Abstract
“Candidatus Methylomirabilis oxyfera” is a newly discovered anaerobic methanotroph that, surprisingly, oxidizes methane through an aerobic methane oxidation pathway. The second step in this aerobic pathway is the oxidation of methanol. In Gram-negative bacteria, the reaction is catalyzed by pyrroloquinoline quinone (PQQ)-dependent methanol dehydrogenase (MDH). The genome of “Ca. Methylomirabilis oxyfera” putatively encodes three different MDHs that are localized in one large gene cluster: one so-called MxaFI-type MDH and two XoxF-type MDHs (XoxF1 and XoxF2). MxaFI MDHs represent the canonical enzymes, which are composed of two PQQ-containing large (α) subunits (MxaF) and two small (β) subunits (MxaI). XoxF MDHs are novel, ecologically widespread, but poorly investigated types of MDHs that can be phylogenetically divided into at least five different clades. The XoxF MDHs described thus far are homodimeric proteins containing a large subunit only. Here, we purified a heterotetrameric MDH from “Ca. Methylomirabilis oxyfera” that consisted of two XoxF and two MxaI subunits. The enzyme was localized in the periplasm of “Ca. Methylomirabilis oxyfera” cells and catalyzed methanol oxidation with appreciable specific activity and affinity (Vmax of 10 μmol min−1 mg−1 protein, Km of 17 μM). PQQ was present as the prosthetic group, which has to be taken up from the environment since the known gene inventory required for the synthesis of this cofactor is lacking. The MDH from “Ca. Methylomirabilis oxyfera” is the first representative of type 1 XoxF proteins to be described.
INTRODUCTION
The bacterium “Candidatus Methylomirabilis oxyfera” is a recently discovered player in the methane cycle. This organism, which is the first representative of the novel division of NC10 bacteria with a known physiological function, derives its energy for growth from the anaerobic oxidation of methane coupled to the reduction of nitrate or nitrite into dinitrogen gas (1, 2). “Ca. Methylomirabilis oxyfera” is an extraordinary organism since methane oxidation is performed by the classical oxygen-dependent (aerobic) pathway in which methane oxidation to CO2 proceeds via methanol, formaldehyde, and formate. In this pathway, the first step, activation of methane into methanol, requires oxygen and this oxygen is produced by “Ca. Methylomirabilis oxyfera” itself in concert with nitrite reduction (2). The second step is the oxidation of methanol. This reaction is catalyzed by methanol dehydrogenase (MDH). MDH is a key enzyme widely distributed among aerobic methane-oxidizing (“methanotrophic”) and methyl group-oxidizing (“methylotrophic”) microorganisms. MDHs fall into different classes. Gram-positive microorganisms harbor nicotinoprotein MDHs in their cytoplasm that use NAD(P)H as the electron acceptor for methanol oxidation (3, 4). In Gram-negative bacteria, a phylogenetically unrelated protein is present in the periplasm that possesses pyrroloquinoline quinone (PQQ) as the catalytic center (5–7). PQQ-containing MDHs are members of a quinoprotein family that are able to oxidize a broad range of alcohol and aldehyde substrates, and each member is more or less tuned for a specific substrate (5–8).
Within genomes, a large variety of protein sequences have been annotated as (putative) quinoprotein MDHs. Functionally and structurally best understood are the so-called MxaFI MDHs. These enzymes are well characterized by the resolution of their atomic structures of six different bacterial species (9–16). MxaFI MDHs are heterotetrameric (α2β2) enzymes composed of two large (α, MxaF) and two small (β, MxaI) subunits. Each large subunit contains one noncovalently bound PQQ prosthetic group and one Ca2+ ion at its active site, both of which are essential for enzyme activity (6). The highly basic small subunit, which is not found in other quinoproteins, tightly wraps against the large subunit, but its function is not fully clear. Active MxaF MDHs, thus lacking the β subunit, have been purified before, suggesting that this subunit is not always essential for enzyme activity per se (17, 18). The functional expression of MxaFI MDH requires the action of up to 30 genes. In Methylobacterium and Methylococcus species, these genes are spread over five gene clusters (mxa, mxb, mxc, pqqABCDE, and pqqFG) (19–21). In the mxa cluster, mxaF and maxI are the structural genes. The mxaG gene codes for cytochrome cL, the cognate electron acceptor during methanol oxidation. Occasionally, the mxaJ gene product has been observed as an additional subunit (22, 23), but most of the MxaFI MDH preparations described to date lack this protein, even though the mxaF and mxaI genes are commonly linked to each other in genomes (8). The mxaACKLD gene products are essential for Ca2+ insertion into the apoprotein (19, 24), whereas the products of the mxaRSEH genes are thought to play a role in further MDH maturation (25–27). MxaB is a DNA-binding protein involved in transcriptional regulation, as are the two two-component regulators encoded by the mxb and mxc clusters (28). The gene products of the pqqABCDE and pqqEF clusters are involved in PQQ biosynthesis for which the 23- to 24-amino-acid (aa) gene product of pqqA is the proposed precursor of PQQ (29–31).
Genome sequencing projects revealed the presence of MxaF homologs, termed XoxF proteins, that showed, at most, 50% sequence identity to known MxaFs (32–34). xoxF genes can be detected in many genomes, not only of methanotrophic and methylotrophic species, but also of microorganisms that have not been implicated in such a life-style (8, 34). Phylogenetic analysis divides XoxF proteins into at least five different clades (XoxF1 to XoxF5) (8, 34). Despite their widespread occurrence, the function of XoxF proteins has been enigmatic for some time. Unlike mxaF, xoxF genes were hardly expressed during growth under laboratory conditions and their deletion did not result in a clear phenotype (32). In striking contrast, xoxF genes were highly expressed in the plant phyllosphere (35) and by communities of nutrient-limited costal ocean waters (36). Certain methanotrophs, such as the Verrucomicrobia (37), lack genes coding for an MxaFI MDH, and an XoxF protein would be the only candidate for methanol oxidation. In agreement with this, the deletion of xoxF from Rhodobacter sphaeroides resulted in the loss of this phototroph's ability to use methanol for photorespiration and aerobic respiration (38). Moreover, the purification of XoxF proteins from different bacterial species established their function as MDHs (39–43). These studies also shed light on their elusive nature. The proteins were specifically induced in the presence of rare earth elements (REEs) like La3+ or Ce3+ (40–42). The recent resolution of the crystal structure of the XoxF MDH from the Verrucomicrobia phylum member Methylacidiphilum fumariolicum SolV showed that this REE took the position near the PQQ catalytic site usually occupied by calcium in MxaFI MDHs (43). Importantly, all of the XoxF MDHs described thus far are homodimeric proteins lacking a small subunit.
The genome of “Ca. Methylomirabilis oxyfera” codes for three MxaF homologs that have been annotated as MxaF1 to MxaF3 and that are encoded by one long gene cluster (2). However, key genes known to be required for the biosynthesis of PQQ are absent from the genome of “Ca. Methylomirabilis oxyfera” (44). A closer analysis described in this paper affiliates these three “MxaFs” with an MxaF MDH and two different XoxF MDHs. To address the question of which of these proteins is functionally expressed, we purified MDH from the organism. Curiously, the only active MDH that could be isolated was a heterotetrameric enzyme composed of two XoxF1-type large subunits and two small subunits, and it contained PQQ as its prosthetic group. Using specific antibodies raised against the XoxF1 large subunit, we localized the enzyme to the periplasm of “Ca. Methylomirabilis oxyfera” cells.
MATERIALS AND METHODS
“Ca. Methylomirabilis oxyfera” enrichment culture.
“Ca. Methylomirabilis oxyfera” strain Ooij was enriched from a sediment sample taken from a ditch draining agricultural land in the Ooij polder, a floodplain of the River Rhine in The Netherlands, by the sequencing batch reactor technique (45). Enrichment, growth, and culture maintenance in a 15-liter bioreactor were performed as described before (1, 2, 46–49). Growth took place in an atmosphere of 95% CH4–5% CO2 (vol/vol) with nitrite as the electron acceptor. The trace elements present in the mineral medium were as specified in reference 48 and lacked any additional REEs. Here, we also added boiled cell extract from the methanotroph M. fumariolicum SolV (43, 50). The addition of the extract (10 ml weekly) resulted in an immediate increase in the methane and nitrite conversion rates; cellular activities had increased no less than 10-fold after 2 months. In the biomass that was collected from the enrichment culture, which was also used in parallel studies (2, 46–49), “Ca. Methylomirabilis oxyfera” made up about 80% of the population, as shown in these studies by fluorescent in situ hybridization and metagenome analysis. The residual community (about 20%) was highly diverse and evenly distributed.
Sequence and expression analyses.
Sequences of the “Ca. Methylomirabilis oxyfera” MDH gene clusters and homologues of PQQ genes were retrieved from the NCBI genome database (BioProject accession number PRJNA161981). Sequences of MDH and PQQ gene clusters of M. extorquens AM1 were extracted from GenBank and NCBI databases. Protein sequence analyses and comparisons used the BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi), ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/), and HHpred homology detection and structure prediction (http://toolkit.tuebingen.mpg.de/hhpred) tools. The SignalP 4.1 program (http://www.cbs.dtu.dk/services/SignalP/) (51) was used to predict N-terminal leader sequences and cleavage sites. Levels of transcription are expressed as transcriptome coverage by short-read Illumina sequences (32 nucleotides [nt]) of reverse-transcribed RNA obtained from the Sequence Read Archive (BioProject accession number SRR022748.2) (2).
