Abstract
Stenotrophomonas maltophilia is a Gram-negative bacterial pathogen of increasing concern to human health. Most clinical isolates of S. maltophilia efficiently form biofilms on biotic and abiotic surfaces, making this bacterium resistant to a number of antibiotic treatments and therefore difficult to eliminate. To date, very few studies have investigated the molecular and regulatory mechanisms responsible for S. maltophilia biofilm formation. Here we constructed a random transposon insertion mutant library of S. maltophilia ATCC 13637 and screened 14,028 clones. A total of 46 nonredundant genes were identified. Mutants of these genes exhibited marked changes in biofilm formation, suggesting that multiple physiological pathways, including extracellular polysaccharide production, purine synthesis, transportation, and peptide and lipid synthesis, are involved in bacterial cell aggregation. Of these genes, 20 putatively contributed to flagellar biosynthesis, indicating a critical role for cell motility in S. maltophilia biofilm formation. Genetic and biochemical evidence demonstrated that an orphan response regulator, FsnR, activated transcription of at least two flagellum-associated operons by directly binding to their promoters. This regulatory protein plays a fundamental role in controlling flagellar assembly, cell motility, and biofilm formation. These results provide a genetic basis to systematically study biofilm formation of S. maltophilia.
INTRODUCTION
Stenotrophomonas maltophilia (formerly Pseudomonas maltophilia or Xanthomonas maltophilia) is an aerobic, Gram-negative bacterium belonging to the family Xanthomonadaceae of the Gammaproteobacteria. S. maltophilia is ubiquitous across a diverse range of ecological niches, including foods, soil, plant roots and stems, and aqueous environments. In clinical settings, S. maltophilia is an opportunistic nosocomial pathogen, predominantly colonizing the skin, the respiratory tract, urinary catheters, and breathing tubes, such as endotracheal tubes (1, 2). Infection generally results in pneumonia, bacteremia, urinary tract infection, or meningitis (3, 4). Over the past 3 decades, S. maltophilia has become an increasing threat to public health, in particular to immunosuppressed or immunocompetent patients in intensive care units (ICU) who are undergoing prolonged mechanical ventilation, tracheostomy, or broad-spectrum antibiotic therapy (5). The reported mortality rates for S. maltophilia-infected patients range from 23% to 77% (4, 6). Treatment of S. maltophilia infection has proven increasingly difficult because of the bacterium's intrinsic multidrug resistance. S. maltophilia is resistant to treatment with most aminoglycosides, quinolones, β-lactams, and β-lactamase inhibitors (7). Worryingly, research has shown that the susceptibility of bacterial isolates to the recommended drug for S. maltophilia infection, trimethoprim-sulfamethoxazole, decreased from >98% to 30 to 40% within a decade (8). Therefore, the development of more effective antibiotic agents targeting critical processes of S. maltophilia pathogenesis that are nonessential to survival and with minimal selective pressure will be a challenge faced by researchers in the future.
Biofilms are slime-enclosed bacterial aggregates affording protection against antibiotics, host immune responses, and multiple stresses (9). Most clinical strains of S. maltophilia form biofilms efficiently on various abiotic and biotic surfaces, including Teflon, plastic, glass, host tissue, and indwelling intravascular devices (10, 11). Environmental factors, including phosphate, pH, temperature, metal ions, and antibiotics, were shown to have a profound effect on S. maltophilia biofilm formation (3). To date, however, only a small number of genes or biochemical cascades associated with S. maltophilia biofilm formation have been reported, and these include the polysaccharide synthesis genes rmlA, rmlC, and xanB (12), diffusible signal factor (DSF)-mediated cell-cell communication (13), the small RNA modulator Hfq (14), and the ABC-type efflux pump MacABC (15). Hence, it is critical to undertake a systematic approach to elucidate the molecular and regulatory properties of S. maltophilia biofilm development.
In this work, we constructed a large-scale S. maltophilia transposon insertion mutant library which was used to screen for S. maltophilia mutants with altered biofilm-forming ability (16). From 14,208 clones, we identified 46 genes belonging to mutants exhibiting remarkable changes in biofilm development. Almost half of these genes (20 genes [43.5%]) were involved in bacterial flagellar or pilus biosynthesis, which suggested that cell motility plays a critical role in S. maltophilia biofilm development. We confirmed that FsnR, an orphan response regulator of the two-component signal transduction system, binds directly to the promoter regions of two gene clusters and activates their transcription. These results provide genetic information to further investigate the molecular process of S. maltophilia biofilm formation.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in the present study are described in Table 1. Escherichia coli DH5α was used as the host in molecular cloning procedures and was routinely cultured at 37°C in Luria-Bertani (LB) medium. S. maltophilia ATCC 13637, obtained from stocks of the China General Microbiological Culture Collection Center (CGMCC), was cultured at 28°C in NYG medium (5 g/liter tryptone, 3 g/liter yeast extract, 20 g/liter glycerol; pH 7.0). Competent cells of S. maltophilia were prepared by culture in 210 medium (4 g/liter yeast extract, 8 g/liter casein enzymatic hydrolysate, 5 g/liter sucrose, 3 g/liter K2HPO4, 0.3 g/liter MgSO4 · 7H2O; pH 7.0) and washed in ice-cold glycerol (10%). Antibiotics were used at the following concentrations: ampicillin, 100 μg/ml; kanamycin, 50 μg/ml; spectinomycin, 100 μg/ml; and streptomycin, 200 μg/ml. Electroporation conditions for the transformation of both S. maltophilia and E. coli were set at 18 kV cm−1, 25 μF, and 200 Ω, and experiments were conducted in a Bio-Rad Pulser XCell electroporation system (Bio-Rad, Hercules, CA).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| S. maltophilia ATCC 13637 | WT strain | CGMCC |
| E. coli strain DH5α | Host strain for molecular cloning | Lab collection |
| E. coli strain BL21(DE3) | Host strain for protein expression; Kanr | Lab collection |
| S. maltophilia SM001 | ΔfsnR; in-frame deletion mutant of fsnR | This study |
| S. maltophilia SM002 | ΔfsnR-fsnR; genetic complementary strain of SM001 mutant; Strr | This study |
| S. maltophilia SM003 | CK; SM001 containing a blank pHM1 vector; Strr | This study |
| Clones from S. maltophilia mutant library | EZ::TN transposon mutants of WT strain with biofilm deficiency; Kanr | See Table S1 in the supplemental material |
| Plasmids | ||
| pK18mobsacB | Suicide vector to create mutant by double-crossover recombination; Kanr | 20 |
| pET30a | Protein expression vector; Kanr | Novagen |
| pHM1 | Broad-host-range vector for genetic complementation; Strr | Lab collection |
| pHM2299 | pHM1::fsnR; genetically complementary vector; Strr | This study |
| pET2299 | pET30a::fsnR; vector for expression of FsnR; Kanr | This study |
Kanr, kanamycin resistant; Strr, streptomycin resistant.
Construction of insertion mutant library and genetic manipulation of bacterial strains.
