Abstract
The main pathogenic enterohemorrhagic Escherichia coli (EHEC) strains are defined as Shiga toxin (Stx)-producing E. coli (STEC) belonging to one of the following serotypes: O157:H7, O26:H11, O103:H2, O111:H8, and O145:H28. Each of these five serotypes is known to be associated with a specific subtype of the intimin-encoding gene (eae). The objective of this study was to evaluate the prevalence of bovine carriers of these “top five” STEC in the four adult cattle categories slaughtered in France. Fecal samples were collected from 1,318 cattle, including 291 young dairy bulls, 296 young beef bulls, 337 dairy cows, and 394 beef cows. A total of 96 E. coli isolates, including 33 top five STEC and 63 atypical enteropathogenic E. coli (aEPEC) isolates, with the same genetic characteristics as the top five STEC strains except that they lacked an stx gene, were recovered from these samples. O157:H7 was the most frequently isolated STEC serotype. The prevalence of top five STEC (all serotypes included) was 4.5% in young dairy bulls, 2.4% in young beef bulls, 1.8% in dairy cows, and 1.0% in beef cows. It was significantly higher in young dairy bulls (P < 0.05) than in the other 3 categories. The basis for these differences between categories remains to be elucidated. Moreover, simultaneous carriage of STEC O26:H11 and STEC O103:H2 was detected in one young dairy bull. Lastly, the prevalence of bovine carriers of the top five STEC, evaluated through a weighted arithmetic mean of the prevalence by categories, was estimated to 1.8% in slaughtered adult cattle in France.
INTRODUCTION
Enterohemorrhagic Escherichia coli (EHEC) strains are responsible for severe clinical symptoms, such as hemorrhagic colitis (HS) and hemolytic uremic syndrome (HUS) (1). EHEC are of serious public health concern because HUS is the leading cause of acute renal failure in children that is potentially fatal (2). Cattle are known to be the reservoir of EHEC strains, and bovine carriers do not show signs of clinical disease (3). Human infection occurs mainly through consumption of contaminated food and water (4). Monitoring EHEC strains is required at all steps along the food chain, from the area of primary production through the area of food production and handling and to foodstuffs, in order to identify and prevent contamination of food. Moreover, quantitative data on the prevalence of EHEC strains should be provided for quantitative microbial risk assessment, which should help in risk management. The identification of EHEC strains during the bacterial examination of food or environmental samples, conducted outside a clinical context involving humans, is challenging. In fact, it is the detection of the different virulence factors within the same strain that enables to estimate its pathogenicity.
The main virulence factor of EHEC contributing to pathogenicity is Shiga toxin (Stx) (5). But not all Shiga toxin-producing E. coli (STEC) strains are able to induce illness, as accessory EHEC genes may also contribute to human disease. Besides the stx gene, typical EHEC strains possess the eae gene, coding for the intimin protein, implicated in attaching and effacing lesions in the intestinal cells (6). Moreover, epidemiological studies have shown that five serotypes are more frequently involved in outbreaks than others (7). Therefore, the French Agency for Food Safety defined five major EHEC strains as STEC, belonging to serotypes O157:H7, O26:H11, O145:H28, O103:H2, and O111:H8 (8). More precisely, the serotypes O157:H7 and O145:H28 are known to be associated with the eae-γ1 subtype, whereas STEC O26:H11, O103:H2, and O111:H8 harbor eae-β1, eae-ε, and eae-θ subtypes, respectively (9, 10). When isolated from the food chain, these “top five” STEC are considered to be highly pathogenic for humans.
The ISO 13136:2012 technical specification (TS) describes a real-time PCR-based approach for the detection of the top five STEC, which should be used to monitor these STEC along the food chain, in order to harmonize the results (11). At the farm level, few studies have focused on the specific detection of the five main pathogenic STEC. Regarding the literature on the prevalence of STEC in cattle, the majority of studies focused only on serotype O157:H7 (12, 13). Recent studies focused on the specific detection of the top five STEC in cattle feces using PCR screening approaches. Nevertheless, the low number of cattle screened and the fact that serogroup-specific strain isolation was restricted to only a few samples did not allow to obtain a reliable estimate of the prevalence of the top five EHEC (14–18).
Therefore, the objective of the present study was to evaluate the prevalence of the five main pathogenic STEC in the different categories of slaughtered cattle used for the production of ground beef in France (young dairy and beef bulls and dairy and beef cows). The aims of our study were (i) to isolate and characterize the top five STEC strains from fecal samples containing EHEC-associated genetic markers, (ii) to evaluate the prevalence of bovine carriers of STEC per cattle category at the time of their slaughter, and finally (iii) to estimate the prevalence of the top five STEC bovine carriers in slaughtered adult cattle in France.
MATERIALS AND METHODS
Sampling plan.
Samples were collected from cattle slaughtered in six of the French abattoirs with the highest slaughter capacities for adult cattle. They produced from 20,000 to 46,000 tons carcass weight equivalent of adult cattle per year. They were selected for inclusion in this study on the basis of their geographical location covering the French cattle production area. Eleven sampling campaigns were conducted from 19 October 2010 to 28 June 2011. The sampling plan was devised in order to enable a good estimation of prevalence among the four categories of slaughtered cattle used for the production of ground beef in France, i.e., young dairy bulls, young beef bulls, dairy cows, and beef cows. Young bulls are defined by the European legislation (Council Regulation [EC] No. 1234/2007) as animals slaughtered before the age of 24 months. A minimum sample size of about 300 animals by category was calculated with the assumption of a 2% prevalence of carriage of the top five STEC and a targeted precision of 1.6%. This assumption was based on data published for STEC O157:H7 shedding in French and European cattle (19–21). Cattle was randomly sampled throughout the slaughter period to avoid as much as possible the sampling of animals from the same batch/herd or farm and to include the requested number of animals in each of the four categories. We checked before the analysis the animal origin and considered only a single positive animal per farm for prevalence calculation if several were detected. In all, feces were collected from 1,318 animals, including 291 young dairy bulls, 296 young beef bulls, 337 dairy cows, and 394 beef cows (Table 1). Fecal samples were obtained by opening the terminal rectum after evisceration. They were kept chilled and sent to the laboratory by overnight courier for analysis.
TABLE 1.
Distribution of sampled cattle by campaign, abattoir, and category
| Sampling campaign ID | Abattoir ID | No. of animals in category |
|||
|---|---|---|---|---|---|
| Young dairy bulls (n = 291) | Young beef bulls (n = 296) | Dairy cows (n = 337) | Beef cows (n = 394) | ||
| B | 2 | 0 | 5 | 3 | 112 |
| C | 1 | 8 | 0 | 88 | 24 |
| D | 3 | 1 | 21 | 32 | 66 |
| E | 4 | 7 | 52 | 1 | 3 |
| F | 5 | 19 | 64 | 9 | 33 |
| G | 6 | 24 | 47 | 46 | 13 |
| H | 5 | 28 | 70 | 6 | 26 |
| I | 1 | 57 | 18 | 28 | 27 |
| J | 3 | 0 | 14 | 46 | 70 |
| K | 1 | 115 | 5 | 0 | 0 |
| L | 5 | 32 | 0 | 78 | 20 |
E. coli control strains.
Seven reference strains were used as positive controls in PCR analysis: Sakaï (O157:H7, stx1, stx2, eae-γ1, ehxA, espK), PMK5 (O103:H2, stx1, eae-ε), H19 (O26:H11, stx1, eae-β1), 95NR1 (O111:H8, stx1, stx2, eae-θ), ED-28 (O145:H28, stx1, eae-γ1), E2348/69 (O127:H6, bfpA, EAF), and EDL933 (O157:H7, stx2, eae-γ1, pagC, nleB, efa1) (22–25). ED-28 was provided by the Istituto Superiore di Sanita (Rome). Laboratory nonpathogenic E. coli strain MG1655 was used as a negative control for all virulence factors investigated.