Enzyme purification.
Approximately 26 g (wet weight) of cells from the “Ca. Methylomirabilis oxyfera” enrichment culture were harvested by centrifugation (6,000 × g, 10 min, 4°C). This and all subsequent steps were performed aerobically. Purification was monitored by measuring MDH activity immediately after each step. Pelleted cells were washed twice in 20 mM potassium phosphate (KPi) buffer (pH 7.0) and resuspended in 20 ml of KPi buffer containing 50 mM sodium pyrophosphate and a few grains of solid DNase. Cells were broken by sonication. Cell debris was removed by centrifugation (6,000 × g, 10 min, 4°C), and the supernatant (1.2 mg of protein ml−1) was collected as cell extract. The fraction containing soluble proteins was obtained as the supernatant after ultracentrifugation of the cell extract (143,000 × g, 60 min, 4°C). To this clear supernatant, saturated ammonium sulfate was added dropwise at 4°C. MDH activity precipitated between 2.0 and 2.8 M (NH4)2SO4. Following centrifugation (6,000 × g, 15 min, 4°C), the precipitated protein fraction was resuspended in 10 ml of 20 mM KPi buffer and applied to a column (1 ml) packed with CHT Ceramic Hydroxyapatite (Bio-Rad, Hercules, CA). The column was equilibrated by washing with 5 column volumes of KPi buffer. Elution was performed with a linear phosphate (pH 7.0) gradient (20 to 500 mM at a flow rate of 1 ml min−1). The only fraction containing MDH activity was eluted at 125 mM phosphate. This fraction was concentrated with a spin column with a 5-kDa cutoff (Sartorius Stedim Biotech, Aubagne, France). Fast protein liquid chromatography (FPLC) column purification was performed at room temperature with an ÄKTA purifier (GE Healthcare, Uppsala, Sweden).
Protein concentrations were determined by the method of Bradford (52) with the Bio-Rad protein assay kit (Bio-Rad, Hercules, CA) with bovine serum albumin (BSA) as the standard. Protein fractions obtained after each step and the purified MDH were analyzed by SDS and native PAGE. Benchmark protein standards (LC0725 NativeMark [Invitrogen Life Technologies, Bleiswijk, The Netherlands] for native PAGE, SM0761 prestained PAGE ruler [Fermentas-Thermo Fisher Scientific, Waltham, MA] for SDS-PAGE) were used to estimate molecular masses. Gels were stained with Coomassie brilliant blue G250 (Bio-Rad).
Enzyme assays.
MDH activity was measured spectrophotometrically at 30°C by monitoring the methanol-dependent and phenazine methosulfate (PMS)-mediated reduction of 2,6-dichlorophenol indophenol (DCPIP) at 600 nm (ε600 of 21,500 M- l cm-l) essentially as described by Antony and Zatman (53) with 100 mM Tris-HCl (pH 8.0) as the buffer and increased concentrations of methanol (20 mM) and NH4Cl (45 mM). Reaction mixtures (0.5 ml) contained 1 mM KCN to suppress the reoxidation of reduced DCPIP. Enzyme kinetic parameters were assessed by nonlinear regression analysis with the Origen 8.5.1 program (OrigenLab Corporation, Northampton, MA) by using Michaelis-Menten kinetics.
Analytical ultracentrifugation.
Analytical equilibrium ultracentrifugation was performed at 20°C in a Beckman XL-I Proteomelab ultracentrifuge (Beckman Coulter, Fullerton, CA) equipped with an An-60 Ti rotor and cells with a 1.2-cm path length. Prior to ultracentrifugation, the protein was equilibrated in 25 mM HEPES-KOH buffer (pH 7.5) containing 25 mM KCl and 1 mM methanol and adjusted to an A280 (1 cm) of 0.75. The speed used was 30,000 rpm. Equilibrium data were evaluated for 300 scans with the SEDFIT program (54) and assuming a frictional ratio (f/f0) of 1.05 for a slightly ellipsoid protein.
UV-visible light absorption spectroscopy and ICP-MS.
The absorption spectrum of the purified enzyme was recorded in a cuvette with a 1-cm path length at room temperature on a Cary 50 spectrophotometer (Agilent, Santa Clara, CA). The concentration and content of the PQQ prosthetic group were calculated on the basis of a molar absorption coefficient of 9,620 M−1 cm−1 at 342 nm for PQQ (55). Inductively coupled plasma mass spectrometry (ICP-MS) was performed to analyze and quantify the metal content of purified protein with the setup and calibration mixture described by Pol et al. (43).
Protein MS.
SDS-gel slices containing the purified MDH were subjected to tryptic in-gel digestion according to Wilm et al. (56). Peptides were extracted and prepared for matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) MS as described by Farhoud et al. (57). MS of the tryptic digests was performed on a Bruker Biflex III spectrometer (Bruker Daltonics, Fremont, CA) operated in reflectron mode. Spectra (500 to 4,000 m/z) were analyzed with the Mascot Peptide Mass Fingerprint search program (Matrix Science, London, United Kingdom) against the “Ca. Methylomirabilis oxyfera” database, with oxidation (M) as variable modification, 0.2 Da peptide tolerance, and a maximum of one missed cleavage. Molecular masses of the native large and small subunits were determined by MALDI-TOF MS operating in the linear mode. These analyses of the as-isolated enzyme were performed by using the [M+H]+, [2M+H]+, [M+2H]+, and [M+3H]+ peaks for determination of molecular masses.
To verify the presence of PQQ, tandem MS was employed. A protein sample was analyzed by C18 reversed-phase nanoflow liquid chromatography (Easy nano-LC; Proxeon, Thermo Fisher Scientific, Waltham, MA) coupled online via a nanoflow electrospray ionization source (Proxeon) to a 7T linear ion trap Fourier transform ion cyclotron resonance (ICR) mass spectrometer (LTQ FT Ultra; Thermo Fisher Scientific). Samples were loaded at a flow rate of 1.2 μl min−1 directly onto the analytical column with 5% acetonitrile. After sample application, peptides and PQQ were eluted from the column with a 15-min linear gradient of 5 to 30% acetonitrile at a flow rate of 0.3 μl min−1. The mass spectrometer was operated in negative-ion mode and optimized for PQQ detection by direct infusion of 1 μM PQQ (Sigma-Aldrich, St. Louis, MO) in 0.5% acetic acid. The ICR cell was programmed to acquire selected ion monitoring spectra of 314 to 344 m/z. The linear ion trap was set to acquire fragmentation scans of m/z 329 with the following parameters: 3E4 ions, a 3Th isolation width, 30% normalized collision energy, a 30-ms activation time, and an activation q of 0.25. Analysis of the MDH sample was performed first and followed by a blank run. Here, the PQQ standard was analyzed. This order was chosen to prevent carryover effects. A simulated (deprotonated) precursor ion spectrum of PQQ was generated with Thermo Scientific Qual browser software.
Antiserum production.
Polyclonal antiserum against the “Ca. Methylomirabilis oxyfera” MDH large (α) subunit (α-XoxF1) was raised by injection of rabbits with the synthetic peptide NQYDPELRSGRWDNK (aa 317 to 331). This antiserum target region was selected on the basis of a unique protein surface peptide sequence deduced from BLAST and ClustalW analyses (see Fig. S1A in the supplemental material). Prior to immunization, an extra amino-terminal cysteine was added to the peptide sequence to enable conjugation to keyhole limpet hemocyanin (Eurogentec, Seraing, Belgium). Two rabbits were immunized by using a 3-month immunization protocol. The antisera from both rabbits were pooled and affinity purified (Eurogentec). This affinity-purified antiserum (α-XoxF1) was used as the primary antiserum for immunoblot analysis and immunogold labeling as described below.
Antiserum specificity.
Antiserum specificity was tested by immunoblot analysis. “Ca. Methylomirabilis oxyfera” cell extracts (30 μg of protein per lane) and purified MDH (10 μg of protein per lane) were separated by SDS–10% PAGE and transferred to a Protran nitrocellulose membrane (Whatman plc, Maidstone, United Kingdom). Immunoblotting and testing of antiserum specificity were performed as described previously (58).
Immunogold labeling and transmission electron microscopy.
Chemical fixation and gelatin embedding of cells from the “Ca. Methylomirabilis oxyfera” enrichment culture, cryosectioning, and subsequent immunogold labeling were done by established protocols (58, 59). The primary antiserum was diluted 50-fold in phosphate-buffered saline containing 1% BSA. Carbon-Formvar-coated grids (copper, hexagonal 100 mesh) containing ultrathin cryosections of “Ca. Methylomirabilis oxyfera” cells were investigated in a transmission electron microscope at 60 kV (JEOL 1010; JEOL Ltd., Akishima-Tokyo, Japan) operating under the iTEM software (Olympus Soft Imaging Solutions, Münster, Germany). Images were recorded with a charge-coupled device camera (MegaView; Olympus Soft Imaging Solutions).
