Abstract
Xylans, including methylglucuronoxylans (MeGXn) and methylglucuronoarabinoxylans (MeGAXn), are the predominant polysaccharides in hemicellulose fractions of dicots and monocots available for conversion to biofuels and chemicals. Paenibacillus sp. strain JDR-2 (Pjdr2) efficiently depolymerizes MeGXn and MeGAXn and assimilates the generated oligosaccharides, resulting in efficient saccharification and subsequent metabolism of these polysaccharides. A xylan utilization regulon encoding a cell-associated GH10 (glycoside hydrolase family 10) endoxylanase, transcriptional regulators, ABC (ATP binding cassette) transporters, an intracellular GH67 α-glucuronidase, and other glycoside hydrolases contributes to complete metabolism. This GH10/GH67 system has been proposed to account for preferential utilization of xylans compared to free oligo- and monosaccharides. To identify additional genes contributing to MeGXn and MeGAXn utilization, the transcriptome of Pjdr2 has been sequenced following growth on each of these substrates as well as xylose and arabinose. Increased expression of genes with different substrates identified pathways common or unique to the utilization of MeGXn or MeGAXn. Coordinate upregulation of genes comprising the GH10/GH67 xylan utilization regulon is accompanied with upregulation of genes encoding a GH11 endoxylanase and a GH115 α-glucuronidase, providing evidence for a novel complementary pathway for processing xylans. Elevated expression of genes encoding a GH43 arabinoxylan arabinofuranohydrolase and an arabinose ABC transporter on MeGAXn but not on MeGXn supports a process in which arabinose may be removed extracellularly followed by its rapid assimilation. Further development of Pjdr2 for direct conversion of xylans to targeted products or introduction of these systems into fermentative strains of related bacteria may lead to biocatalysts for consolidated bioprocessing of hemicelluloses released from lignocellulose.
INTRODUCTION
Plant biomass represents a source of lignocellulosic materials for production of alternative sources of energy. This renewable resource is primarily composed of polymeric cellulose, hemicelluloses, and lignin and therefore does not compete with agricultural commodities used for human and animal nutrition. Lignocelluloses in their native state are recalcitrant to bioprocessing and require suitable pretreatment followed by enzyme-mediated saccharification to generate fermentable sugars (1). Microbial fermentation of carbohydrates derived from lignocellulose generates biofuels and chemicals (2–6), thereby reducing the need for nonrenewable sources of energy. Strains of the yeast Saccharomyces cerevisiae and bacteria, e.g., Escherichia coli, Klebsiella oxytoca, and Zymomonas mobilis, have been developed for the production of ethanol from hexoses and pentoses derived from cellulose and hemicellulose fractions comprising lignocellulosic biomass (2–4, 7). Strains of E. coli, K. oxytoca, and Bacillus coagulans have been developed for commercial production of chemical feedstocks, e.g., d- or l-lactic acid, for production of bioplastics (2, 6, 8). Processes using these biocatalysts requires the addition of cellulases and hemicellulases for saccharification to release fermentable sugars, with the enzymes representing a major cost for production of desired biofuels and chemicals (2). Consolidated bioprocessing (CBP) by cellulolytic Clostridium thermocellum has provided an approach for direct conversion of cellulose to useful products without the addition of commercial enzymes and its associated cost (9). Systems for CBP involve a single biocatalyst to process both cellulose and xylan or participation of cocultures of cellulolytic and xylanolytic bacteria. With respect to this approach, cellulolytic Caldicellulosiruptor and xylanolytic Thermoanaerobacter species have been evaluated for consolidated bioprocessing of lignocellulose (10, 11).
Hemicelluloses of angiosperms are predominantly complex polysaccharides referred to as xylans which differ in composition depending upon their source. Methylglucuronoxylans (MeGXn) comprise the xylans in dicots, including hardwoods, whereas methylglucuronoarabinoxylans (MeGAXn) comprise the xylans in monocots, including grasses. Hardwood MeGXn is typically a linear polymer of β-1,4-linked xylopyranose units variably modified with α-1,2-linked 4-O-methylglucuronate and acetyl esters at the C-2 and C-3 positions on xylose. Depending on the species, from 6 to 10% of the xylose residues may be modified with methylglucuronate (MeG). MeGAXn from grasses is also modified with methylglucuronates although to a lesser extent than the MeGXn from hardwoods. MeGAXn commonly has 10% or more of the xylose residues substituted with α-1,2- or α-1,3-linked l-arabinofuranose units, some of which are modified with ferulate and p-coumarate esters. These ester linkages may be hydrolyzed during alkaline pretreatment that is used to solubilize the MeGAXn and render it accessible to endoxylanases for digestion (12–16). The mild thermochemical pretreatment (17) followed by enzymatic saccharification involving endoxylanases, β-xylosidases, α-glucuronidases, α-l-arabinofuranosidases, and acetyl esterases complementing each other may efficiently and completely convert the complex xylans to monomeric sugars (12, 13, 18–20). These sugars can be completely metabolized by suitable bacterial biocatalysts for fermentation to produce biofuels and chemicals.
An aggressively xylanolytic bacterium, Paenibacillus sp. strain JDR-2 (Pjdr2), with a sequenced genome (21) has a defined system for MeGXn and MeGAXn utilization (19, 20, 22, 23). This system includes a xyn10A1 gene (earlier referred to as xynA1 [23]) encoding a cell-associated multimodular GH10 (glycoside hydrolase family 10) endoxylanase (Xyn10A1) and an agu67A gene (earlier referred to as aguA [19]) encoding an intracellular GH67 α-glucuronidase (Agu67A). The xyn10A1 gene along with an aldouronate utilization gene cluster encoding transcriptional regulators, ATP binding cassette (ABC) transporters, and intracellular glycoside hydrolases, including a GH10 xylanase (Xyn10A2) (earlier referred to as XynA2 [19]), a GH43 β-xylosidase (Xyn43B1) (earlier referred to as XynB [19]), and a GH67 α-glucuronidase (Agu67A) collectively comprise a xylan utilization regulon (22). A distally located abf51B gene (earlier referred to as abfB [20]) encoding a GH51 α-l-arabinofuranosidase (Abf51B) along with neighboring genes encoding transcriptional regulators is preferentially upregulated in response to growth on MeGAXn compared to growth on MeGXn (20). Recombinant forms of Xyn10A1, Agu67A, Xyn10A2, and Abf51B from Pjdr2 have been assigned functional roles based on biochemical characterization. To further define this GH10/GH67 xylan utilization system, this study investigates the transcriptome of Pjdr2 cultured on polysaccharides from a representative dicot and monocot, sweetgum MeGXn and sorghum MeGAXn, respectively, and also their constituent monosaccharides, including arabinose and xylose. The genome of this bacterium includes several genes encoding carbohydrate active enzymes (CAZy) (24) that contribute to xylan depolymerization, such as glycoside hydrolases (GHs) and carbohydrate esterases (CEs), as well as ABC transporters for assimilation of a variety of oligosaccharides, including oligoxylosides (XOS), oligoarabinoxylosides (AXOS), and aldouronates (MeG-linked XOS). These studies have identified additional genes that contribute to systems for xylan depolymerization and assimilation of the generated oligosaccharides for intracellular metabolism and conversion to desired products.
MATERIALS AND METHODS
Preparation of xylans.
MeGXn from sweetgum (Liquidambar styraciflua) wood and MeGAXn from stalks of sorghum [Sorghum bicolor (L.) Moench] were prepared by alkaline extraction as previously described (20, 23). The phenol-sulfuric acid assay (25) was used for determination of total carbohydrate concentration.
Growth studies of Paenibacillus sp. strain JDR-2.
Paenibacillus sp. strain JDR-2 (Pjdr2) was previously isolated from decaying sweetgum wood in our laboratory (19, 21–23). Pjdr2 was stored, resuscitated, and cultured to prepare initial inocula as described previously (20). To carry out growth studies, Pjdr2 cells were harvested and suspended as 2% inocula into 20 ml ZH medium (Zucker-Hankin minerals [pH 7.4]) (26) containing 0.5% yeast extract with either 0.5% sweetgum MeGXn, sorghum MeGAXn, arabinose, xylose, or no carbohydrate and cultured at 30°C in 250-ml baffled culture flasks with shaking at 220 rpm using a G-2 gyratory shaker (New Brunswick Scientific). Aliquots of cultures were sampled at regular intervals, and growth was determined by measuring the optical density at 600 nm (OD600) using a 1.00-cm cuvette and Beckman series DU500 spectrophotometer. The cultures were diluted to obtain an OD600 between 0.2 and 0.8 and corrected for dilution to generate the growth curves. Culture supernatants were recovered by centrifugation for determination of substrate utilization by measuring total carbohydrate remaining using the phenol-sulfuric acid assay (25) with xylose as the standard.
Preparation of RNA.
Pjdr2 was cultured on different substrates as described above. Cells for RNA isolation and purification were harvested from triplicate cultures at estimated early mid-exponential growth phase by centrifugation at 4,300 × g for 10 min at 4°C. Culture purity was verified by streaking onto xylan agar plates. The procedure derived from the RNAprotect Bacteria Reagent Handbook (Qiagen) (27) was used for preparing RNA from cell lysates. RNeasy column elution was followed by RNA treatment with DNase using the TURBO-DNA free kit following the prescribed protocol (Ambion). RNA concentrations were determined by absorbance at 260 nm (A260), and the A260/A280 ratio was determined to estimate the purity of the RNA. RNA quantification was carried out using the Qubit fluorescence dye-based system (Life Technologies). Analysis of RNA quality was performed with an Agilent bioanalyzer available through the Interdisciplinary Center for Biotechnology Research (ICBR) at the University of Florida. Quantitative reverse transcription-PCR (qRT-PCR) was performed using the QuantiTect SYBR green RT-PCR kit (Qiagen) as described earlier (20) to characterize the RNA preparation with or without reverse transcriptase.