Sample enrichment and DNA extraction.
Upon arrival, each sample (10 g) was diluted 10-fold (wt/vol) in 90 ml of modified tryptone soya broth (Oxoid, Dardilly, France) supplemented with novobiocin (Oxoid, Dardilly, France) at 16 mg · liter−1 and incubated overnight at 37°C. Bacterial DNA was extracted from 1 ml of each enriched broth using lysis tubes (Pall GeneDisc Technologies, Bruz, France), as already described (26).
PCR-based screenings for EHEC-associated genetic markers.
DNA extracts were subjected to a sequential real-time PCR-based approach for the detection of EHEC-associated genetic markers. This PCR-based strategy used to detect suspect samples was the same as the one described in the ISO 13136:2012 technical specification, to which a screening of the eae subtypes associated with the five major EHEC was added (11, 26). An initial screening step was performed for the detection of stx, eae genes, and eae subtypes γ1, β1, ε, and θ. The detection of stx1, stx2, and an internal control was performed as already described (27), and two additional assays allowed the detection of the eae gene and eae subtypes as previously published (28, 29). A second screening step was performed on stx- and eae-positive samples for the detection of the five O group markers. This second step included two simplex real-time PCR assays to detect rfbEO157 and wbd1O111 genes and one triplex real-time PCR assay for the screening of wzxO26, wzxO103, and ihp1O145 genes, with primers and probes described elsewhere (30, 31). All the PCR experiments were performed using a CFX96 instrument (Bio-Rad), except those targeting eae subtypes, which were performed on a LightCycler 480 instrument (Roche Diagnostics).
Isolation procedures.
Isolations of E. coli strains belonging to the five targeted serotypes were performed for samples that tested positive by PCR for the simultaneous presence of an stx gene, an eae subtype, and its associated O group marker. Three isolation procedures were used in parallel, except for two campaigns, in order to maximize the recovery of strains. The first isolation procedure consisted of immunomagnetic separation (IMS) assays using Dynabeads (Invitrogen, Cergy Pontoise, France), as recommended by the manufacturer. Ten microliters of immunoconcentrated bacteria was plated onto cefixime-tellurite-sorbitol-MacConkey agar (Oxoid, Dardilly, France) for E. coli O157, O103, and O111, onto cefixime-tellurite-rhamnose-MacConkey agar for E. coli O26, and onto cefixime-tellurite-raffinose-MacConkey agar for E. coli O145 (32). A total of 10 plates were used for each serogroup, and all media were incubated for 18 to 24 h at 37°C. Up to 10 suspect colonies were tested by slide agglutination with serogroup-specific antisera (Statens Serum Institut, Copenhagen, Denmark), and each serogroup was confirmed by real-time PCR as described above. In the second procedure, E. coli O157 strains were isolated using the Vidas Immuno-Concentration E. coli O157 (ICE) kit (bioMérieux, Marcy l'Etoile, France) according to the manufacturer's instructions. Automated immunoconcentration of E. coli O26 was performed using the E. coli Serogroup (EES) kit (bioMérieux), adapted for immunoconcentration. IMS-based isolations (Dynabeads; Invitrogen, Cergy Pontoise, France) were performed to isolate E. coli strains belonging to serogroups O103, O111, and O145 according to the manufacturer's instructions. In the third procedure, isolation of E. coli strains belonging to serogroups O157, O26, O103, O111, and O145 was performed using the Vidas UP E. coli serogroups (ESPT) kits under development (bioMérieux). For the second and third procedures, 50 μl of immunoconcentrated bacteria was inoculated on two selective isolation media, Rapid'E. coli O157:H7 (Bio-Rad, Marne la Coquette, France) and ChromoID O157:H7 (bioMérieux), supplemented with cefixime and tellurite for E. coli O157, with sorbitol-MacConkey agar and rhamnose-MacConkey agar (Oxoid) for E. coli O26, or with sorbitol-MacConkey agar and a selective differential medium for E. coli O103, O111, and O145 (32). All media were incubated for at 18 to 24 h at 37°C. Up to 10 suspect colonies per each serogroup screened were tested by real-time PCR, as described above.
For the three isolation procedures, once the targeted serogroup was confirmed by PCR, isolates were characterized for the presence of stx genes and eae subtypes by real-time PCR, as described above. The presence of the fliCH alleles (fliCH2, fliCH7, fliCH8, fliCH11, and fliCH28) was also investigated as previously described (29). Isolates were also confirmed as E. coli using an API 20E test (bioMérieux, Marcy l'Etoile, France). Based on PCR results, E. coli isolates positive for stx genes were classified as Shiga toxin-producing E. coli (STEC). E. coli isolates positive for eae gene and negative for stx genes were classified as enteropathogenic E. coli (EPEC).
Virulence profiles.
Subtyping of stx1 and stx2 genes was performed as described previously (33). The presence of additional EHEC virulence markers (ehxA gene and OI-122-associated genes, namely, pagC, nleB, and efa1 genes) was screened by PCR as described previously (34, 35). The presence of the virulence marker espK was screened by PCR for STEC O26:H11 (36). The presence of typical EPEC markers, i.e., bfpA and EPEC adherence factor (EAF) genes, was also tested by PCR (37, 38). E. coli isolates positive for the eae gene and negative for the bfpA gene and EAF plasmid were classified as atypical enteropathogenic E. coli (aEPEC) (1).
PFGE typing of the top five STEC strains.
STEC strains were typed using the Standard PulseNet pulsed-field gel electrophoresis (PFGE) protocol for E. coli O157 (39). Agarose-embedded DNAs were digested overnight at 37°C with 20 U of XbaI enzyme (Promega Corp., Madison, WI, USA). XbaI-digested DNA of Salmonella enterica serotype Braenderup strain H9812 (Centers for Disease Control and Prevention, Atlanta, GA) was used as a universal molecular size marker. Restriction fragments were resolved at 14°C in 0.5× Tris-borate-EDTA (TBE) buffer on 1% Seakem gold agarose gels (FMC Bioproducts, Rockland, ME, USA) using a pulsed-field Chef-DR-III system (Bio-Rad laboratories, Munich, Germany). After being stained with ethidium bromide (10 μg ml−1), gels were visualized on gel image digitization by Easy RH equipment (Herolab GmBH, Germany) and an image analyzer (VisioCapt-Bio1D; Fisher Bioblock Scientific, Illrisch, France), and the PFGE profiles were analyzed using GelCompar II software version 6.5 (Applied Maths, Ghent, Belgium). A dendrogram was generated using the band-based Dice similarity coefficient with a 1.5% band position tolerance and the unweighted pair group method with arithmetic mean clustering.
Statistical analysis.
Statistical analyses were conducted using R software (R Foundation for Statistical Computing, Vienna, Austria [http://www.R-project.org/]). Comparisons of detection of EHEC-associated genetic markers among the different cattle categories were done using logistic models (the response variable was presence or absence of the specific gene; the explicative variable was animal category). The prevalence of bovine carriers of the top five STEC per cattle category was calculated considering in the numerator one single positive animal per farm (as the animals of a same farm are correlated). Comparisons of prevalences between cattle categories were done using the chi-square test or Fisher's exact test if the number of animals was too small. For all the analyses, the significance level was set to 0.05. The prevalence of the top five STEC (all serotypes included) among French slaughtered adult cattle was calculated through the weighted mean prevalence, using the proportion of animals of each category slaughtered in France as weights. These proportions were given by the French Cattle and Meat Association (Interbev) for the 2010-2011 period.