The S. maltophilia transposon insertion mutant library was constructed using an EZ::TN <KAN-2> Tnp transposome kit (Epicentre) as described previously and in accordance with the manufacturer's protocol (17, 18). A total of 14,208 kanamycin-resistant transformants were selected and stored in 37 384-well plates at −80°C until use. Southern blot hybridization was used to determine the transgene copy number. In brief, total DNAs were extracted from bacterial strains, and the electrophoresed fragments were transferred to a Hybond N+ membrane and hybridized with an [α-32P]dCTP-labeled PCR probe (Prime-A-Gene; Promega). PCR primers used to amplify the kanamycin resistance sequence and used as DNA probes are listed in Table 2.
TABLE 2.
Primers used in this study
| Primer | Direction of primer | Sequence (5′–3′)a | Purpose |
|---|---|---|---|
| SP1 | GATAGATTGTCGCACCTGATTG | Specific primer 1 for TAIL-PCR | |
| SP2 | AAGACGTTTCCCGTTGAATATG | Specific primer 2 for TAIL-PCR | |
| SP3 | GCAATGTAACATCAGAGATTTTGAG | Specific primer 3 for TAIL-PCR | |
| AD1 | NTCGASTWTSGWGTT | Arbitrary primer for TAIL-PCR | |
| AD4 | TGWGNANCASAGA | Arbitrary primer for TAIL-PCR | |
| AD5 | AGWGNAGWANCAWAGG | Arbitrary primer for TAIL-PCR | |
| AD6 | CAWCGICNGAIASGAA | Arbitrary primer for TAIL-PCR | |
| AD8 | STTGNTASTNCTNTGC | Arbitrary primer for TAIL-PCR | |
| Kan | Sense | GCAATCAGGTGCGACAATC | DNA probe used in Southern blotting |
| Antisense | AAGTCAGCGTAATGCTCTGC | ||
| IFD2299 | Sense-W | GAATTCTTACTCCAGCTCGTGCTG | ΔfsnR mutant construction |
| Antisense-X | ACTAGTAGCAACAAGGAAATCGCC | ||
| Sense-Y | ACTAGTGTCCATCAGGACCACGTC | ||
| Antisense-Z | AAGCTTGTGCGAGTTCTCATCGTC | ||
| C2299 | Sense | AAGCTTGTGCGAGTTCTCATCGTC | ΔfsnR-fsnR strain construction |
| Antisense | GAATTCTTACTCCAGCTCGTGCTG | ||
| P2299 | Sense | CGCCATATGCGAGTTCTCATCGTCGAC | FsnR protein expression |
| Antisense | AAGCTTCTCCAGCTCGTGCTGATG | ||
| Pb2303 | Sense | GTGATTCTCCTGGATCC | EMSA probe for Smlt2303 |
| Antisense | CTCTTTCAGCGCTGTACCT | ||
| Pb2318 | Sense | GGCTGCTGTATCGGCGGGGT | EMSA probe for Smlt2318 |
| Antisense | GGCGCCTCCTGTCGATGG | ||
| 0706 | Sense | GTGCCGTTGCCGGTCGTAC | Semiquantitative RT-PCR analysis of Smlt0706 |
| Antisense | GGACCTGGTTGTTCGAGC | ||
| 0710 | Sense | CATCAACCGGCCGTGGCC | Semiquantitative RT-PCR analysis of Smlt0710 |
| Antisense | CTACGGTCTGCTGCAGGG | ||
| 2274 | Sense | GTGGGAGCATTCATGGTC | Semiquantitative RT-PCR analysis of Smlt2274 |
| Antisense | GAAATGAAGGAGAGCGAG | ||
| 2283 | Sense | GAGAACAGCATCAGCAGG | Semiquantitative RT-PCR analysis of Smlt2283 |
| Antisense | CTTCACCCAGTCCAAGCAC | ||
| 2290 | Sense | GACCATCACCTTGGCGAG | Semiquantitative RT-PCR analysis of Smlt2290 |
| Antisense | CTGCCGGGCACGGTGCTG | ||
| 2297 | Sense | GACGGTGGACTCGTGCATG | Semiquantitative RT-PCR analysis of Smlt2297 |
| Antisense | AGTACGAACAGGCGCTGG | ||
| 2303 | Sense | GCTGACGTAGCCGTCGAC | Semiquantitative RT-PCR analysis of Smlt2303 |
| Antisense | TGGGACTGCAGACGCGTG | ||
| 2306 | Sense | GAGATCTGCAGGCCGGAG | Semiquantitative RT-PCR analysis of Smlt2306 |
| Antisense | TCCTTCACCAGCCAGCTG | ||
| 2318 | Sense | GTTCTCGCCTGAGGCGTG | Semiquantitative RT-PCR analysis of Smlt2318 |
| Antisense | GTGCTCGATCCGGCGATG | ||
| 2319 | Sense | CAACACCACGGTGGAGGT | Semiquantitative RT-PCR analysis of Smlt2319 |
| Antisense | CCGGGCAGCGTCACGCTG | ||
| 2324 | Sense | TCGTCTTCCTGCAGGGAG | Semiquantitative RT-PCR analysis of Smlt2324 |
| Antisense | CAGCTTCCATGCCGGCCTG | ||
| tmRNA | Sense | GGGGGTGCACTGGTTTCG | Semiquantitative RT-PCR analysis of tmRNA |
| Antisense | TGGTGGAGGTGGGCGGAAT |
The letters used are standard IUB/IUPAC nucleic acid codes.
General molecular cloning procedures, including PCR, DNA extraction and ligation, restriction enzyme digestion, and Southern blotting, were conducted per the methods of Sambrook and Russell (19). All primers used for mutagenesis, protein expression, and reverse transcription-PCR (RT-PCR) are listed in Table 2. The in-frame deletion mutant of fsnR was constructed by homologous double-crossover recombination using the suicide vector pk18mobsacB (20). Correction of the in-frame deletion mutant was verified by PCR and sequencing. A genetically complementary strain was constructed as outlined below. EcoRI and HindIII restriction sites were introduced into full-length fsnR, the gene sequence was amplified by PCR, and the amplicon was ligated with the pHM1 broad-host-range vector and electroporated into S. maltophilia strains. In this construct, fsnR is under the control of the lacZp promoter.
Biofilm assay and quantification.
The crystal violet staining method for the quantification of biofilm formation was adapted from a previously described method (21, 22). In brief, S. maltophilia cultures were inoculated quantitatively into NYG medium within 96-well polystyrene plates and incubated at 28°C for 8 h without shaking. The optical density at 600 nm (OD600) for each well was measured on a Tecan Infinite 200 Pro microplate reader. The wells were washed rigorously with water prior to staining with 0.1% crystal violet solution for 20 min. The wells were washed, the crystal violet stain was solubilized in absolute ethanol, and biofilm formation was determined by measuring the OD590 on a microplate reader. Each data point was the average for at least four replicates.
Swimming motility and transmission electron microscopy.
For the swimming motility assay, bacterial strains were inoculated into 0.1% NYG agar with a toothpick and cultured for 36 h at 28°C, and the diameters of the swimming zones were measured.
