19204 | 1357 | 11 |
| CHAOS | 5635 | 3060 | 70 | 31 |
| GLASS | 56445 | 17437 | 984 | 2866 |
| MUMmer − | 18457 | 5634 | 0 | 24 |
| Chimp | ||||
| AVID | 852611 | 0 | 0 | 36 |
| BLASTZ | 881206 | 0 | 0 | 11330 |
| BLASTZ (chaining) | 854023 | 0 | 0 | 45 |
| CHAOS | −680491 | 0 | 0 | 4471 |
| GLASS | * | * | * | * |
| MUMmer − | 752809 | 0 | 0 | 5 |
| Cow | ||||
| AVID | 1195913 | 26359 | 10777 | 89 |
| BLASTZ | 1288895 | 27911 | 13505 | 79 |
| BLASTZ (chaining) | 1150323 | 25814 | 10434 | 57 |
| CHAOS | 317660 | 22551 | 6633 | 198 |
| GLASS | 1131693 | 25187 | 12860 | 10115 |
| MUMmer | 996019 | 22775 | 9621 | 135 |
| Dog | ||||
| AVID | 1015426 | 26278 | 11406 | 84 |
| BLASTZ | 1123496 | 27643 | 15321 | 91 |
| BLASTZ (chaining) | 973670 | 26131 | 11386 | 50 |
| CHAOS | 258696 | 22298 | 6182 | 173 |
| GLASS −− | 741066 | 20565 | 10364 | 7635 |
| MUMmer | 790831 | 22217 | 6848 | 116 |
| Pig | ||||
| AVID | 2163592 | 60903 | 22932 | 180 |
| BLASTZ | 2288008 | 65757 | 28016 | 168 |
| BLASTZ (chaining) | 2075031 | 61988 | 25900 | 110 |
| CHAOS | 574740 | 53854 | 9863 | 398 |
| GLASS − | 1951056 | 58207 | 23318 | 13689 |
| MUMmer | 1715068 | 55250 | 20505 | 235 |
| Rat | ||||
| AVID | 1686932 | 82920 | 38973 | 209 |
| BLASTZ | 2115917 | 90662 | 42267 | 447 |
| BLASTZ (chaining) | 1618195 | 83338 | 39099 | 101 |
| CHAOS | 456152 | 59918 | 14415 | 403 |
| GLASS −−−− − | 1076219 | 62978 | 29376 | 28993 |
| MUMmer | 850710 | 65361 | 31006 | 258 |
Coverage of the human genome using the mouse genome is described in O. . An asterisk indicates that the program was not able to successfully align the sequences. A minus sign indicates that the program crashed on one sequence pair. Multiple minus signs are used for multiple crashes. RefSeq annotations are based on the human December 2001 hg10 freeze.
It is reassuring to note that the BLASTZ coverage with chaining on is very similar to the AVID coverage. It is important to note, however, that the coverage is not always at the same place. We observed that roughly 7% of the alignments were unique to AVID and another 7% to BLASTZ. This is similar to results obtained on the whole genome alignments (Waterston et al. 2002).
A comparison of coverage results for the different organisms shows that the different programs are sensitive to the evolutionary distances of the sequences. The chimp sequences were alignable with all of the programs except GLASS, although CHAOS and BLASTZ (without chaining) took much longer than the other programs. Coding exon coverage results were not reported because they are not informative for highly similar sequences.
AVID currently has worse results on chicken in comparison to the other organisms because of the low similarity between human and chicken on the nucleotide level. We are currently trying different approaches to optimize the use of both protein and DNA matches for increasing sensitivity on coding regions in highly divergent organisms.
The running time of the methods was also measured because this measure is important in practice, especially if the alignment methods are to be used on a whole genome scale. All the programs were fast, except for GLASS. Of the three global alignment programs, AVID was, in general, the fastest.