RESULTS
Detection of EHEC-associated genetic markers in bovine feces.
A total of 1,318 fecal bovine samples were screened for the presence of stx1, stx2, eae, and eae subtypes β1, ε, γ1, and θ (Table 2). Whatever the category studied, the most frequently detected EHEC marker was stx. The percentage of stx-positive fecal samples was significantly higher in young dairy bulls than in the three other categories (P < 0.001). Moreover, the percentage of eae-positive fecal samples was significantly higher in young dairy bulls and in young beef bulls than in dairy and beef cows (P < 0.001). For the four cattle categories, the most frequently detected eae subtypes were eae-β1 and eae-θ, followed by eae-γ1 and eae-ε. Regarding the association of EHEC markers, samples positive for both stx and eae genes were more frequently detected in young dairy bulls than in the three other categories (P < 0.001).
TABLE 2.
Detection of stx1, stx2, eae, and eae subtypes in cattle categories
| EHEC marker targeted by real-time PCR | No. (%) of samples positive for EHEC-associated genetic markers in: |
|||
|---|---|---|---|---|
| Young dairy bulls (n = 291) | Young beef bulls (n = 296) | Dairy cows (n = 337) | Beef cows (n = 394) | |
| stxa | 245 (84.2) | 208 (70.3) | 235 (69.7) | 278 (70.6) |
| stx1 | 122 (41.9) | 81 (27.4) | 136 (40.4) | 165 (41.9) |
| stx2 | 219 (75.3) | 182 (61.5) | 201 (59.6) | 247 (62.7) |
| eaeb | 199 (68.4) | 190 (64.2) | 155 (46.0) | 229 (58.1) |
| eae-β1 | 100 (34.4) | 98 (33.1) | 70 (20.8) | 111 (28.2) |
| eae-γ1 | 34 (11.7) | 21 (7.1) | 12 (3.6) | 28 (7.1) |
| eae-ε | 33 (11.3) | 40 (13.5) | 23 (6.8) | 52 (13.2) |
| eae-θ | 80 (27.5) | 71 (24.0) | 76 (22.6) | 101 (25.6) |
| stx-eae | 177 (60.8) | 145 (49.0) | 125 (37.1) | 182 (46.2) |
Samples positive for stx were positive for stx1 and/or stx2.
Detection of the eae gene with the universal primers/probe.
In all, 629 samples were stx and eae positive (47.7%) and were subjected to a screening for the five EHEC O group markers. The results showed that 555 (42.1%) of the samples were positive for at least one of the five stx-eae-O group marker combinations. The stx-eae-O group marker combinations were detected 1,217 times, as several samples contained more than one O group marker, and the most frequently detected combination was stx-eae-ihp1O145 (Table 3). When taking into account the results of the detection of the eae subtypes, 235 (17.8%) samples were positive for at least one of five targeted stx-eae subtype-O group marker combinations. The most frequently identified combinations were stx–eae-β1–wzxO26, stx–eae-ε–wzxO103, and stx–eae-γ1–ihpO145 (Table 3). In all, the stx-eae subtype/associated O group marker combinations were detected 363 times, as several samples contained more than one combination of EHEC-associated genetic markers.
TABLE 3.
Number of combinations of the top five EHEC-associated genetic markers detected in 1,318 bovine fecal samples and results of isolation assays targeting the top five EHEC serogroups
| Combinations of EHEC markers | No. of combinations (% of positive samples) | Results of isolation assays |
||
|---|---|---|---|---|
| No. of strains isolated | No. of STEC | No. of aEPEC | ||
| stx-eae-O group marker | 1,217 (NAa) | |||
| stx-eae subtype/associated O group marker | 363 (NA) | 96 | 33 | 63 |
| stx-eae-rfbEO157 | 216 (16.4) | |||
| stx–eae-γ1–rfbEO157 | 47 (3.6) | 20 | 18 | 2 |
| stx-eae-wzxO26 | 202 (15.3) | |||
| stx–eae-β1–wzxO26 | 129 (9.8) | 37 | 3 | 34 |
| stx-eae-wzxO103 | 262 (19.9) | |||
| stx–eae-ε–wzxO103 | 93 (7.1) | 27 | 8 | 19 |
| stx-eae-wbd1O111 | 27 (2.0) | |||
| stx–eae-θ–wbd1O111 | 14 (1.1) | 2 | 2 | 0 |
| stx-eae-ihp1O145 | 510 (38.7) | |||
| stx–eae-γ1–ihp1O145 | 80 (6.1) | 10 | 2 | 8 |
NA, not applicable, as several samples contained more than one combination of EHEC-associated genetic markers.
Isolation of STEC and aEPEC strains belonging to the five EHEC serotypes.
Samples positive for one or more stx-eae subtype/associated O group marker combinations were subjected to isolation assays using three procedures in parallel (see Materials and Methods). An E. coli strain belonging to one of the five targeted EHEC serotypes was isolated for 96 of the 363 assays (Table 3). Of the 96 strains isolated, 33 were the top five STEC and belonged to serotypes O157:H7 (n = 18), O26:H11 (n = 3), O103:H2 (n = 8), O111:H8 (n = 2), and O145:H28 (n = 2). The 63 other E. coli strains isolated belonged to serotypes O157:H7, O103:H2, O145:H28, and O26:H11 and harbored the corresponding eae subtype but lacked an stx gene. These strains were classified as aEPEC, as they were negative for the bfpA gene and the EAF plasmid (1). No aEPEC was isolated for the serotype O111:H8. Finally, the proportion of STEC among the obtained E. coli isolates varied between the five serotypes. Concerning the O157:H7 serotype, isolation procedures led to the isolation of STEC strains rather than aEPEC strains, whereas the reverse was observed for serotypes O26:H11 and O103:H2.
Virulence profiles of the top five STEC strains.
The 33 STEC belonging to the top five serotypes were further characterized (Table 4). All these isolates harbored the eae subtype known to be specifically associated with its serotype. Concerning the 18 STEC O157:H7 strains, they were all positive for the stx2 gene, and the stx2c subtype was the most frequently detected (n = 14). The stx2a subtype was detected in 3 STEC O157:H7, and the simultaneous presence of stx2a and stx2c subtypes was detected in one STEC O157:H7 strain (B3-O157-1). In addition to the stx2 gene(s), an stx1 gene (stx1a subtype) was present in two STEC O157:H7 strains, isolated from cows (C61-O157-1 and B3-O157-1). The stx1a subtype was detected in all the non-O157 STEC strains, whereas the stx2a subtype was detected in only one STEC O103:H2 strain (L154-O103-3). The screening of additional EHEC virulence markers showed that all STEC strains possessed the ehxA gene, except for the O26:H11 strain I92-O26-1. Moreover, all STEC strains of serotypes O157:H7, O103:H2, and O111:H8 were positive for pagC, nleB, and efa1 genes. STEC strains of serotype O26:H11 were positive only for nleB and efa1 genes, and STEC strains of serotype O145:H28 were negative for these three OI-122-associated genes. Finally, all STEC O26:H11 strains harbored the espK gene.
TABLE 4.
