Abstract
Lactobacillus is the largest genus within the lactic acid bacteria (LAB), with almost 180 species currently identified. Motility has been reported for at least 13 Lactobacillus species, all belonging to the Lactobacillus salivarius clade. Motility in lactobacilli is poorly characterized. It probably confers competitive advantages, such as superior nutrient acquisition and niche colonization, but it could also play an important role in innate immune system activation through flagellin–Toll-like receptor 5 (TLR5) interaction. We now report strong evidence of motility in a species outside the L. salivarius clade, Lactobacillus curvatus (strain NRIC 0822). The motility of L. curvatus NRIC 0822 was revealed by phase-contrast microscopy and soft-agar motility assays. Strain NRIC 0822 was motile at temperatures between 15°C and 37°C, with a range of different carbohydrates, and under varying atmospheric conditions. We sequenced the L. curvatus NRIC 0822 genome, which revealed that the motility genes are organized in a single operon and that the products are very similar (>98.5% amino acid similarity over >11,000 amino acids) to those encoded by the motility operon of Lactobacillus acidipiscis KCTC 13900 (shown for the first time to be motile also). Moreover, the presence of a large number of mobile genetic elements within and flanking the motility operon of L. curvatus suggests recent horizontal transfer between members of two distinct Lactobacillus clades: L. acidipiscis in the L. salivarius clade and L. curvatus in the L. sakei clade. This study provides novel phenotypic, genetic, and phylogenetic insights into flagellum-mediated motility in lactobacilli.
INTRODUCTION
Motility in bacterial species is often mediated by a sophisticated molecular structure called the flagellum. This chief organelle of bacterial motility is self-assembled using dozens of different proteins and rotates to propel the cell forward (1). The filament of the bacterial flagellum is composed of one or more flagellin proteins, a microbe-associated molecular pattern (MAMP) which is recognized by the host Toll-like receptor 5 (TLR5) (2) and which, via activation of the nuclear factor kappa B (NF-κB) signaling pathway, engages defense responses both systemically and at epithelial surfaces (3). Several flagellate bacterial pathogens (alphaproteobacteria and epsilonproteobacteria) have evolved flagellin proteins with sequence changes that avoid TLR5 recognition while maintaining motility (4). In an ecosystem, flagellum-mediated motility may confer a competitive advantage on motile species over nonmotile species with respect to niche colonization, biofilm formation, and the secretion of virulence proteins by pathogenic bacteria (5).
Lactobacillus spp. constitute a very diverse group and the largest genus within the lactic acid bacteria (LAB). Lactobacilli are associated mainly with food production and probiotics (6). Some Lactobacillus species colonize the gastrointestinal, oral, and genital tracts of humans, making them important members of the human microbiota (7). The genus Lactobacillus has been widely researched because of its importance for health and food applications. However, to date, motility in lactobacilli is poorly characterized. The motility of flagellate species has not attracted much scientific consideration, leading to the continued perception that the Lactobacillus genus is nonmotile (8–10). To date, at least 13 motile species have been officially recognized in the genus Lactobacillus (5, 11), and all of them belong to the Lactobacillus salivarius clade (Table 1) (12). The motility of L. ruminis has been particularly well studied previously in strain ATCC 27782 at both the phenotypic and genomic levels (5). In this strain, all the genes required to produce a fully functional flagellar apparatus have been identified and have been shown to be organized in a single operon (5, 13).
TABLE 1.
Origin and phylogeny of motile Lactobacillus species described to date
| Species | Strain | Source | Type strain | Cladea | Reference(s) for motility |
|---|---|---|---|---|---|
| L. acidipiscis | KCTC 13900 | Halloumi cheese made from ovine milk | Yesb | L. salivarius | This study |
| L. agilis | DSM 20509T | Municipal sewage | Yes | L. salivarius | 5, 48 |
| L. aquaticus | DSM 21051T | Korean freshwater pond | Yes | L. salivarius | 5, 49 |
| L. cacaonum | DSM 21116T | Cocoa bean heap fermentation | Yes | L. salivarius | This study |
| L. capillatus | DSM 19910T | Fermented stinky tofu brine | Yes | L. salivarius | 5, 50 |
| L. curvatus | NRIC 0822 | Kabura-zushi | No | L. sakei | This study |
| L. ghanensis | DSM 18630T | Cocoa fermentation | Yes | L. salivarius | 5, 51 |
| L. hordei | DSM 19519T | Malted barley | Yes | L. salivarius | This study |
| L. mali | DSM 20444T | Apple juice from cider press | Yes | L. salivarius | 5, 52 |
| L. nagelii | DSM 13675T | Commercial red wine | Yes | L. salivarius | 5, 53 |
| L. oeni | DSM 19972T | Bobal grape wine | Yes | L. salivarius | 5, 54 |
| L. ruminis | ATCC 27782 | Bovine rumen | No | L. salivarius | 5, 55 |
| L. satsumensis | DSM 16230T | Mashes of shochu | Yes | L. salivarius | 5, 56 |
| L. sicerae | CECT 8227T | Ropy natural cider | Yes | L. salivarius | 11 |
| L. sucicola | DSM 21376T | Sap of oak tree | Yes | L. salivarius | 5, 57 |
| L. uvarum | DSM 19971T | Bobal grape musts | Yes | L. salivarius | 5, 58 |
| L. vini | DSM 20605T | Grape must, fermenting at high temp | Yes | L. salivarius | 5, 59 |
L. curvatus is one of the LAB most commonly associated with fermented meat goods, vacuum-packaged refrigerated meat, and, to a lesser extent, ready-to-eat meat, fish, and poultry products (14). L. curvatus is a member of the L. sakei clade (based on 16S rRNA gene phylogeny) and is phylogenetically related to the species L. sakei, L. fuchuensis, and L. graminis (12), all of which are associated with meat environments. To date, the only genome sequence available for L. curvatus is that of strain CRL705 (14), isolated from an Argentinean artisanal fermented sausage and well known for bacteriocin production (15).
In this study, we provide strong evidence of motility in a Lactobacillus species outside the L. salivarius clade, L. curvatus (strain NRIC 0822). We assessed this motility behavior under different environmental conditions and confirmed the presence of motility genes organized in a single operon. We also identified L. acidipiscis KCTC 13900 as a motile strain, which had not been shown before, and we identified two other Lactobacillus species (L. cacaonum and L. hordei), also belonging to the L. salivarius clade, as potentially motile. The gaps in 16 motility loci that were present on different contigs in the draft genome assemblies of Lactobacillus spp. were closed by PCR and sequencing to allow a global comparison of the motility loci of this genus. We also analyzed the phylogeny of Lactobacillus motility proteins and found it to be congruent with the 16S rRNA gene-based phylogeny of the species in the L. salivarius clade.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and motility evaluation.
Lactobacillus strains (Table 1) were routinely cultured in MRS medium at 37°C or 30°C either anaerobically, aerobically, or under 5% CO2, depending on the strain.
Strain NRIC 0822, isolated in Japan from kabura-zushi, a fermented archetype of sushi (16), was identified as L. curvatus by Gram staining, catalase testing, CO2 production from glucose, lactic acid isomer production, and carbohydrate fermentation, as well as by sequencing of the 16S rRNA gene. This identification was confirmed again after genome sequencing, (i) by BLAST analysis of the nucleotide sequences of the pheS, rpoA, tuf, atpA, and hsp60 genes against the NCBI nonredundant (nr) database and (ii) by whole draft genome comparisons using two DNA-DNA hybridization (DDH) prediction models: average nucleotide identity (ANI [17]) and genome-to-genome distance calculator (GGDC [18]) values.