RESULTS
Genomic organization of three MDH systems of “Ca. Methylomirabilis oxyfera.”
The genome of “Ca. Methylomirabilis oxyfera” codes for three different MDH systems (MDH-1, MDH-2, and MDH-3) that are located on the same strand of a cluster of partly overlapping genes (DAMO_0112 to DAMO_0138) (Fig. 1). The cluster is made up of three subclusters. Each subcluster is preceded by genes encoding proteins (DAMO_0112, DAMO_0124, and DAMO_0134) showing 40 to 74% amino acid sequence identity both to each other and to the large subunits (MxaF, XoxF) of well-defined MxaFI and XoxF MDHs (see next). In all three subclusters, genes coding for the putative MDH large subunits are linked to mxaJ and mxaG homologs (termed xoxJ and xoxG in the case of XoxF systems). Herein, distinct MxaG/XoxG proteins with the typical CXXCH motif for heme c binding would represent the cognate physiological electron acceptor for methanol oxidation, cytochrome cL. The function of the MxaJ/XoxJ-like proteins (DAMO_0113, DAMO_0125, DAMO_0136) is not known, but sequence analysis identified it as a member of the family 3 extracellular solute-binding proteins (COG0834, pfam13531), suggesting a role in the binding of methanol or the release of the toxic reaction product formaldehyde. Only the MDH-1 subcluster harbors a gene (DAMO_0115) coding for the small (β) subunit of canonical heterotetrameric MxaFI MDHs. Similarly, only the MDH-1 subcluster comprises the nearly complete set of genes coding for proteins involved in PQQ and Ca2+ insertion (mxaACKLD; DAMO_0118 to DAMO_0121) and in MDH maturation (mxaRSE; DAMO_0116 to DAMO_0117, DAMO_0122) (Fig. 1). In “Ca. Methylomirabilis oxyfera,” the mxaL and mxaD genes are fused (DAMO_0121). The gene order in the MDH-1 subcluster is the same as that found in M. extorquens (19) and in Mxa systems in other organisms (8). However, homologs of mxaH and the regulatory mxaB are absent, which is not unusual (8).
FIG 1.
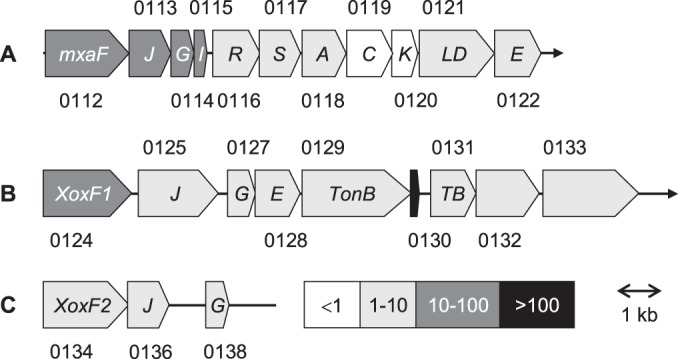
Three MDH systems in the genome of “Ca. Methylomirabilis oxyfera.” The numbers above and below the illustrations are (DAMO) gene identifiers. Levels of transcription expressed as the transcriptome coverage by short read sequences (32 nt) of reverse-transcribed RNA (2) are represented by the gray scale shown in the right half of panel C. Gene sizes and their intergenic regions are drawn to scale. TB, TonB-like protein.
MDH-2 and MDH-3 subclusters lack nearly all of the genes coding for Ca2+ insertion and maturation proteins, which seems to be a common property of XoxF systems (8). In this respect, the presence of an MxaE homolog (DAMO_0128) in the MDH-2 subcluster is remarkable. The two subclusters are separated by five genes (DAMO_0129 to DAMO_0133) (Fig. 1), of which DAMO_0129 and DAMO_0131 encode putative TonB-dependent and TonB-like transporter proteins, respectively. It is interesting that homologs of DAMO_0129 and DAMO_0131 are widely detected in genomes of XoxF-containing methylotrophs (data not shown). DAMO_0132 and DAMO_0133 code for “Ca. Methylomirabilis oxyfera”-specific exported surface proteins that are structurally characterized by β-propeller strands. Deep RNA sequencing indicated that the DAMO_0130 coding region is one of the most highly expressed parts of the genome (Fig. 1), but it remains to be established whether its transcript is translated into an unknown protein (as annotated) or it is a small noncoding RNA that has some regulatory function.
As described below, all of the amino acids involved in the binding of the PQQ prosthetic group are conserved in all three MDH large subunits from “Ca. Methylomirabilis oxyfera.” Moreover, in this work, the presence of this cofactor in one of these MDHs could be confirmed by direct purification. Therefore, it was puzzling that the genome of the organism possessed only three of the seven or eight genes required for PQQ biosynthesis (44), i.e., DAMO_0005 (ppqF), DAMO_0004 (ppqG), and DAMO_0982 (pqqE) (2, 44). However, reexamination of the gene products revealed only low sequence similarity to the particular PQQ biosynthesis enzymes. Like validated PpqF (760 aa) (30), DAMO_0005 (427 aa) and DAMO_0004 (448 aa) belong to the Zn-dependent M16 peptidase superfamily. DAMO_0982 (371 aa) and PqqE (∼380 aa) are members of the radical S-adenosylmethionine superfamily. Significant differences in protein length and low sequence similarities (<20%) with respect to known PQQ biosynthesis proteins suggest other functions for these three “Ca. Methylomirabilis oxyfera” proteins, if they are expressed. Hence, “Ca. Methylomirabilis oxyfera” seems to be devoid of PQQ biosynthesis machinery, at least as far as we know.
Sequence analysis of the MDH large and small subunits of “Ca. Methylomirabilis oxyfera.”
The sequences of the three MDH large subunits (DAMO_0112, DAMO_0124, and DAMO_0134) are 42 to 50% identical, and nearly all of the amino acids that had been implicated in MDH crystal structures with the binding of PQQ and metal (calcium) prosthetic groups are conserved in these subunits (see Fig. S1A in the supplemental material). Major differences included deletions and/or insertions of small amino acid stretches. More-detailed sequence comparison and phylogenetic analysis assigned those large subunits to three different phylogenetic clades (see Fig. S2 in the supplemental material). DAMO_0112 was affiliated with the MxaF MDHs. At the amino acid level, DAMO_0112 (referred to here as MxaFMo) displayed 69 to 74% sequence identity to MxaF MDHs with validated function and known crystal structures. According to the numbering system of Chistoserdova (34), DAMO_0124 and DAMO_0134 are members of the XoxF1 and XoxF2 MDHs, respectively. XoxF1 from “Ca. Methylomirabilis oxyfera” (DAMO_0124; XoxF1Mo) showed the highest sequence identity (67 to 74%) to putative MDHs from methanotrophic Methylotenera, Methyloferula, and Methylocella species, as well as Hyphomicrobium and Xanthomonas representatives. XoxF2 from “Ca. Methylomirabilis oxyfera” (DAMO_0134; XoxF2Mo) showed the highest sequence identity (62 to 63%) to tentative MDHs from the thermoacidophilic methanotrophic Verrucomicrobia (37), including the XoxF-type MDH from M. fumariolicum SolV (XoxFMf), the crystal structure of which has been resolved (43). A particular feature of XoxFMf is the presence of a lanthanide REE (La3+, Ce3+) at its catalytic site instead of Ca2+. This REE is coordinated in close proximity to PQQ by three highly conserved amino acids (Glu172, Arg256, and Asp299; numbering according to that of processed XoxFMf) that are also involved in the coordination of Ca2+ (8, 43). However, the proper coordination of the REE requires one more amino acid, Asp301. Besides other sequence characteristics, the presence of Asp301 is a highly diagnostic property of XoxF MDHs (8). Next, two amino acid substitutions (Pro259→Thr, Ala171→Gly) are observed to accommodate the larger REE in the XoxFMf structure (43). Sequence comparison showed that Asp301 was present in both XoxF1Mo and XoxF2Mo (see Fig. S1A in the supplemental material). In addition, XoxF2Mo displayed the same proline-to-threonine and alanine-to-glycine substitutions seen in XoxFMf, but these substitutions were different in XoxF1Mo (Pro→Asp, Thr→His) (see Fig. S1A). These observations might indicate that both XoxF MDHs from “Ca. Methylomirabilis oxyfera” possess a REE instead of calcium.
As mentioned before, only the MDH-1 subcluster contained a gene coding for a small subunit, namely, DAMO_0115 (Fig. 1). DAMO_0115 was translated as a polypeptide (MxaIMo) of 94 aa. SignalP analysis (51) suggested a 25-aa leader sequence, pointing to the translocation of the processed protein across the cytoplasmic membrane. The processed protein (69 aa; theoretical molecular mass, 8,075.11 Da) showed 61 to 65% sequence identity to the small subunit of MDHs with known crystal structures (see Fig. S1B in the supplemental material). In these structures, the small subunit tightly binds to the large subunit through a conserved set of large-subunit amino acids (see Fig. S1A). To quite an extent, these amino acids are also conserved in XoxFs, even though the latter may not contain such a small subunit. Like MxaIMo, all three MDH large subunits from “Ca. Methylomirabilis oxyfera” were predicted to contain N-terminal leader sequences, again suggesting a periplasmic localization of the processed enzymes. After cleavage of the N-terminal 29 or 30 aa, this would result MxaFMo, XoxF1Mo, and XoxF2Mo apoprotein molecular masses of 63,526, 67,660, and 63,713 Da, respectively.