Origin and characterization of STEC O157:H7, O26:H11, O103:H2, O111:H8, and O145:H28 isolated from bovine feces
| Serotypea | Origin | Strainb | Presencec of gene: |
Farm ID | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| stx1 (subtype) | stx2 (subtype) | eae (subtype) | ehxA | pagC | nleB | efa1 | espK | ||||
| O157:H7 | Young dairy bull | C117-O157-1 | − | + (stx2c) | + (γ1) | + | + | + | + | ND | 4 |
| O157:H7 | Young dairy bull | H105-O157-1 | − | + (stx2a) | + (γ1) | + | + | + | + | ND | 10 |
| O157:H7 | Young dairy bull | I113-O157-1 | − | + (stx2c) | + (γ1) | + | + | + | + | ND | 15 |
| O157:H7 | Young dairy bull | I114-O157-1 | − | + (stx2c) | + (γ1) | + | + | + | + | ND | 15 |
| O157:H7 | Young dairy bull | K118-O157-1 | − | + (stx2c) | + (γ1) | + | + | + | + | ND | 23 |
| O157:H7 | Young dairy bull | K143-O157-1 | − | + (stx2c) | + (γ1) | + | + | + | + | ND | 24 |
| O157:H7 | Young dairy bull | L81-O157-1 | − | + (stx2c) | + (γ1) | + | + | + | + | ND | 27 |
| O157:H7 | Young beef bull | G15-O157-1 | − | + (stx2c) | + (γ1) | + | + | + | + | ND | 6 |
| O157:H7 | Young beef bull | H13-O157-1 | − | + (stx2c) | + (γ1) | + | + | + | + | ND | 9 |
| O157:H7 | Young beef bull | H37-O157-1 | − | + (stx2a) | + (γ1) | + | + | + | + | ND | 11 |
| O157:H7 | Young beef bull | J28-O157-1 | − | + (stx2c) | + (γ1) | + | + | + | + | ND | 16 |
| O157:H7 | Dairy cow | C29-O157-1 | − | + (stx2c) | + (γ1) | + | + | + | + | ND | 2 |
| O157:H7 | Dairy cow | C61-O157-1 | + (stx1a) | + (stx2c) | + (γ1) | + | + | + | + | ND | 3d |
| O157:H7 | Dairy cow | J49-O157-1 | − | + (stx2c) | + (γ1) | + | + | + | + | ND | 3bisd |
| O157:H7 | Dairy cow | L71-O157-1 | − | + (stx2a) | + (γ1) | + | + | + | + | ND | 26 |
| O157:H7 | Beef cow | B3-O157-1 | + (stx1a) | + (stx2a, 2c) | + (γ1) | + | + | + | + | ND | 1 |
| O157:H7 | Beef cow | G79-O157-1 | − | + (stx2c) | + (γ1) | + | + | + | + | ND | 8 |
| O157:H7 | Beef cow | J76-O157-1 | − | + (stx2c) | + (γ1) | + | + | + | + | ND | 18 |
| O26:H11 | Young dairy bull | I92-O26-1 | + (stx1a) | − | + (β1) | − | − | + | + | + | 14 |
| O26:H11 | Young dairy bull | K106-O26-1 | + (stx1a) | − | + (β1) | + | − | + | + | + | 22 |
| O26:H11 | Beef cow | J77-O26-1 | + (stx1a) | − | + (β1) | + | − | + | + | + | 19 |
| O103:H2 | Young dairy bull | K56-O103-1 | + (stx1a) | − | + (ε) | + | + | + | + | ND | 21 |
| O103:H2 | Young dairy bull | K106-O103-1 | + (stx1a) | − | + (ε) | + | + | + | + | ND | 22 |
| O103:H2 | Young dairy bull | K146-O103-1 | + (stx1a) | − | + (ε) | + | + | + | + | ND | 24 |
| O103:H2 | Young dairy bull | L24-O103-1 | + (stx1a) | − | + (ε) | + | + | + | + | ND | 25 |
| O103:H2 | Young beef bull | F63-O103-1 | + (stx1a) | − | + (ε) | + | + | + | + | ND | 5 |
| O103:H2 | Young beef bull | G22-O103-1 | + (stx1a) | − | + (ε) | + | + | + | + | ND | 7 |
| O103:H2 | Young beef bull | H115-O103-1 | + (stx1a) | − | + (ε) | + | + | + | + | ND | 13 |
| O103:H2 | Dairy cow | L154-O103-3 | + (stx1a) | + (stx2a) | + (ε) | + | + | + | + | ND | 28 |
| O111:H8 | Young dairy bull | K50-O111-1 | + (stx1a) | − | + (θ) | + | + | + | + | ND | 20 |
| O111:H8 | Dairy cow | J43-O111-1 | + (stx1a) | − | + (θ) | + | + | + | + | ND | 17 |
| O145:H28 | Young dairy bull | H99-O145-1 | + (stx1a) | − | + (γ1) | + | − | − | − | ND | 12 |
| O145:H28 | Young dairy bull | H101-O145-1 | + (stx1a) | − | + (γ1) | + | − | − | − | ND | 12 |
The serotype was determined by PCR.
The first letter of the name of the strain corresponds to the sampling campaign.
+, detected by PCR; −, not detected by PCR; ND, not determined.
Fecal samples were collected from cattle coming from farm 3 on two different campaigns (3bis, second campaign).
Origin and genetic diversity of the top five STEC strains.
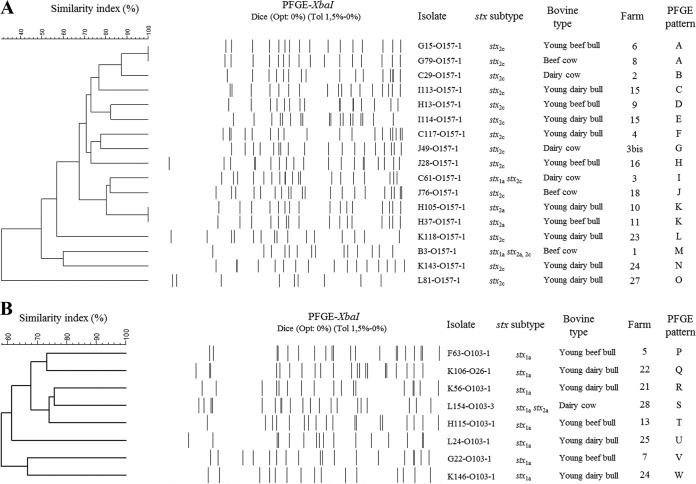
The 33 STEC strains were isolated from 32 distinct animals, as the young dairy bull K106 harbored both STEC O26:H11 and STEC O103:H2. Besides, it was noteworthy that young dairy bulls K143 and K146, coming from the same farm (farm 24), carried STEC O157:H7 and STEC O103:H2, respectively (Table 4). In order to explore the genetic relatedness of STEC within a same serotype, the 33 top five STEC strains were subjected to PFGE analysis. The strain L71-O157-1 could not be typed. The dendrograms of STEC O157:H7 and STEC O103:H2 are shown in Fig. 1.Within the same serotype, a high diversity was observed. Nevertheless, on two occasions, STEC O157:H7 strains isolated from animals coming from different farms but sampled during the same campaign showed an identical PFGE pattern (PFGE patterns A and K). A unique PFGE pattern was also observed for the two STEC O145:H28 carried by two young dairy bulls coming from the same farm and sampled at the same campaign (data not shown). In contrast, it was noteworthy that STEC O157:H7 strains isolated from cattle coming from the same farm (farms 3 and 15), sampled or not during the same campaign, showed different PFGE patterns.