Transmission electron microscopy was used to observe flagellar morphology. Briefly, bacterial suspensions prepared from cultures of S. maltophilia were placed directly on glow-discharged carbon-coated grids, stained with 2.0% uranyl acetate for 2 min, washed twice with water, and air dried. Bacterial flagella were then visualized using a JEDL JEM-1400 transmission electron microscope (JEOL).
RNA extraction and semiquantitative RT-PCR.
Total bacterial RNA was extracted using TRIzol (Invitrogen) according to the manufacturer's instructions. The extracted RNA was quantified with a NanoDrop spectrophotometer (Thermo Fisher). Total RNA was treated with DNA-free DNase (Life Technologies) to remove contaminating DNA. The first strand of cDNA was synthesized using random primers (Promega) and Superscript III reverse transcriptase (Invitrogen). Controls used in the semiquantitative RT-PCR assay were as follows: DNA templates of the wild-type (WT) strain were included as the positive control for PCR, transfer-messenger RNA (tmRNA) amplification was the loading control for RT-PCR, and samples that lacked reverse transcriptase during cDNA synthesis were included as negative controls to evaluate potential DNA contamination. Primers used for the amplification of sample genes and tmRNA genes are listed in Table 2.
Protein expression and electrophoretic mobility shift assay (EMSA).
Recombinant FsnR-His6 protein was expressed in E. coli BL21(DE3) cells transformed with a recombinant pET30a (Novagen) expression vector. PCR primers used for the construction of the recombinant vector are listed in Table 2. The FsnR-His6 protein was purified by Ni-nitrilotriacetic acid (Ni-NTA) affinity chromatography according to the manufacturer's instructions (Novagen). The purified protein was concentrated using Centricon YM-10 columns (Millipore, Australia), and the buffer was exchanged with storage buffer (50 mM Tris-HCl, 0.5 mM EDTA, 50 mM NaCl, 5% glycerol; pH 8.0) for further use.
For EMSA, promoter regions of the Smlt2303 and Smlt2318 operons were amplified by PCR (see Table 2 for primer sequences). The PCR products (299 bp and 267 bp, respectively) were labeled with [α-32P]dCTP by use of a Prime-A-Gene kit (Promega), and free [α-32P]dCTP was removed by use of a ProbeQuant G-50 column (GE). DNA probes (4 fmol) and proteins (1 μg) were coincubated in reaction buffer containing 10 mM Tris (pH 7.0), 50 mM KCl, 1 mM dithiothreitol (DTT) (pH 7.5), 2.5% glycerol, 5 μl MgCl2, 50 ng/μl poly(dI-dC), 0.05% NP-40, and 10 mM EDTA for 20 min at room temperature. A 4-μl aliquot of DNA loading buffer (0.25% bromophenol blue, 40% sucrose) was added to stop the reaction, and the samples were loaded into a 5% native PAGE gel. Electrophoresis was performed at 120 V for about 40 min, using 0.5× Tris-borate-EDTA (TBE) buffer. The gel was placed in a ziplock bag, and phosphorimaging screens were used for signal detection. Unlabeled probes were used in the competitive experiments.
RESULTS
Construction and screening of random insertion mutant library of S. maltophilia.
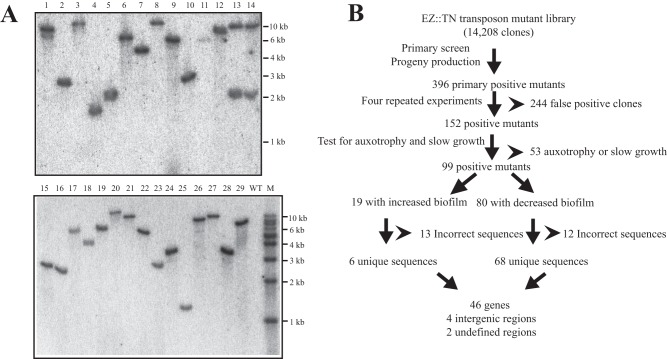
S. maltophilia ATCC 13637 is a clinical strain that was isolated from the oropharyngeal region of a patient with mouth cancer. It was chosen for use in the current study because it rapidly forms biofilms, within several hours. A random Tn5 transposon insertion mutant library was constructed with the EZ::TN transposome system (16). The library comprises 37 384-well plates and 14,208 kanamycin-resistant transformants. To evaluate the randomness and proportion of single-copy insertions, total DNAs from 29 randomly chosen clones were extracted, digested with SmaI, and analyzed by Southern blotting. As shown in Fig. 1A, 27 clones (93%) contained single-copy insertions, while 2 clones (7%) (lanes 13 and 14) simultaneously received two EZ::TN transposon insertions. Additionally, six clones cultivated in NYGB medium in the absence of kanamycin for 20 generations maintained their resistance to kanamycin, confirming the stability of the transposon inserts (see Fig. S1 in the supplemental material).
FIG 1.
Construction and screening of a random transposon insertion mutant library of S. maltophilia. (A) Determination of randomness and insertion copy numbers of mutants. Twenty-nine randomly chosen colonies of EZ::TN transposon insertion transformants and the WT strain were analyzed by Southern blotting. Total DNAs were extracted from the bacterial strains, digested with SmaI, and fractionated in a 1% agarose gel. A PCR product of the EZ::TN transposon was labeled with [α-32P]dCTP and used as the DNA hybridization probe. M, 1-kb DNA ladder. (B) Pipeline of screening for mutants with biofilm deficiency. The number of mutants obtained after each round of screening is shown. Details of the screens are described in Materials and Methods.
The library was then screened for mutants with altered biofilm-forming capabilities by using the crystal violet staining assay. Bacterial growth was also quantified to eliminate potential growth-associated contributions to changes in biofilm formation. The primary screening round identified a total of 396 clones exhibiting changes in the quantity of biofilm on the polypropylene surface (Fig. 1B). These clones were selected, propagated in 96-well plates, and subjected to four additional rounds of screening by biofilm assay. Of the 396 clones selected, 244 were identified as being false positive, because their phenotypic change failed to be replicated stably throughout the screening process. Additionally, 53 clones with poor or no growth were excluded from subsequent analyses because the decrease in biofilm formation may have been caused by auxotrophy or slow growth. Thermal asymmetric interlaced PCR (TAIL-PCR) was used to identify genomic sequences flanking the EZ::TN transposon insertion sites for 99 clones exhibiting decreased (80 clones) and increased (19 clones) biofilm formation.
Identification of genes responsible for biofilm formation emphasizes the role of flagellar biosynthesis.