RNA sequencing.
RNA sequencing was carried out by the members of the Joint Genome Institute (JGI), U.S. Department of Energy, Walnut Creek, CA, using the following protocols. The rRNA was removed from 1 μg of total RNA using Ribo-Zero rRNA removal kit (Epicentre). The cDNA libraries were generated using an Illumina mRNA sample preparation kit. The rRNA-depleted RNA was fragmented using divalent cations and high temperature. The fragmented RNA was reverse transcribed using random hexamers and Superscript II (Invitrogen) followed by second-strand synthesis. The fragmented cDNA was treated to allow end pair A-tailing adapter ligation and 10 cycles of PCR. Libraries were quantified using KAPA Biosystem's next-generation sequencing library qPCR kit and run on a Roche LightCycler 480 real-time PCR instrument. The quantified libraries were then prepared for sequencing on the Illumina HiSeq sequencing platform utilizing a TruSeq paired-end cluster kit, v3, and Illumina's cBot instrument to generate clustered flow cells for sequencing. Sequencing of the flow cells was performed on the Illumina HiSeq2000 sequencer using a TruSeq SBS sequencing kit, v3, for 200 cycles, following a 2×100 indexed run recipe.
Transcriptomic analysis.
The filtered raw data files containing RPKM (reads per kilobase per million reads sequenced) values were analyzed using ArrayStar software from DNAStar. The results from all growth conditions (in triplicate cultures) studied were mapped to the annotated genome and normalized together for comparisons. The mapped and normalized conditions were then grouped and averaged to represent the corresponding growth conditions. Analysis of variance (ANOVA) was performed to access data quality per gene over all data sets. Statistical analysis for comparisons of two conditions was performed with the moderated t test, and adjusted P values were calculated using the false discovery rate (FDR) (Benjamini-Hochberg) method (28). Growth on 0.5% yeast extract without carbohydrate served as a control (YE control) for growth on different carbohydrates with 0.5% yeast extract. The expression of genes discussed in this study is based upon fold difference relative to YE control. Data with P values less than 0.05 were considered within the significant levels. Protein BLAST from NCBI (http://www.ncbi.nlm.nih.gov/) and IMG database (http://img.jgi.doe.gov/) were used to evaluate the candidate genes of interest and assign functional roles and names. In this study, all the genes in Pjdr2 are identified by their locus tags referenced as Pjdr2_XXXX, where XXXX represents a four-digit gene number. The raw data files for this study are available for access from the JGI database using the following link: http://genome.jgi.doe.gov/Paespnscriptome/Paespnscriptome.info.html.
RESULTS AND DISCUSSION
Growth of Pjdr2.
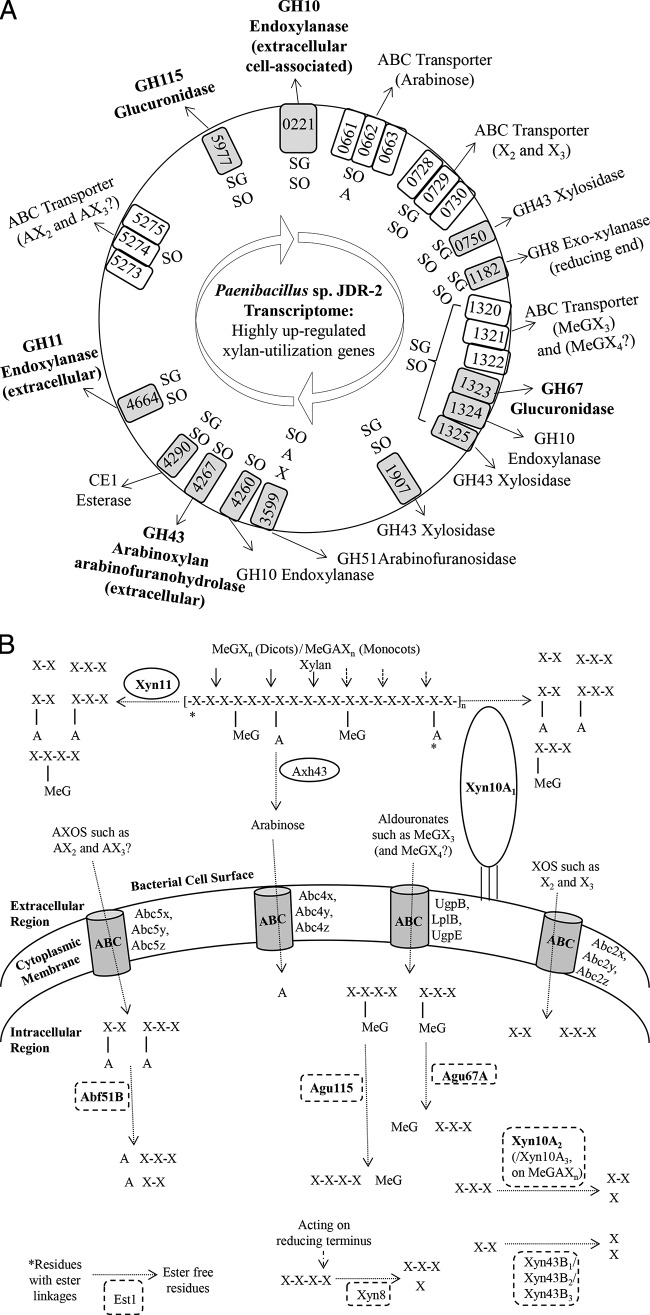
Rapid and efficient growth of Pjdr2 was observed when polysaccharides such as sweetgum MeGXn or sorghum MeGAXn were used as the substrates compared to monosaccharides such as arabinose or xylose (Fig. 1). The growth yields and growth rates on the polymeric xylans were on average 1.7- and 3.1-fold greater than on monosaccharides, indicating that these polysaccharides are preferred growth substrates for Pjdr2 compared to arabinose or xylose. The substrates were almost completely utilized with approximately 10% remaining in the medium as observed in previous studies (20, 23). Phase-contrast microscopy confirmed sporulation by Pjdr2 upon reaching stationary phase (data not shown). The transcriptome of Pjdr2 following growth on these substrates has confirmed the presence of a xylan utilization regulon comprising the GH10/GH67 system previously defined by qRT-PCR studies (20, 22) and identified additional genes and gene cassettes that likely contribute to the utilization of MeGXn and/or MeGAXn, as shown in Table 1 and Fig. 2A.
FIG 1.
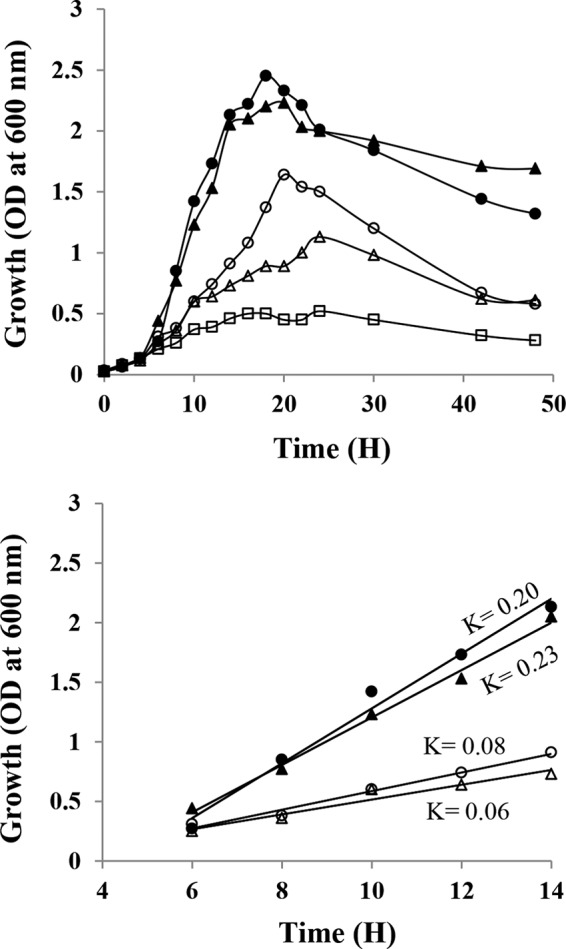
Growth of Paenibacillus sp. strain JDR-2 (Pjdr2) on xylans and monosaccharides. Growth studies were performed by determining the optical density (OD) at 600 nm of 20-ml cultures in 250-ml baffled flasks at 30°C with gyratory agitation at 220 rpm. ZH medium containing 0.5% yeast extract was supplemented with 0.5% of either sorghum MeGAXn (closed circles), sweetgum MeGXn (closed triangles), arabinose (open circles), or xylose (open triangles) or without carbohydrate as YE control (open squares). Comparison of Pjdr2 growth yields (top) and growth rates during exponential phase (bottom) on different carbohydrate substrates. The rate of growth was determined as the slope (K) with R2 values within the range 0.9400 to 0.9950. Time is shown in hours.
TABLE 1.