FIG 1.
XbaI PFGE patterns and origins of 17 STEC O157:H7 strains (A) and 8 STEC O103:H2 strains (B) isolated from 1,318 bovine feces, in France, in 2010-2011. The dendrograms were generated using the band-based Dice similarity coefficient with a 1.5% band position tolerance and the unweighted pair group method with arithmetic mean clustering.
Prevalence of bovine carriers of the top five STEC per cattle category.
The prevalence rates of the top five STEC were 4.5%, 2.4%, 1.8%, and 1.0% in young dairy bulls, young beef bulls, dairy cows, and beef cows, respectively (Table 5). Young dairy bulls harbored significantly more STEC strains than other categories (P < 0.01). The prevalence of STEC serotype O157:H7 was significantly higher in young dairy bulls than in other categories (P < 0.05). Finally, taking into account the proportion of animals of each category slaughtered in France during the sampling period, the weighted mean prevalence of STEC with the five targeted serotypes combined was estimated at 1.8% in adult slaughtered cattle. It was estimated at 1.2% for STEC O157:H7.
TABLE 5.
Prevalence of bovine carriers of STEC O157:H7, O26:H11, O103:H2, O111:H8, and O145:H28 per cattle category
| STEC serotype(s) | Prevalence (%) of bovine carriers of top five STEC in: |
|||
|---|---|---|---|---|
| Young dairy bulls (n = 291) | Young beef bulls (n = 296) | Dairy cows (n = 337) | Beef cows (n = 394) | |
| Top five STEC | 4.5 | 2.4 | 1.8 | 1.0 |
| STEC O157:H7 | 2.1 | 1.4 | 1.2 | 0.8 |
| STEC O26:H11 | 0.3 | 0.0 | 0.0 | 0.3 |
| STEC O103:H2 | 1.0 | 1.0 | 0.3 | 0.0 |
| STEC O111:H8 | 0.3 | 0.0 | 0.3 | 0.0 |
| STEC O145:H28 | 0.3 | 0.0 | 0.0 | 0.0 |
| STEC O26:H11 and O103:H2 | 0.3 | 0.0 | 0.0 | 0.0 |
DISCUSSION
The main objective of our study was to evaluate the prevalence of the five main pathogenic STEC in French cattle per category of cattle that are slaughtered for the production of ground beef. The PCR-based strategy used to detect suspect samples was that described in the ISO 13136:2012 technical specification, to which we added a screening of the eae subtypes associated with the five major EHEC (11, 26). Indeed, we previously showed that this additional screening step helped to be more discriminating for the specific detection of suspect samples likely to contain the five major STEC in cattle feces. As confirmed in this study, identification of samples positive for stx-eae subtype/associated O group marker combinations rather than stx-eae-O group marker combinations allowed to narrow down the number of potential positive samples that should be subjected to isolation assays for confirmation. The proportion of isolation assays leading to the isolation of one of the top five STEC strains was relatively low, as previously observed in various studies (14, 15, 17). The reasons for this discrepancy, including PCR-based strategy and isolation assays limitations, have been already discussed elsewhere (26, 40–42). In this study, three isolation procedures were used in parallel and allowed us to improve the isolation fraction of the top five STEC strains. In addition to STEC strains, we isolated stx-negative aEPEC belonging to the same top five serotypes and harboring eae subtypes associated with the targeted serotypes. It was noteworthy that the proportion of STEC among the recovered E. coli strains depended on the targeted serotype and was high for serotype O157:H7 but much lower for serotypes O26:H11 and O103:H2. These results are in agreement with previous studies evaluating stx carriage in top five serogroups of E. coli strains isolated from Irish beef slaughter chains and Scottish and Swiss cattle (14, 16, 43). Whether these results reflect a higher stability of stx bacteriophages within strains of serotype O157:H7 remains to be investigated.
In all, 33 STEC strains belonging to the top five serotypes were isolated from the 1,318 bovine fecal samples screened. They all harbored the expected eae subtypes specifically associated with the five targeted serotypes. According to their virulence genetic profiles, these top five STEC should be considered highly pathogenic for humans (8, 9, 23, 44). All STEC O157:H7 strains were positive for the stx2 gene. The stx2c subtype was the most frequently detected in those strains. This is in agreement with previous studies showing that STEC O157 isolated from cattle were dominated by subtype stx2c (45–47). In contrast, the non-O157:H7 STEC strains were all stx1a positive. Another epidemiological study also showed that stx1 predominated in STEC O26 isolated from Scotland cattle (43). Moreover, stx1 predominated in STEC O26, O111, and O103 isolated from humans, food, and cattle in Belgium, whereas isolates of STEC O145 displayed a heterogeneous distribution of stx genes (48). Finally, EHEC additional virulence genes ehxA and OI-122-associated genes were detected in the top five STEC strains isolated from cattle feces, as well as espK in the case of STEC O26:H11. These EHEC virulence markers have already been shown to be associated with the top five STEC strains that cause severe diseases and outbreaks (36, 49–51).
When looking at STEC bovine carriers, one of the most striking features is the fact that a young dairy bull carried STEC of two different serotypes. Moreover, the identification of farms harboring STEC bovine carriers highlighted the fact that STEC of a given serotype could be carried by several animals belonging to the same farm. We also identified two young bulls that came from the same farm and carried STEC of different serotypes. Within each serotype, PFGE analysis showed that the genetic diversity of the top five STEC was high, as already observed by others (52, 53). STEC of a given serotype carried by cattle coming from the same farm were either genetically related (for serotype O145:H28) or not (for serotype O157:H7). Moreover, within a same sampling campaign, identical PFGE patterns could be observed for STEC O157:H7 harbored by cattle coming from different farms. We can assume that the same clone might be present at a given time in several farms. Alternatively, we can also assume that these bovine carriers of a same clone might have been batched into the same facilities for fattening before slaughter.
Finally, the main objective of the present study was to obtain a reliable estimate of the prevalence of bovine carriers of the top five STEC in slaughtered adult cattle in France and thus new elements for assessment of human exposure to the top five STEC through consumption of beef. It is noteworthy that cattle prevalence studies have rarely addressed the question of the reliability of their results. Indeed, screening strategies, analytical methods, and sampling strategies might lead to certain limitations. The limitations of the screening strategy and the analytical method used in the present study have been discussed above. Concerning the sampling strategy, it was optimized in order to obtain a reliable estimate of the prevalence of each of the five STEC serotypes per slaughtered cattle category. This allowed to identify differences in carriage between categories. All serotypes combined, young dairy bulls harbored significantly more STEC strains than other categories. The prevalence of STEC O157:H7 was also significantly higher in this category. These results are in agreement with the results of PCR screenings showing that the simultaneous presence of stx and eae genes was significantly more frequently detected in feces from young dairy bulls than in other categories. Overall, these results are consistent with the results of previous studies evaluating the influence of type of production and age of animal on STEC fecal shedding. Concerning the effect of type of production on the prevalence of STEC, a survey conducted on 180 Belgium farms showed that the highest prevalence of E. coli O157:H7 was found in dairy cattle farms (61.2%) compared to beef farms (22.7%) or mixed dairy and beef farms (44.4%) (54). Concerning the effect of age on STEC shedding, a Scotland investigation on 14,856 cattle fecal samples showed that an increased probability of a sampling group containing a STEC O157:H7-shedding animal was associated with larger numbers of 12- to 30-month-old finished cattle (55). They also showed that a higher maximum age of animals in the sampling group was significantly associated with a lower prevalence of STEC O157:H7. Moreover, a review of published farm prevalence surveys had already shown that 0.5 to 1% of sampled animals were E. coli O157:H7 carriers, and this proportion was raised to 5% for later-weaned calves and heifers (56). Nevertheless, it should be noted that a seasonality of production exists for young bulls, with a peak of production observed around the summer, and this seasonality has been observed in France (http://www.agrireseau.qc.ca/bovinsboucherie/documents/pdf_D379-v.pdf). Consequently, in the present study, the feces of this cattle category was largely sampled during this period. It is also worth noting that the season had been shown to have an influence on STEC shedding, the warmer months being associated with a peak in the prevalence of STEC (57). Anyway, the biological basis for either age-related or rearing conditions or seasonal peak shedding by cattle is unknown and remains to be elucidated. Various hypotheses have been advanced to try to explain these variations in STEC carriage; and among them, the seasonal presence of increased numbers of young high shedders might explain a seasonal peak in the prevalence of STEC (58).