Standard motility agar assays were used to evaluate the influence of atmospheric conditions (aerobic, anaerobic, 5% CO2), temperature (10°C, 15°C, 20°C, 25°C, 30°C, 37°C, and 42°C), and carbohydrates (glucose, galactose, fructose, mannose, and maltose, which had been identified as the carbohydrates fermented by strain NRIC 0822 during its identification) on the motility of L. curvatus NRIC 0822. A carbohydrate-free MRS medium (cfMRS), in which the carbohydrate source (glucose) and meat extract were omitted as described previously (19), was used as a basal growth medium. Motility under the different atmospheric conditions was assessed in cfMRS broth supplemented with 0.5% glucose and 0.3% to 0.5% (wt/vol) agar (soft agar) at 30°C. Six-well plates (Corning Incorporated) containing 8 ml of soft agar were inoculated (by stabbing) with 5 μl of an overnight culture of the strain to be tested. The plates were allowed to dry for 3 min and were incubated for a maximum of 48 h. Test tubes containing 17 ml of soft agar were inoculated with a needle previously plunged into an overnight culture. The tubes were allowed to dry for 3 min and were incubated as described above. The same procedure was used to test the motility of L. curvatus NRIC 0822 (i) at different temperatures in cfMRS containing 0.5% glucose under anaerobic conditions and (ii) in the presence of different carbohydrates in cfMRS containing 0.5% of the test carbohydrate at 30°C under 5% CO2.
For testing the motility of L. curvatus NRIC 0822, three control strains were always included. L. ruminis ATCC 27782 was used as a positive control, since its motility and genome sequence have been described well previously (5, 13). The previously sequenced L. acidipiscis strain KCTC 13900 (20) was used because its genome harbors the top BLAST hit for all motility genes in L. curvatus NRIC 0822. L. curvatus DSM 20019T was used as a negative control for motility, because its genome does not contain motility genes.
All the experiments were carried out in biological triplicate. Images of each of the 6-well plates and test tubes were taken using the GeneGenius bioimaging system (Syngene) and an Xperia Z camera (Sony), respectively.
Culture motility was also evaluated by phase-contrast microscopy as described previously (5). Briefly, glass capillary tubes were first filled with an aliquot of the bacterial culture and then placed on a heated microscope stage that was maintained at 30 to 37°C (according to the strain growth conditions) for the evaluation of culture motility. When every bacterium in a field of view of the microscope was either running or tumbling and moving quickly, the culture was considered “motile.” If no motile bacteria were observed in the fields of view examined, the culture was considered “nonmotile.”
Chemotaxis was also assessed by a chemical-in-plug assay. In this assay, a solid (1.5%) agar plug (diameter, about 7 mm) containing the chemical to be tested was placed in the center of a petri dish. A soft (0.2%) MRS agar medium without yeast extract (prewarmed at 48°C) was inoculated (0.05%) with an overnight culture, and 20 ml was poured around the agar plug. The plates were allowed to dry for 10 min and were incubated for a maximum of 48 h at 30°C under 5% CO2. The presence of an outer chemotactic ring indicated positive results for chemotaxis.
Microscopy.
For transmission electron microscopy (TEM), L. curvatus NRIC 0822 cells were added to an equal volume of 5% glutaraldehyde, and the container was gently inverted to mix the contents. Cells were allowed to sediment overnight at 4°C. The supernatant was removed, and each cell pellet was resuspended in 2.5% glutaraldehyde and was flicked briefly to mix. Five microliters of the cell suspension was added to Formvar carbon-coated 200-mesh copper grids and was incubated at room temperature for 5 min. Liquid was wicked away by touching the side of the grid with a sheet of clean filter paper. Grids were washed by quick dipping in filtered sterile water and were dried by wicking. Five microliters of uranyl acetate (0.5%) was added to the grids for 5 min. Liquid was wicked away, and the grids were allowed to dry for 5 min. Grids were imaged by transmission electron microscopy under a vacuum.
For light microscopy, L. curvatus NRIC 0822 cells were prestained with a Flagella Reagent Dropper (Becton Dickinson Microbiology Systems) according to the manufacturer's instructions.
Genome sequencing and annotation of L. curvatus NRIC 0822.
Paired-end reads were obtained using the Illumina HiSeq 2000 reversible dye terminator system (Macrogen, Seoul, South Korea), with read lengths of 101 bp. Sequencing generated 29,761,714 reads (3,005,933,114 bp). De novo genome assembly of the Illumina sequences was performed using Velvet (version 1.2.07 [21]), producing an assembly of 144 contigs These 144 assembled contigs represent 1,417-fold genome coverage based on an estimated genome size of 1.94 Mb. The N50 score for the assembly was 25,925 bp. Automated gene calling was performed using Glimmer, version 3 (22). tRNA genes were identified using tRNAscan (version 1.23 [23]). All predicted proteins were searched (BLASTP) against the NCBI nonredundant (nr) protein database.
DNA extraction and gap closure.
After 24 to 48 h of culture, the genomic DNA of Lactobacillus species positive for motility genes was isolated by using the Qiagen DNeasy blood and tissue kit according to the manufacturer's instructions for Gram-positive bacteria. The genomic DNA was quantified using a spectrophotometer (NanoDrop 2000; Thermo Scientific) and was checked for integrity on a 0.8% agarose gel.
A PCR-based strategy was adopted for gap closure in the motility operons. Contig-contig gaps were closed using primers designed at the beginnings (reverse strand) and ends (forward strand) of contigs, and these regions were amplified using Phusion Hot Start II polymerase (Thermo Scientific). Contigs were ordered and oriented by PCR. A two-step walking PCR method (24) was also used to amplify the upstream or downstream contig regions when they were unknown, in order to check for the presence or absence of mobile genetic elements flanking the motility operon. Primers were designed using the Primer3Plus Web tool. Purified PCR products for both closing gaps and walking PCRs were sequenced by GATC Biotech (Cologne, Germany). Once the gaps were closed, genes in the motility operons were predicted as described above.
In silico confirmation and identification of motile Lactobacillus species.
Protein sequence motifs for each of five motility components were identified using the available draft genomes of L. curvatus NRIC 0822, L. ruminis ATCC 27782, L. acidipiscis KCTC 13900, L. mali DSM 20444T, and L. vini DSM 20605T. The motifs identified corresponded to different regions of the motility operons and encompassed the flagellin FliC, the flagellar hook-basal body complex protein FliE, the flagellar motor switch protein FliG, the flagellar biosynthesis protein FlhA, and the chemotaxis protein MotA (see Table S1 in the supplemental material). A TBLASTN search for these 5 motifs was performed against all publicly available Lactobacillus sp. complete and draft genomes, as well as against the Lactobacillus sp. draft genomes that were recently sequenced as part of the Lactobacillus genome-sequencing initiative (Z. Sun, H. M. B. Harris, A. McCann, X. Yang, S. Argimon, W. Zhang, C. Guo, I. B. Jeffery, J. C. Cooney, T. F. Kagawa, W. Liu, Y. Song, E. Salvetti, A. Wrobel, P. Rasinkangas, J. Parkhill, M. C. Rea, O. O'Sullivan, J. Ritari, F. P. Douillard, R. P. Ross, R. Yang, A. Briner, G. Felis, W. M. de Vos, R. Barrangou, T. R. Klaenhammer, P. W. Caufield, Y. Cui, H. Zhang, and P. W. O'Toole, submitted for publication), for a total of 349 genomes. The L. sicerae species, described as motile (11), was not included in this study, because this new species was described too recently, and the partial draft genome did not allow us to identify all the motility genes.
Alignments and phylogenetic analyses.
The sequence of the 16S rRNA gene of the type strain of each Lactobacillus species was downloaded from the NCBI GenBank database. A maximum likelihood (ML) phylogeny was constructed in MEGA6 (25) from a MUSCLE alignment with the appropriate substitution model for the ML option selected (26) and 1,000 bootstrap replications. The phylogenetic tree shown in Fig. 5 was made online using the Interactive Tree Of Life (27).
FIG 5.
16S rRNA gene phylogenetic tree and motility of lactobacilli. The tree, based on 16S rRNA gene sequence analysis, depicts the phylogenetic relationships among species of the genus Lactobacillus. The 16S rRNA gene sequences were aligned with MUSCLE, and the tree was inferred with MEGA6 software (25). The evolutionary history was inferred by using the maximum likelihood method based on the general time-reversible model. Statistical support was estimated with bootstraps (1,000 replicates). The main clades are indicated according to the last taxonomic update of the lactobacilli (12). Motile Lactobacillus species (Table 1) described previously are shown in black, and species newly recognized as motile in this study are shown in red.