Deep RNA sequencing (2) indicated that mxaF was most highly expressed at the mRNA level (transcriptome coverage: mxaFMo, 62-fold; xoxF1Mo, 14-fold; xoxF2Mo, 6-fold) (Fig. 1). Strikingly, XoxF1Mo was most highly expressed in the proteome of “Ca. Methylomirabilis oxyfera” strain Ooij, which was used in the present study, whereas XoxF2Mo was most abundant in the proteome of closely related “Ca. Methylomirabilis oxyfera” strain Twente (2), despite the fact that both strains were enriched and cultured under similar conditions. The above considerations raised the questions of which of the three MDHs is (are) functionally expressed, whether their large subunit contains PQQ or an alternative cofactor, which metal is present at the catalytic site, and where the MDH is localized. To address these questions, we purified the MDH from “Ca. Methylomirabilis oxyfera” and assessed its cellular localization by immunogold labeling.
Purification of a heterotetrameric XoxF MDH from “Ca. Methylomirabilis oxyfera.”
MDH was purified with a 32% yield by the two-step purification procedure summarized in Table 1. The preparation obtained after the hydroxyapatite step represented the predominant MDH activity (>95%); a few other fractions displayed negligible activity and were not further investigated. After ultracentrifugation of the cell extract, 93% of the MDH activity was recovered in the supernatant. Hence, MDH is soluble or only loosely membrane associated. The purification factor (8.4-fold) suggested that MDH is a major protein. The purified enzyme catalyzed methanol oxidation by simple Michaelis-Menten kinetics with apparent Vmax and Km values of 10 μmol min−1 mg of protein−1 and 17 μM, respectively. The presence of ammonium strongly stimulated its activity; in the absence of ammonium, it was 10-fold less active.
TABLE 1.
Purification of MDH from “Ca. Methylomirabilis oxyfera” enrichment culture
| Purification step | Total protein (mg) | Sp act (μmol min−1 mg−1) | Total activity (μmol min−1) | Yield (%) |
|---|---|---|---|---|
| Cell extract | 24.5 | 1.10 ± 0.06 | 26.4 | 100 |
| Soluble fraction | 19.7 | 0.97 ± 0.05 | 19.2 | 73 |
| Membranes | 3.4 | 0.53 ± 0.03 | 1.8 | 7 |
| (NH4)2SO4 | 5.7 | 2.94 ± 0.23 | 16.9 | 64 |
| Hydroxyapatite | 0.9 | 9.66 ± 1.52 | 8.4 | 32 |
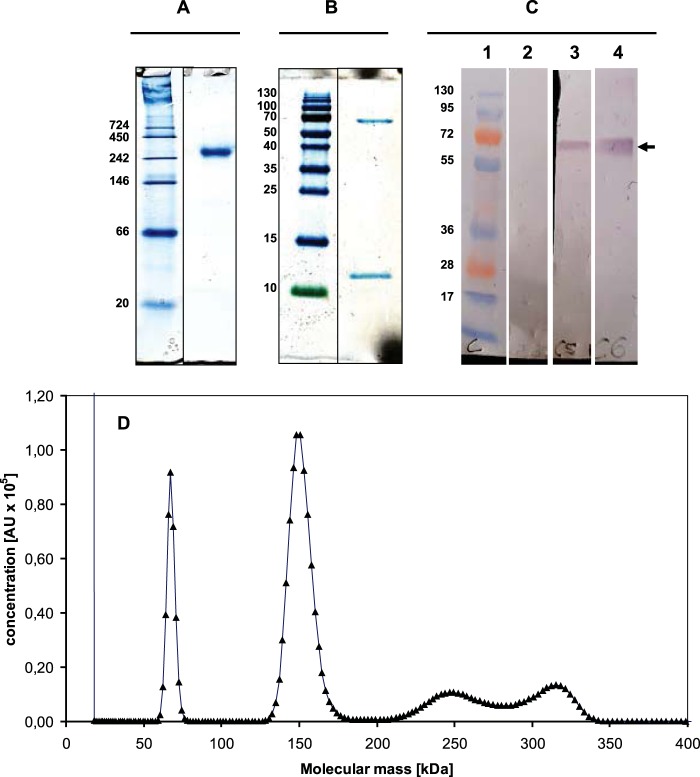
On native PAGE, purified MDH showed one single band with an apparent molecular mass of approximately 260 kDa (Fig. 2A). SDS-PAGE displayed two bands of 67 and 11 kDa (Fig. 2B). Linear-mode MALDI-TOF MS analysis applied to the as-isolated MDH revealed an Mr of 8,200 ± 66 (n = 5) for the small subunit, which was significantly less than that judged by SDS-PAGE (see Fig. S3 in the supplemental material). However, this value was close to the calculated molecular mass (8,075.11 Da) of MxaIMo, which is encoded by a gene (DAMO_0115) in the MDH-1 subcluster, after cleavage of the predicted signal peptide. In agreement with this, trypsin cleavage of the MDH small subunit and subsequent reflectron mode MALDI-TOF MS analysis verified the presence of several peptides that were to be expected in DAMO_0115 (see Fig. S1B). The large subunit was resistant to proteolytic cleavage, and only a few peptides were recovered within the 500-to-4,000 m/z frame used for MS analysis. These peptides uniquely mapped to DAMO_0124 (XoxF1Mo) (see Fig. S1A). Linear MALDI-TOF MS gave a molecular mass of 67.3 ± 0.27 kDa (n = 7) for the large subunit (see Fig. S3), which agrees with both that estimated by SDS-PAGE and that expected for N-terminally cleaved XoxF1Mo (DAMO_0124; 67.66 kDa). It should be noted that the molecular masses of the other two large subunits are ∼4 kDa less (MxaFMo, 63,526 Da; XoxF2Mo, 63,713 Da), differences that are incompatible with MS. Upon analytical equilibrium ultracentrifugation of native MDH, the dominant 280-nm-absorbing band could be modeled (SEDFIT) to an ∼150-kDa protein (Fig. 2D), which is clearly less than that observed by native PAGE. This molecular mass of 150 kDa is consistent with an α2β2 protein composed of two 67-kDa large subunits and two 8-kDa small subunits. Besides this major band, three more bands were obtained during analytical ultracentrifugation of the apparently pure protein preparation (Fig. 2B). These bands sedimented at ∼67, ∼240, and ∼310 kDa. The molecular mass of the first band (67 kDa) again agreed with that expected for XoxF1Mo. Both high-molecular-mass bands might represent higher aggregates (α3β3, α4β4). Taken together, the observations concluded that the MDH purified from “Ca. Methylomirabilis oxyfera” was predominantly a heterotetrameric (α2β2) enzyme composed of two MxaIMo small subunits and two XoxF1Mo large subunits.
FIG 2.
PAGE, immunoblotting, and analytical ultracentrifugation of MDH from “Ca. Methylomirabilis oxyfera.” (A) Native 10% PAGE of purified MDH (6 μg). (B) SDS–15% PAGE of the purified MDH (4 μg). The upper band corresponds to XoxF1, and lower band corresponds to MxaI. Marker proteins and their corresponding molecular masses are shown in the left lanes of panels A to C. (C) Immunoblot analysis of the affinity-purified antiserum (α-XoxF1) directed against XoxF1 of “Ca. Methylomirabilis oxyfera.” For SDS–10% PAGE, gels were loaded with cell extract from “Ca. Methylomirabilis oxyfera” (30 μg of protein) or with purified MDH (10 μg) and blotted onto a nitrocellulose membrane. Lanes: 1, marker proteins; 2, cell extract blot incubated with only secondary antiserum; 3, cell extract blot incubated with α-XoxF1 antiserum; 4, purified MDH blot incubated with α-XoxF1 antiserum. The expected target size (∼67 kDa) is indicated by the arrow. The values to the left are molecular sizes in kilodaltons. (D) Analytical ultracentrifugation of purified MDH. Analytical ultracentrifugation was performed as described in Materials and Methods, and equilibrium data were fitted by the SEDFIT program (54). Concentrations are expressed in arbitrary units (AU) of absorbance at 280 nm.
The XoxF MDH from “Ca. Methylomirabilis oxyfera” contains PQQ.