Lastly, the prevalence per category was weighted by the number of slaughtered cattle within each category, and the prevalence of the top five STEC (all five serotypes included) was estimated to 1.8% in slaughtered adult cattle in France. The weighted mean prevalence of the most prevalent serotype, O157:H7, was estimated at 1.2%. STEC O157:H7 was detected in the four cattle categories. These values concerning STEC O157:H7 shedding are in agreement with previous results of European prevalence studies. The average proportion of STEC O157-positive samples, based on the investigation of a high number of feces or hides from animals sampled either at the farm or at slaughter, ranged from 0.2% to 2.3% for the 2009-2011 period (59, 60). Concerning the non-O157:H7 STEC serotypes, prevalence data directly comparable to our results are lacking in the literature. Recent studies focused on the detection of the top five STEC in cattle feces, but none of them led to the estimation of their prevalence, due either to a limited number of serogroup-specific strain isolations performed (14, 15) or to a sampling strategy that did not allow a national estimation of the prevalence (16, 18). In our study, the four non-O157:H7 STEC serotypes were detected in slaughtered categories at a low prevalence, ranging from 0.0% to 1.0%.
In conclusion, an estimation of a reliable value of the prevalence of STEC bovine carriers in slaughtered adult cattle in France was attempted here. To this end, the top five STEC strains considered highly pathogenic were isolated, and prevalence weighted by the number of slaughtered animals within each category was calculated. This study also allowed to identify differences in STEC carriage, which need to be clarified. Factors affecting STEC carriage are under investigation; notably, in the farms from which the bovine carriers came.
ACKNOWLEDGMENTS
This work was supported by funds from the French Cattle and Meat Association (Interbev) and the French National Authority for Agriculture and Sea Products (FranceAgriMer). PFGE experimentations performed by VetAgro Sup were supported by an additional fund from the French Ministry of Agriculture (n°2011-303/2100616738 and 2013-304/2101231963).
We thank bioMérieux Industry for providing kits under development for automated immunoconcentration assays.
We are grateful to A. A. Diallo (ISRA/LNERV, Dakar-Hann, Senegal), D. Boudjerba (Université de Jijel, Algérie), T. Méheut (Anses), and S. Ganet (VetAgro Sup) for their involvement in the study.
REFERENCES
- 1.Kaper JB, Nataro JP, Mobley HL. 2004. Pathogenic Escherichia coli. Nat Rev Microbiol 2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 2.Tarr PI, Gordon CA, Chandler WL. 2005. Shiga-toxin-producing Escherichia coli and haemolytic uraemic syndrome. Lancet 365:1073–1086. doi: 10.1016/S0140-6736(05)71144-2. [DOI] [PubMed] [Google Scholar]
- 3.Naylor SW, Gally DL, Low JC. 2005. Enterohaemorrhagic E. coli in veterinary medicine. Int J Med Microbiol 295:419–441. doi: 10.1016/j.ijmm.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 4.Karmali MA, Gannon V, Sargeant JM. 2010. Verocytotoxin-producing Escherichia coli (VTEC). Vet Microbiol 140:360–370. doi: 10.1016/j.vetmic.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 5.Croxen MA, Finlay BB. 2010. Molecular mechanisms of Escherichia coli pathogenicity. Nat Rev Microbiol 8:26–38. doi: 10.1038/nrmicro2265. [DOI] [PubMed] [Google Scholar]
- 6.Dean-Nystrom EA, Bosworth BT, Moon HW, O'Brien AD. 1998. Escherichia coli O157:H7 requires intimin for enteropathogenicity in calves. Infect Immun 66:4560–4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson KE, Thorpe CM, Sears CL. 2006. The emerging clinical importance of non-O157 Shiga toxin-producing Escherichia coli. Clin Infect Dis 43:1587–1595. doi: 10.1086/509573. [DOI] [PubMed] [Google Scholar]
- 8.Anses. 2010. French Agency for Food, Environmental and Occupational Health and Safety. Opinion of the French Food Safety Agency on the advisability of revising the definition of pathogenic STEC, specified in AFSSA's Opinion of 15 July 2008. Anses, Maisons-Alfort, France. [Google Scholar]
- 9.Tarr CL, Whittam TS. 2002. Molecular evolution of the intimin gene in O111 clones of pathogenic Escherichia coli. J Bacteriol 184:479–487. doi: 10.1128/JB.184.2.479-487.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adu-Bobie J, Frankel G, Bain C, Goncalves AG, Trabulsi LR, Douce G, Knutton S, Dougan G. 1998. Detection of intimins alpha, beta, gamma, and delta, four intimin derivatives expressed by attaching and effacing microbial pathogens. J Clin Microbiol 36:662–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.International Organization for Standardization. 2012. ISO/TS 13136:2012. Microbiology of food and animal feed. Real-time polymerase chain reaction (PCR)-based method for the detection of food-borne pathogens. Horizontal method for the detection of Shiga toxin-producing Escherichia coli (STEC) and the determination of O157, O111, O26, O103 and O145 serogroups. International Organization for Standardization, Geneva, Switzerland. [Google Scholar]
- 12.Hussein HS. 2007. Prevalence and pathogenicity of Shiga toxin-producing Escherichia coli in beef cattle and their products. J Anim Sci 85:E63–E72. doi: 10.2527/jas.2006-421. [DOI] [PubMed] [Google Scholar]
- 13.Hussein HS, Sakuma T. 2005. Prevalence of shiga toxin-producing Escherichia coli in dairy cattle and their products. J Dairy Sci 88:450–465. doi: 10.3168/jds.S0022-0302(05)72706-5. [DOI] [PubMed] [Google Scholar]
- 14.Hofer E, Stephan R, Reist M, Zweifel C. 2012. Application of a real-time PCR-based system for monitoring of O26, O103, O111, O145 and O157 Shiga toxin-producing Escherichia coli in cattle at slaughter. Zoonoses Public Health 59:408–415. doi: 10.1111/j.1863-2378.2012.01468.x. [DOI] [PubMed] [Google Scholar]
- 15.Barlow RS, Mellor GE. 2010. Prevalence of enterohemorrhagic Escherichia coli serotypes in Australian beef cattle. Foodborne Pathog Dis 7:1239–1245. doi: 10.1089/fpd.2010.0574. [DOI] [PubMed] [Google Scholar]
- 16.Thomas KM, McCann MS, Collery MM, Logan A, Whyte P, McDowell DA, Duffy G. 2012. Tracking verocytotoxigenic Escherichia coli O157, O26, O111, O103 and O145 in Irish cattle. Int J Food Microbiol 153:288–296. doi: 10.1016/j.ijfoodmicro.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 17.Lynch MJ, Fox EM, O'Connor L, Jordan K, Murphy M. 2012. Surveillance of verocytotoxigenic Escherichia coli in Irish bovine dairy herds. Zoonoses Public Health 59:264–271. doi: 10.1111/j.1863-2378.2011.01443.x. [DOI] [PubMed] [Google Scholar]