Start codons were manually corrected by using the average length of the sequences for each protein to indicate the most likely start site and by looking for the presence of a ribosome binding site (RBS) motif upstream. This was a necessary step, because gene prediction software is often inaccurate at predicting start sites for genes. Motility protein sequences were then concatenated. Alignment and phylogenetic tree construction were carried out as described above.
Protein identity and similarity scores were obtained from the LALIGN Web tool (28), using alignment default values.
Nucleotide sequence accession numbers.
The BioProject identification code (ID) for this study is PRJNA265031. This whole-genome shotgun project for L. curvatus NRIC 0822 has been deposited at DDBJ/EMBL/GenBank under accession number JTJV00000000. The version described in this paper is version JTJV01000000. The GenBank accession number for the full-length 16S rRNA gene of L. curvatus NRIC 0822 is KM676454. The GenBank accession numbers for the 16 individual motility operons are KM886858 to KM886873 (KM886858 for L. acidipiscis KCTC 13900, KM886859 for L. agilis DSM 20509T, KM886860 for L. aquaticus DSM 21051T, KM886861 for L. cacaonum DSM 21116T, KM886862 for L. capillatus DSM 19910T, KM886863 for L. curvatus NRIC 0822, KM886864 for L. ghanensis DSM 18630T, KM886865 for L. hordei DSM 19519T, KM886866 for L. mali DSM 20444T, KM886867 for L. nagelii ATCC 700692T, KM886868 for L. oeni DSM 19972T, KM886869 for L. ruminis ATCC 27782, KM886870 for L. satsumensis DSM 16230T, KM886871 for L. sucicola DSM 21376T, KM886872 for L. uvarum DSM 19971T, and KM886873 for L. vini DSM 20605T).
RESULTS
L. curvatus NRIC 0822 employs flagellum-mediated motility.
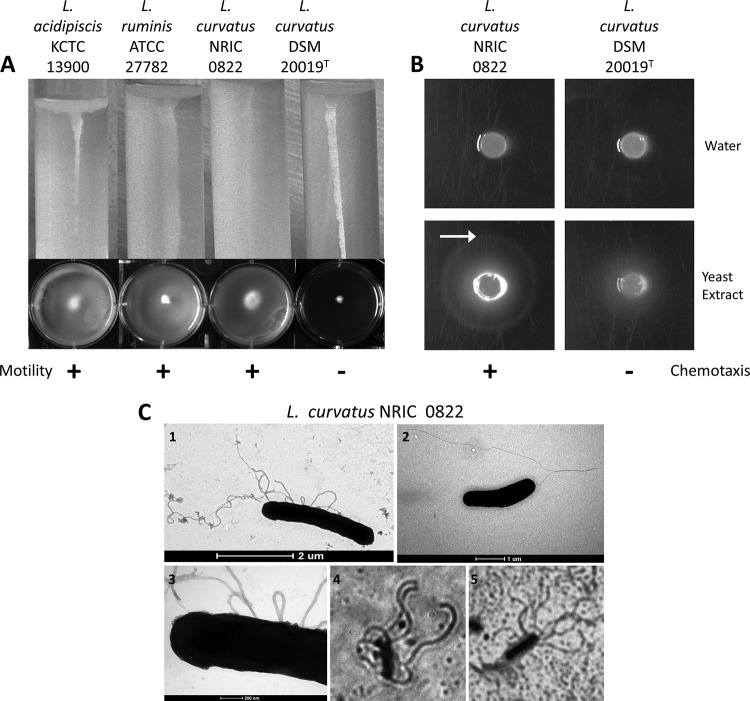
Strain NRIC 0822 was isolated from kabura-zushi, a Japanese fermented food made from rice, turnip, and fish. This strain was observed to be motile in our routine lab tests and was identified as L. curvatus by 16S rRNA gene sequencing. To investigate the motility of L. curvatus NRIC 0822, phase-contrast microscopy and standard motility agar assays were conducted. Under microscopy, this strain displayed moderate motility in the exponential phase in MRS broth, demonstrating swimming and tumbling motilities. L. curvatus NRIC 0822 was also observed to be motile in both soft agar assays used (test tubes and 6-well plates) (Fig. 1A). The cultures of L. curvatus NRIC 0822 were nonmotile in the stationary phase. The two positive controls, L. ruminis ATCC 27782 and L. acidipiscis KCTC 13900, were also motile, whereas L. curvatus DSM 20019T, the type strain of the species, was nonmotile. Preliminary investigation demonstrated positive chemotaxis for L. curvatus NRIC 0822 but not for the nonmotile L. curvatus strain DSM 20019T (Fig. 1B). As expected, enhanced growth was observed for both motile and nonmotile L. curvatus strains around the plug containing yeast extract. However, a sharp ring of concentrated cells (a typical chemotactic ring) specific for the medium components in the plug and plate was present only with L. curvatus NRIC 0822, and not with water. Thus, this strain sensed and responded positively chemotactically to the chemical stimulus. To further investigate the flagellate phenotype of L. curvatus NRIC 0822, TEM and flagellar staining were performed, revealing the presence of flagella with a peritrichous organization (Fig. 1C). The peritrichous flagella are more visible with the light microscope after flagellar staining than on the TEM images, where the flagella look sparse. This is probably due to fixation during the preparation of cells for TEM, which altered the fragile flagellar filaments. In addition, the effects of different environmental conditions (atmospheric conditions, temperature, and carbohydrate sources) on the motility of L. curvatus NRIC 0822 were investigated, and the results were consistent throughout the triplicate experiments. L. curvatus NRIC 0822 was motile under 5% CO2, as well as under anaerobic and aerobic conditions, in both the 6-well plates and the test tubes (Table 2). Motility was also observed at temperatures ranging from 15 to 37°C (Table 2). L. curvatus NRIC 0822 grew at 10°C and 42°C, but no motility was observed. L. curvatus NRIC 0822 was also motile when grown with all five carbohydrates tested in both test tubes and 6-well plates (Table 2).
FIG 1.
Motility and flagella of L. curvatus NRIC 0822. (A) Soft-agar assays in tubes (top) and 6-well plates (bottom) showing the motility of L. curvatus NRIC 0822. Three controls were included: L. ruminis ATCC 27782 (positive control for motility), L. acidipiscis KCTC 13900 (top BLAST hit for all the motility genes in L. curvatus NRIC 0822), and L. curvatus DSM 20019T (negative control for motility). (B) Chemotaxis phenotype of L. curvatus NRIC 0822 for yeast extract. Chemotaxis was assessed by a chemical-in-plug assay, with sterile water or yeast extract (2.5%) as the test compound. The arrow indicates the location of the outer chemotactic ring, indicating a positive result for chemotaxis. (C) Images of an L. curvatus NRIC 0822 whole cell and flagella. (Panels 1 to 3) Transmission electron micrographs (magnifications, ×8,200, ×11,500, and ×60,000, respectively); (panels 4 and 5) light microscopy pictures after flagellum staining (magnification, ×1,000).
TABLE 2.
Effects of growth conditions on the motility of Lactobacillus curvatus NRIC 0822
| Growth conditions | Motilitya |
|---|---|
| Atmospheric conditionsb | |
| Aerobic | + |
| 5% CO2 | + |
| Anaerobic | + |
| Temp (°C)c | |
| 10 | − |
| 15 | + |
| 20 | + |
| 25 | + |
| 30 | + |
| 37 | + |
| 42 | − |
| Carbohydrated | |
| Glu | + |
| Gal | + |
| Fru | + |
| Man | + |
| Mal | + |
+, motile; −, nonmotile.
With glucose at 30°C.
With glucose under anaerobic conditions.
At 30°C under 5% CO2. Glu, glucose; Gal, galactose; Fru, fructose; Man, mannose; Mal, maltose.
Genome sequencing of L. curvatus NRIC 0822 confirms the presence of motility genes organized in a single operon.