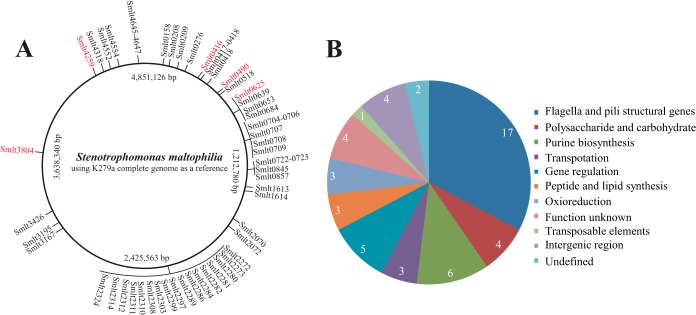
TAIL-PCR analysis successfully identified the transposon insertion sites in each of the clones. A total of 46 nonredundant genes, 4 intergenic regions, and 2 undefined insertion sites within a multicopy locus were identified (Fig. 2A; see Table S1 in the supplemental material). Because the genome of S. maltophilia ATCC 13637 has not been sequenced, the inserted genes were mapped onto the complete genome of S. maltophilia reference strain K279a (1). The mutant genes can be functionally classified into the following nine categories: (i) flagellum and pilus assembly; (ii) polysaccharide biosynthesis, e.g., xanA; (iii) peptide and lipid synthesis; (iv) purine biosynthesis, e.g., purE, purK, guaA, and purC; (v) transmembrane transportation; (vi) gene expression regulation; (vii) oxidoreduction; (viii) transposable elements; and (ix) unknown function (Fig. 2B; see Table S1). Overall, mutations identified in 5 genes were associated with an increase in biofilm formation, while the remaining 41 genes showed deficits. Southern blotting revealed that all the above-mentioned mutants contained single-copy transposons (see Fig. S2).
FIG 2.
Mapping of insertion sites on the genome of S. maltophilia. (A) Schematic view of insertion sites on the genome of S. maltophilia, using the complete genome of K279a as a reference. Coding sequences (CDSs) were labeled per the genomic annotation for K279a. Genes carrying mutations which resulted in decreases and increases in biofilm formation are depicted in black and red, respectively. (B) Functional classification of mutated genes. Detailed information on the mutated genes in both panels is provided in Table S1 in the supplemental material.
Almost half of the genes identified (20 genes [43.5%]) were associated with flagellar structure and regulation, which is highly suggestive of the flagellum playing a prominent role in S. maltophilia biofilm formation. As shown in Fig. 3, these genes were organized into 10 putative operons in the genome of S. maltophilia reference strain K279a. These operons encode structural and assembly proteins associated with different stages of flagellar assembly. FlhA, FliO, FliN, FliM, FliI, FliF, FlgI, FlgH, and FlgG are structural components of the basal body. FliD is the flagellar filament capping protein, while FliK controls flagellar hook length during assembly. FlhF is a putative GTPase that is often necessary for assembly and placement of polar flagella. FliJ acts as a chaperone, exporting flagellar proteins from the cytoplasm into the periplasm. FlgL, FlgK, FlgJ, and FlgE are components of the flagellar hook and assist in connecting the basal body to the filament.
FIG 3.
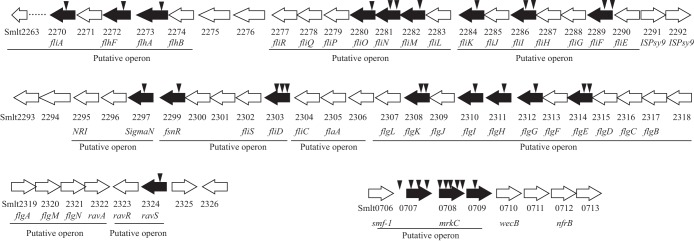
Transposon insertions in gene clusters associated with flagellar or pilus assembly. White arrows indicate CDSs and the direction in which they are transcribed. Black arrows indicate genes disrupted by transposon insertions. Black triangles indicate independent EZ::TN transposon insertions. Virtual Institute of Microbial Stress and Survival (VIMSS) operon predictions are illustrated below.
It is noteworthy that five regulatory genes were identified within or in the vicinity of flagellar gene clusters. Smlt2270 (fliA) and Smlt2297 encode σ28 and σ54, respectively. These alternative sigma factors are involved in regulating flagellar gene transcription. Smlt0158 encodes a histidine kinase (NRII or GlnL) which may phosphorylate the putative cognate response regulator Smlt0159 (NRI or GlnG). Smlt2324 encodes a PAS/PAC domain-containing histidine kinase that is orthologous to RavS of Xanthomonas campestris (23). In the vicinity of ravS, there is another histidine kinase gene (ravA or Smlt2322) and a GGDEF/EAL domain-containing response regulator gene, Smlt2323 (an ortholog of ravR). Additionally, a response regulator gene, Smlt2299, was also identified. Smlt2299 encodes a putative protein product with 210 amino acid residues. The protein contains an N-terminal receiver domain for accepting phosphoryl groups and a C-terminal LuxR-type DNA-binding domain. In the absence of a histidine kinase gene near Smlt2299, its product, Smlt2299, was considered an “orphan” response regulator. To our knowledge, Smlt2299 and its orthologs have not been reported previously. As this gene is not an ortholog of genes for known flagellum-associated response regulators, such as the FlgR proteins of Campylobacter jejuni and Helicobacter pylori, we proposed “FsnR,” for flagellum biosynthesis regulator, as a more suitable name. The regulatory role of FsnR was investigated further in the current study.
FsnR controls biofilm development and flagellar biosynthesis.
Since fsnR is putatively located in an operon and transposon insertion may lead to polar effects to complicate genetic analysis, we constructed a nonpolar, in-frame deletion mutant of fsnR via homologous double-crossover recombination. This mutant strain was labeled SM001 (ΔfsnR). To generate the complementary gene mutation, the full-length sequence of fsnR was amplified by PCR, cloned into the broad-host-range vector pHM1, and transformed into SM001, giving rise to strain SM002 (ΔfsnR-fsnR). In this construct, fsnR was under the control of the lacZp promoter. As shown in Fig. 4A and B, SM001 biofilm production decreased significantly, to 27.5% of the WT level, while genetic complementation substantially restored the bacterium's biofilm-forming ability (to 77.6% of the WT level). Similar growth curves were observed for the SM001 and WT strains, while SM002 grew more slowly (Fig. 4C). The disparity in growth curves may have been caused by overexpression of the fsnR gene for SM002. These results demonstrated that mutations in the fsnR gene were responsible for deficiencies in bacterial biofilm formation and that the phenotypic change was not due to changes in growth rate.
FIG 4.