Summarized candidate genes in xylan utilization systems
| Location or function and family | LTa | Assigned protein productb | Assigned namec | SPd | Suggested function |
|---|---|---|---|---|---|
| Extracellular | |||||
| GH11 | 4664 | Endo-1,4-β-xylanase | Xyn11 | Yes | Depolymerization of xylans to produce MeGX4, XOS, and AXOS |
| GH10 | 0221 | Endo-1,4- β-xylanase | Xyn10A1 | Yes | Depolymerization of xylans to produce MeGX3, XOS, and AXOS |
| GH43 | 4267 | Arabinoxylan arabinofuranohydrolase | Axh43 | Yes | Release certain arabino linkages from the backbone of MeGAXn |
| Transport/assimilation | |||||
| ABC | 1320 | Extracellular SBP | UgpB | Yes | ABC transport of aldouronates such as MeGX3 (and MeGX4?) |
| 1321 | BPD transport system IMP | LplB | No | ABC transport of aldouronates such as MeGX3 (and MeGX4?) | |
| 1322 | BPD transport system IMP | UgpE | No | ABC transport of aldouronates such as MeGX3 (and MeGX4?) | |
| ABC | 0728 | Extracellular SBP | Abc2x | Yes | ABC transport of XOS such as X2 and X3 |
| 0729 | BPD transport system IMP | Abc2y | No | ABC transport of XOS such as X2 and X3 | |
| 0730 | BPD transport system IMP | Abc2z | No | ABC transport of XOS such as X2 and X3 | |
| ABC | 1809 | BPD transport system IMP | Abc3x | No | Multiple sugar transport system? |
| 1810 | BPD transport system IMP | Abc3y | No | Multiple sugar transport system? | |
| 1811 | Extracellular SBP | Abc3z | Yes | Multiple sugar transport system? | |
| ABC | 0661 | Extracellular SBP | Abc4x | Yes | ABC transport of arabinose |
| 0662 | ABC ATPase | Abc4y | No | ABC transport of arabinose | |
| 0663 | IMP | Abc4z | No | ABC transport of arabinose | |
| ABC | 5273 | BPD transport system IMP | Abc5x | No | ABC transport of AXOS such as AX2 and AX3? |
| 5274 | BPD transport system IMP | Abc5y | No | ABC transport of AXOS such as AX2 and AX3? | |
| 5275 | Extracellular SBP | Abc5z | Yes | ABC transport of AXOS such as AX2 and AX3? | |
| ABC | 5314 | BPD transport system IMP | Abc6x | No | Multiple sugar transport system? |
| 5315 | BPD transport system IMP | Abc6y | No | Multiple sugar transport system? | |
| 5316 | Extracellular SBP | Abc6z | Yes | Multiple sugar transport system? | |
| ABC | 5596 | BPD transport system IMP | Abc7x | No | Unknown carbohydrate transport system? |
| 5597 | BPD transport system IMP | Abc7y | No | Unknown carbohydrate transport system? | |
| 5598 | Extracellular SBP | Abc7z | Yes | Unknown carbohydrate transport system? | |
| Intracellular | |||||
| GH115 | 5977 | α-Glucuronidase | Agu115 | No | Aldouronate (MeGX4) processing |
| GH67 | 1323 | α-Glucuronidase | Agu67A | No | Aldouronate (MeGX3) processing |
| GH10 | 1324 | Endo-1,4-β-xylanase | Xyn10A2 | No | XOS (X3) processing |
| GH43 | 1325 | Xylan 1,4-β-xylosidase | Xyn43B1 | No | XOS (X2) processing |
| GH43 | 0750 | Xylan 1,4-β-xylosidase | Xyn43B2 | No | XOS (X2) processing |
| GH43 | 1907 | Xylan 1,4-β-xylosidase | Xyn43B3 | No | XOS (X2) processing |
| GH51 | 3599 | α-N-Arabinofuranosidase | Abf51B | No | AXOS (AX2/AX3) processing |
| GH10 | 4260 | Endo-1,4-β-xylanase | Xyn10A3 | No | AXOS (X3) processing |
| GH8 | 1182 | Exo-oligoxylanase (reducing end) | Xyn8 | No | XOS processing from reducing end |
| CE1 | 4290 | Esterase | Est1 | No | Cleaving ester linkages |
LT, locus tag referenced as Pjdr2_XXXX with XXXX being a four-digit number. Only the four-digit number is shown in the table.
Annotation for assignments derived from analysis of proteins and domains in the IMG database (http://img.jgi.doe.gov/) and protein BLAST from NCBI (http://www.ncbi.nlm.nih.gov/). SBP, solute binding protein; IMP, inner membrane protein; BPD, binding protein dependent.
The protein name assigned to the xylan utilization gene with those studied in our laboratory shown in bold type.
SP, signal peptide.
FIG 2.
(A) Summary of genes that are highly expressed with assigned functional roles contributing to the xylan utilization systems in the genome of Pjdr2. Genes included on the basis of results of transcriptomic analysis following growth of Pjdr2 on either sweetgum MeGXn (SG), sorghum MeGAXn (SO), arabinose (A), or xylose (X). The substrates preferred for upregulation are indicated adjacent to the corresponding gene or gene clusters inside the circle. Genes are identified as Pjdr2_XXXX with XXXX being the four-digit number shown in the gray or white boxes; the genes encoding catalytic enzymes are shown in gray boxes, and the genes encoding ABC transporters are shown in white boxes. The genes represented in the figure are not drawn to scale. (B) Model displaying the GH10/GH67 and GH11/GH115 systems in Pjdr2 for bioconversion of MeGXn from dicots or MeGAXn from monocots. X, β-1,4-linked xylopyranosyl units (X, xylose; X2, xylobiose, X3, xylotriose; X4, xylotetraose); MeG, α-1,2-linked 4-O-methyl-d-glucuronopyranosyl residues (MeG, methylglucuronate; MeGX3, aldotetrauronate; MeGX4, aldopentauronate); A, α-1,2- and/or 1,3-linked l-arabinofuranosyl residues (A, arabinose; AX2, arabino-linked X2; AX3, arabino-linked X3). AX2 and AX3 are generated significantly only in the case of MeGAXn. Enzyme candidates assigned a functional role based on biochemical characterization are indicated in bold type.
Expression of genes encoding glycoside hydrolases and a carbohydrate esterase during xylan utilization by Pjdr2.
Following growth of Pjdr2 on sweetgum MeGXn and sorghum MeGAXn, certain genes encoding glycoside hydrolases and a single carbohydrate esterase were upregulated. These genes encode glycoside hydrolases belonging to families GH10, GH11, GH67, GH115, GH43, and GH51 that are expected to play a role in xylan utilization in line with their reported functions. Candidate genes that belong to the potential xylan utilization systems were identified by increased transcript levels relative to the YE control condition and are listed in Table 2. Included in these findings are all genes previously assigned a functional role in xylan utilization by Pjdr2 (19, 20, 22, 23) as well as numerous other genes identified by transcriptomic analysis. The xylan-inducible genes Pjdr2_0221 encoding the extracellular multimodular cell-associated GH10 Xyn10A1, Pjdr2_1323 encoding the GH67 Agu67A, Pjdr2_1324 encoding the GH10 xylanase Xyn10A2, and also Pjdr2_3599 encoding the GH51 Abf51B (upregulated only on MeGAXn) have functional roles defined by biochemical characterization (19, 20, 22, 23). The Pjdr2_1325 gene encoding a putative GH43 β-xylosidase Xyn43B1 has also been previously shown to be highly upregulated on xylans and is part of the aldouronate utilization gene cluster (22). The expression of abf51B, which was previously shown to be enhanced by qRT-PCR when grown on MeGAXn versus MeGXn supplemented with 0.1% yeast extract (20), could not be interpreted due to high P values (Table 2).
TABLE 2.
Expression of candidate genes encoding glycoside hydrolases and carbohydrate esterase by growth on xylans
| Family | LTa | Protein product | Nameb | SPc | Fold changed |
Linear RPKM valuee |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SG/YE | SO/YE | A/YE | X/YE | SG | SO | A | X | YE | |||||
| CE1 | 4290 | Esterase | Est1 | No | 3.6 | 8.5 | 1.2* | 1.6 | 84.43 | 199.50 | 27.18 | 37.51 | 23.40 |
| GH8 | 1182 | Reducing end exo-oligoxylanase | Xyn8 | No | 37.2 | 65.9 | 0.8 | 0.8* | 280.68 | 498.05 | 5.69 | 5.80 | 7.56 |
| GH10 | 0221 | Endo-1,4-β-xylanase | Xyn10A1 | Yes | 17.0 | 9.6 | 0.2 | 0.2 | 100.79 | 57.13 | 0.91 | 1.01 | 5.93 |
| GH10 | 1324 | Endo-1,4-β-xylanase | Xyn10A2 | No | 40.1 | 45.6 | 0.4 | 0.6 | 309.01 | 352.19 | 2.92 | 4.60 | 7.72 |
| GH10 | 4260 | Endo-1,4-β-xylanase | Xyn10A3 | No | 0.4 | 2.7 | 0.6 | 0.8 | 6.37 | 39.98 | 8.64 | 11.46 | 15.05 |
| GH11 | 4664 | Endo-1,4-β-xylanase | Xyn11 | Yes | 7.1 | 50.9 | 0.2 | 0.2 | 14.65 | 104.76 | 0.39 | 0.46 | 2.06 |
| GH43 | 0750 | Xylan 1,4-β-xylosidase | Xyn43B2 | No | 104.4 | 107.8 | NS | 2.1 | 163.56 | 168.96 | 1.46 | 3.24 | 1.57 |
| GH43 | 1325 | Xylan 1,4-β-xylosidase | Xyn43B1 | No | 44.8 | 48.4 | 0.6 | 0.7 | 507.81 | 548.19 | 6.89 | 8.45 | 11.32 |
| GH43 | 1907 | Xylan 1,4-β-xylosidase | Xyn43B3 | No | 258.9 | 126.3 | 0.4 | NS | 3276.93 | 1597.93 | 5.35 | 13.11 | 12.66 |
| GH43 | 4267 | Arabinoxylan arabinofuranohydrolase | Axh43 | Yes | 0.1 | 7.2 | 0.3 | 0.3 | 2.79 | 196.78 | 9.15 | 7.86 | 27.35 |
| GH51 | 3599 | α-N-Arabinofuranosidase | Abf51B | No | NS | 1.4* | 2.6 | 1.4* | 13.06 | 19.04 | 34.08 | 19.03 | 13.18 |
| GH51 | 3612 | α-l-Arabinofuranosidase | No | 0.3 | 2.3 | 0.7 | NS | 1.35 | 12.65 | 3.87 | 4.91 | 5.38 | |
| GH67 | 1323 | α-Glucuronidase | Agu67A | No | 32.9 | 43.7 | 0.4 | 0.6 | 191.78 | 254.73 | 2.35 | 3.22 | 5.82 |
| GH115 | 5977 | α-Glucuronidase | Agu115 | No | 52.0 | 32.8 | 0.8 | NS | 268.82 | 169.43 | 4.06 | 4.46 | 5.17 |
| GH2 | 2523 | Mannosidase | No | 8.7 | 11.1 | 0.3 | 0.5 | 201.48 | 259.35 | 6.19 | 10.70 | 23.27 | |
| GH26 | 1350 | Mannosidase | Yes | 3.0 | 3.2 | 3.3 | 4.8 | 27.76 | 30.35 | 30.88 | 45.20 | 9.36 | |
| GH13 | 0774 | α-Amylase | Amy13A1 | Yes | 28.7 | 24.2 | 0.2 | 0.3 | 283.56 | 239.33 | 1.53 | 2.92 | 9.88 |
| GH13 | 0783 | α-Amylase | Amy13A3 | No | 69.1 | 53.9 | 1.4 | 1.3 | 359.64 | 280.47 | 7.31 | 7.00 | 5.21 |
| GH13 | 5200 | α-Amylase | Amy13A2 | Yes | 4.1 | 2.7 | 0.2 | 0.3 | 28.60 | 19.13 | 1.30 | 1.82 | 6.97 |
| GH73 | 3505 | Glucosamidase | No | 4.3 | NS | 1.8 | 1.6 | 21.74 | 5.18 | 9.04 | 7.99 | 5.11 | |
| GH25 | 4070 | 1,4-β-N-Acetylmuramidase | No | 15.6 | NS | 7.7 | 3.3 | 6.58 | 0.65 | 3.26 | 1.39 | 0.42 | |
LT, locus tag referenced as Pjdr2_XXXX with XXXX being a four-digit number.