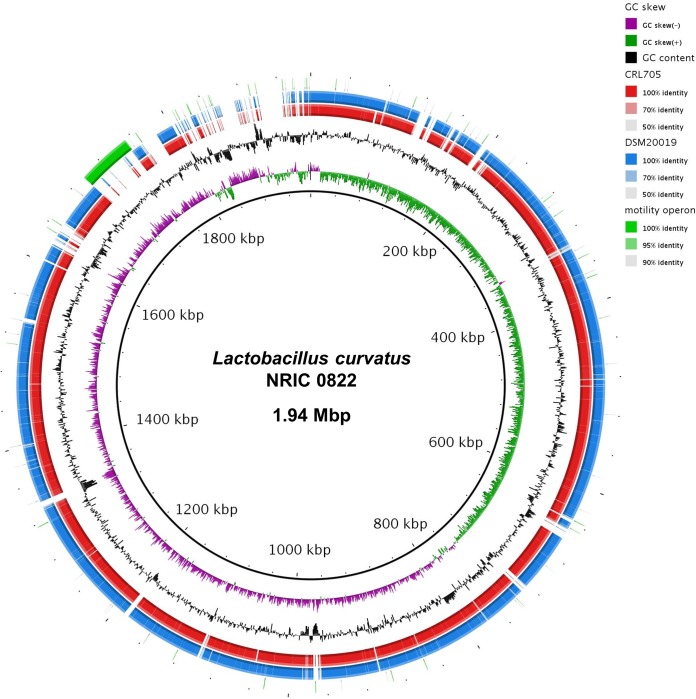
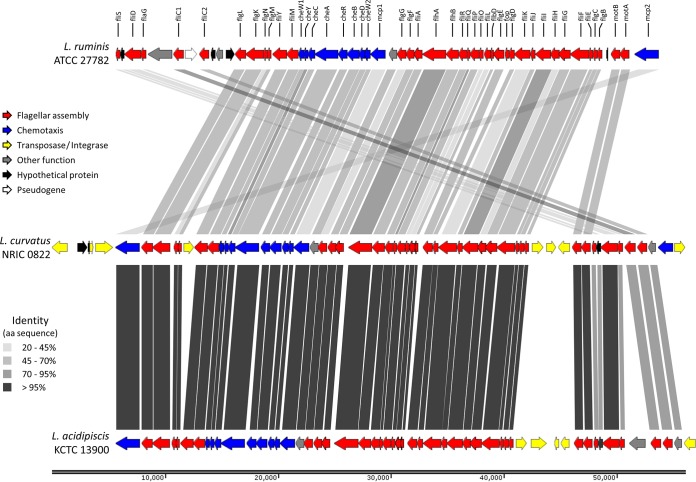
The draft genome of L. curvatus NRIC 0822 comprised 144 contigs, for a total assembly size of 1.94 Mbp, an N50 value of 25,925 bp, and 1,417-fold coverage (see Table S2 in the supplemental material). The draft genome of L. curvatus NRIC 0822 includes 1,944,912 bases (GC content, 41.70%). It comprises 2,060 predicted genes or coding DNA sequences (CDS), and 56 predicted tRNAs, representing all 20 amino acids, were identified in the genome. The genome of L. ruminis ATCC 27782 had been sequenced in our laboratory previously (13) and was used to direct our study of L. curvatus NRIC 0822 and the organization of its motility genes. Annotation of the L. curvatus NRIC 0822 draft genome identified motility genes spread over 8 contigs in the draft assembly. The sequences of these contigs were joined by PCR and sequencing. Walking PCR was also used on the flanking regions in order to confirm the limit of motility-related (or mobile genetic element-related) sequences. Among the genome sequences of L. curvatus available so far, the presence of motility genes is a unique trait of strain NRIC 0822 (Fig. 2). The genome of NRIC 0822 is otherwise nearly identical to the two other L. curvatus draft genomes (Fig. 2; see also Tables S2 and S3 in the supplemental material). The motility genes in L. curvatus NRIC 0822 are organized in a single operon of 49.3 kb, as in L. ruminis ATCC 27782 (Fig. 3). This motility operon is flanked by transposases, which bring the motility locus to 56 kb when included. Among the 46 motility proteins (involved in flagellum assembly, export, and chemotaxis) of L. curvatus NRIC 0822, 45 have a top BLASTP hit in the predicted proteins of L. acidipiscis KCTC 13900, and only the MCP3 protein of L. curvatus NRIC 0822 has no homolog (but this predicted gene seems to have been disrupted by an integrase [Fig. 3]). The sequence of the motility operon of L. acidipiscis KCTC 13900 (4 contigs in the publicly available draft genome in the NCBI database [GenBank Assembly ID GCA_000260635.1]) was also closed by PCR in order to allow the overall comparison of motility operons between strains. The organization of the L. curvatus NRIC 0822 motility operon was compared to that of the well-studied L. ruminis ATCC 27782 motility operon and its closest relative based on BLAST hits, L. acidipiscis KCTC 13900 (Fig. 3; see also Table S4 in the supplemental material). The organization of the L. curvatus NRIC 0822 motility operon is quite similar to that of L. ruminis ATCC 27782, with a major central block showing the same gene positions, whereas the beginning and end of the operon are inverted between the two strains (Fig. 3). The organization and gene content of the L. curvatus NRIC 0822 and L. acidipiscis KCTC 13900 motility operons are very similar (98.8% identity between the concatenated proteins). Over the individual proteins, similarity ranges from 96% to 100% and identity from 93% to 100% (see Table S4). The GC content was also more similar between the motility operons of these two strains (0.10% difference) than over their whole genomes (>2% difference), supporting the idea of a recent horizontal gene transfer. The L. ruminis ATCC 27782 and L. curvatus NRIC 0822 motility proteins have an average similarity of 83% (minimum, 59%; maximum, 98%) and an average identity of 54% (minimum, 23%; maximum, 87%). With regard to the gene composition of the motility operons, the LRC_16020 gene, encoding a flagellar operon protein (FOP), is present only in L. ruminis ATCC 27782. The L. curvatus NRIC 0822 genome harbors a gene encoding an additional methyl-accepting chemotaxis protein (MCP), MCP3, at the end of its motility operon (Fig. 3).
FIG 2.
BLAST Ring Image Generator representation of three L. curvatus draft genomes. The draft reference genome of L. curvatus NRIC 0822 is compared to the draft genomes of the nonmotile strains L. curvatus DSM 20019T (blue) and CRL 705 (red) by using the BLAST Ring Image Generator (46). The reference genome is an ordered set of contigs, based on an alignment with the closest complete genome, L. sakei 23K (BioProject accession no. PRJNA13435), by use of Mauve (47). The innermost rings show the GC skew (purple/green) and GC content (black). The red and blue rings show BLAST comparisons of the other two L. curvatus draft genomes against the L. curvatus NRIC 0822 draft genome assembly. The outermost arc, shown in green, highlights the motility operon of L. curvatus NRIC 0822.
FIG 3.
Comparison of motility locus organization in L. curvatus NRIC 0822, L. ruminis ATCC 27782, and L. acidipiscis KCTC 13900. Motility operon representations of L. ruminis ATCC 27782, L. curvatus NRIC 0822, and L. acidipiscis KCTC 13900 were built in SnapGene Viewer, version 2.4.3. Identity scores between protein sequences were obtained from the LALIGN Web tool by using alignment default values (see Table S4 in the supplemental material for percentages of identity).
Identification of motility genes in L. salivarius clade genomes.
Motility is described solely for members of the L. salivarius clade of lactobacilli. We used 5 motility protein sequence motifs (ranging from 63 to 325 amino acids) to search for homologs in the draft genomes of additional species in this clade whose genomes were recently sequenced as part of the Lactobacillus genome-sequencing initiative (Sun et al., submitted). All 12 species previously described as motile in the L. salivarius clade (Table 1) returned significant hits, based on TBLASTN searches, with all 5 motifs. In addition to the genomes of the 4 strains belonging to the L. salivarius clade used to design the motifs (i.e., L. ruminis, L. mali, L. vini, and L. acidipiscis strains; the latter is newly described as motile in this study), the motility motif search confirmed the presence of genes involved in motility in L. agilis DSM 20509T, L. aquaticus DSM 21051T, L. capillatus DSM 19910T, L. ghanensis DSM 18630T, L. nagelii DSM 13675T, L. oeni DSM 19972T, L. satsumensis DSM 16230T, L. sucicola DSM 21376T, and L. uvarum DSM 19971T. Two additional strains, L. cacaonum DSM 21116T and L. hordei DSM 19519T, belonging to species that are also part of the L. salivarius clade, were identified as likely to be motile, because BLAST analysis using motility protein search motifs returned significant hits. The sequences of motility contigs in all of the draft genomes were joined by PCR and sequencing to allow a global comparison of the 16 motility operons.