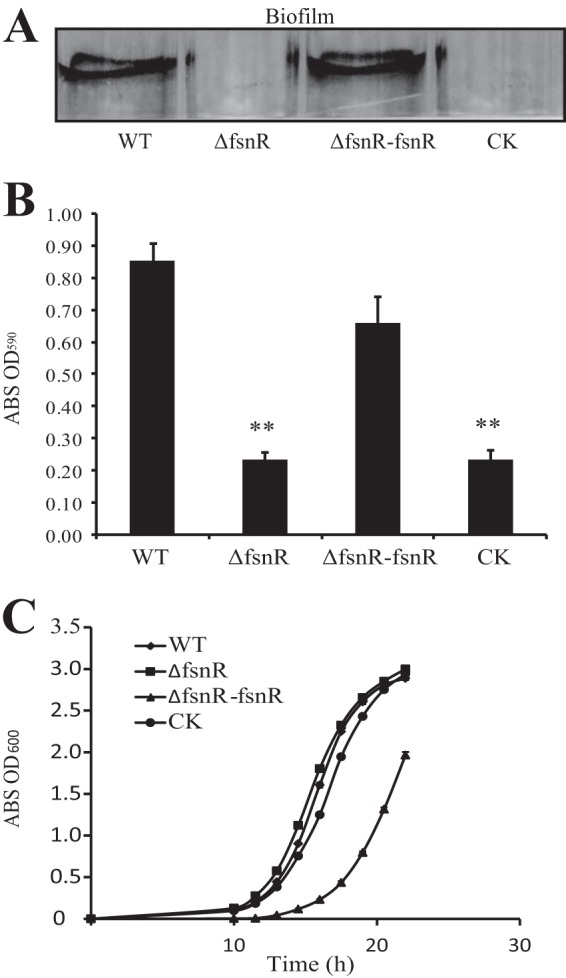
Mutations in fsnR substantially reduced biofilm formation. (A) Crystal violet-stained biofilms formed by bacterial strains. (B) Quantification of bacterial biofilm formation by OD590 measurements. Error bars represent standard deviations (n = 4). **, P < 0.01, calculated by Student's t test. WT, wild-type strain; ΔfsnR, in-frame fsnR deletion mutant SM001; ΔfsnR-fsnR, complementary strain SM002; CK, ΔfsnR strain containing blank pHM1 vector (SM003). ABS, absorbance. (C) Growth curves for bacterial strains in rich NYG medium. Error bars represent standard deviations (n = 4).
Because fsnR resides in an operon with fliS, fliD, and two as yet functionally unknown genes, Smlt2300 and Smlt2301, we surmised that Smlt2299 may be required for the regulation of flagellar biosynthesis and bacterial motility. As such, swimming motility and flagellar morphology were investigated for each of the bacterial strains. The SM001 mutant was found to be nonmotile on semisolid NYG agar plates containing 0.1% agar, while genetic complementation partially restored the swimming ability of SM002 to 50.5% of the WT strain level (Fig. 5A). Transmission electron microscopy confirmed the presence of clearly visible polar, single flagella for most WT and SM002 cells, while SM001 cells lacked flagella (Fig. 5B). These results demonstrate that the response regulator FsnR controls flagellar biosynthesis and cell motility.
FIG 5.
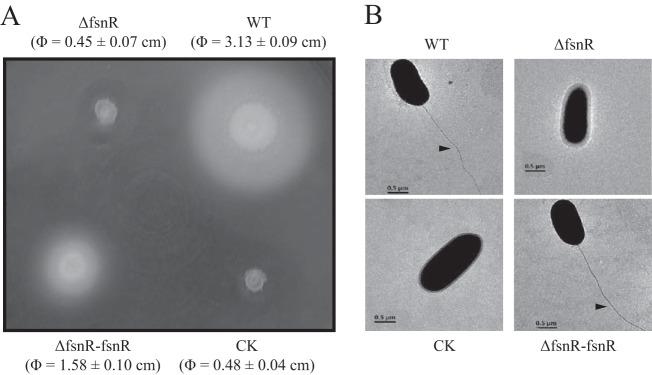
fsnR mutation resulted in deficiencies in swimming motility and flagellar development. (A) Swimming motilities of bacterial strains. All strains were inoculated into 0.1% NYG agar and grown for 36 h. WT, wild-type strain; ΔfsnR, in-frame fsnR deletion mutant SM001; ΔfsnR-fsnR, complementary strain SM002; CK, ΔfsnR strain containing blank pHM1 vector (SM003). (B) fsnR mutation caused abnormalities in flagellar assembly. Bacterial morphology was observed with a scanning transmission electron microscope. Magnification, ×4,000. Black arrows indicate the presence of flagella.
FsnR directly binds to promoters of flagellar genes and activates their transcription.
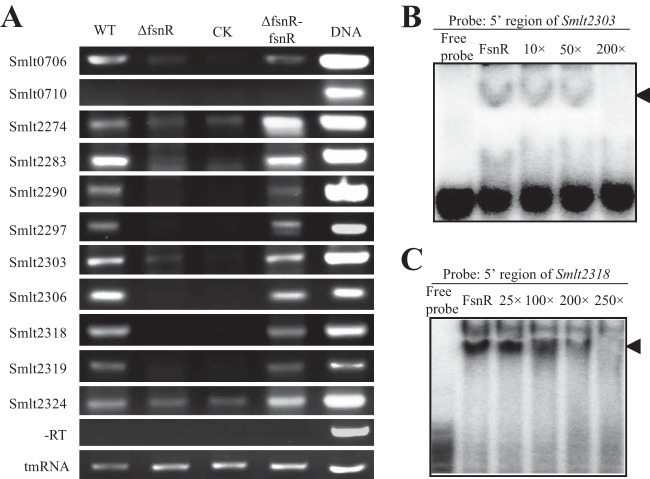
Semiquantitative RT-PCR was used to establish whether FsnR is a critical transcription factor in controlling flagellar gene expression. The transcription levels of 11 genes located in the 5′ regions of operons associated with flagellar assembly (Fig. 3) were measured. As shown in Fig. 6A, with the exception of Smlt0710, whose expression was undetectable, the quantities of mRNA for these representative genes were markedly reduced in the fsnR mutant. In contrast, the transcriptional levels of all genes were either restored or increased to levels greater than those for the WT for the complementary strain SM002. This result suggests that FsnR is a canonical positive regulator directly or indirectly controlling the transcription of most flagellar genes.
FIG 6.
FsnR controls expression of flagellar genes and directly binds to promoter regions. (A) Semiquantitative RT-PCR assays of flagellum-associated gene transcription. The bands represent RT-PCR amplification of cDNAs synthesized from total RNA of bacterial strains. −RT, negative control in which reverse transcriptase was absent during cDNA synthesis. Amplification using bacterial DNA and cDNA for tmRNA as templates were used as the positive control and loading control, respectively. WT, wild-type strain; ΔfsnR, in-frame fsnR deletion mutant; ΔfsnR-fsnR, complementary strain. (B and C) EMSAs detected FsnR binding directly to promoter regions of the Smlt2303 (B) and Smlt2318 (C) operons. Four femtomoles of [α-32P]dCTP-labeled double-stranded DNA corresponding to the Smlt2303 or Smlt2318 promoter region was used in each reaction mixture. Increasing amounts of unlabeled probes were used as competitors (10× to 250×) for binding to 1 μg of FsnR-His6 recombinant protein. Black triangles indicate the protein-DNA binding complexes.