The name assigned to gene candidates with enzymes characterized in our laboratory shown in bold type.
SP, signal peptide.
Transcript levels of candidate genes that were upregulated (underlined) and those that were expressed 4-fold greater (bold) than the yeast extract without carbohydrate (YE control) growth are indicated. The substrates are shown as follows: SG, sweetgum MeGXn; SO, sorghum MeGAXn; A, arabinose; X, xylose. Significance of fold change data is judged by having a P value no more than 0.01. Data with P values between 0.01 and 0.05 are denoted with an asterisk, and those with P values greater than 0.05 are designated as not significant (NS).
RPKM (reads per kilobase per million reads sequenced) values for cultures grown on yeast extract without carbohydrate (YE control) or cultures grown on the following substrates: SG, sweetgum MeGXn; SO, sorghum MeGAXn; A, arabinose; X, xylose.
Following growth of Pjdr2 on xylans compared to the YE controls, significant upregulation of genes encoding endoxylanases with signal peptides for secretion was observed. Pjdr2_0221 (encoding cell-associated GH10 Xyn10A1) and Pjdr2_4664 (encoding GH11 Xyn11) were expressed, respectively, at levels 17- and 7.1-fold higher on MeGXn and 9.6- and 50.9-fold higher on MeGAXn than the YE controls, supporting their contribution to the extracellular depolymerization of xylan (Table 2). The secreted endoxylanase Xyn11 shares 81% identity with the GH11 endoxylanase XynA from Bacillus subtilis 168 which is well defined and characterized for depolymerization of xylan to MeGX4 (aldopentauronate) and oligoxylosides (15). This extracellular Xyn11 may depolymerize xylan to MeGX4 and oligoxylosides (15, 29, 30) (Table 1 and Fig. 2B). MeGX4, in which MeG is α-1,2 linked to the penultimate xylose at the nonreducing terminus of the β-1,4 xylotetraose, is not a substrate for the GH67 α-glucuronidase (12, 14, 31, 32).
Another gene, Pjdr2_4267 (encoding a putative GH43 arabinoxylan arabinofuranohydrolase Axh43) shows 7.2-fold-higher relative expression on sorghum MeGAXn while repressed on sweetgum MeGXn (Table 2). Axh43 shares 53% amino acid identity with the characterized enzyme XynD, arabinoxylan arabinofuranohydrolase, from Bacillus subtilis 168 (18) and has a signal peptide sequence and a carbohydrate binding module family 6 (CBM6), as predicted by BLAST. The XynD from Bacillus subtilis catalyzes the release of free arabinose from sorghum MeGAXn (unpublished data). XynD has higher activity on arabinoxylans compared to oligoarabinoxylosides (18) which are preferred substrates for processing by intracellular GH51 Abf2 from B. subtilis 168 (33). This suggests that similar to B. subtilis 168, Pjdr2 may secrete Axh43 to bind to arabinoxylans and release certain arabinose units from the backbone (Table 1 and Fig. 2B). In Pjdr2, the AXOS that are generated by Xyn10A1 are processed by intracellular Abf51B (20), and AXOS products generated by Xyn11 may be intracellularly processed by Abf51B as well (Table 1 and Fig. 2B).
In addition to genes encoding enzymes for intracellular processing described earlier, there are other genes of interest that encode enzymes lacking a signal peptide. Pjdr2_5977 encodes a GH115 α-glucuronidase Agu115 which is 45% identical to that from Bacteroides ovatus (34). The expression of this gene is 52-fold higher on MeGXn and 32.8-fold higher on MeGAXn than on YE control (Table 2) and may be involved in processing aldouronates such as MeGX4 generated by GH11 digestion of xylan (34). Coregulated with the expression of genes comprising the GH10/GH67 system is the expression of xyn11 and also agu115. The agu115 gene is highly expressed on xylans to a level similar to that of agu67A, and neither of these genes include sequences corresponding to a signal peptide. GH115 α-glucuronidases from the prokaryotic Gram-negative bacterium Bacteroides ovatus ATCC 8483 (34) and eukaryotic Pichia stipitis CBS 6054 (35) have been shown to have a preference for the aldopentauronate generated by GH11 endoxylanases. Agu115 from the Gram-positive bacterium Pjdr2 also shows activity on MeGX4 generated by Xyn11 digestion of MeGXn (unpublished data). A GH11/GH115 system in Pjdr2 may complement the defined GH10/GH67 system for complete xylan utilization (Table 1 and Fig. 2).
Pjdr2_0750 encodes GH43 Xyn43B2 and Pjdr2_1907 encodes Xyn43B3, and both are expressed more than 100-fold on both xylans (Table 2). These enzymes may contribute to oligoxyloside processing. Pjdr2_4260 encodes GH10 endoxylanase Xyn10A3 expressed 2.7-fold higher only in the case of MeGAXn (Table 2). This enzyme may work synergistically with intracellular arabinofuranosidases to generate oligoxylosides from AXOS (Table 1 and Fig. 2B). In addition to these genes, the translated sequence of Pjdr2_3612 encoding GH51 arabinofuranosidase, with 2.3-fold-higher expression only on MeGAXn (Table 2) as the substrate shows 23% identity to a putative arabinofuranosidase from Geobacillus stearothermophilus T6. The selective upregulation of this gene on MeGAXn but not on MeGXn or arabinose suggests a role in the processing of AXOS yet to be defined. Pjdr2_1182 encoding a GH8 xylanase (Xyn8) which shares 62% identity with a characterized GH8 reducing end exo-oligoxylanase from Bacillus halodurans C-125 (36) was expressed 35-fold greater on both xylans (Table 2). The GH8 enzyme from B. halodurans C-125 has been shown to hydrolyze xylose from the reducing terminus of xylooligosaccharides (36). This suggests a role for the Pjdr2 Xyn8 in hydrolyzing oligoxylosides and aldouronates to xyloses and smaller aldouronates by acting from the reducing terminus (Table 1 and Fig. 2B).
Other genes relevant to the xylan utilization systems include those encoding esterases. Pjdr2 has 21 genes encoding putative carbohydrate esterases of CE1, -4, -7, -9, -12, -14, and -15 families; of these 21 genes, only the Pjdr2_4290 gene (est1) encoding a CE1 esterase was upregulated 8.5-fold higher with MeGAXn as the substrate, and it showed 3.6-fold-greater expression when grown on MeGXn and much lower expression when grown on arabinose and xylose (Table 2). Est1 may be involved in cleaving coumaroyl or feruloyl ester linkages from xylans which are more prominent in the case of MeGAXn than MeGXn. The translated sequence of est1 shares 61% identity with the characterized feruloyl esterase from Thermobacillus xylanilyticus (GenBank accession no. ADK73591) (37). The feruloyl esterase from T. xylanilyticus exhibited activity against methyl esters of hydrocinnamic acids and feruloylated arabinoxylotetraose (37). It should be noted that the preparation of MeGXn and MeGAXn for these studies included alkaline extraction that was expected to hydrolyze feruloyl as well as acetyl esters from these sources prior to the use as the substrates for growth, although some may have been more resistant to hydrolysis and therefore may have remained and were processed by Pjdr2.
Other genes encoding glycoside hydrolases upregulated during growth on MeGXn and MeGAXn include Pjdr2_2523 encoding a putative GH2 mannosidase or β-galactosidase/β-glucuronidase orthologous to a putative GH from Thermoanaerobacter mathranii, and Pjdr2_1350 encoding a GH26 mannosidase orthologous to putative GHs from Paenibacillus, Bacillus, and Bacteroides species (Table 2). Genes expressed on xylans also include Pjdr2_0774 (encoding extracellular amylase Amy13A1) and Pjdr2_5200 (encoding extracellular multimodular cell-associated amylase Amy13A2) sharing 29% and 36% amino acid identity with extracellular amylase, AmyE (BSU03040), from Bacillus subtilis, and the translated sequence of Pjdr2_0783 (encoding intracellular amylase Amy13A3) that is 47% orthologous to intracellular amylase (BSU34620) from B. subtilis (Table 2). Although these genes are expressed on xylans, their translated products are primary candidates for starch utilization in Pjdr2 that will be evaluated further.