- 18.Joris MA, Pierard D, De Zutter L. 2011. Occurrence and virulence patterns of E. coli O26, O103, O111 and O145 in slaughter cattle. Vet Microbiol 151:418–421. doi: 10.1016/j.vetmic.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 19.EFSA. 2010. The Community Summary Report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in the European Union in 2008. EFSA J 8(1):410. doi: 10.2903/j.efsa.2010.1496. [DOI] [Google Scholar]
- 20.Andral B, Aspan A, Perelle S, Fach P. 2004. PCR detection of virulence genes and molecular epidemiology of STEC O157 isolates from French abattoirs. Vet Rec 155:365–368. doi: 10.1136/vr.155.12.365. [DOI] [PubMed] [Google Scholar]
- 21.Rogerie F, Marecat A, Gambade S, Dupond F, Beaubois P, Lange M. 2001. Characterization of Shiga toxin producing E. coli and O157 serotype E. coli isolated in France from healthy domestic cattle. Int J Food Microbiol 63:217–223. doi: 10.1016/S0168-1605(00)00422-0. [DOI] [PubMed] [Google Scholar]
- 22.Hayashi T, Makino K, Ohnishi M, Kurokawa K, Ishii K, Yokoyama K, Han CG, Ohtsubo E, Nakayama K, Murata T, Tanaka M, Tobe T, Iida T, Takami H, Honda T, Sasakawa C, Ogasawara N, Yasunaga T, Kuhara S, Shiba T, Hattori M, Shinagawa H. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res 8:11–22. doi: 10.1093/dnares/8.1.11. [DOI] [PubMed] [Google Scholar]
- 23.Oswald E, Schmidt H, Morabito S, Karch H, Marches O, Caprioli A. 2000. Typing of intimin genes in human and animal enterohemorrhagic and enteropathogenic Escherichia coli: characterization of a new intimin variant. Infect Immun 68:64–71. doi: 10.1128/IAI.68.1.64-71.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levine MM, Nataro JP, Karch H, Baldini MM, Kaper JB, Black RE, Clements ML, O'Brien AD. 1985. The diarrheal response of humans to some classic serotypes of enteropathogenic Escherichia coli is dependent on a plasmid encoding an enteroadhesiveness factor. J Infect Dis 152:550–559. doi: 10.1093/infdis/152.3.550. [DOI] [PubMed] [Google Scholar]
- 25.Perna NT, Plunkett G III, Burland V, Mau B, Glasner JD, Rose DJ, Mayhew GF, Evans PS, Gregor J, Kirkpatrick HA, Posfai G, Hackett J, Klink S, Boutin A, Shao Y, Miller L, Grotbeck EJ, Davis NW, Lim A, Dimalanta ET, Potamousis KD, Apodaca J, Anantharaman TS, Lin J, Yen G, Schwartz DC, Welch RA, Blattner FR. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529–533. doi: 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- 26.Bibbal D, Loukiadis E, Kerouredan M, Peytavin de Garam C, Ferre F, Cartier P, Gay E, Oswald E, Auvray F, Brugere H. 2014. Intimin gene (eae) subtype-based real-time PCR strategy for specific detection of Shiga toxin-producing Escherichia coli serotypes O157:H7, O26:H11, O103:H2, O111:H8, and O145:H28 in cattle feces. Appl Environ Microbiol 80:1177–1184. doi: 10.1128/AEM.03161-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Auvray F, Lecureuil C, Dilasser F, Tache J, Derzelle S. 2009. Development of a real-time PCR assay with an internal amplification control for the screening of Shiga toxin-producing Escherichia coli in foods. Lett Appl Microbiol 48:554–559. doi: 10.1111/j.1472-765X.2009.02561.x. [DOI] [PubMed] [Google Scholar]
- 28.Nielsen EM, Andersen MT. 2003. Detection and characterization of verocytotoxin-producing Escherichia coli by automated 5′ nuclease PCR assay. J Clin Microbiol 41:2884–2893. doi: 10.1128/JCM.41.7.2884-2893.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Madic J, Peytavin de Garam C, Vingadassalon N, Oswald E, Fach P, Jamet E, Auvray F. 2010. Simplex and multiplex real-time PCR assays for the detection of flagellar (H-antigen) fliC alleles and intimin (eae) variants associated with enterohaemorrhagic Escherichia coli (EHEC) serotypes O26:H11, O103:H2, O111:H8, O145:H28 and O157:H7. J Appl Microbiol 109:1696–1705. doi: 10.1111/j.1365-2672.2010.04798.x. [DOI] [PubMed] [Google Scholar]
- 30.Perelle S, Dilasser F, Grout J, Fach P. 2004. Detection by 5′-nuclease PCR of Shiga-toxin producing Escherichia coli O26, O55, O91, O103, O111, O113, O145 and O157:H7, associated with the world's most frequent clinical cases. Mol Cell Probes 18:185–192. doi: 10.1016/j.mcp.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 31.Perelle S, Dilasser F, Grout J, Fach P. 2005. Detection of Escherichia coli serogroup O103 by real-time polymerase chain reaction. J Appl Microbiol 98:1162–1168. doi: 10.1111/j.1365-2672.2005.02545.x. [DOI] [PubMed] [Google Scholar]
- 32.Posse B, De Zutter L, Heyndrickx M, Herman L. 2008. Novel differential and confirmation plating media for Shiga toxin-producing Escherichia coli serotypes O26, O103, O111, O145 and sorbitol-positive and -negative O157. FEMS Microbiol Lett 282:124–131. doi: 10.1111/j.1574-6968.2008.01121.x. [DOI] [PubMed] [Google Scholar]
- 33.Scheutz F, Teel LD, Beutin L, Pierard D, Buvens G, Karch H, Mellmann A, Caprioli A, Tozzoli R, Morabito S, Strockbine NA, Melton-Celsa AR, Sanchez M, Persson S, O'Brien AD. 2012. Multicenter evaluation of a sequence-based protocol for subtyping Shiga toxins and standardizing Stx nomenclature. J Clin Microbiol 50:2951–2963. doi: 10.1128/JCM.00860-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paton AW, Paton JC. 1998. Detection and characterization of Shiga toxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic E. coli hlyA, rfbO111, and rfbO157. J Clin Microbiol 36:598–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Valat C, Haenni M, Saras E, Auvray F, Forest K, Oswald E, Madec JY. 2012. CTX-M-15 extended-spectrum beta-lactamase in a Shiga toxin-producing Escherichia coli isolate of serotype O111:H8. Appl Environ Microbiol 78:1308–1309. doi: 10.1128/AEM.06997-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bugarel M, Beutin L, Scheutz F, Loukiadis E, Fach P. 2011. Identification of genetic markers for differentiation of Shiga toxin-producing, enteropathogenic, and avirulent strains of Escherichia coli O26. Appl Environ Microbiol 77:2275–2281. doi: 10.1128/AEM.02832-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Franke J, Franke S, Schmidt H, Schwarzkopf A, Wieler LH, Baljer G, Beutin L, Karch H. 1994. Nucleotide sequence analysis of enteropathogenic Escherichia coli (EPEC) adherence factor probe and development of PCR for rapid detection of EPEC harboring virulence plasmids. J Clin Microbiol 32:2460–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gunzburg ST, Tornieporth NG, Riley LW. 1995. Identification of enteropathogenic Escherichia coli by PCR-based detection of the bundle-forming pilus gene. J Clin Microbiol 33:1375–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, Swaminathan B, Barrett TJ. 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog Dis 3:59–67. doi: 10.1089/fpd.2006.3.59. [DOI] [PubMed] [Google Scholar]