The UV-visible light absorption spectrum of the purified MDH from “Ca. Methylomirabilis oxyfera” exhibited the characteristics of a quinoprotein (Fig. 3), having an absorption maximum at 345 to 350 nm and a wide shoulder at 375 to 400 nm (60, 61). The spectrum of the enzyme (6.2 mg ml−1; 82 μM calculated on the basis of an Mr of 75,700 for an αβ heterodimer) had an absorbance at 342 nm of 0.91. Assuming a PQQ molar coefficient of absorption at 342 nm of 9,620 M−1.cm−1 (55), the quinone concentration was 95 μM, which indicated that the protein bound PQQ in an about 1:1 stoichiometry. The prominent absorption peak around 345 nm was also indicative of the presence of a metal ion in the active site of the enzyme (62, 63). Incubation of the enzyme preparation with EGTA had no effect on the overall spectrum of the enzyme, indicating that the metal would be tightly bound. To assess the nature of the metal, ICP-MS was used as described before (43). The analysis revealed the presence of calcium only; rare earth metals were below the limit of detection (<0.1 mol%). Since column fractions lacking MDH contained calcium as well, presumably derived from hydroxyapatite [Ca10(PO4)6] used for purification, it was not possible to determine the content of this compound in the protein.
FIG 3.
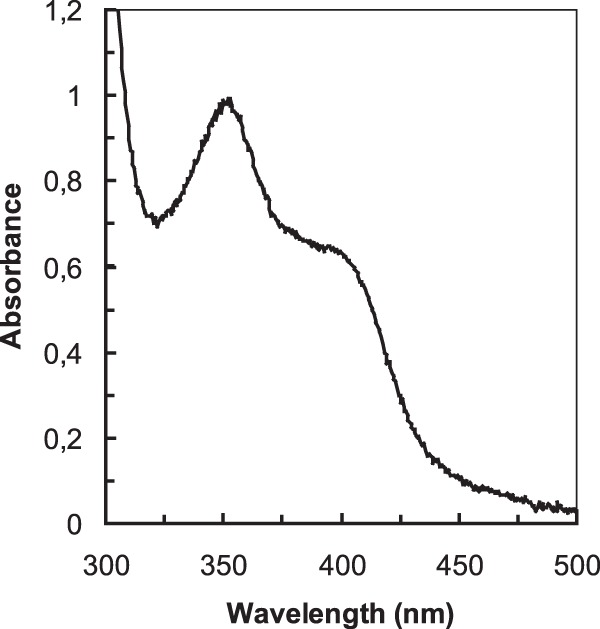
UV-visible light absorption spectrum of purified MDH of “Ca. Methylomirabilis oxyfera.” The spectrum was recorded in 150 mM phosphate buffer (pH 7.0) at 30°C. The protein concentration was 6.2 mg ml−1.
The presence of PQQ in the purified MDH from “Ca. Methylomirabilis oxyfera” was confirmed by MS with both precursor ion and fragmentation level data. The acquired precursor ion spectra from the PQQ standard and the purified MDH fully compared the simulated spectrum of PQQ. The monoisotopic mass of the [M-H]-PQQ ion from both the standard MDH (m/z 329.0053; Δ = 4.0 ppm) and the purified MDH (m/z 329.0052; Δ = 3.6 ppm) accurately matched the calculated (deprotonated) monoisotopic mass of PQQ (m/z 329.0040) (see Fig. S4A in the supplemental material). Moreover, the relative abundance of the 13C isotope peak in precursor spectra from both the PQQ standard (17%) and the purified MDH (16%) was in good agreement with the simulated spectrum (15%). The isotope abundance spectrum was simulated according to de Hoffmann and Stroobant (64). The presence of PQQ in the purified MDH was also confirmed by comparing the collision-induced dissociation fragmentation spectra of m/z 329 from the PQQ standard and the purified enzyme (see Fig. S4B). The two spectra were nearly identical and showed the presence of two abundant fragment ions corresponding to the loss of CO2 (m/z 285 [M-H-CO2]−) and 2CO2 (m/z 241 [M-H-2CO2]−) from the precursor ion. The findings agreed with those of Noji et al. (65), except that in neither the standard nor the purified enzyme was a third diagnostic fragment ion (m/z 197 [M-H-3CO2]−) detectable, which was probably due to different instrument characteristics and/or conditions.
Cellular localization of the XoxF MDH in “Ca. Methylomirabilis oxyfera.”
The N-terminal signal sequences in XoxF1Mo suggested a periplasmic localization of the processed protein (see Fig. S1A in the supplemental material). To investigate this, we generated primary antiserum with a synthetic peptide targeting a sequence of XoxF1Mo (α-XoxF1) that was specific for this protein (see Fig. S1A). The specificity of the derived antiserum was confirmed with both cell extracts of “Ca. Methylomirabilis oxyfera” and the purified enzyme by SDS-PAGE and immunoblot analysis (Fig. 2C). No bands were detected in blots incubated with the secondary antiserum only. These results showed that the derived α-XoxF1 antiserum was specific and suited for the intracellular localization of XoxF1Mo in ultrathin cryosections of “Ca. Methylomirabilis oxyfera” cells. Ultrathin sections of “Ca. Methylomirabilis oxyfera” cells incubated with colloidal gold alone showed no gold particles associated with the atypical polygon-shaped “Ca. Methylomirabilis oxyfera” cells (negative control; data not shown) (58, 59). When ultrathin sections were incubated with α-XoxF1, very little background was detected and most of the gold particles were observed near the outer side of the cytoplasmic membrane, as expected for a periplasmic enzyme (Fig. 4).
FIG 4.
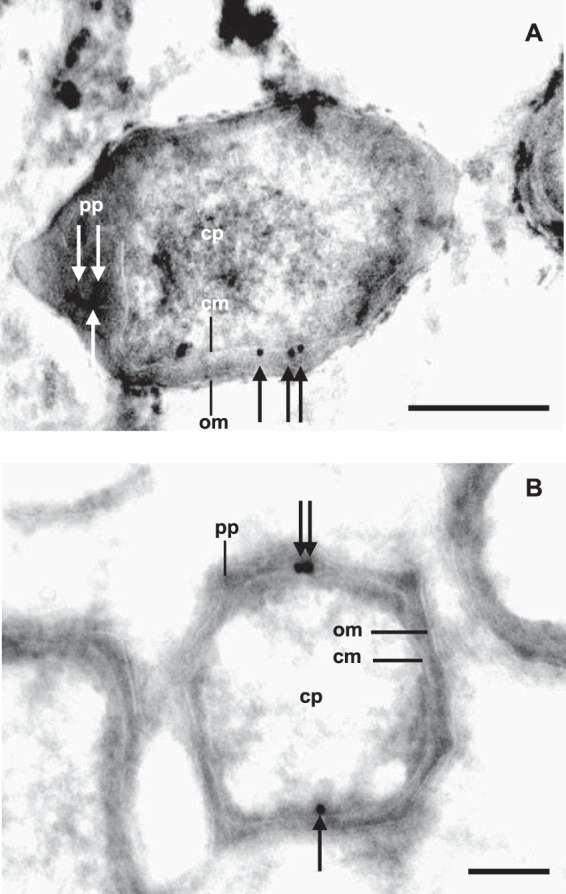
Transmission electron micrographs of chemically fixed and cryosectioned “Ca. Methylomirabilis oxyfera” cells. The immunogold localization of affinity-purified antiserum directed against XoxF1 is shown (black and white arrows). Longitudinal (A) and cross (B) sections were blocked with 1% BSA and treated with 50-fold-diluted α-XoxF1 serum. Abbreviations: cp, cytoplasm; pp, periplasm; cm, cytoplasmic membrane; om, outer membrane. Scale bars, 200 nm.
DISCUSSION
The genome of the anaerobic methanotroph “Ca. Methylomirabilis oxyfera” codes for three different MDH systems that are localized in one large gene cluster, which is highly unusual: one MxaFI MDH and two XoxF MDHs (Fig. 1). Here, we purified the predominant MDH that is functionally expressed under the growth conditions used (Fig. 2 and Table 1). The enzyme was composed of two small and two large subunits, as is common among MxaFI MDHs (5–8). MALDI-TOF MS identified the small subunit as the N-terminally cleaved gene product of DAMO_0115 (MxaIMo). Five lines of evidence (SDS-PAGE, MALDI-TOF MS of the native and trypsin-cleaved protein, analytical ultracentrifugation, antibody specificity) consistently indicated that the large subunit with a size of 67 kDa was XoxF1Mo (DAMO_0124). Immunogold labeling localized the protein in the periplasm (Fig. 4), as is also known for MDHs from other methanotrophs and methylotrophs. In the established (PMS and DCPIP) dye-coupled assay (6, 53), “Ca. Methylomirabilis oxyfera” MDH catalyzed the oxidation of methanol with high affinity (Km of 17 μM) and a high Vmax of 10 μmol min−1 mg −1. The apparent Km for methanol is comparable to those of well-investigated MxaFI MDHs (2 to 20 μM), while the Vmax of the “Ca. Methylomirabilis oxyfera” enzyme was substantially higher than those of known MxaFI MDHs (0.8 to 1.0 μmol min−1 mg−1) (60, 63, 66). In addition, MDH was a major enzyme in “Ca. Methylomirabilis oxyfera.” Taking into account that the very slowly growing bacterium “Ca. Methylomirabilis oxyfera” oxidizes methane at an extremely low rate (1.7 nmol min−1 mg of protein−1) (2, 46), the abundance of MDH and its favorable catalytic efficiency (Vmax/Km) allow the conversion of methanol, the product of methane activation, down to low concentrations.