The Pjdr2_3505 and Pjdr2_4070 genes are upregulated significantly following growth on MeGXn compared to other carbohydrates (Table 2). The Pjdr2_3505 gene encodes a GH73 mannosyl-glycoprotein endo-β-N-acetylglucosaminidase that shares 29% identity with GH73 glucosamidase from Bacillus subtilis 168 (BSU31120) which affects peptidoglycan structure during vegetative growth (38). The function of this gene is not established, although its similarity to genes for which encoded proteins have been determined suggests that it contributes to cell division- and growth-related activities. The Pjdr2_4070 gene shares 30% amino acid sequence identity with characterized GH25 proteins from Bacillus anthracis (39) encoding a putative GH25 1,4-β-N-acetylmuramidase. The role of this gene is hypothetical and suggests involvement in cell wall modification.
Expression of genes encoding xylan-inducible ABC transporters in Pjdr2.
Following growth of Pjdr2 on sweetgum MeGXn, sorghum MeGAXn, arabinose, or xylose, several gene sets encoding ABC transporters show a significant transcript increase relative to the YE controls (Table 3). The contribution of genes adjacent to agu67A in the aldouronate utilization gene cluster, Pjdr2_1320 (encoding UgpB), Pjdr2_1321 (encoding LplB), and Pjdr2_1322 (encoding UgpE) have been studied previously on MeGXn and MeGAXn by qRT-PCR (20, 22) establishing their role in the assimilation of aldotetrauronate MeGX3 generated by cell-associated Xyn10A1 and therefore contributing to the GH10/GH67 system (Table 1 and Fig. 2). Transcriptomic analysis with xylans as the substrates also revealed significant upregulation of gene clusters, Pjdr2_0728, -0729, and -0730 (abc2x, abc2y, and abc2z) were expressed 26.3- and 5.9-fold higher on MeGXn and MeGAXn, respectively, and Pjdr2_1809, -1810, and -1811 (abc3x, abc3y, and abc3z) were expressed 10.6- and 34.7-fold higher on MeGXn and MeGAXn, respectively (average fold changes shown) (Table 3). These clusters of genes encode ATP binding cassettes (ABC) and are predicted to be involved with sugar transport systems that may include transport of the different oligosaccharides generated as products of xylan depolymerization. The well-defined solute binding protein specific for xylobiose (X2) and xylotriose (X3) from Streptomyces thermoviolaceus OPC-520 (40) and Geobacillus stearothermophilus (41) share 30% and 49% identity, respectively, with amino acid sequences encoded by gene clusters abc2x, abc2y, and abc2z. This cassette in Pjdr2 is likely involved with transport of oligoxylosides such as xylobiose and xylotriose (Table 1 and Fig. 2B).
TABLE 3.
Expression of candidate genes encoding ABC transport proteins by growth on xylans
| LTa | Protein productb | Fold changec |
Linear RPKM valued |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SG/YE | SO/YE | A/YE | X/YE | SG | SO | A | X | YE | ||
| 0661 | Extracellular SBP | 0.2 | 58.3 | 1,917.5 | NS | 0.87 | 226.77 | 7,460.49 | 4.82 | 3.89 |
| 0662 | ABC transporter | 0.2 | 27.6 | 1,267.8 | NS | 0.43 | 73.54 | 3,374.44 | 2.07 | 2.66 |
| 0663 | Inner membrane translocator | 0.2 | 30.3 | 1,301.2 | 0.6* | 0.52 | 101.12 | 4,343.48 | 2.04 | 3.34 |
| 0664 | ABC periplasmic component | 0.3 | NS | 27.8 | 0.5 | 1.5 | 6.6 | 165.8 | 2.8 | 6.0 |
| 0728 | Extracellular SBP | 27.9 | 6.2 | 0.4 | 0.5 | 669.39 | 147.82 | 9.12 | 10.95 | 23.99 |
| 0729 | BPD transport system IMP | 21.3 | 4.8 | 0.6 | 0.6 | 295.87 | 67.04 | 8.47 | 8.89 | 13.87 |
| 0730 | BPD transport system IMP | 29.8 | 6.8 | 0.8 | 0.8* | 453.36 | 103.99 | 11.49 | 12.81 | 15.21 |
| 0771 | Extracellular SBP | 13.2 | 9.3 | 0.1 | 0.1 | 842.81 | 595.64 | 3.29 | 4.79 | 63.96 |
| 0772 | BPD transport system IMP | 14.2 | 9.9 | 0.1 | 0.1 | 177.17 | 122.81 | 1.29 | 1.40 | 12.46 |
| 0773 | BPD transport system IMP | 22.0 | 17.1 | 0.1 | 0.2 | 227.01 | 176.51 | 1.11 | 1.89 | 10.31 |
| 1320 | Extracellular SBP (UgpB) | 170.4 | 147.2 | 0.2 | 0.5 | 3,171.57 | 2,740.67 | 3.23 | 8.48 | 18.61 |
| 1321 | BPD transport system IMP (LplB) | 158.8 | 126.3 | 0.3 | 0.4 | 791.00 | 629.36 | 1.41 | 2.12 | 4.98 |
| 1322 | BPD transport system IMP (UgpE) | 141.1 | 149.9 | 0.6* | 0.7 | 1,040.91 | 1,105.83 | 4.30 | 5.23 | 7.38 |
| 1809 | BPD transport system IMP | 15.3 | 31.1 | 0.5 | 0.5 | 22.26 | 45.11 | 0.75 | 0.76 | 1.45 |
| 1810 | BPD transport system IMP | 24.5 | 53.2 | NS | NS | 18.87 | 40.91 | 0.54 | 0.69 | 0.77 |
| 1811 | Extracellular SBP | 10.1 | 19.9 | NS | NS | 86.58 | 170.32 | 7.69 | 9.31 | 8.56 |
| 3245 | Periplasmic binding protein | 1.6 | 12.7 | 12.3 | 14.0 | 72.49 | 581.88 | 560.14 | 638.30 | 45.72 |
| 4270 | Extracellular SBP | 0.1 | 16.4 | 0.5 | 0.4 | 1.68 | 194.94 | 5.62 | 5.01 | 11.89 |
| 5273 | BPD transport system IMP | 0.6* | 6.0 | NS | NS | 0.83 | 8.21 | 1.09 | 1.36 | 1.38 |
| 5274 | BPD transport system IMP | NS | 4.8 | NS | 1.6* | 1.34 | 6.96 | 1.81 | 2.31 | 1.44 |
| 5275 | Extracellular SBP | 0.2 | 8.1 | 0.4 | 0.5 | 0.32 | 11.61 | 0.51 | 0.67 | 1.44 |
| 5314 | BPD transport system IMP | 31.2 | 34.1 | 0.4 | 0.5 | 333.79 | 364.75 | 4.66 | 5.11 | 10.69 |
| 5315 | BPD transport system IMP | 32.5 | 30.7 | 0.4 | 0.4 | 292.54 | 277.08 | 3.53 | 3.55 | 9.02 |
| 5316 | Extracellular SBP | 28.2 | 29.4 | 0.2 | 0.2 | 723.90 | 755.00 | 4.82 | 5.74 | 25.65 |
| 5596 | BPD transport system IMP | 61.4 | 24.6 | 0.5 | 0.7 | 385.74 | 154.77 | 3.18 | 4.69 | 6.29 |
| 5597 | BPD transport system IMP | 42.5 | 15.7 | 0.3 | NS | 178.95 | 66.13 | 1.20 | 3.08 | 4.21 |
| 5598 | Extracellular SBP | 72.1 | 22.8 | 0.1 | 0.3* | 654.34 | 206.97 | 1.02 | 3.10 | 9.08 |
LT, locus tag referenced as Pjdr2_XXXX with XXXX being a four-digit number.
SBP, solute binding protein; IMP, inner membrane protein; BPD, binding protein dependent. The name of the candidates previously evaluated in our laboratory is shown in bold type.
Transcript levels of candidate genes upregulated with 4-fold-greater expression (underlined) or 10-fold-greater expression (bold) than the yeast extract without carbohydrate (YE control) growth are indicated. The substrates are shown as follows: SG, sweetgum MeGXn; SO, sorghum MeGAXn; A, arabinose; X, xylose. The Results and Discussion section refers to the data in this table as the average fold increases (average of expression levels of all three component genes of the ABC cluster on each substrate being considered). Significance of fold change data is judged by having a P value no more than 0.01. Data with P values between 0.01 and 0.05 are denoted with an asterisk, and those with P values greater than 0.05 are designated as not significant (NS).
RPKM values for cultures grown on yeast extract without carbohydrate (YE control) or cultures grown on the substrates shown in footnote c.
Upregulation of genes Pjdr2_5273, -5274, and -5275 (abc5x, abc5y, and abc5z) following growth of Pjdr2 on sorghum MeGAXn compared to all other carbohydrates suggests that these genes play a role in the transport of oligoarabinoxylosides. The abc5z gene, which is expressed 8.1-fold higher on MeGAXn (Table 3) and encodes a solute binding protein, has a translated sequence 23% and 25% identical to arabino-linked oligosaccharide binding protein of ABC transporters from Bacillus subtilis (AraN) (33) and Geobacillus stearothermophilus (AbnE) (42), respectively. These genes are also predicted to be involved with sugar transport systems. The ABC transporter Abc5x, Abc5y, and Abc5z may contribute to oligoarabinoxyloside transport into the cell (Table 1 and Fig. 2B).