FsnR has a C-terminal LuxR-type DNA-binding domain and may act as a transcription factor. A recombinant FsnR-His6 protein expressed in E. coli and purified by Ni-NTA affinity chromatography was used in EMSAs to establish whether FsnR binds to the 5′ promoters of the Smlt2303 and Smlt2318 operons. The promoter region of the Smlt2303 operon was selected because fsnR is putatively located within this operon. If FsnR binds to the cis-regulatory elements of the Smlt2303 operon, it will form an autoregulation loop. Conversely, the 5′ promoter sequence of the Smlt2318 operon was selected because the operon contains 12 genes (Smlt2307 to Smlt2318) and is the largest gene cluster of all flagellum-associated operons. FsnR-His6 formed stable protein-DNA complexes with the promoter sequences of both Smlt2303 (Fig. 6B) and Smlt2318 (Fig. 6C). The addition of unlabeled probes effectively competed with the binding of FsnR-His6 to isotope-labeled probes. These results demonstrated that FsnR specifically binds to the promoter regions of the Smlt2303 and Smlt2318 operons and activates their transcription, which may be a critical regulatory process during S. maltophilia biofilm development.
DISCUSSION
The molecular process of S. maltophilia biofilm formation is important in defending this emergent pathogen (3). Here we identified 46 genes containing mutations associated with changed biofilm formation (Fig. 2; see Table S1 in the supplemental material). As these genes are involved in multiple biochemical and regulatory cascades, we surmised that S. maltophilia biofilm formation is a tightly controlled and highly complex process. With the exception of a few genes, such as xanA, which is involved in polysaccharide biosynthesis (12), all remaining genes had not previously been reported, and hence their roles in biofilm development require further investigation. We provided evidence that an orphan response regulator, FsnR, directly binds to promoters of at least two flagellum-associated operons, activating their transcription (Fig. 6). Inactivation of fsnR resulted in deficiencies in flagellar assembly, swimming motility, and biofilm formation (Fig. 4 and 5). These results identify FsnR as a pivotal modulator of S. maltophilia biofilm formation and, as such, a desirable target in the development of novel antibacterial agents.
In the current study, we identified 17 structural and 3 regulatory genes associated with flagellar assembly (see Table S1 in the supplemental material). The vital role that bacterial flagella play in biofilm development is well recognized (24). Flagella not only contribute to cell motility during biofilm formation and dispersal but also take part in surface sensing and colonization (25). Consequently, subtle regulation of flagellar biosynthesis and cell motility behavior is important for generating and maintaining the bacterial biofilm matrix. In general, regulatory genes of flagella can be assigned to one of three groups: group I regulates flagellar gene expression and assembly, group II regulates flagellum-associated chemotaxis, and group III regulates modes of flagellar operation (24). To date, these regulatory genes have not been studied in S. maltophilia. Here we identified a number of regulatory genes. fsnR was characterized as a group I transcription regulator responsible for activation of a number of flagellar genes of S. maltophilia. BLASTP sequence alignment revealed that FsnR shared the greatest homology with E. coli UvrY (e = 1e−40; identity = 38%; 100% coverage). Inactivation of uvrY caused deficiencies in biofilm formation and virulence for a number of enterobacterial species. A uvrY gene mutation downregulated type 1 and Pap fimbriae in E. coli (26). Teplitski et al. reported that uvrY (sirA) inactivation caused repression of the master regulator of flagellar genes, flhDC, in Salmonella enterica (27). The present study provides molecular evidence that FsnR is a transcription activator directly binding to the promoter regions of flagellar genes (Fig. 6B). Collectively, these results indicate that a UvrY/FsnR-like response regulator may modulate flagellar gene expression on multiple levels or that these response regulators have experienced functional differentiation of signal rewiring during bacterial evolution. Furthermore, FsnR is an “orphan” response regulator whose cognate histidine kinase remains unknown. Identification of its cognate histidine kinase by use of computational methods for prediction of protein-protein interactions, together with phosphorylation profiling (28, 29), will improve our understanding of the effects that environmental stimuli have on FsnR-mediated flagellar assembly.
Four other regulatory genes (Smlt2270, Smlt0158, Smlt2297, and Smlt2324) associated with S. maltophilia biofilm formation were identified in the current study. Smlt2270 (fliA) encodes the flagellar sigma factor FliA (σ28). The σ28-containing holoenzyme of S. enterica serovar Typhimurium was shown to selectively bind to the −35 regions downstream of fliC and the anti-sigma-factor gene fliM, activating their transcription at a late stage of flagellar assembly (30). Furthermore, σ28 was shown to control type III secretion system gene expression in S. Typhi (31), suggesting that it is a global regulator connecting different physiological pathways. The regulatory gene Smlt2297 resides in the fsnR operon, and hence its transcription is also modulated by FsnR autoregulation. Smlt2297 encodes an alternative sigma factor (σ54) that is known to regulate flagellar gene expression in a diverse range of bacterial species, including Enterococcus faecalis (32), Vibrio fischeri (33), Pseudomonas aeruginosa (34), and Burkholderia cenocepacia (35). Consequently, inactivation of σ28 or σ54 has been shown to result in deficiencies in motility, virulence, and biofilm formation. To date, the consensus binding motifs of σ28 and σ54 in most of the aforementioned bacteria, including S. maltophilia, remain unclear.
As with fsnR, the regulatory genes glnL and ravS encode histidine kinase sensors belonging to two-component signal transduction systems. GlnL (Smlt0158 or NRII) is a putative histidine kinase with a predicted transmembrane domain. The presence of a PAS domain in the N-terminal region of GlnL suggests that GlnL has the capacity to detect intracellular redox potentials, oxygen, or other stimuli. Not surprisingly, glnL is situated with a putative cognate response regulator gene, glnG (Smlt0159). GlnG (NRI) is an NtrC family transcription factor containing the receiver AAA and a helix-turn-helix output domain. Future studies will focus on the GlnL-GlnG regulon and its relationship to S. maltophilia biofilm formation. The fourth regulatory gene identified in the present study was Smlt2324, an ortholog of the ravS gene of X. campestris. In X. campestris, ravS encodes a PAS domain-containing histidine kinase and appears to constitute a “three-component” signal transduction system (RavSAR) with the histidine kinase RavA and the response regulator RavR (23, 36). The RavSAR system seems to regulate bacterial gene expression by affecting turnover of the second messenger c-di-GMP, which is conceivable because RavR contains both putative diguanylate cyclase (GGDEF) and c-di-GMP phosphodiesterase (EAL) domains (23). RavSAR gene inactivation markedly attenuated the virulence of X. campestris (23, 36). However, comparatively little is known of RavSAR regulation, and it would be worthwhile to investigate the role of RavS during RavA-RavR phosphotransfer, as well as the nature of the signals detected by RavS and RavA, and to establish how these three proteins constitute a dynamic protein complex during regulation.
Gene mutations causing increased biofilm formation have been identified in other bacterial species, including lipopolysaccharide-related genes in E. coli (37), an S-ribosylhomocysteinase gene (luxS) in Listeria monocytogenes (38), and two-component signaling system genes in P. aeruginosa (39). We identified five genes whose mutants exhibited substantial increases in biofilm formation compared to that of the WT strain (Fig. 2A; see Table S1 in the supplemental material). Four of these genes, Smlt3804, Smlt4259, Smlt0490, and Smlt0625, encode enzymes, while Smlt0416 encodes a protein of unknown function. Insufficient information was obtained in the current study to elucidate a relationship between biofilm formation and the functionality of these genes. For example, Smlt3804 encodes a glyceraldehyde-3-phosphate dehydrogenase (GAPDH) which is involved in glycolysis. Studies have suggested a role for GAPDH in bacterial biofilm formation. GAPDH expression was altered in mutants with biofilm deficiencies for Streptococcus suis and Streptococcus mutans (40, 41). In Streptococcus oralis, cell surface GAPDH acts as the binding site for biofilm formation by the periodontopathic bacterium Porphyromonas gingivalis (42, 43). These studies indicated that GAPDH has a role in bacterial biofilms. However, further experimental evidence is needed to elucidate why mutations in the five genes listed above resulted in increases in S. maltophilia biofilm production.