“Ca. Methylomirabilis oxyfera” MDH contains PQQ as its prosthetic group. While the unambiguous presence of this cofactor in an XoxF-type large subunit was established only quite recently (43), the finding was expected. Except for nicotinoprotein methanol/alcohol dehydrogenase (3, 4), all of the MDHs known to date rely on PQQ as the catalytic center and PQQ-binding motifs have been identified before in XoxF MDHs (see Fig. S1A) (8, 32). However, the genome of “Ca. Methylomirabilis oxyfera” lacks known PQQ biosynthesis machinery. So, either the organism has evolved a novel biosynthetic pathway, which is unlikely considering the highly specific chemistry of the (oxygen-dependent) reactions involved (67), or the microorganism derives the cofactor from producers in the enrichment culture. Our finding that the supply of boiled cells of M. fumariolicum strongly stimulated “Ca. Methylomirabilis oxyfera” culture activity and growth supports the latter view. Otherwise, the dependence on exogenous PQQ is not without precedent. For example, Escherichia coli and other enteric bacteria are incapable of PQQ synthesis, but the organisms readily incorporate the cofactor present in growth medium into their glucose dehydrogenase quinoproteins (68, 69). In addition, it has been known for quite some time that microorganisms that are capable of PQQ biosynthesis may excrete the compound, thereby stimulating the growth of other organisms (70). Still, it would be astonishing if “Ca. Methylomirabilis oxyfera,” with its unique life-style, depended on other microorganisms in the environment for the supply of an essential cofactor of one of its key enzymes.
The large subunit of purified MDH from “Ca. Methylomirabilis oxyfera” belongs to the XoxF MDHs. XoxF MDHs likely represent an ecologically highly relevant but grossly overlooked group of MDHs (8). Genes coding for these proteins are widely found in the genomes of methylotrophic and methanotrophic bacteria and can be phylogenetically differentiated into at least five lineages (see Fig. S2 in the supplemental material) (8, 34). While hardly expressed under laboratory growth conditions (32), xoxF genes are among the most highly expressed genes in natural systems (35, 36). The reason for their long-time elusiveness is that XoxF apparently harbors lanthanide REEs, compounds that are rarely included in laboratory trace element solutions, at its catalytic site (40–43). The presence of a REE may confer on the proteins a catalytic efficiency that is superior to that of their commonly studied calcium-containing MxaFI MDH counterparts (8, 43). Typically, the XoxF MDHs isolated thus far lack a small subunit (39–43). In agreement with this, XoxF genes are not associated with genes encoding a small subunit (8). In these respects, MDH from “Ca. Methylomirabilis oxyfera,” being the first member of the XoxF1 MDH family to be documented, already might represent a variation on a still only emerging theme. First, enzyme preparations did not contain appreciable amounts of REEs. However, calcium was present, although part of it may have stemmed from the hydroxyapatite used during purification. Nonetheless, the UV-visual light spectrum of “Ca. Methylomirabilis oxyfera” MDH, having an absorbance maximum at 445 nm (Fig. 3), is typical of a calcium-containing quinoprotein rather than a REE-containing MDH, in which the PQQ absorbance band is shifted to a longer wavelength (454 nm) (43). These considerations suggest that “Ca. Methylomirabilis oxyfera” MDH possesses calcium. A second striking difference between the “Ca. Methylomirabilis oxyfera” MDH and the XoxF MDHs known thus far is the presence in the former of a small subunit that is derived from the MxaFIMo gene cluster (MDH-1) (Fig. 1). The function of the small subunit is not precisely known, but it has been suggested by us (8) that it enables MxaFI MDHs to properly coordinate calcium. Also in the XoxF MDH described here, the small subunit might have such a function. Obviously, the concerted action of different proteins from different gene clusters requires tight regulation. The mechanisms underlying regulation are a wide-open field of research, regarding not only “Ca. Methylomirabilis oxyfera” but also in other methylotrophs and methanotrophs that harbor multiple MxaFI and XoxF MDH systems in their genomes.
The presence of multiple MDH systems offers a methylotroph or methanotroph the means to adapt its metabolism in an optimal way to prevailing environmental conditions (availability of substrates and essential cofactors, like metals). The as yet poorly explored XoxF MDHs add to this by their (presumably) superior catalytic properties, yet utilizing difficult-to-access catalytic REEs (8). The hybrid heterotetrameric MDH described here provides “Ca. Methylomirabilis oxyfera” with a most efficient means to oxidize methanol in the pathway from methane to CO2.
Supplementary Material
ACKNOWLEDGMENTS
We thank Katinka van de Pas-Schoonen for support in the maintenance of the enrichment cultures and Geert-Jan Janssen from the General Instruments Department for his help in operating the transmission electron microscope. Thomas Barends of the Max Planck Institute for Biomedical Research (Heidelberg, Germany) is acknowledged for the analytical ultracentrifugation analysis. We thank Wouter Maalcke and Naomi de Almeida for their support in operating the FPLC system.
The research by M.L.W. was funded by a Horizon grant (050-71-058), L.V.N. was funded by the Netherlands Organization for Scientific Research (VENI grant 863.09.009), and M.S.M.J. was funded by two European Union Advanced Research Grants (ERC 232937, ERC 339880) and by Zwaartekrachtsubsidie (gravitation grant) SIAM 024 002 002 from the Dutch Government to the Soehngen Institute for Anaerobic Microbiology.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03292-14.
REFERENCES
- 1.Raghoebarsing AA, Pol A, van de Pas-Schoonen KT, Smolders AJ, Ettwig KF, Rijpstra WI, Schouten S, Sinninghe Damsté JS, Op den Camp HJM, Jetten MSM, Strous M. 2006. A microbial consortium couples anaerobic methane oxidation to denitrification. Nature 440:918–921. doi: 10.1038/nature04617. [DOI] [PubMed] [Google Scholar]
- 2.Ettwig KF, Butler MK, Le Paslier D, Pelletier E, Mangenot S, Kuypers MMM, Schreiber F, Dutilh BE, Zedelius J, de Beer D, Gloerich J, Wessels HJ, van Alen T, Luesken F, Wu ML, van de Pas-Schoonen KT, Op den Camp HJM, Janssen-Megens EM, Francoijs KJ, Stunnenberg H, Weissenbach J, Jetten MSM, Strous M. 2010. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 464:543–548. doi: 10.1038/nature08883. [DOI] [PubMed] [Google Scholar]
- 3.Hektor HJ, Kloosterman H, Dijkhuizen L. 2000. Nicotinoprotein methanol dehydrogenase enzymes in Gram-positive methylotrophic bacteria. J Mol Catal B 8:103–109. doi: 10.1016/S1381-1177(99)00073-9. [DOI] [Google Scholar]
- 4.Hektor HJ, Kloosterman H, Dijkhuizen L. 2002. Identification of a magnesium-dependent NAD(P)(H)-binding domain in the nicotinoprotein methanol dehydrogenase from Bacillus methanolicus. J Biol Chem 277:46966–46973. doi: 10.1074/jbc.M207547200. [DOI] [PubMed] [Google Scholar]
- 5.Anthony C, Ghosh M. 1998. The structure and function of the PQQ-containing quinoprotein dehydrogenases. Prog Biophys Mol Biol 69:1–21. doi: 10.1016/S0079-6107(97)00020-5. [DOI] [PubMed] [Google Scholar]
- 6.Anthony C, Williams P. 2003. The structure and mechanism of methanol dehydrogenase. Biochim Biophys Acta 1647:18–23. doi: 10.1016/S1570-9639(03)00042-6. [DOI] [PubMed] [Google Scholar]
- 7.Anthony C. 2004. The quinoprotein dehydrogenases for methanol and glucose. Arch Biochem Biophys 428:2–9. doi: 10.1016/j.abb.2004.03.038. [DOI] [PubMed] [Google Scholar]
- 8.Keltjens JT, Pol A, Reimann J, Op den Camp HJM. 2014. PQQ-dependent methanol dehydrogenases: rare-earth elements make a difference. Appl Microbiol Biotechnol 98:6163–6183. doi: 10.1007/s00253-014-5766-8. [DOI] [PubMed] [Google Scholar]