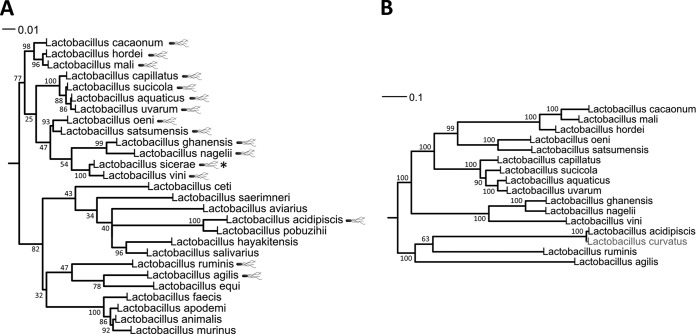
Concordant phylogeny of motility proteins in the L. salivarius clade.
Phylogenetic analysis was performed on the motility operons of the 16 strains listed in Table 1 (excluding L. sicerae), which included a total of 749 predicted genes. Three genes presented a frameshift due to a stop codon, and the corresponding proteins were corrected manually before running the alignment in order to avoid the bias that truncated proteins would have introduced into the phylogenetic analyses. These three genes were fliI in L. vini DSM 20605T, fliM in L. hordei DSM 19519T, and fliP in L. agilis DSM 20509T. This phylogenetic analysis of the 16 motility operons demonstrated strong concordance with the 16S rRNA gene-based phylogenetic tree (Fig. 4). The L. salivarius clade can be divided into two subclades, represented by L. mali and L. ruminis (the latter subclade includes L. salivarius). All 13 species in the L. mali subclade are motile, whereas only 3 of the 15 species in the L. ruminis subclade are motile (Fig. 4A). The ML tree based on the concatenated motility proteins is in accordance with this division into subclades and also confirmed the strong relationship between the L. curvatus NRIC 0822 and L. acidipiscis KCTC 13900 motility operons (Fig. 4B). Moreover, the clustering of the motile Lactobacillus species based on the concatenated motility proteins is concordant with the 16S rRNA phylogeny (Fig. 4). This concordance between the 16S rRNA gene and concatenated motility proteins was also observed in most of the individual motility protein trees (see Fig. S1 in the supplemental material). The global identities (see Table S5 in the supplemental material) and similarities (see Table S6 in the supplemental material) between the motility operons also demonstrated the same close relationships within the L. salivarius clade that were shown by the 16S rRNA phylogenetic analysis. All these data (see Fig. S1 and Tables S4, S5, and S6 in the supplemental material) confirmed again the close phylogenetic relationship between the motility operons of L. curvatus NRIC 0822 and L. acidipiscis KCTC 13900.
FIG 4.
Concordant phylogeny of rRNA genes and motility proteins in the L. salivarius clade. (A) 16S rRNA gene ML subtree displaying only the L. salivarius clade.  , motile Lactobacillus species; *, the motile species L. sicerae (11) was not included in this study because this new species was described too recently, and the partial draft genome did not allow us to identify all the motility proteins. (B) Phylogenetic analysis of motility proteins in lactobacilli. This phylogenetic tree is based on concatenated protein sequence analysis of the predicted motility genes and depicts the phylogenetic relationships among motile species of the genus Lactobacillus. The sequences were aligned with MUSCLE, and the trees were inferred with MEGA6 software (25). The evolutionary history was inferred by using the maximum likelihood method based on the LG model. Statistical support was estimated with bootstraps (1,000 replicates).
, motile Lactobacillus species; *, the motile species L. sicerae (11) was not included in this study because this new species was described too recently, and the partial draft genome did not allow us to identify all the motility proteins. (B) Phylogenetic analysis of motility proteins in lactobacilli. This phylogenetic tree is based on concatenated protein sequence analysis of the predicted motility genes and depicts the phylogenetic relationships among motile species of the genus Lactobacillus. The sequences were aligned with MUSCLE, and the trees were inferred with MEGA6 software (25). The evolutionary history was inferred by using the maximum likelihood method based on the LG model. Statistical support was estimated with bootstraps (1,000 replicates).
Motility in Lactobacillus spp.
We also used the 5 motility protein sequence motifs to search against the draft genomes of all the Lactobacillus species that were recently sequenced as part of the Lactobacillus genome-sequencing initiative (Sun et al., submitted). No new species returned significant hits. Thus, apart from L. curvatus NRIC 0822, motility in lactobacilli is confined to the L. salivarius clade (Fig. 5). Our study resulted in a new total of 17 motile species (15 species with a motile phenotype and motility genes and 2 potentially motile species, carrying motility genes). Examination of the motility gene composition reveals that 42 genes are present in all 16 operons (the number excludes L. sicerae, because it has been described too recently). This set of genes was searched for correspondence with the KEGG flagellar assembly and bacterial chemotaxis pathways (see Fig. S2 in the supplemental material). With regard to the flagellar assembly pathway, 27 of the 33 genes returned a positive hit (see Fig. S2A). All of the genes involved in the flagellum structure are present except, unsurprisingly, the L and P rings encoded by the flgI and flgH genes, specific to Gram-negative bacteria. These rings are also missing in Bacillus subtilis (29). The same explanation likely holds for the absence of some expression regulators (FlgA, FliT, FhlC, and FhlD) with no matches on the KEGG flagellar assembly map (see Fig. S2A). For the bacterial chemotaxis map, 13 of the 17 genes returned a positive hit; only the Aer, CheV, CheX, and CheZ proteins were not found (see Fig. S2B). Few differences in motility gene composition were observed between the 16 motility operons, and these are also in accordance with the phylogenetic analysis. Indeed, the gene encoding the flagellar protein FlaG is absent only in L. cacaonum DSM 21116T and L. mali DSM 20444T (at the top of the ML tree in Fig. 4B). The gene encoding the flagellar operon protein FOP is absent in the L. acidipisicis, L. curvatus, and L. agilis motility operons (Fig. 4B, bottom) but is present in L. ruminis and in all the species in the L. mali subclade. The gene encoding the flagellin FliC is duplicated in all the species of the L. ruminis subclade: L. acidipisicis, L. curvatus, L. ruminis, and L. agilis (Fig. 4B, bottom). Phylogenetic analysis of this flagellin protein, which is potentially important for interaction with the immune system via TLR5, showed a different organization than the 16S rRNA phylogeny (see Fig. S3A in the supplemental material). The two FliC proteins of L. ruminis ATCC 27782 and L. agilis DSM 20509T were clustered together, and the L. ruminis proteins were clustered with those of L. acidipiscis and L. curvatus, whereas L. agilis FliC was clustered with the L. vini, L. ghanensis, and L. nagelii FliC proteins. The two FliC proteins of L. acidipiscis and L. curvatus were clustered with homologs (see Fig. S3A). Almost all the amino acids important for the interaction with the TLR5 receptor are well conserved. A few substitutions were evident and involved similar amino acids (see Fig. S3B). From an evolutionary point of view, across all 16 motility operons, the CheY protein (a chemotaxis regulator transmitting a signal to a flagellar motor component) presents the highest residue identity, at 68%, and the lowest minimum evolution number, with 0.133 ± 0.020 amino acid difference per site, from averaging over all sequence pairs (see Table S7 in the supplemental material). The most divergent proteins are FliK (1.49% identity and 0.783 amino acid difference per site) and FlaG (4.43% identity and 0.593 amino acid difference per site). Two other predicted proteins were present in all 16 operons: the cell division ATP-binding protein FtsE and a hypothetical protein that might be involved in motility. The sizes of the motility operons range from 39 kb for L. ghanensis DSM 18630T and L. nagelii DSM 13675T to 70 kb for L. capillatus DSM 19910T. Mobile genetic elements were found in 10 of the 16 motility operons, either inside the operon or flanking it. Some of the lactobacillus motility operons have a very simple organization, harboring only motility genes, but other motility loci are interrupted by genes with no known role in motility.