Supplementary Material
ACKNOWLEDGMENTS
This study was financially supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (grant XDB11040700), the National Basic Research Program of the Ministry of Science and Technology of China (grant 2011CB100700), the National Science Foundation of China (grants 31370127, 31070081, and 31400071), and the China Ocean Mineral Resources R&D Association (grant DY125-15-T-07).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03408-14.
REFERENCES
- 1.Crossman LC, Gould VC, Dow JM, Vernikos GS, Okazaki A, Sebaihia M, Saunders D, Arrowsmith C, Carver T, Peters N, Adlem E, Kerhornou A, Lord A, Murphy L, Seeger K, Squares R, Rutter S, Quail MA, Rajandream MA, Harris D, Churcher C, Bentley SD, Parkhill J, Thomson NR, Avison MB. 2008. The complete genome, comparative and functional analysis of Stenotrophomonas maltophilia reveals an organism heavily shielded by drug resistance determinants. Genome Biol 9:R74. doi: 10.1186/gb-2008-9-4-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ryan RP, Monchy S, Cardinale M, Taghavi S, Crossman L, Avison MB, Berg G, van der Lelie D, Dow JM. 2009. The versatility and adaptation of bacteria from the genus Stenotrophomonas. Nat Rev Microbiol 7:514–525. doi: 10.1038/nrmicro2163. [DOI] [PubMed] [Google Scholar]
- 3.Brooke JS. 2012. Stenotrophomonas maltophilia: an emerging global opportunistic pathogen. Clin Microbiol Rev 25:2–41. doi: 10.1128/CMR.00019-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Looney WJ, Narita M, Muhlemann K. 2009. Stenotrophomonas maltophilia: an emerging opportunist human pathogen. Lancet Infect Dis 9:312–323. doi: 10.1016/S1473-3099(09)70083-0. [DOI] [PubMed] [Google Scholar]
- 5.Brooke JS. 2014. New strategies against Stenotrophomonas maltophilia: a serious worldwide intrinsically drug-resistant opportunistic pathogen. Expert Rev Anti Infect Ther 12:1–4. doi: 10.1586/14787210.2014.864553. [DOI] [PubMed] [Google Scholar]
- 6.Safdar A, Rolston KV. 2007. Stenotrophomonas maltophilia: changing spectrum of a serious bacterial pathogen in patients with cancer. Clin Infect Dis 45:1602–1609. doi: 10.1086/522998. [DOI] [PubMed] [Google Scholar]
- 7.Nicodemo AC, Paez JI. 2007. Antimicrobial therapy for Stenotrophomonas maltophilia infections. Eur J Clin Microbiol Infect Dis 26:229–237. doi: 10.1007/s10096-007-0279-3. [DOI] [PubMed] [Google Scholar]
- 8.Toleman MA, Bennett PM, Bennett DM, Jones RN, Walsh TR. 2007. Global emergence of trimethoprim/sulfamethoxazole resistance in Stenotrophomonas maltophilia mediated by acquisition of sul genes. Emerg Infect Dis 13:559–565. doi: 10.3201/eid1304.061378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pompilio A, Piccolomini R, Picciani C, D'Antonio D, Savini V, Di Bonaventura G. 2008. Factors associated with adherence to and biofilm formation on polystyrene by Stenotrophomonas maltophilia: the role of cell surface hydrophobicity and motility. FEMS Microbiol Lett 287:41–47. doi: 10.1111/j.1574-6968.2008.01292.x. [DOI] [PubMed] [Google Scholar]
- 10.de Oliveira-Garcia D, Dall'Agnol M, Rosales M, Azzuz AC, Alcantara N, Martinez MB, Giron JA. 2003. Fimbriae and adherence of Stenotrophomonas maltophilia to epithelial cells and to abiotic surfaces. Cell Microbiol 5:625–636. doi: 10.1046/j.1462-5822.2003.00306.x. [DOI] [PubMed] [Google Scholar]
- 11.Jucker BA, Harms H, Zehnder AJ. 1996. Adhesion of the positively charged bacterium Stenotrophomonas (Xanthomonas) maltophilia 70401 to glass and Teflon. J Bacteriol 178:5472–5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang TP, Somers EB, Wong AC. 2006. Differential biofilm formation and motility associated with lipopolysaccharide/exopolysaccharide-coupled biosynthetic genes in Stenotrophomonas maltophilia. J Bacteriol 188:3116–3120. doi: 10.1128/JB.188.8.3116-3120.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ryan RP, Fouhy Y, Garcia BF, Watt SA, Niehaus K, Yang L, Tolker-Nielsen T, Dow JM. 2008. Interspecies signalling via the Stenotrophomonas maltophilia diffusible signal factor influences biofilm formation and polymyxin tolerance in Pseudomonas aeruginosa. Mol Microbiol 68:75–86. doi: 10.1111/j.1365-2958.2008.06132.x. [DOI] [PubMed] [Google Scholar]
- 14.Roscetto E, Angrisano T, Costa V, Casalino M, Forstner KU, Sharma CM, Di Nocera PP, De Gregorio E. 2012. Functional characterization of the RNA chaperone Hfq in the opportunistic human pathogen Stenotrophomonas maltophilia. J Bacteriol 194:5864–5874. doi: 10.1128/JB.00746-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin YT, Huang YW, Liou RS, Chang YC, Yang TC. 19 August 2014. MacABCsm, an ABC-type tripartite efflux pump of Stenotrophomonas maltophilia involved in drug resistance, oxidative and envelope stress tolerances and biofilm formation. J Antimicrob Chemother doi: 10.1093/jac/dku317. [DOI] [PubMed] [Google Scholar]
- 16.Goryshin IY, Jendrisak J, Hoffman LM, Meis R, Reznikoff WS. 2000. Insertional transposon mutagenesis by electroporation of released Tn5 transposition complexes. Nat Biotechnol 18:97–100. doi: 10.1038/72017. [DOI] [PubMed] [Google Scholar]
- 17.Qian W, Jia Y, Ren SX, He YQ, Feng JX, Lu LF, Sun Q, Ying G, Tang DJ, Tang H, Wu W, Hao P, Wang L, Jiang BL, Zeng S, Gu WY, Lu G, Rong L, Tian Y, Yao Z, Fu G, Chen B, Fang R, Qiang B, Chen Z, Zhao GP, Tang JL, He C. 2005. Comparative and functional genomic analyses of the pathogenicity of phytopathogen Xanthomonas campestris pv. campestris. Genome Res 15:757–767. doi: 10.1101/gr.3378705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun Q, Wu W, Qian W, Hu J, Fang R, He C. 2003. High-quality mutant libraries of Xanthomonas oryzae pv. oryzae and X. campestris pv. campestris generated by an efficient transposon mutagenesis system. FEMS Microbiol Lett 226:145–150. doi: 10.1016/S0378-1097(03)00583-4. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook JR, Russell DW. 2001. Molecular cloning. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- 20.Schafer A, Tauch A, Jager W, Kalinowski J, Thierbach G, Puhler A. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 21.Musken M, Di Fiore S, Romling U, Haussler S. 2010. A 96-well-plate-based optical method for the quantitative and qualitative evaluation of Pseudomonas aeruginosa biofilm formation and its application to susceptibility testing. Nat Protoc 5:1460–1469. doi: 10.1038/nprot.2010.110. [DOI] [PubMed] [Google Scholar]