- 9.Xia ZX, Dai WW, Xiong JP, Hao ZP, Davidson VL, White S, Mathews FS. 1992. The three-dimensional structures of methanol dehydrogenase from two methylotrophic bacteria at 2.6-Å resolution. J Biol Chem 267:22289–22297. [PubMed] [Google Scholar]
- 10.Ghosh M, Anthony C, Harlos K, Goodwin MG, Blake C. 1995. The refined structure of the quinoprotein methanol dehydrogenase from Methylobacterium extorquens at 1.94 A. Structure 3:177–187. [DOI] [PubMed] [Google Scholar]
- 11.Xia Z, Dai W, Zhang Y, White SA, Boyd GD, Mathews FS. 1996. Determination of the gene sequence and the three-dimensional structure at 2.4 angstroms resolution of methanol dehydrogenase from Methylophilus W3A1. J Mol Biol 259:480–501. doi: 10.1006/jmbi.1996.0334. [DOI] [PubMed] [Google Scholar]
- 12.Xia ZX, He YN, Dai WW, White SA, Boyd GD, Mathews FS. 1999. Detailed active site configuration of a new crystal form of methanol dehydrogenase from Methylophilus W3A1 at 1.9 Å resolution. Biochemistry 38:1214–1220. doi: 10.1021/bi9822574. [DOI] [PubMed] [Google Scholar]
- 13.Xia ZX, Dai WW, He YN, White SA, Mathews FS, Davidson VL. 2003. X-ray structure of methanol dehydrogenase from Paracoccus denitrificans and molecular modeling of its interactions with cytochrome c-551i. J Biol Inorg Chem 8:843–854. doi: 10.1007/s00775-003-0485-0. [DOI] [PubMed] [Google Scholar]
- 14.Nojiri M, Hira D, Yamaguchi K, Okajima T, Tanizawa K, Suzuki S. 2006. Crystal structures of cytochrome cL and methanol dehydrogenase from Hyphomicrobium denitrificans: structural and mechanistic insights into interactions between the two proteins. Biochemistry 45:3481–3492. doi: 10.1021/bi051877j. [DOI] [PubMed] [Google Scholar]
- 15.Williams P, Coates L, Mohammed F, Gill R, Erskine P, Bourgeois D, Wood SP, Anthony C, Cooper JB. 2006. The 1.6 Å X-ray structure of the unusual c-type cytochrome, cytochrome cL, from the methylotrophic bacterium Methylobacterium extorquens. J Mol Biol 357:151–162. doi: 10.1016/j.jmb.2005.12.055. [DOI] [PubMed] [Google Scholar]
- 16.Choi JM, Kim HG, Kim JS, Youn HS, Eom SH, Yu SL, Kim SW, Lee SH. 2011. Purification, crystallization and preliminary X-ray crystallographic analysis of a methanol dehydrogenase from the marine bacterium Methylophaga aminisulfidivorans MP(T). Acta Crystallogr Sect F Struct Biol Cryst Commun 67:513–516. doi: 10.1107/S1744309111006713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grosse S, Wendlandt KD, Kleber HP. 1997. Purification and properties of methanol dehydrogenase from Methylocystis sp. GB 25 J Basic Microbiol 37:269–279. doi: 10.1002/jobm.3620370406. [DOI] [PubMed] [Google Scholar]
- 18.Grosse S, Voigt C, Wendlandt K-D, Kleber H-P. 1998. Purification and properties of methanol dehydrogenase from Methylosinus sp. WI 14. J Basic Microbiol 38:189–196. doi:. [DOI] [PubMed] [Google Scholar]
- 19.Chistoserdova L, Chen SW, Lapidus A, Lidstrom ME. 2003. Methylotrophy in Methylobacterium extorquens AM1 from a genomic point of view. J Bacteriol 185:2980–2987. doi: 10.1128/JB.185.10.2980-2987.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ward N, Larsen Ø Sakwa J, Bruseth L, Khouri H, Durkin AS, Dimitrov G, Jiang L, Scanlan D, Kang KH, Lewis M, Nelson KE, Methé B, Wu M, Heidelberg JF, Paulsen IT, Fouts D, Ravel J, Tettelin H, Ren Q, Read T, DeBoy RT, Seshadri R, Salzberg SL, Jensen HB, Birkeland NK, Nelson WC, Dodson RJ, Grindhaug SH, Holt I, Eidhammer I, Jonasen I, Vanaken S, Utterback T, Feldblyum TV, Fraser CM, Lillehaug JR, Eisen JA. 2004. Genomic insights into methanotrophy: the complete genome sequence of Methylococcus capsulatus (Bath). PLoS Biol 2:e303. doi: 10.1371/journal.pbio.0020303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chistoserdova L, Lapidus A, Han C, Goodwin L, Saunders L, Brettin T, Tapia R, Gilna P, Lucas S, Richardson PM, Lidstrom ME. 2007. Genome of Methylobacillus flagellatus, molecular basis for obligate methylotrophy, and polyphyletic origin of methylotrophy. J Bacteriol 189:4020–4027. doi: 10.1128/JB.00045-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsushita K, Takahashi K, Adachi O. 1993. A novel quinoprotein methanol dehydrogenase containing an additional 32-kilodalton peptide purified from Acetobacter methanolicus: identification of the peptide as a MoxJ product. Biochemistry 32:5576–5582. doi: 10.1021/bi00072a012. [DOI] [PubMed] [Google Scholar]
- 23.Kim HG, Han GH, Kim D, Choi JS, Kim SW. 2012. Comparative analysis of two types of methanol dehydrogenase from Methylophaga aminisulfidivorans MPT grown on methanol. J Basic Microbiol 52:141–149. doi: 10.1002/jobm.201000479. [DOI] [PubMed] [Google Scholar]
- 24.Richardson IW, Anthony C. 1992. Characterization of mutant forms of the quinoprotein methanol dehydrogenase lacking an essential calcium ion. Biochem J 287:709–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anderson DJ, Morris CJ, Nunn DN, Anthony C, Lidstrom ME. 1990. Nucleotide sequence of the Methylobacterium extorquens AM1 moxF and moxJ genes involved in methanol oxidation. Gene 90:173–176. doi: 10.1016/0378-1119(90)90457-3. [DOI] [PubMed] [Google Scholar]
- 26.Nunn DN, Lidstrom ME. 1986. Isolation and complementation analysis of 10 methanol oxidation mutant classes and identification of the methanol dehydrogenase structural gene for Methylobacterium sp. strain AM1. J Bacteriol 166:581–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nunn DN, Lidstrom ME. 1986. Phenotypic characterization of 10 methanol oxidation mutant classes in Methylobacterium sp. strain AM1. J Bacteriol 166:591–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang M, Lidstrom ME. 2003. Promoters and transcripts for genes involved in methanol oxidation in Methylobacterium extorquens AM1. Microbiology 149:1033–1040. doi: 10.1099/mic.0.26105-0. [DOI] [PubMed] [Google Scholar]
- 29.Goosen N, Huinen RGM, Vandeputte P. 1992. A 24-amino-acid polypeptide is essential for the biosynthesis of the coenzyme pyrrolo-quinoline-quinone. J Bacteriol 174:1426–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Puehringer S, Metlitzky M, Schwarzenbacher R. 2008. The pyrroloquinoline quinone biosynthesis pathway revisited: a structural approach. BMC Biochem 9:8. doi: 10.1186/1471-2091-9-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gliese N, Khodaverdi V, Görisch H. 2010. The PQQ biosynthetic operons and their transcriptional regulation in Pseudomonas aeruginosa. Arch Microbiol 192:1–14. doi: 10.1007/s00203-009-0523-6. [DOI] [PubMed] [Google Scholar]
- 32.Chistoserdova L, Lidstrom ME. 1997. Molecular and mutational analysis of a DNA region separating two methylotrophy gene clusters in Methylobacterium extorquens AM1. Microbiology 143:1729–1736. doi: 10.1099/00221287-143-5-1729. [DOI] [PubMed] [Google Scholar]
- 33.Kalyuzhnaya MG, Hristova KR, Lidstrom ME, Chistoserdova L. 2008. Characterization of a novel methanol dehydrogenase in representatives of Burkholderiales: implications for environmental detection of methylotrophy and evidence for convergent evolution. J Bacteriol 190:3817–3823. doi: 10.1128/JB.00180-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chistoserdova L. 2011. Modularity of methylotrophy, revisited. Environ Microbiol 13:2603–2622. doi: 10.1111/j.1462-2920.2011.02464.x. [DOI] [PubMed] [Google Scholar]
- 35.Delmotte N, Knief C, Chaffron S, Innerebner G, Roschitzki B, Schlapbach R, von Mering C, Vorholt JA. 2009. Community proteogenomics reveals insights into the physiology of phyllosphere bacteria. Proc Natl Acad Sci U S A 106:16428–16433. doi: 10.1073/pnas.0905240106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sowell SM, Abraham PE, Shah M, Verberkmoes NC, Smith DP, Barofsky DF, Giovannoni SJ. 2011. Environmental proteomics of microbial plankton in a highly productive coastal upwelling system. ISME J 5:856–865. doi: 10.1038/ismej.2010.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Op den Camp HJM, Islam T, Stott MB, Harhangi HR, Hynes A, Schouten S, Jetten MSM, Birkeland N-K, Pol A, Dunfield PF. 2009. Environmental, genomic and taxonomic perspectives on methanotrophic Verrucomicrobia. Environ Microbiol Rep 1:293–306. doi: 10.1111/j.1758-2229.2009.00022.x. [DOI] [PubMed] [Google Scholar]