The genes Pjdr2_0661, -0662, and -0663 (abc4x, abc4y, and abc4z) are expressed 1,495.5- and 58.1-fold (averages) higher following growth of Pjdr2 on arabinose and sorghum MeGAXn, respectively, but not on sweetgum MeGXn (Table 3). The linked gene Pjdr2_0664 is also upregulated on arabinose although expression on MeGAXn cannot be interpreted due to a high P value. These genes are predicted to belong to the ATP-dependent d-xylose and l-arabinose uptake systems. The solute binding protein, Abc4x, shares 71% amino acid identity with arabinose binding protein and 37% amino acid identity with arabinan-derived oligosaccharide binding protein of ABC transporters from Geobacillus stearothermophilus (AraE and AbnE, respectively) (42). The high sequence identity with the arabinose binding protein and the greater upregulation by arabinose of the genes in this cluster compared to MeGAXn indicate that the expression of this cluster may be inducible by arabinose. This is the only set of genes commonly expressed on both MeGAXn and arabinose (Table 3). The Abc4x, Abc4y, and Abc4z transporter may transport arabinose generated outside the cell by action of Axh43 on MeGAXn (Table 1 and Fig. 2B).
Gene cassettes Pjdr2_0771, -0772, and -0773 are predicted to encode a maltose/maltodextrin transport system and are upregulated slightly more on MeGXn than MeGAXn (Table 3). These genes are located adjacent to amy13A1 encoding an extracellular amylase which is also upregulated on xylans (Table 2). On the basis of its annotation and location adjacent to amy13A1, this gene cluster may encode an ABC transporter that may be associated with transport of starch-derived oligosaccharides and will be considered for further investigation. Cassettes Pjdr2_5314, -5315, and -5316 (abc6x, abc6y, and abc6z) and Pjdr2_5596, -5597, and -5598 (abc7x, abc7y, and abc7z) are also expressed on xylans (Table 3) and are predicted to be involved with transport of multiple or unknown carbohydrates, respectively. The functional roles of these cassettes are yet to be investigated.
Pjdr2_4270 encoding an extracellular solute binding protein was expressed 16.4-fold greater following growth on sorghum MeGAXn but not on sweetgum MeGXn (Table 3), suggesting its potential role in transport of oligoarabinoxylosides. Pjdr2_3245 which encodes a predicted periplasmic binding protein was upregulated with an average 13-fold higher following growth of Pjdr2 on MeGAXn, arabinose, and xylose (Table 3) and is predicted to be involved with an iron complex transport system and may be important for cell growth-related activities. Neither of these two genes is found in a cluster encoding other ABC components.
It has been proposed for Gram-positive bacteria such as B. subtilis that one ATPase (also sometimes referred to as nucleotide binding domain [NBD]) of an ABC transporter may be shared by more than one ABC transport system (22, 43). The Pjdr2_0262 gene encoding an ABC transporter ATPase component sharing 28% amino acid sequence identity with a putative ABC transporter ATPase from B. subtilis 168 (BSU03730) may be recruited to provide ATP to other ABC transporters for assimilation of oligosaccharides. Relative to YE controls, this gene has a transcript level that is 2.6-fold higher on MeGXn, 2.8-fold higher on MeGAXn, 7.3-fold higher on arabinose, and 4.8-fold higher on xylose.
Expression of genes encoding transcriptional regulators during xylan utilization in Pjdr2.
Most prominently upregulated genes encoding transcriptional regulators (Table 4) were identified. These genes include the Pjdr2_0332 encoding GntR family protein, which showed 9.7- and 6.0-fold higher expression on MeGXn and MeGAXn, respectively. The Pjdr2_1270 gene encoding Fur family protein was 11.2-, 13.4-, and 9.6-fold higher on MeGAXn, arabinose, and xylose, respectively, and the Pjdr2_3244 gene encoding TetR family protein was 6.8-, 9.4-, and 16.2-, fold higher on MeGAXn, arabinose, and xylose, respectively. The gene Pjdr2_4272 encoding an AraC family protein with 5.2- and 17.0-fold-higher expression on MeGXn and MeGAXn, respectively, was also identified (Table 4). The Pjdr2_5182 gene encoding a ROK (repressor, open reading frame, kinase) family protein showed 5.4-, 7.3-, and 6.8-fold-higher expression on MeGXn, MeGAXn, and xylose, respectively, and the Pjdr2_5888 gene encoding Fur family protein showed a 4.3- and 5.7-fold-higher expression on MeGAXn and arabinose, respectively. In Geobacillus stearothermophilus, the glucuronic acid utilization gene cluster includes a regulatory gene encoding a product predicted to be a member of the GntR family (44) which is highly expressed on xylans in Pjdr2, supporting a role in the regulatory control of the xylan utilization systems. When screened on the basis of their location on the genome and synteny with other genes in the xylan utilization systems, the level of expression of the Pjdr2_1318 gene encoding YesN, AraC family, was 2.4-fold on MeGXn and 1.5-fold on MeGAXn, and the Pjdr2_1319 gene encoding YesM, the HAMP (histidine kinase, adenylyl cyclase, methyl binding protein, phosphatase) sensor domain, was 1.9-fold on MeGXn and an undetermined value on MeGAXn due to a high P value. Both the yesN and yesM genes as well as other genes in the aldouronate utilization gene cluster showed coordinate upregulation when determined by qRT-PCR in Pjdr2 (22).
TABLE 4.
Summarized candidate genes encoding transcriptional regulators expressed by growth on xylans
| LTa | Neighboring gene(s) | Protein productb | Fold changec |
Linear RPKM valued |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SG/YE | SO/YE | A/YE | X/YE | SG | SO | A | X | YE | |||
| 0332 | GntR family TR | 9.7 | 6.0 | NS | NS | 35.81 | 22.31 | 3.19 | 3.47 | 3.69 | |
| 0665 | abc4x, abc4y, abc4z | Sensor with HAMP domain | 0.2 | NS | 19.9 | 0.5 | 1.74 | 7.60 | 149.38 | 3.44 | 7.51 |
| 0666 | AraC family TR | 0.4 | NS | 11.8 | 0.7 | 5.44 | 16.87 | 168.52 | 9.22 | 14.29 | |
| 0667 | LysR family TR | 0.7* | 1.7* | 7.2 | NS | 14.15 | 33.98 | 145.04 | 21.88 | 20.05 | |
| 1270 | Fur family ferric uptake regulator | 1.6 | 11.2 | 13.4 | 9.6 | 144.98 | 1,012.65 | 1,216.09 | 873.80 | 90.71 | |
| 1318 | Belongs to GH10/GH67 regulon | AraC family TR (YesN) | 2.4 | 1.5* | 0.2 | 0.3 | 34.98 | 21.95 | 3.52 | 3.63 | 14.64 |
| 1319 | Sensor with HAMP domain (YesM) | 1.9 | NS | 0.3 | 0.2 | 11.61 | 7.03 | 1.63 | 1.45 | 5.98 | |
| 1442 | LuxR family TR | 7.1 | 0.6 | 2.5 | 11.8 | 229.43 | 18.14 | 80.42 | 378.98 | 32.17 | |
| 1806 | Pjdr2_1809, -1810, -1811 (encoding ABC) | HxlR family TR | 1.2* | 2.7 | 1.9 | 2.5 | 119.27 | 263.88 | 184.06 | 236.80 | 96.22 |
| 1808 | AraC family TR | 2.9 | 7.9 | NS | 1.9 | 29.48 | 79.99 | 11.22 | 19.70 | 10.19 | |
| 3244 | TetR family TR | NS | 6.8 | 9.4 | 16.2 | 27.82 | 156.13 | 216.67 | 372.79 | 22.96 | |
| 3598 | abf51B | ArsR family TR (ArsR) | NS | 1.8 | 1.0* | 1.5* | 5.97 | 8.52 | 4.82 | 6.91 | 4.77 |
| 4254 | xyn10A3 and xyn43B4 | AraC family TR | 0.2* | 32.0 | 0.4* | 0.6 | 0.34 | 49.90 | 0.56 | 0.99 | 1.56 |
| 4266 | AraC family TR | 0.1 | 2.4* | 0.2 | 0.2 | 0.65 | 15.84 | 1.33 | 1.37 | 6.55 | |
| 4268 | AraC family TR | 0.1 | 7.5 | 0.4 | 0.3 | 2.06 | 163.94 | 8.34 | 6.25 | 21.86 | |
| 4269 | Sensor with HAMP domain | 0.1 | 10.1 | 0.5 | 0.4 | 1.61 | 136.44 | 6.08 | 5.38 | 13.54 | |
| 4272 | AraC family TR | 5.2 | 17.0 | 0.7 | 0.8 | 55.35 | 179.91 | 7.38 | 8.27 | 10.56 | |
| 5181 | XRE family TR | NS | 1.8 | 1.3 | 1.8 | 30.78 | 44.19 | 31.18 | 46.09 | 25.00 | |
| 5182 | ROK family protein | 5.4 | 7.3 | 1.2* | 6.8 | 115.10 | 157.00 | 24.56 | 144.62 | 21.40 | |
| 5887 | Winged helix family two-component TR | NS | 4.0 | 5.6 | 3.1 | 32.61 | 175.80 | 249.47 | 136.09 | 44.45 | |
| 5888 | Fur family ferric uptake regulator | NS | 4.3 | 5.7 | 3.1 | 28.27 | 119.64 | 158.95 | 86.81 | 28.13 | |
LT, locus tag referenced as Pjdr2_XXXX with XXXX being a four-digit number.
TR, transcriptional regulator. Name of the candidates previously evaluated in our laboratory are shown in bold type.
Transcript levels of summarized genes upregulated (underlined) and those expressed 4- fold greater (bold) than the yeast extract without carbohydrate (YE control) growth are indicated. The substrates are shown as follows: SG, sweetgum MeGXn; SO, sorghum MeGAXn; A, arabinose; X, xylose. Significance of fold change data is judged by having a P value no more than 0.01. Data with P values between 0.01 and 0.05 are denoted with an asterisk, and those with P values greater than 0.05 are designated as not significant (NS).