DISCUSSION
The property of motility is biologically important for bacteria and may potentially confer competitive advantages, such as nutrient acquisition and niche colonization. This would justify the established metabolic costs associated with the assembly and energization of flagella (5). The number of Lactobacillus species that have been reported to be motile makes up only a small proportion of the 179 species that currently belong to the genus Lactobacillus, but that number has been increasing in recent years and merits further investigation. From an evolutionary perspective, the almost complete restriction of flagellate species to the L. salivarius clade is also noteworthy (5). To date, 17 Lactobacillus species have been historically described as motile. Origins of isolation differ greatly, from fermented food products (cheese, cider, wine, cocoa) to environmental sources (sap of oak tree, sewage, and freshwater pond) and animal sources (bovine rumen) (Table 1). This great diversity of sources for motile Lactobacillus species is indicative of the importance of flagellum-meditated motility and the advantages that likely accompany this trait, such as niche colonization or biofilm formation. Interestingly, the species of the L. salivarius clade isolated from vertebrates are not motile (except for L. ruminis), and the species isolated from the environment are motile (except for L. pobuzihii) (see Fig. S4 in the supplemental material). We hypothesize that motility genes were selected against while the lactobacillus was in contact with a host. This hypothesis seems to agree with the study of Cullender et al. (30) showing that the development of flagellin-specific adaptive immune responses can downregulate or select against the production of flagella by the gut microbiome.
This study presents unequivocal evidence for the existence of a motile strain/species outside the L. salivarius clade, which, until now, comprised all known motile species. These findings were supported by both phenotypic and genomic data. L. curvatus NRIC 0822 was nonmotile in the stationary phase, which is in accordance with a previous study on L. ruminis ATCC 27782 and suggests that nutrient depletion may influence the motility phenotype (5). A previous study also reported that the chemotaxis/motility operon of L. ruminis L5 was upregulated in the “late” growth phase (optical density at 600 nm [OD600], ∼1.0) in comparison to the very “early” growth phase (OD600, ∼0.1) (31), so the growth phase must be considered when one is deciding on motility phenotypes. Likewise, we noted three genes (in motile Lactobacillus species) that harbored frameshifts that should abolish motility. Selection against motility in laboratory culture can lead to such mutations, as in the case of the fliP mutation in Helicobacter pylori strain 26695 (32). Other studies have described motile species of Lactobacillus, but often these studies are quite old, the strains are not available for testing, and the species-level identification of the isolates or strains in question was probably not robust (33). For example, Torriani et al. reported that some strains of L. curvatus are motile but lose their motility upon subculture. This motile phenotype may be unreliable because the conditions for testing were not reported, and there was no molecular investigation of motility genes (34). Another recent study from Cullender et al. described the motility of L. brevis DSM 20054T and L. sakei NRRL B-1917, but these strains did not stimulate TLR5, and the Western blot experiment targeting the flagellin proteins did not identify a reacting protein (L. brevis) or displayed a protein with a molecular mass too high (≈250 kDa for L. sakei) for it to be a flagellin protein (the mean molecular mass of the predicted lactobacillus flagellin proteins in this study is 33.01 ± 3.72 kDa) (30). In addition, the genome of L. brevis DSM 20054T is available online (and was also sequenced in the Lactobacillus genome-sequencing initiative), and none of the motility motifs returned significant hits in this genome during our analysis. More recently, L. koreensis DCY50T was described as motile (35), but the motility agar used contained only 0.15% agar (very soft, compared to the usual 0.3 to 0.5% agar for motility tests), and the TEM or atomic force microscopy (AFM) micrographs failed to show any flagella, leading us to question the motility of this strain. L. koreensis strain DCY50T was retested in our study and was not shown to be motile, and none of the motility motifs returned significant sequence matches (data not shown). In the future, when a new Lactobacillus sp. is suspected to be motile, it would be desirable to test the presence of motility genes by PCR to confirm the observed motile phenotype.
TEM and light microscopy images of the motile L. curvatus NRIC 0822 cell showed the presence of peritrichous flagella, also observed in L. ruminis ATCC 27782 (5) but different from the polar flagellum observed in L. sicerae CECT 8227T (11) and L. ruminis L5 (31). The organization and genes of the L. curvatus NRIC 0822 and L. acidipiscis KCTC 13900 motility operons are very similar (98.8% identity between the concatenated proteins), suggesting that the L. curvatus NRIC 0822 motility genes were horizontally acquired recently. This is strongly supported by the presence of mobile genetic elements inside and flanking the L. curvatus NRIC 0822 motility operon. This proposed horizontal transfer would therefore have occurred between members of two distinct Lactobacillus clades: L. acidipiscis in the L. salivarius clade and L. curvatus in the L. sakei clade. In addition, the motility of L. acidipiscis was newly described in this study. L. acidipiscis KCTC 13900 is motile at 30°C but not at 37°C (data not shown), which can explain the nonmotile phenotype recorded for this strain to date, since all the culture for this species was previously performed at 37°C (20, 36). In kabura-zushi, L. acidipiscis was not detected (37), whereas this species has been found together with L. curvatus in a similar fermented food, narezushi, produced in the same area of Japan (38).
The motility operon structure in Lactobacillus spp. seems to be relatively conserved, with identical gene blocks. This selection presumably results in efficient flagellum assembly, allowing the flagellar substructures serving as checkpoints to coordinate flagellar gene expression with assembly (39). Some of the Lactobacillus motility operons have a very simple organization, but other motility loci are interrupted by genes with no known role in motility, leading to the size variability observed between the lactobacillus motility operons (from 39 to 70 kb). For example, the inclusion of genes for rhamnose utilization in the motility loci of L. mali DSM 20444T, L. cacaonum DSM 21116T, and L. sucicola DSM 21376T, and of glycosyltransferase genes in the motility loci of L. ruminis ATCC 22782, L. agilis DSM 20509T, and L. vini DSM 20605T, may indicate that the flagellin proteins of these species are modified by glycosylation, since rhamnose has been found as a posttranslational modification of flagellin in other bacteria (40).
In the future, it will be necessary to investigate the expression level of the motility operon in vivo in the gut and to determine whether the flagellum is assembled in this environment. A previous study suggested that some control occurs in the gut to render commensal gut bacteria generally nonmotile, explaining the low levels of flagellin protein in a healthy gut despite the capacity of the gut microbiome to produce flagella (41, 42). This control might be due to the ability of flagellin-specific IgA to inhibit bacterial motility and downregulate flagellar gene expression in vitro (30).
In the mammalian gut, the microbe-associated molecular patterns (MAMPs) of commensal microbiota are recognized by host pattern recognition receptors (PRRs), such as the TLRs. This interaction may contribute to homeostasis and protection from injury in the gut (43). The flagellin-TLR5 interaction leads to a proinflammatory response (44), also shown with L. ruminis TH14 (45) and with flagellated cells and flagellin proteins of L. ruminis ATCC 27782 (5). The relatively high conservation of the TLR5 interaction amino acid motif among all the motile Lactobacillus species suggests that all flagellin proteins in this species are able to trigger this immune response via TLR5.
This study provides global phenotypic, genetic, and phylogenetic insights into flagellum-mediated motility in lactobacilli. The characterization of the in vivo expression level and the immune response to flagellated lactobacilli may become particularly relevant in the context of probiotic, pharmaceutical, or vaccination applications.
Supplementary Material
ACKNOWLEDGMENTS
Work in the laboratory of P.W.O. was supported by Science Foundation Ireland through a Centre award to the Alimentary Pharmabiotic Centre.
TEM imaging was performed by Dimitri Scholtz at the University College Dublin Biological Imaging Facility. We also thank Ceara Clancy and Michelle O'Donnell for advice.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03594-14.