- 38.Wilson SM, Gleisten MP, Donohue TJ. 2008. Identification of proteins involved in formaldehyde metabolism by Rhodobacter sphaeroides. Microbiology 154:296–305. doi: 10.1099/mic.0.2007/011346-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmidt S, Christen P, Kiefer P, Vorholt JA. 2010. Functional investigation of methanol dehydrogenase-like protein XoxF in Methylobacterium extorquens AM1. Microbiology 156:2575–2586. doi: 10.1099/mic.0.038570-0. [DOI] [PubMed] [Google Scholar]
- 40.Fitriyanto NA, Fushimi M, Matsunaga M, Pertiwiningrum A, Iwama T, Kawai K. 2011. Molecular structure and gene analysis of Ce3+-induced methanol dehydrogenase of Bradyrhizobium sp. MAFF211645. J Biosci Bioeng 111:613–617. doi: 10.1016/j.jbiosc.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 41.Hibi Y, Asai K, Arafuka H, Hamajima M, Iwama T, Kawai K. 2011. Molecular structure of La3+-induced methanol dehydrogenase-like protein in Methylobacterium radiotolerans. J Biosci Bioeng 111:547–549. doi: 10.1016/j.jbiosc.2010.12.017. [DOI] [PubMed] [Google Scholar]
- 42.Nakagawa T, Mitsui R, Tani A, Sasa K, Tashiro S, Iwama T, Hayakawa T, Kawai K. 2012. A catalytic role of XoxF1 as La3+-dependent methanol dehydrogenase in Methylobacterium strain AM1. PLoS One 7:e50480. doi: 10.1371/journal.pone.0050480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pol A, Barends TRM, Dietl A, Khadem AF, Eygensteyn J, Jetten MSM, Op den Camp HJM. 2014. Rare earth metals are essential for methanotrophic life in volcanic mudpots. Environ Microbiol 16:255–264. doi: 10.1111/1462-2920.12249. [DOI] [PubMed] [Google Scholar]
- 44.Wu ML, Ettwig KF, Jetten MSM, Strous M, Keltjens JT, van Niftrik L. 2011. A new intra-aerobic metabolism in the nitrite-dependent anaerobic methane-oxidizing bacterium Candidatus ‘Methylomirabilis oxyfera'. Biochem Soc Trans 39:243–248. doi: 10.1042/BST0390243. [DOI] [PubMed] [Google Scholar]
- 45.Strous M, Heijnen JJ, Kuenen JG, Jetten MSM. 1998. The sequencing batch reactor as a powerful tool for the study of slowly growing anaerobic ammonium-oxidizing microorganisms. Appl Microbiol Biotechnol 50:589–596. doi: 10.1007/s002530051340. [DOI] [Google Scholar]
- 46.Ettwig KF, Shima S, van de Pas-Schoonen KT, Kahnt J, Medema MH, Op den Camp HJM, Jetten MSM, Strous M. 2008. Denitrifying bacteria anaerobically oxidize methane in the absence of Archaea. Environ Microbiol 10:3164–3173. doi: 10.1111/j.1462-2920.2008.01724.x. [DOI] [PubMed] [Google Scholar]
- 47.Ettwig KF, van Alen T, van de Pas-Schoonen KT, Jetten MS, Strous M. 2009. Enrichment and molecular detection of denitrifying methanotrophic bacteria of the NC10 phylum. Appl Environ Microbiol 75:3656–3662. doi: 10.1128/AEM.00067-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luesken FA, Wu ML, Op den Camp HJM, Keltjens JT, Stunnenberg H, Francoijs K-J, Strous M, Jetten MSM. 2012. Effect of oxygen on the anaerobic methanotroph ‘Candidatus Methylomirabilis oxyfera’: kinetic and transcriptional analysis. Environ Microbiol 14:1024–1034. doi: 10.1111/j.1462-2920.2011.02682.x. [DOI] [PubMed] [Google Scholar]
- 49.Rasigraf O, Kool DM, Jetten MS, Sinninghe Damsté JS, Ettwig KF. 2014. Autotrophic carbon dioxide fixation via the Calvin-Benson-Bassham cycle by the denitrifying methanotroph “Candidatus Methylomirabilis oxyfera.” Appl Environ Microbiol 80:2451–2460. doi: 10.1128/AEM.04199-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pol A, Heijmans K, Harhangi HR, Tedesco D, Jetten MSM, Op den Camp HJM. 2007. Methanotrophy below pH 1 by a new Verrucomicrobia species. Nature 450:874–878. doi: 10.1038/nature06222. [DOI] [PubMed] [Google Scholar]
- 51.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 52.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 53.Anthony C, Zatman LJ. 1967. Microbial oxidation of methanol—prosthetic group of alcohol dehydrogenase Pseudomonas sp. M27—a new oxidoreductase prosthetic group. Biochem J 104:960–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schuck P. 2000. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys J 78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Duine JA, Frank J, Jongejan JA. 1987. Enzymology of quinoproteins. Adv Enzymol Relat Areas Mol Biol 59:169–212. [DOI] [PubMed] [Google Scholar]
- 56.Wilm M, Shevchenko A, Houthaeve T, Breit S, Schweigerer L, Fotsis T, Mann M. 1996. Femtomole sequencing of proteins from polyacrylamide gels by nano-electrospray mass spectrometry. Nature 379:466–469. [DOI] [PubMed] [Google Scholar]
- 57.Farhoud MH, Wessels HJ, Steenbakkers PJ, Mattijssen S, Wevers RA, van Engelen BG, Jetten MSM, Smeitink JA, van den Heuvel LP, Keltjens JT. 2005. Protein complexes in the archaeon Methanothermobacter thermautotrophicus analyzed by blue native/SDS-PAGE and mass spectrometry. Mol Cell Proteomics 4:1653–1663. doi: 10.1074/mcp.M500171-MCP200. [DOI] [PubMed] [Google Scholar]
- 58.Wu ML, van Alen TA, van Donselaar EG, Strous M, Jetten MSM, van Niftrik L. 2012. Co-localization of particulate methane monooxygenase and cd1 nitrite reductase in the denitrifying methanotroph ‘Candidatus Methylomirabilis oxyfera.’ FEMS Microbiol Lett 334:49–56. doi: 10.1111/j.1574-6968.2012.02615.x. [DOI] [PubMed] [Google Scholar]
- 59.Wu ML, van Teeseling MC, Willems MJ, van Donselaar EG, Klingl A, Rachel R, Geerts WJ, Jetten MSM, Strous M, van Niftrik L. 2012. Ultrastructure of the denitrifying methanotroph “Candidatus Methylomirabilis oxyfera,” a novel polygon-shaped bacterium. J Bacteriol 194:284–291. doi: 10.1128/JB.05816-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Miyazaki SS, Toki S-I, Izumi Y, Yamada H. 1987. Purification and characterization of methanol dehydrogenase of a serine-producing methylotroph, Hyphomicrobium methylovorum. J Ferment Technol 65:371–377. [Google Scholar]
- 61.Anthony C, Holzenburg A, Scrutton N. 2002. Methanol dehydrogenase, a PQQ-containing quinoprotein dehydrogenase, p 73-117. In Harris JR, Biswas BB, Quinn P (ed), Enzyme-catalyzed electron and radical transfer. Springer US, New York, NY. [Google Scholar]
- 62.Goodwin MG, Anthony C. 1996. Characterization of a novel methanol dehydrogenase containing a Ba2+ ion at the active site. Biochem J 318:673–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Goodwin MG, Avezoux A, Dales SL, Anthony C. 1996. Reconstitution of the quinoprotein methanol dehydrogenase from inactive Ca2+-free enzyme with Ca2+, Sr2+ or Ba2+. Biochem J 319:839–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.de Hoffmann E, Stroobant V. 2007. Mass spectrometry: principles and applications, 3rd edition John Wiley & Sons Ltd., Hoboken, NJ. [Google Scholar]
- 65.Noji N, Nakamura T, Kitahata N, Taguchi K, Kudo T, Yoshida S, Tsujimoto M, Sugiyama T, Asami T. 2007. Simple and sensitive method for pyrroloquinoline quinone (PQQ) analysis in various foods using liquid chromatography/electrospray-ionization tandem mass spectrometry. J Agric Food Chem 55:7258–7263. doi: 10.1021/jf070483r. [DOI] [PubMed] [Google Scholar]
- 66.Page MD, Anthony C. 1986. Regulation of formaldehyde oxidation by the methanol modifier proteins of Methylophilus methylotrophus and Pseudomonas AM1. J Gen Microbiol 132:1553–1563. [Google Scholar]
- 67.Shen YQ, Bonnot F, Imsand EM, RoseFigura JM, Sjölander K, Klinman JP. 2012. Distribution and properties of the genes encoding the biosynthesis of the bacterial cofactor, pyrroloquinoline quinone. Biochemistry 51:2265–2275. doi: 10.1021/bi201763d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Neijssel OM. 1987. PQQ-linked enzymes in enteric bacteria. Microbiol Sci 4:87–90. [PubMed] [Google Scholar]
- 69.Oubrie A, Rozeboom HJ, Kalk KH, Olsthoorn AJ, Duine JA, Dijkstra BW. 1999. Structure and mechanism of soluble quinoprotein glucose dehydrogenase. EMBO J 18:5187–5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ameyama M, Matsushita K, Shinagawa E, Hayashi M, Adachi O. 1988. Pyrroloquinoline quinone: excretion by methylotrophs and growth stimulation for microorganisms. Biofactors 1:51–53. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.