RPKM values for cultures grown on yeast extract without carbohydrate (YE control) or cultures grown on the substrates shown in footnote c.
Xylan utilization as previously defined in Pjdr2 utilizes a GH10/GH67 system for the efficient processing of xylans such as sweetgum MeGXn and sorghum MeGAXn (19, 20, 22, 23). An advantage of this system is its ability to assimilate MeGX3 generated by the action of the extracellular cell-associated GH10 xylanase, Xyn10A1, on both MeGXn and MeGAXn and process MeGX3 with an intracellular GH67 α-glucuronidase, Agu67A, to release xylotriose, which is then further processed to xylose (19), as depicted in Fig. 2B. Key to this GH10/GH67 system is the specificity of the GH67 α-glucuronidase for the GH10 Xyn10A1-generated MeGX3 product in which the nonreducing terminal xylose of β-1,4-xylotriose is modified with an α-1,2-linked 4-O-methylglucuronate. This allows the assimilation and subsequent metabolism of all of the oligoxylosides and aldouronates generated by the cell-associated Xyn10A1. In the case of MeGAXn, oligoarabinoxylosides released by Xyn10A1 are assimilated by transporters for processing, presumably by Abf51B, to release xylobiose and xylotriose for further metabolism (20). This allows maximal utilization of both arabinose as well as xylose in the hemicellulose fractions of both dicots and monocots (20, 22). The surface layer homology (SLH) domains on Xyn10A1 contribute to association with the bacterial cell surface. The carbohydrate binding modules (CBMs) through their interactions with cellulose and xylans may contribute to the generation of oligosaccharides at the cell surface (Fig. 2B). Proximity to the substrate binding domain of the ABC transporter complex may enable rapid assimilation of oligosaccharides without diffusion into the medium (19, 23). GH10/GH67 systems have been identified in other bacteria, including Thermotoga maritima (45–47), and Thermoanaerobacterium sp. strain JW/SL-YS485 (48, 49). Both of these organisms are under investigation as biocatalysts for consolidated bioprocessing of lignocellulosic biomass. Earlier studies carried out following growth of Pjdr2 on xylose supplemented with 0.1% yeast extract led to the expression of the GH10/GH67 system (20), which was lower when 0.5% yeast extract was used in this study. This suggests that larger amounts of yeast extract drive the growth of Pjdr2 without the expression of xylan utilization genes on xylose as a substrate.
The metabolic potential of Pjdr2 for direct conversion of xylans and its ability to process aldouronates supports its further development as a biocatalyst for direct conversion of the hemicelluloses to targeted products. The previously defined GH10/GH67 MeGXn and MeGAXn utilization systems (19–23) has here been extended to a GH11/GH115 system for the utilization of xylans in Pjdr2 (Table 1 and Fig. 2). An alternative system containing GH11 and GH30 endoxylanases occurs in the Bacillus subtilis 168 and related species that processes xylan to xylobiose and xylotriose, and MeGX3. In this system, only the oligoxylosides are assimilated and metabolized, and the aldouronate MeGX3 accumulates in the medium (15, 29, 30). In contrast, Pjdr2 contains genes encoding ABC transporters for assimilation of aldouronates (MeGX3 and MeGX4) and genes encoding GH67 and GH115 α-glucuronidases for intracellular processing, which make this bacterium uniquely efficient for xylan utilization (19, 22, 23). These genes of interest may be introduced into fermentative strains of related bacteria for complete conversion of xylans to useful products.
The sequenced genome of Pjdr2 includes genes for efficient utilization of a variety of polysaccharides, including soluble β-glucans, starch, and arabinans, as well as xylans. For each of these substrates, cell-associated enzymes with SLH domains and CBMs for binding to polysaccharides are thought to play a role and contribute to the preferential utilization of the polysaccharides compared to oligosaccharides derived from depolymerization. Preliminary studies have demonstrated the formation of lactate, acetate, and ethanol from starch and xylans under oxygen-limiting conditions (unpublished results). The efficient utilization of these polysaccharides by Pjdr2 supports its further development as a biocatalyst for the direct conversion of the hemicellulose fraction of energy crops and agricultural residues to targeted products.
ACKNOWLEDGMENTS
We thank Christa Pennacchio for managing the RNA sequencing project and also the other members of the Joint Genome Institute, Walnut Creek, CA, for conducting the RNA sequencing work. We appreciate the assistance provided by K. T. Shanmugam, L. O. Ingram, John D. Rice, Virginia Chow, and Yinping Guo, all at the Department of Microbiology and Cell Science, University of Florida, Gainesville, FL. We also thank Daniel Cullen for his expertise and helpful discussions regarding transcriptome studies and Diane Dietrich for professional support, both at the USDA, U.S. Forest Service, Forest Products Laboratory, Madison, WI.
This research was supported by Biomass Research & Development Initiative competitive grant 2011-10006-30358 from the USDA National Institute of Food and Agriculture and by Florida Energy Systems Consortium, State University System of Florida, project 00077818. The RNA sequencing work at the Joint Genome Institute, Walnut Creek, CA, was supported by the Office of Science of the U.S. Department of Energy under contract DE-AC02-05CH11231.
REFERENCES
- 1.Chundawat SP, Beckham GT, Himmel ME, Dale BE. 2011. Deconstruction of lignocellulosic biomass to fuels and chemicals. Annu Rev Chem Biomol Eng 2:121–145. doi: 10.1146/annurev-chembioeng-061010-114205. [DOI] [PubMed] [Google Scholar]
- 2.Ingram LO, Aldrich HC, Borges AC, Causey TB, Martinez A, Morales F, Saleh A, Underwood SA, Yomano LP, York SW, Zaldivar J, Zhou S. 1999. Enteric bacterial catalysts for fuel ethanol production. Biotechnol Prog 15:855–866. doi: 10.1021/bp9901062. [DOI] [PubMed] [Google Scholar]
- 3.Jarboe LR, Grabar TB, Yomano LP, Shanmugan KT, Ingram LO. 2007. Development of ethanologenic bacteria. Adv Biochem Eng Biotechnol 108:237–261. doi: 10.1007/10_2007_068. [DOI] [PubMed] [Google Scholar]
- 4.Jeffries TW, Jin YS. 2004. Metabolic engineering for improved fermentation of pentoses by yeasts. Appl Microbiol Biotechnol 63:495–509. doi: 10.1007/s00253-003-1450-0. [DOI] [PubMed] [Google Scholar]
- 5.Lynd LR, Wyman CE, Gerngross TU. 1999. Biocommodity engineering. Biotechnol Prog 15:777–793. doi: 10.1021/bp990109e. [DOI] [PubMed] [Google Scholar]
- 6.Shanmugam KT, Ingram LO. 2008. Engineering biocatalysts for production of commodity chemicals. J Mol Microbiol Biotechnol 15:8–15. doi: 10.1159/000111988. [DOI] [PubMed] [Google Scholar]
- 7.Lau MW, Gunawan C, Balan V, Dale BE. 2010. Comparing the fermentation performance of Escherichia coli KO11, Saccharomyces cerevisiae 424A(LNH-ST) and Zymomonas mobilis AX101 for cellulosic ethanol production. Biotechnol Biofuels 3:11. doi: 10.1186/1754-6834-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ou MS, Ingram LO, Shanmugam KT. 2011. l(+)-Lactic acid production from non-food carbohydrates by thermotolerant Bacillus coagulans. J Ind Microbiol Biotechnol 38:599–605. doi: 10.1007/s10295-010-0796-4. [DOI] [PubMed] [Google Scholar]
- 9.Zhang YH, Lynd LR. 2005. Cellulose utilization by Clostridium thermocellum: bioenergetics and hydrolysis product assimilation. Proc Natl Acad Sci U S A 102:7321–7325. doi: 10.1073/pnas.0408734102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Svetlitchnyi VA, Kensch O, Falkenhan DA, Korseska SG, Lippert N, Prinz M, Sassi J, Schickor A, Curvers S. 2013. Single-step ethanol production from lignocellulose using novel extremely thermophilic bacteria. Biotechnol Biofuels 6:31. doi: 10.1186/1754-6834-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao GL, Zhao L, Wang AJ, Wang ZY, Ren NQ. 2014. Single-step bioconversion of lignocellulose to hydrogen using novel moderately thermophilic bacteria. Biotechnol Biofuels 7:82. doi: 10.1186/1754-6834-7-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Biely P, Vrsanska M, Tenkanen M, Kluepfel D. 1997. Endo-beta-1,4-xylanase families: differences in catalytic properties. J Biotechnol 57:151–166. doi: 10.1016/S0168-1656(97)00096-5. [DOI] [PubMed] [Google Scholar]
- 13.Kuhad RC, Singh A, Eriksson KE. 1997. Microorganisms and enzymes involved in the degradation of plant fiber cell walls. Adv Biochem Eng Biotechnol 57:45–125. [DOI] [PubMed] [Google Scholar]
- 14.Preston JF, Hurlbert JC, Rice JD, Ragunathan A, St. John FJ. 2003. Microbial strategies for the depolymerization of glucuronoxylan: leads to the biotechnological applications of endoxylanases. ACS Symp Ser 855:191–210. doi: 10.1021/bk-2003-0855.ch012. [DOI] [Google Scholar]