The Celera and EDGP assemblies of the tip of the X chromosome from Drosophila melanogaster were used to test the efficiency of AVID in aligning assemblies. The alignment took 30 sec on a 2-GHz Linux machine, and used less than 600 MB of RAM. Results are posted at http://baboon.math.berkeley.edu/∼syntenic/avid/benos/. The alignment compares favorably with the MUMmer alignment performed in the original paper (Benos et al. 2001). The process of aligning the assemblies is completely automatic, requiring no manual intervention (as was needed with MUMmer), and the subsequent visualization is easy to construct and examine (see Web site). Aside from MUMmer, none of the other programs in this paper were able to align assemblies. AVID was also used to align the Celera mouse chromosome 16 sequence to the public mouse chromosome 16. This comparison represents the largest assembly comparison to date (the Celera chromosome is 92 Mb long). This alignment required the division of the sequences into 10 pieces, but should be doable on a large memory machine in one pass. Results can be viewed at http://baboon.math.berkeley.edu/∼syntenic/avid/celera/.
Results on the whole genome alignment of human and mouse using AVID are reported in another paper (O. Couronne et al. 2003).
DISCUSSION
The problem of alignment has expanded and become substantially more complicated during the past two decades. Whereas the original alignment problem asked for a comparison of two short sequences according to a straightforward evolutionary model, the current sequences being analyzed demand robust alignment algorithms that satisfy many criteria and requirements. Among the desirable attributes a program should have are speed (in order to be able to process whole genomes); the ability to deal with rearrangements, duplications, and other large-scale genomic events; sensitivity for the detection of remote homologies and short coding exons; accurate results regardless of the evolutionary distance of the sequences; functionality for dealing with draft sequence, seamless integration with visualization tools; and methods for incorporating phylogenetic information in multiple alignment.
Our results show that AVID is a very effective and practical alignment tool that addresses many, although not all, of these problems. The critical assumption that the sequences being aligned have the property that their functional elements are preserved in order and orientation is used to reduce false alignments, but, at the same time, is a weakness in that it restricts the possible applications of the program. This latter problem can be remedied by combining AVID with a local alignment program, as we have done for the whole human and mouse genome alignment (O. Couronne et al. 2003). Nevertheless, it remains an unsolved problem (and in our opinion an important one) to develop a sophisticated yet efficient alignment program that combines the best of both local and global alignment algorithms.
Our comparison of alignment programs on several BAC-sized regions reveals that coding exons are relatively straightforward to identify, and are correctly aligned by most methods (global and local). Weaker homologies are more difficult to detect, and differences emerge between local and global alignment programs. The extra alignments that result from local alignments that are not required to be order and orientation preserving in regions in which gene order is conserved are strong indicators that local alignment methods may align regions that are similar in sequence, but are not necessarily biologically significant. Perhaps most important, the fact that different alignment programs do not return the exact same alignments, even for conserved regions, indicates that users may want to try all the available methods when aligning their sequences.
The comparison of alignment programs is a non-trivial issue; in particular, we were not successful in running all the available programs on a modestly sized data set. We hope that any omissions resulting from errors on our part will be rectified by further investigations that lead to detailed comparative studies of alignment methods analogous to those that have been undertaken for gene finding. In order to facilitate such comparisons, we have made all our format conversion tools available at http://baboon.math.berkeley.edu/avid/.
The draft mode of AVID has proved to be extremely useful to us, and we use it routinely to order and orient GenBank draft sequence by alignment with finished sequence on the VISTA server http://www-gsd.lbl.gov/vista/ (Mayor et al. 2000). AVID is also used in the genome VISTA server at http://pipeline.lbl.gov/cgi-bin/GenomeVista (O. Couronne et al. 2003), which is useful for locating and aligning user-submitted sequences to whole genomes (human or mouse at the present time).
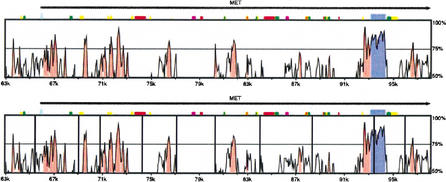
AVID can be used online at http://bio.math.berkeley.edu/avid/. AVID integrates well with the VISTA visualization tool (Mayor et al. 2000). Figure 4 shows an example of a VISTA visualization of an AVID alignment of cat and mouse sequence. Coding exons are displayed in blue and easy to identify. Noncoding highly conserved regions are displayed in red. A server for running AVID and displaying alignments using VISTA is operational at http://www-gsd.lbl.gov/vista/. These two servers are currently aligning almost 2000 sequence pairs per month, with submissions originating from over 30 countries. The bio.math.berkely.edu server is set up to handle requests up to 2 Mb in size. The program is also available for download (free for nonprofit use) at http://www-gsd.lbl.gov/vista/VISTAdownload2.html. Executables are available for Solaris, Mac OS X, Linux, and Alpha, and the source code is available on request.