- 22.O'Toole GA, Kolter R. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol 28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- 23.He YW, Boon C, Zhou L, Zhang LH. 2009. Co-regulation of Xanthomonas campestris virulence by quorum sensing and a novel two-component regulatory system RavS/RavR. Mol Microbiol 71:1464–1476. doi: 10.1111/j.1365-2958.2009.06617.x. [DOI] [PubMed] [Google Scholar]
- 24.Guttenplan SB, Kearns DB. 2013. Regulation of flagellar motility during biofilm formation. FEMS Microbiol Rev 37:849–871. doi: 10.1111/1574-6976.12018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Belas R. 2014. Biofilms, flagella, and mechanosensing of surfaces by bacteria. Trends Microbiol 22:517–527. doi: 10.1016/j.tim.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 26.Mitra A, Palaniyandi S, Herren CD, Zhu X, Mukhopadhyay S. 2013. Pleiotropic roles of uvrY on biofilm formation, motility and virulence in uropathogenic Escherichia coli CFT073. PLoS One 8:e55492. doi: 10.1371/journal.pone.0055492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teplitski M, Al-Agely A, Ahmer BM. 2006. Contribution of the SirA regulon to biofilm formation in Salmonella enterica serovar Typhimurium. Microbiology 152:3411–3424. doi: 10.1099/mic.0.29118-0. [DOI] [PubMed] [Google Scholar]
- 28.Skerker JM, Perchuk BS, Siryaporn A, Lubin EA, Ashenberg O, Goulian M, Laub MT. 2008. Rewiring the specificity of two-component signal transduction systems. Cell 133:1043–1054. doi: 10.1016/j.cell.2008.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skerker JM, Prasol MS, Perchuk BS, Biondi EG, Laub MT. 2005. Two-component signal transduction pathways regulating growth and cell cycle progression in a bacterium: a system-level analysis. PLoS Biol 3:e334. doi: 10.1371/journal.pbio.0030334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schaubach OL, Dombroski AJ. 1999. Transcription initiation at the flagellin promoter by RNA polymerase carrying sigma28 from Salmonella typhimurium. J Biol Chem 274:8757–8763. doi: 10.1074/jbc.274.13.8757. [DOI] [PubMed] [Google Scholar]
- 31.Eichelberg K, Galan JE. 2000. The flagellar sigma factor FliA (σ28) regulates the expression of Salmonella genes associated with the centisome 63 type III secretion system. Infect Immun 68:2735–2743. doi: 10.1128/IAI.68.5.2735-2743.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iyer VS, Hancock LE. 2012. Deletion of sigma(54) (rpoN) alters the rate of autolysis and biofilm formation in Enterococcus faecalis. J Bacteriol 194:368–375. doi: 10.1128/JB.06046-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolfe AJ, Millikan DS, Campbell JM, Visick KL. 2004. Vibrio fischeri sigma54 controls motility, biofilm formation, luminescence, and colonization. Appl Environ Microbiol 70:2520–2524. doi: 10.1128/AEM.70.4.2520-2524.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thompson LS, Webb JS, Rice SA, Kjelleberg S. 2003. The alternative sigma factor RpoN regulates the quorum sensing gene rhlI in Pseudomonas aeruginosa. FEMS Microbiol Lett 220:187–195. doi: 10.1016/S0378-1097(03)00097-1. [DOI] [PubMed] [Google Scholar]
- 35.Saldias MS, Lamothe J, Wu R, Valvano MA. 2008. Burkholderia cenocepacia requires the RpoN sigma factor for biofilm formation and intracellular trafficking within macrophages. Infect Immun 76:1059–1067. doi: 10.1128/IAI.01167-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tao J, Li C, Luo C, He C. 2014. RavA/RavR two-component system regulates Xanthomonas campestris pathogenesis and c-di-GMP turnover. FEMS Microbiol Lett 358:81–90. doi: 10.1111/1574-6968.12529. [DOI] [PubMed] [Google Scholar]
- 37.Nakao R, Ramstedt M, Wai SN, Uhlin BE. 2012. Enhanced biofilm formation by Escherichia coli LPS mutants defective in Hep biosynthesis. PLoS One 7:e51241. doi: 10.1371/journal.pone.0051241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sela S, Frank S, Belausov E, Pinto R. 2006. A mutation in the luxS gene influences Listeria monocytogenes biofilm formation. Appl Environ Microbiol 72:5653–5658. doi: 10.1128/AEM.00048-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang L, Fritsch M, Hammond L, Landreville R, Slatculescu C, Colavita A, Mah TF. 2013. Identification of genes involved in Pseudomonas aeruginosa biofilm-specific resistance to antibiotics. PLoS One 8:e61625. doi: 10.1371/journal.pone.0061625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Palmer SR, Crowley PJ, Oli MW, Ruelf MA, Michalek SM, Brady LJ. 2012. YidC1 and YidC2 are functionally distinct proteins involved in protein secretion, biofilm formation and cariogenicity of Streptococcus mutans. Microbiology 158:1702–1712. doi: 10.1099/mic.0.059139-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Y, Zhang W, Wu Z, Zhu X, Lu C. 2011. Functional analysis of luxS in Streptococcus suis reveals a key role in biofilm formation and virulence. Vet Microbiol 152:151–160. doi: 10.1016/j.vetmic.2011.04.029. [DOI] [PubMed] [Google Scholar]
- 42.Maeda K, Nagata H, Kuboniwa M, Ojima M, Osaki T, Minamino N, Amano A. 2013. Identification and characterization of Porphyromonas gingivalis client proteins that bind to Streptococcus oralis glyceraldehyde-3-phosphate dehydrogenase. Infect Immun 81:753–763. doi: 10.1128/IAI.00875-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nagata H, Iwasaki M, Maeda K, Kuboniwa M, Hashino E, Toe M, Minamino N, Kuwahara H, Shizukuishi S. 2009. Identification of the binding domain of Streptococcus oralis glyceraldehyde-3-phosphate dehydrogenase for Porphyromonas gingivalis major fimbriae. Infect Immun 77:5130–5138. doi: 10.1128/IAI.00439-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.