- 15.St. John FJ, Rice JD, Preston JF. 2006. Characterization of XynC from Bacillus subtilis subsp. subtilis strain 168 and analysis of its role in depolymerization of glucuronoxylan. J Bacteriol 188:8617–8626. doi: 10.1128/JB.01283-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Verbruggen MA, Spronk BA, Schols HA, Beldman G, Voragen AG, Thomas JR, Kamerling JP, Vliegenthart JF. 1998. Structures of enzymically derived oligosaccharides from sorghum glucuronoarabinoxylan. Carbohydr Res 306:265–274. [DOI] [PubMed] [Google Scholar]
- 17.Mosier N, Wyman C, Dale B, Elander R, Lee YY, Holtzapple M, Ladisch M. 2005. Features of promising technologies for pretreatment of lignocellulosic biomass. Bioresour Technol 96:673–686. doi: 10.1016/j.biortech.2004.06.025. [DOI] [PubMed] [Google Scholar]
- 18.Bourgois TM, Van Craeyveld V, Van Campenhout S, Courtin CM, Delcour JA, Robben J, Volckaert G. 2007. Recombinant expression and characterization of XynD from Bacillus subtilis subsp. subtilis ATCC 6051: a GH 43 arabinoxylan arabinofuranohydrolase. Appl Microbiol Biotechnol 75:1309–1317. doi: 10.1007/s00253-007-0956-2. [DOI] [PubMed] [Google Scholar]
- 19.Nong G, Rice JD, Chow V, Preston JF. 2009. Aldouronate utilization in Paenibacillus sp. strain JDR-2: physiological and enzymatic evidence for coupling of extracellular depolymerization and intracellular metabolism. Appl Environ Microbiol 75:4410–4418. doi: 10.1128/AEM.02354-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sawhney N, Preston JF. 2014. GH51 arabinofuranosidase and its role in the methylglucuronoarabinoxylan utilization system in Paenibacillus sp. strain JDR-2. Appl Environ Microbiol 80:6114–6125. doi: 10.1128/AEM.01684-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chow V, Nong G, St. John FJ, Rice JD, Dickstein E, Chertkov O, Bruce D, Detter C, Brettin T, Han J, Woyke T, Pitluck S, Nolan M, Pati A, Martin J, Copeland A, Land ML, Goodwin L, Jones JB, Ingram LO, Shanmugam KT, Preston JF. 2012. Complete genome sequence of Paenibacillus sp. strain JDR-2. Stand Genomic Sci 6(1):1–10. doi: 10.4056/sigs.2374349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chow V, Nong G, Preston JF. 2007. Structure, function, and regulation of the aldouronate utilization gene cluster from Paenibacillus sp. strain JDR-2. J Bacteriol 189:8863–8870. doi: 10.1128/JB.01141-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.St. John FJ, Rice JD, Preston JF. 2006. Paenibacillus sp. strain JDR-2 and XynA1: a novel system for methylglucuronoxylan utilization. Appl Environ Microbiol 72:1496–1506. doi: 10.1128/AEM.72.2.1496-1506.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. 2014. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dubois M, Gilles KA, Hamilton JK, Rebers PA, Smith F. 1956. Colorimetric method for the determination of sugars and related substances. Anal Chem 28:350–356. doi: 10.1021/ac60111a017. [DOI] [PubMed] [Google Scholar]
- 26.Zucker M, Hankin L. 1970. Regulation of pectate lyase synthesis in Pseudomonas fluorescens and Erwinia carotovora. J Bacteriol 104:13–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qiagen. 2005. RNAprotect bacteria reagent handbook, 2nd ed. Qiagen, Hilden, Germany. [Google Scholar]
- 28.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B 57:289–300. [Google Scholar]
- 29.Rhee MS, Wei L, Sawhney N, Rice JD, St. John FJ, Hurbert JC, Preston JF. 2014. Engineering the xylan utilization system in Bacillus subtilis for production of acidic xylooligosaccharides. Appl Environ Microbiol 80:917–927. doi: 10.1128/AEM.03246-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishitani K, Nevins DJ. 1991. Glucuronoxylan xylanohydrolase. A unique xylanase with the requirement for appendant glucuronosyl units. J Biol Chem 266:6539–6543. [PubMed] [Google Scholar]
- 31.Bronnenmeier K, Meissner H, Stocker S, Staudenbauer WL. 1995. Alpha-d-glucuronidases from the xylanolytic thermophiles Clostridium stercorarium and Thermoanaerobacterium saccharolyticum. Microbiology 141:2033–2040. doi: 10.1099/13500872-141-9-2033. [DOI] [PubMed] [Google Scholar]
- 32.Nagy T, Nurizzo D, Davies GJ, Biely P, Lakey JH, Bolam DN, Gilbert HJ. 2003. The alpha-glucuronidase, GlcA67A, of Cellvibrio japonicus utilizes the carboxylate and methyl groups of aldobiouronic acid as important substrate recognition determinants. J Biol Chem 278:20286–20292. doi: 10.1074/jbc.M302205200. [DOI] [PubMed] [Google Scholar]
- 33.Inácio JM, Correia IL, de Sá-Nogueira I. 2008. Two distinct arabinofuranosidases contribute to arabino-oligosaccharide degradation in Bacillus subtilis. Microbiology 154:2719–2729. doi: 10.1099/mic.0.2008/018978-0. [DOI] [PubMed] [Google Scholar]
- 34.Rogowski A, Baslé A, Farinas CS, Solovyova A, Mortimer JC, Dupree P, Gilbert HJ, Bolam DN. 2014. Evidence that GH115 α-glucuronidase activity, which is required to degrade plant biomass, is dependent on conformational flexibility. J Biol Chem 289:53–64. doi: 10.1074/jbc.M113.525295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kolenová K, Ryabova O, Vrsanská M, Biely P. 2010. Inverting character of family GH115 α-glucuronidases. FEBS Lett 584:4063–4068. doi: 10.1016/j.febslet.2010.08.031. [DOI] [PubMed] [Google Scholar]
- 36.Honda Y, Kitaoka M. 2004. A family 8 glycoside hydrolase from Bacillus halodurans C-125 (BH2105) is a reducing end xylose-releasing exo-oligoxylanase. J Biol Chem 279:55097–55103. doi: 10.1074/jbc.M409832200. [DOI] [PubMed] [Google Scholar]
- 37.Rakotoarivonina H, Hermant B, Chabbert B, Touzel JP, Remond C. 2011. A thermostable feruloyl-esterase from the hemicellulolytic bacterium Thermobacillus xylanilyticus releases phenolic acids from non-pretreated plant cell walls. Appl Microbiol Biotechnol 90:541–552. doi: 10.1007/s00253-011-3103-z. [DOI] [PubMed] [Google Scholar]
- 38.Horsburgh GJ, Atrih A, Williamson MP, Foster SJ. 2003. LytG of Bacillus subtilis is a novel peptidoglycan hydrolase: the major active glucosaminidase. Biochemistry 42:257–264. doi: 10.1021/bi020498c. [DOI] [PubMed] [Google Scholar]
- 39.Martinez-Fleites C, Korczynska JE, Davies GJ, Cope MJ, Turkenburg JP, Taylor EJ. 2009. The crystal structure of a family GH25 lysozyme from Bacillus anthracis implies a neighboring-group catalytic mechanism with retention of anomeric configuration. Carbohydr Res 344:1753–1757. doi: 10.1016/j.carres.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 40.Tsujibo H, Kosaka M, Ikenishi S, Sato T, Miyamoto K, Inamori Y. 2004. Molecular characterization of a high-affinity xylobiose transporter of Streptomyces thermoviolaceus OPC-520 and its transcriptional regulation. J Bacteriol 186:1029–1037. doi: 10.1128/JB.186.4.1029-1037.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shulami S, Zaide G, Zolotnitsky G, Langut Y, Feld G, Sonenshein AL, Shoham Y. 2007. A two-component system regulates the expression of an ABC transporter for xylo-oligosaccharides in Geobacillus stearothermophilus. Appl Environ Microbiol 73:874–884. doi: 10.1128/AEM.02367-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shulami S, Raz-Pasteur A, Tabachnikov O, Gilead-Gropper S, Shner I, Shoham Y. 2011. The l-arabinan utilization system of Geobacillus stearothermophilus. J Bacteriol 193:2838–2850. doi: 10.1128/JB.00222-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Quentin Y, Fichant G, Denizot F. 1999. Inventory, assembly and analysis of Bacillus subtilis ABC transport systems. J Mol Biol 287:467–484. doi: 10.1006/jmbi.1999.2624. [DOI] [PubMed] [Google Scholar]
- 44.Shulami S, Gat O, Sonenshein AL, Shoham Y. 1999. The glucuronic acid utilization gene cluster from Bacillus stearothermophilus T-6. J Bacteriol 181:3695–3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Conners SB, Montero CI, Comfort DA, Shockley KR, Johnson MR, Chhabra SR, Kelly RM. 2005. An expression-driven approach to the prediction of carbohydrate transport and utilization regulons in the hyperthermophilic bacterium Thermotoga maritima. J Bacteriol 187:7267–7282. doi: 10.1128/JB.187.21.7267-7282.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Winterhalter C, Liebl W. 1995. Two extremely thermostable xylanases of the hyperthermophilic bacterium Thermotoga maritima MSB8. Appl Environ Microbiol 61:1810–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liebl W, Winterhalter C, Baumeister W, Armbrecht M, Valdez M. 2008. Xylanase attachment to the cell wall of the hyperthermophilic bacterium Thermotoga maritima. J Bacteriol 190:1350–1358. doi: 10.1128/JB.01149-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shao W, Deblois S, Wiegel J. 1995. A high-molecular-weight, cell-associated xylanase isolated from exponentially growing Thermoanaerobacterium sp. strain JW/SL-YS485. Appl Environ Microbiol 61:937–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu SY, Gherardini FC, Matuschek M, Bahl H, Wiegel J. 1996. Cloning, sequencing, and expression of the gene encoding a large S-layer-associated endoxylanase from Thermoanaerobacterium sp. strain JW/SL-YS 485 in Escherichia coli. J Bacteriol 178:1539–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]