Figure 4.
Cat versus mouse: A VISTA picture showing an AVID alignment of the 5′ region of the MET gene in cat and mouse. The top panel shows the alignment of the two finished sequences with the X-axis showing coordinates in the mouse, and the Y-axis showing the %identity in a 100-bp window. The bottom panel shows the results of a draft placement simulation: the cat sequence has been sliced at locations corresponding to the vertical black lines. The resulting contigs were permuted (order and orientation changed randomly) and realigned to the mouse.
WEB SITE REFERENCES
http://baboon.math.berkeley.edu/avid/; AVX alignment format, test sets for comparing programs, Celera mouse chromosome 16 to public mouse chromosome 16 alignment, Drosophila assembly comparison, AVID syntenic map whole-genome human–mouse alignment.
http://bibiserv.techfak.uni-bielefeld.de/dialign/; DIALIGN.
http://bio.cse.psu.edu/; BLASTZ.
http://bio.math.berkeley.edu/avid/; AVID.
http://crossspecies.lcs.mit.edu/; GLASS.
http://ftp.genome.washington.edu/cgi-bin/RepeatMasker; RepeatMasker, Smit, A. and Green, P.
http://pipeline.lbl.gov/cgi-bin/GenomeVista; VISTA/AVID.
http://www.cse.ucsc.edu/∼kent/xenoAli/; WABA.
http://www-gsd.lbl.gov/vista/; VISTA/AVID.
http://www-gsd.lbl.gov/vista/VISTAdownload2.html; VISTA/AVID download.
http://www.sanger.ac.uk/Software/Wise2/dbaform.shtml; DBA.
http://www.stanford.edu/∼brudno/chaos/; CHAOS.
http://www.tigr.org/software/mummer/; MUMmer.
Acknowledgments
We thank Alex Poliakov for helping in setting up the AVID Web servers and provided extensive debugging support and assistance. We also thank Jody Schwartz for help in testing and debugging AVID and Jim Lord who helped in developing overlap identification methods for draft contigs. Thanks also to the Mouse Sequencing Consortium for generating whole genome mouse sequence, which helped greatly in refining and streamlining AVID. Some of the sequence data used to benchmark the alignment programs were generated by the NIH Intramural Sequencing Center (www.nisc.nih.gov). This project was supported in part by a Program in Genomic Applications grant (PGA) from the National Heart Lung and Blood Institute and a grant from the NIH (ROI-HG02362-01).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
E-MAIL lpachter@math.berkeley.edu; FAX (510) 642-8204.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.789803. Article published online before print in December 2002.
REFERENCES
- 1.Altschul S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403-410. [DOI] [PubMed] [Google Scholar]
- 2.Batzoglou S., Pachter, L., Mesirov, J.P., Berger, B., and Lander, E.S. 2000. Human and mouse gene structure: Comparative analysis and application to exon prediction. Genome Res. 10: 950-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benos P.V., Gatt, M.K., Murphy, L., Harris, D., Barrell, B., Ferraz, C., Vidal, S., Brun, C., Demaille, J., Cadieu, E., et al. 2001. From first base: The sequence of the tip of the X chromosome of Drosophila melanogaster, a comparison of two sequencing strategies. Genome Res. 11: 710-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brudno M. and Morgenstern, B., 2002. Fast and sensitive alignment of large genomic sequences. In Proceedings of the First IEEE Computer Society Conference on Bioinformatics. IEEE Computer Society Press. [PubMed]
- 5.Burset M. and Guigo, R. 1996. Evaluation of gene structure prediction programs. Genomics 34: 353-357. [DOI] [PubMed] [Google Scholar]
- 6.Chain, P. 2001. Examining the problems of whole genome comparison: A review. http://cmgm.stanford.edu/biochem218/Projects2001/Chain.pdf.