REFERENCES
- 1.Macnab RM. 2003. How bacteria assemble flagella. Annu Rev Microbiol 57:77–100. doi: 10.1146/annurev.micro.57.030502.090832. [DOI] [PubMed] [Google Scholar]
- 2.Hayashi F, Smith KD, Ozinsky A, Hawn TR, Yi EC, Goodlett DR, Eng JK, Akira S, Underhill DM, Aderem A. 2001. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 410:1099–1103. doi: 10.1038/35074106. [DOI] [PubMed] [Google Scholar]
- 3.Smith KD, Ozinsky A. 2002. Toll-like receptor-5 and the innate immune response to bacterial flagellin. Curr Top Microbiol Immunol 270:93–108. doi: 10.1007/978-3-642-59430-4_6. [DOI] [PubMed] [Google Scholar]
- 4.Andersen-Nissen E, Smith KD, Strobe KL, Barrett SLR, Cookson BT, Logan SM, Aderem A. 2005. Evasion of Toll-like receptor 5 by flagellated bacteria. Proc Natl Acad Sci U S A 102:9247–9252. doi: 10.1073/pnas.0502040102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neville BA, Forde BM, Claesson MJ, Darby T, Coghlan A, Nally K, Ross RP, O'Toole PW. 2012. Characterization of pro-inflammatory flagellin proteins produced by Lactobacillus ruminis and related motile lactobacilli. PLoS One 7:e40592. doi: 10.1371/journal.pone.0040592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Claesson MJ, van Sinderen D, O'Toole PW. 2007. The genus Lactobacillus—a genomic basis for understanding its diversity. FEMS Microbiol Lett 269:22–28. doi: 10.1111/j.1574-6968.2006.00596.x. [DOI] [PubMed] [Google Scholar]
- 7.Turroni F, Ventura M, Butto LF, Duranti S, O'Toole PW, Motherway MOC, van Sinderen D. 2014. Molecular dialogue between the human gut microbiota and the host: a Lactobacillus and Bifidobacterium perspective. Cell Mol Life Sci 71:183–203. doi: 10.1007/s00018-013-1318-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herbel SR, Vahjen W, Wieler LH, Guenther S. 2013. Timely approaches to identify probiotic species of the genus Lactobacillus. Gut Pathog 5:27. doi: 10.1186/1757-4749-5-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paco RS, Leme IL, Bottino JA, Ferreira AJP. 2003. Identification of Lactobacillus spp. from broiler litter in Brazil. Braz J Microbiol 34:236–237. doi: 10.1590/S1517-83822003000300010. [DOI] [Google Scholar]
- 10.Goyal R, Dhingra H, Bajpai P, Joshi N. 2012. Characterization of the Lactobacillus isolated from different curd samples. Afr J Biotechnol 11:14448–14452. doi: 10.5897/AJB11.310. [DOI] [Google Scholar]
- 11.Puertas AI, Arahal DR, Ibarburu I, Elizaquivel P, Aznar R, Dueñas MT. 2014. Lactobacillus sicerae sp. nov., a new lactic acid bacterium isolated from Spanish natural cider. Int J Syst Evol Microbiol 64:2949–2955. doi: 10.1099/ijs.0.059980-0. [DOI] [PubMed] [Google Scholar]
- 12.Salvetti E, Torriani S, Felis GE. 2012. The genus Lactobacillus: a taxonomic update. Probiotics Antimicrob Proteins 4(4):217–226. doi: 10.1007/s12602-012-9117-8. [DOI] [PubMed] [Google Scholar]
- 13.Forde BM, Neville BA, O'Donnell MM, Riboulet-Bisson E, Claesson MJ, Coghlan A, Ross RP, O'Toole PW. 2011. Genome sequences and comparative genomics of two Lactobacillus ruminis strains from the bovine and human intestinal tracts. Microb Cell Fact 10:S13. doi: 10.1186/1475-2859-10-S1-S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hebert EM, Saavedra L, Taranto MP, Mozzi F, Magni C, Nader MEF, de Valdez GF, Sesma F, Vignolo G, Raya RR. 2012. Genome sequence of the bacteriocin-producing Lactobacillus curvatus strain CRL705. J Bacteriol 194:538–539. doi: 10.1128/JB.06416-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Castellano P, Raya R, Vignolo G. 2003. Mode of action of lactocin 705, a two-component bacteriocin from Lactobacillus casei CRL705. Int J Food Microbiol 85:35–43. doi: 10.1016/S0168-1605(02)00479-8. [DOI] [PubMed] [Google Scholar]
- 16.Irisawa T, Tanaka N, Takano K, Okada S. 2010. Isolation and identification of lactic acid bacteria inhabited in Kaburazushi. Food Preserv Sci 36:83–87. [Google Scholar]
- 17.Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM. 2007. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol 57:81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- 18.Meier-Kolthoff JP, Auch AF, Klenk HP, Goker M. 2013. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O'Donnell MM, Forde BM, Neville B, Ross PR, O'Toole PW. 2011. Carbohydrate catabolic flexibility in the mammalian intestinal commensal Lactobacillus ruminis revealed by fermentation studies aligned to genome annotations. Microb Cell Fact 10:S12. doi: 10.1186/1475-2859-10-S1-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim D-S, Choi S-H, Kim D-W, Kim RN, Nam S-H, Kang A, Kim A, Park H-S. 2011. Genome sequence of Lactobacillus cypricasei KCTC 13900. J Bacteriol 193:5053–5054. doi: 10.1128/JB.05659-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delcher AL, Bratke KA, Powers EC, Salzberg SL. 2007. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23:673–679. doi: 10.1093/bioinformatics/btm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25:955–964. doi: 10.1093/nar/25.5.0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pilhofer M, Bauer AP, Schrallhammer M, Richter L, Ludwig W, Schleifer KH, Petroni G. 2007. Characterization of bacterial operons consisting of two tubulins and a kinesin-like gene by the novel two-step gene walking method. Nucleic Acids Res 35:e135. doi: 10.1093/nar/gkm836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hall BG. 2013. Building phylogenetic trees from molecular data with MEGA. Mol Biol Evol 30:1229–1235. doi: 10.1093/molbev/mst012. [DOI] [PubMed] [Google Scholar]
- 27.Letunic I, Bork P. 2011. Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res 39:W475–W478. doi: 10.1093/nar/gkr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McWilliam H, Li W, Uludag M, Squizzato S, Park YM, Buso N, Cowley AP, Lopez R. 2013. Analysis tool web services from the EMBL-EBI. Nucleic Acids Res 41:W597–W600. doi: 10.1093/nar/gkt376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kubori T, Okumura M, Kobayashi N, Nakamura D, Iwakura M, Aizawa S-I. 1997. Purification and characterization of the flagellar hook–basal body complex of Bacillus subtilis. Mol Microbiol 24:399–410. doi: 10.1046/j.1365-2958.1997.3341714.x. [DOI] [PubMed] [Google Scholar]
- 30.Cullender TC, Chassaing B, Janzon A, Kumar K, Muller CE, Werner JJ, Angenent LT, Bell ME, Hay AG, Peterson DA, Walter J, Vijay-Kumar M, Gewirtz AT, Ley RE. 2013. Innate and adaptive immunity interact to quench microbiome flagellar motility in the gut. Cell Host Microbe 14:571–581. doi: 10.1016/j.chom.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lawley B, Sims IM, Tannock GW. 2013. Whole-transcriptome shotgun sequencing (RNA-seq) screen reveals upregulation of cellobiose and motility operons of Lactobacillus ruminis L5 during growth on tetrasaccharides derived from barley β-glucan. Appl Environ Microbiol 79:5661–5669. doi: 10.1128/AEM.01887-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Josenhans C, Eaton KA, Thevenot T, Suerbaum S. 2000. Switching of flagellar motility in Helicobacter pylori by reversible length variation of a short homopolymeric sequence repeat in fliP, a gene encoding a basal body protein. Infect Immun 68:4598–4603. doi: 10.1128/IAI.68.8.4598-4603.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McGroarty JA. 1994. Cell-surface appendages of lactobacilli. FEMS Microbiol Lett 124:405–409. doi: 10.1111/j.1574-6968.1994.tb07316.x. [DOI] [PubMed] [Google Scholar]