- 40.Beutin L, Jahn S, Fach P. 2009. Evaluation of the ‘GeneDisc’ real-time PCR system for detection of enterohaemorrhagic Escherichia coli (EHEC) O26, O103, O111, O145 and O157 strains according to their virulence markers and their O- and H-antigen-associated genes. J Appl Microbiol 106:1122–1132. doi: 10.1111/j.1365-2672.2008.04076.x. [DOI] [PubMed] [Google Scholar]
- 41.Verstraete K, De Zutter L, Messens W, Herman L, Heyndrickx M, De Reu K. 2010. Effect of the enrichment time and immunomagnetic separation on the detection of Shiga toxin-producing Escherichia coli O26, O103, O111, O145 and sorbitol positive O157 from artificially inoculated cattle faeces. Vet Microbiol 145:106–112. doi: 10.1016/j.vetmic.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 42.Jenkins C, Pearce MC, Smith AW, Knight HI, Shaw DJ, Cheasty T, Foster G, Gunn GJ, Dougan G, Smith HR, Frankel G. 2003. Detection of Escherichia coli serogroups O26, O103, O111 and O145 from bovine faeces using immunomagnetic separation and PCR/DNA probe techniques. Lett Appl Microbiol 37:207–212. doi: 10.1046/j.1472-765X.2003.01379.x. [DOI] [PubMed] [Google Scholar]
- 43.Pearce MC, Evans J, McKendrick IJ, Smith AW, Knight HI, Mellor DJ, Woolhouse ME, Gunn GJ, Low JC. 2006. Prevalence and virulence factors of Escherichia coli serogroups O26, O103, O111, and O145 shed by cattle in Scotland. Appl Environ Microbiol 72:653–659. doi: 10.1128/AEM.72.1.653-659.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karmali MA, Mascarenhas M, Shen S, Ziebell K, Johnson S, Reid-Smith R, Isaac-Renton J, Clark C, Rahn K, Kaper JB. 2003. Association of genomic O island 122 of Escherichia coli EDL 933 with verocytotoxin-producing Escherichia coli seropathotypes that are linked to epidemic and/or serious disease. J Clin Microbiol 41:4930–4940. doi: 10.1128/JCM.41.11.4930-4940.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Franz E, van Hoek AH, van der Wal FJ, de Boer A, Zwartkruis-Nahuis A, van der Zwaluw K, Aarts HJ, Heuvelink AE. 2012. Genetic features differentiating bovine, food, and human isolates of Shiga toxin-producing Escherichia coli O157 in The Netherlands. J Clin Microbiol 50:772–780. doi: 10.1128/JCM.05964-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shringi S, Schmidt C, Katherine K, Brayton KA, Hancock DD, Besser TE. 2012. Carriage of stx2a differentiates clinical and bovine-biased strains of Escherichia coli O157. PLoS One 7:e51572. doi: 10.1371/journal.pone.0051572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee K, French NP, Hara-Kudo Y, Iyoda S, Kobayashi H, Sugita-Konishi Y, Tsubone H, Kumagai S. 2011. Multivariate analyses revealed distinctive features differentiating human and cattle isolates of Shiga toxin-producing Escherichia coli O157 in Japan. J Clin Microbiol 49:1495–1500. doi: 10.1128/JCM.02640-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Verstraete K, de Reu K, van Weyenberg S, Pierard K, de Zutter L, Herman L, Robyn J, Heyndrickx M. 2013. Genetic characteristics of Shiga toxin-producing E. coli O157, O26, O103, O111 and O145 isolates from humans, food, and cattle in Belgium. Epidemiol Infect 141:2503–2515. doi: 10.1017/S0950268813000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmidt H, Beutin L, Karch H. 1995. Molecular analysis of the plasmid-encoded hemolysin of Escherichia coli O157:H7 strain EDL 933. Infect Immun 63:1055–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schmidt H, Karch H. 1996. Enterohemolytic phenotypes and genotypes of Shiga toxin-producing Escherichia coli O111 strains from patients with diarrhea and hemolytic-uremic syndrome. J Clin Microbiol 34:2364–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ju W, Shen J, Toro M, Zhao S, Meng J. 2013. Distribution of pathogenicity islands OI-122, OI-43/48, and OI-57 and a high-pathogenicity island in Shiga toxin-producing Escherichia coli. Appl Environ Microbiol 79:3406–3412. doi: 10.1128/AEM.03661-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Madic J, Vingadassalon N, de Garam CP, Marault M, Scheutz F, Brugere H, Jamet E, Auvray F. 2011. Detection of Shiga toxin-producing Escherichia coli serotypes O26:H11, O103:H2, O111:H8, O145:H28, and O157:H7 in raw-milk cheeses by using multiplex real-time PCR. Appl Environ Microbiol 77:2035–2041. doi: 10.1128/AEM.02089-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Islam MA, Mondol AS, de Boer E, Beumer RR, Zwietering MH, Talukder KA, Heuvelink AE. 2008. Prevalence and genetic characterization of Shiga toxin-producing Escherichia coli isolates from slaughtered animals in Bangladesh. Appl Environ Microbiol 74:5414–5421. doi: 10.1128/AEM.00854-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cobbaut K, Berkvens D, Houf K, De Deken R, De Zutter L. 2009. Escherichia coli O157 prevalence in different cattle farm types and identification of potential risk factors. J Food Prot 72:1848–1853. [DOI] [PubMed] [Google Scholar]
- 55.Gunn GJ, McKendrick IJ, Ternent HE, Thomson-Carter F, Foster G, Synge BA. 2007. An investigation of factors associated with the prevalence of verocytotoxin producing Escherichia coli O157 shedding in Scottish beef cattle. Vet J 174:554–564. doi: 10.1016/j.tvjl.2007.08.024. [DOI] [PubMed] [Google Scholar]
- 56.Meyer-Broseta S, Bastian SN, Arne PD, Cerf O, Sanaa M. 2001. Review of epidemiological surveys on the prevalence of contamination of healthy cattle with Escherichia coli serogroup O157:H7. Int J Hyg Environ Health 203:347–361. doi: 10.1078/1438-4639-4410041. [DOI] [PubMed] [Google Scholar]
- 57.Ferens WA, Hovde CJ. 2011. Escherichia coli O157:H7: animal reservoir and sources of human infection. Foodborne Pathog Dis 8:465–487. doi: 10.1089/fpd.2010.0673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Garcia A, Fox JG, Besser TE. 2010. Zoonotic enterohemorrhagic Escherichia coli: a One Health perspective. ILAR J 51:221–232. doi: 10.1093/ilar.51.3.221. [DOI] [PubMed] [Google Scholar]
- 59.European Food Safety Authority. 2012. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2010. EFSA J 7(11):2597. doi: 10.2903/j.efsa.2012.2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.European Food Safety Authority. 2013. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2011. EFSA J 11(4):3129. doi: 10.2903/j.efsa.2013.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]