- 7.Couronne, O., Poliakov, A., Bray, N., Ishkhanov, T., Ryaboy, D., Rubin, E., Pachter, L., Dubchak, I. 2003. Strategies and tools for whole-genome alignments. Genome Res. (this issue). [DOI] [PMC free article] [PubMed]
- 8.Delcher A.L., Kasif, S., Fleischmann, R.D., Peterson, J., White, O., and Salzberg, S.L. 1999. Alignment of whole genomes. Nucleic Acids Res. 27: 2369-2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delcher A.L., Phillippy, A., Carlton, J., and Salzberg, S.L. 2002. Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Res. 30: 2478-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dubchak I. and Pachter, L. 2002. The computational challenges of applying comparative-based computational methods to whole genomes. Brief. Bioinform. 3: 18-22. [DOI] [PubMed] [Google Scholar]
- 11.Gusfield D., 1997. Algorithms on strings, trees, and sequences: Computer science and computational biology. Cambridge University Press, Cambridge, UK.
- 12.Jareborg N., Birney, E., and Durbin, R. 1999. Comparative analysis of non-coding regions of 77 orthologous mouse and human gene pairs. Genome Res. 9: 815-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kent J. and Zahler, M. 2000. The Intronerator: Exploring introns and alternative splicing in C. elegans genomic alignment. Genome Res. 10: 1115-1125. [Google Scholar]
- 14.Mayor C., Brudno, M., Schwartz, J.R., Poliakov, A., Rubin, E.M., Frazer, K.A., Pachter, L.S., and Dubchak, I. 2000. VISTA: Visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 16: 1046-1047. [DOI] [PubMed] [Google Scholar]
- 15.Miller W. 2001. Comparison of genomic DNA sequences: Solved and unsolved problems. Bioinformatics 17: 391-397. [DOI] [PubMed] [Google Scholar]
- 16.Morgenstern B., Frech, K., Dress, A., and Werner, T. 1998. DIALIGN: Finding local similarities by multiple sequence alignment. Bioinformatics 14: 290-294. [DOI] [PubMed] [Google Scholar]
- 17.Morgenstern B., Rinner, O., Abdeddaïm, S., Haase, D., Mayer, K., Dress, A., and Mewes, H-W. 2002. Exon discovery by genomic sequence alignment. Bioinformatics 18: 777-787. [DOI] [PubMed] [Google Scholar]
- 18.Mural R., Adams, M.D., Myers, E.W., Smith, H.O., Miklos, G.L., Wides, R., Halpern, A., Li, P.W., Sutton, G.G., Nadeau, J., et al. 2002. A comparison of whole-genome-shotgun-derived mouse chromosome 16 and the human genome. Science 296: 1667-1671. [DOI] [PubMed] [Google Scholar]
- 19.Needleman S.B. and Wunsch, C.D. 1970. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 48: 443-453. [DOI] [PubMed] [Google Scholar]
- 20.Pachter, L. 1999. “Domino tiling, gene recognition, and mice.” Ph.D thesis, MIT, Cambridge, Massachusetts.
- 21.Schwartz S., Zhang, Z., Frazer, K.A., Smit, A., Riemer, C., Bouck, J., Gibbs, R., Hardison, R., and Miller, W. 2000. PipMaker—A web server for aligning two genomic DNA sequences. Genome Res. 10: 577-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith T.F. and Waterman, M.S. 1981. Identification of common molecular subsequences. J. Mol. Biol. 147: 195-197. [DOI] [PubMed] [Google Scholar]
- 23.Waterston R.H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J.F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520-562. [DOI] [PubMed] [Google Scholar]
- 24.Wiehe T., Guigó, R., and Miller, W. 2000. Genome sequence comparisons: Hurdles in the fast lane to functional genomics. Briefings in Bioinformatics 1: 381-388. [DOI] [PubMed] [Google Scholar]