- 34.Torriani S, VanReenen CA, Klein G, Reuter G, Dellaglio F, Dicks LMT. 1996. Lactobacillus curvatus subsp. curvatus subsp. nov. and Lactobacillus curvatus subsp. melibiosus subsp. nov. and Lactobacillus sake subsp. sake subsp. nov. and Lactobacillus sake subsp. carnosus subsp. nov., new subspecies of Lactobacillus curvatus Abo-Elnaga and Kandler 1965 and Lactobacillus sake Katagiri, Kitahara, and Fukami 1934 (Klein et al. 1996, emended descriptions), respectively. Int J Syst Bacteriol 46:1158–1163. [DOI] [PubMed] [Google Scholar]
- 35.Bui TPN, Kim YJ, In JG, Yang DC. 2011. Lactobacillus koreensis sp. nov., isolated from the traditional Korean food kimchi. Int J Syst Evol Microbiol 61:772–776. doi: 10.1099/ijs.0.021386-0. [DOI] [PubMed] [Google Scholar]
- 36.Naser SM, Vancanneyt M, Hoste B, Snauwaert C, Swings J. 2006. Lactobacillus cypricasei Lawson et al. 2001 is a later heterotypic synonym of Lactobacillus acidipiscis Tanasupawat et al. Int J Syst Evol Microbiol 56:1681–1683. doi: 10.1099/ijs.0.64229-0. [DOI] [PubMed] [Google Scholar]
- 37.Koyanagi T, Nakagawa A, Kiyohara M, Matsui H, Yamamoto K, Barla F, Take H, Katsuyama Y, Tsuji A, Shijimaya M, Nakamura S, Minami H, Enomoto T, Katayama T, Kumagai H. 2013. Pyrosequencing analysis of microbiota in Kaburazushi, a traditional medieval sushi in Japan. Biosci Biotechnol Biochem 77:2125–2130. doi: 10.1271/bbb.130550. [DOI] [PubMed] [Google Scholar]
- 38.Koyanagi T, Kiyohara M, Matsui H, Yamamoto K, Kondo T, Katayama T, Kumagai H. 2011. Pyrosequencing survey of the microbial diversity of ‘narezushi’, an archetype of modern Japanese sushi. Lett Appl Microbiol 53:635–640. doi: 10.1111/j.1472-765X.2011.03155.x. [DOI] [PubMed] [Google Scholar]
- 39.Anderson JK, Smith TG, Hoover TR. 2010. Sense and sensibility: flagellum-mediated gene regulation. Trends Microbiol 18:30–37. doi: 10.1016/j.tim.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Merino S, Tomas JM. 2014. Gram-negative flagella glycosylation. Int J Mol Sci 15:2840–2857. doi: 10.3390/ijms15022840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Verberkmoes NC, Russell AL, Shah M, Godzik A, Rosenquist M, Halfvarson J, Lefsrud MG, Apajalahti J, Tysk C, Hettich RL, Jansson JK. 2009. Shotgun metaproteomics of the human distal gut microbiota. ISME J 3:179–189. doi: 10.1038/ismej.2008.108. [DOI] [PubMed] [Google Scholar]
- 42.Neville BA, Sheridan PO, Harris HMB, Coughlan S, Flint HJ, Duncan SH, Jeffery IB, Claesson MJ, Ross RP, Scott KP, O'Toole PW. 2013. Pro-inflammatory flagellin proteins of prevalent motile commensal bacteria are variably abundant in the intestinal microbiome of elderly humans. PLoS One 8:e68919. doi: 10.1371/journal.pone.0068919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. 2004. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 44.Tallant T, Deb A, Kar N, Lupica J, de Veer MJ, DiDonato JA. 2004. Flagellin acting via TLR5 is the major-activator of key signaling pathways leading to NF-κB and proinflammatory gene program activation in intestinal epithelial cells. BMC Microbiol 4:33. doi: 10.1186/1471-2180-4-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taweechotipatr M, Iyer C, Spinler JK, Versalovic J, Tumwasorn S. 2009. Lactobacillus saerimneri and Lactobacillus ruminis: novel human-derived probiotic strains with immunomodulatory activities. FEMS Microbiol Lett 293:65–72. doi: 10.1111/j.1574-6968.2009.01506.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alikhan NF, Petty N, Ben Zakour N, Beatson S. 2011. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Darling ACE, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weiss N, Schillinger U, Laternser M, Kandler O. 1981. Lactobacillus sharpeae sp.nov. and Lactobacillus agilis sp.nov., two new species of homofermentative, meso-diaminopimelic acid-containing lactobacilli isolated from sewage. Zentralbl Bakteriol Mikrobiol Hyg C 2:242–253. [Google Scholar]
- 49.Mañes-Lázaro R, Song J, Pardo I, Cho JC, Ferrer S. 2009. Lactobacillus aquaticus sp. nov., isolated from a Korean freshwater pond. Int J Syst Evol Microbiol 59:2215–2218. doi: 10.1099/ijs.0.008276-0. [DOI] [PubMed] [Google Scholar]
- 50.Chao SH, Tomii Y, Sasamoto M, Fujimoto J, Tsai YC, Watanabe K. 2008. Lactobacillus capillatus sp. nov., a motile bacterium isolated from stinky tofu brine. Int J Syst Evol Microbiol 58:2555–2559. doi: 10.1099/ijs.0.65834-0. [DOI] [PubMed] [Google Scholar]
- 51.Nielsen DS, Schillinger U, Franz C, Bresciani J, Amoa-Awua W, Holzapfel WH, Jakobsen M. 2007. Lactobacillus ghanensis sp. nov., a motile lactic acid bacterium isolated from Ghanaian cocoa fermentations. Int J Syst Evol Microbiol 57:1468–1472. doi: 10.1099/ijs.0.64811-0. [DOI] [PubMed] [Google Scholar]
- 52.Carr JG, Davies PA. 1970. Homofermentative lactobacilli of ciders including Lactobacillus mali nov. spec. J Appl Bacteriol 33:768–774. doi: 10.1111/j.1365-2672.1970.tb02261.x. [DOI] [PubMed] [Google Scholar]
- 53.Edwards CG, Collins MD, Lawson PA, Rodriguez AV. 2000. Lactobacillus nagelii sp. nov., an organism isolated from a partially fermented wine. Int J Syst Evol Microbiol 50:699–702. doi: 10.1099/00207713-50-2-699. [DOI] [PubMed] [Google Scholar]
- 54.Mañes-Lázaro R, Ferrer S, Rossello-Mora R, Pardo I. 2009. Lactobacillus oeni sp. nov., from wine. Int J Syst Evol Microbiol 59:2010–2014. doi: 10.1099/ijs.0.007567-0. [DOI] [PubMed] [Google Scholar]
- 55.Sharpe ME, Latham MJ, Garvie EI, Zirngibl J, Kandler O. 1973. Two new species of Lactobacillus isolated from the bovine rumen, Lactobacillus ruminis sp.nov. and Lactobacillus vitulinus sp.nov. J Gen Microbiol 77:37–49. doi: 10.1099/00221287-77-1-37. [DOI] [PubMed] [Google Scholar]
- 56.Endo A, Okada S. 2005. Lactobacillus satsumensis sp. nov., isolated from mashes of shochu, a traditional Japanese distilled spirit made from fermented rice and other starchy materials. Int J Syst Evol Microbiol 55:83–85. doi: 10.1099/ijs.0.63248-0. [DOI] [PubMed] [Google Scholar]
- 57.Irisawa T, Okada S. 2009. Lactobacillus sucicola sp. nov., a motile lactic acid bacterium isolated from oak tree (Quercus sp.) sap. Int J Syst Evol Microbiol 59:2662–2665. doi: 10.1099/ijs.0.006478-0. [DOI] [PubMed] [Google Scholar]
- 58.Mañes-Lázaro R, Ferrer S, Rossello-Mora R, Pardo I. 2008. Lactobacillus uvarum sp. nov.—a new lactic acid bacterium isolated from Spanish Bobal grape must. Syst Appl Microbiol 31:425–433. doi: 10.1016/j.syapm.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 59.Rodas AM, Chenoll E, Macian MC, Ferrer S, Pardo I, Aznar R. 2006. Lactobacillus vini sp. nov., a wine lactic acid bacterium homofermentative for pentoses. Int J Syst Evol Microbiol 56:513–517. doi: 10.1099/ijs.0.63877-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.