Abstract
Ldl1 is a virulent phage infecting the dairy starter Lactobacillus delbrueckii subsp. lactis LdlS. Electron microscopy analysis revealed that this phage exhibits a large head and a long tail and bears little resemblance to other characterized phages infecting Lactobacillus delbrueckii. In vitro propagation of this phage revealed a latent period of 30 to 40 min and a burst size of 59.9 ± 1.9 phage particles. Comparative genomic and proteomic analyses showed remarkable similarity between the genome of Ldl1 and that of Lactobacillus plantarum phage ATCC 8014-B2. The genomic and proteomic characteristics of Ldl1 demonstrate that this phage does not belong to any of the four previously recognized L. delbrueckii phage groups, necessitating the creation of a new group, called group e, thus adding to the knowledge on the diversity of phages targeting strains of this industrially important lactic acid bacterial species.
INTRODUCTION
Lactobacillus delbrueckii subsp. lactis is a member of the lactic acid bacteria (LAB) and is commonly used in the production of commercial fermented milk products, such as Emmental-like cheeses, where it is employed as a starter culture contributing to the acidification and the organoleptic properties of the final product (1, 2). Bacteriophage (or phage) infection of these starter strains represents a major hurdle to their technological activity, as this may cause lysis of the starter, which in turn may lead to reduced acidification activity and a poor(er)-quality product, with a consequent negative economic impact (3).
The commercial importance of L. delbrueckii fermentations has catalyzed extensive research into the occurrence, diversity, and impact of its infecting phages. Currently, L. delbrueckii phages are organized into four distinct groups (designated groups a, b, c, and d), classified based on DNA homology by hybridization (4–6) and, more recently, by comparative genome analysis (7). L. delbrueckii phages belonging to groups a and c have enjoyed considerable scientific scrutiny, with phage LL-H representing a prototype Lactobacillus phage. LL-H was originally isolated in 1972 (8), and its full genome sequence and transcriptional map, which revealed two distinct phases of transcription, have been determined (9). Furthermore, research on phage-host interactions, culminating in the identification of gp71 as the receptor-binding protein of LL-H (10) and lipoteichoic acids as the recognized receptor, have significantly advanced our understanding of this phage and the way in which it recognizes its host (11, 12). Also, the prolate-head, temperate phage JCL1032, being a member of group c L. delbrueckii phages, has been subjected to considerable scientific characterization, including genome sequencing (13), analysis of genomic integration (14), and identification of lipoteichoic acid as its receptor (12).
Due to their apparent increased frequency of isolation, research has recently focused on group b phages, facilitated by the sequencing of six phages, c5 and LL-Ku (13); Ld3, Ld17, and Ld25A (7); and phiLdb (15). Ld17 is the most comprehensively studied group b phage, with a defined transcriptome consisting of two transcripts (E1 and L1), while structural proteins were identified not only for this phage but also for group b phages Ld3 and Ld25A (7). Finally, group d phages remain significantly understudied, being represented by just a single isolate, the prolate-headed phage 0252, which infects L. delbrueckii subsp. lactis (6).
Here we present the complete genome sequence of Ldl1, a novel phage infecting L. delbrueckii subsp. lactis, isolated in 2010 from a cheese manufacturing facility in Switzerland and representing a fifth, possibly newly emerging group of bacteriophages infecting L. delbrueckii.
MATERIALS AND METHODS
Bacteriophages, bacterial strains, and growth conditions.
Phage Ldl1 was isolated from a Swiss whey sample by employing an Emmental-specific starter culture manufactured by Sacco s.r.l. in 2010. L. delbrueckii subsp. lactis strain LdlS was used for the isolation, propagation, and enumeration of phage Ldl1. Both phage and host were provided by Sacco s.r.l. The strain was grown overnight at 42°C in MRS-LCT (MRS broth [Oxoid] supplemented with 1% lactose, 20 mM CaCl2, and 0.5% tryptone). Phage propagation was performed by picking an individual plaque and infecting a host at an optical density at 600 nm (OD600) of ∼0.15 in MRS-LCT, followed by incubation at 42°C until lysis occurred. The resulting lysate was passed through a 0.45-μm filter to remove cell debris. Phage enumerations, expressed as PFU ml−1, were carried out by employing the double-layer plaque assay method with single-plaque isolates (16), using MRS-LCT agar supplemented with 2% glycine (17).
Electron microscopic analysis.
Bacteriophage lysates were purified on a continuous cesium chloride gradient and were dialyzed three times against TBT buffer (20 mM Tris [pH 7.2], 10 mM NaCl, 20 mM MgSO4). Staining was performed with 2% (wt/vol) uranyl acetate on freshly prepared carbon films. Grids were analyzed with a Tecnai 10 transmission electron microscope (FEI Company) at an acceleration voltage of 80 kV. Micrographs were taken with a MegaView II charge-coupled device camera (Soft Imaging Solutions) at the Max Rubner Institute, Kiel, Germany (18).
One-step growth curve.
A one-step growth experiment was performed to ascertain the burst size and latent, rise, and eclipse phases of Ldl1 by using a modification of the one-step growth curve protocol of Lu et al. (19). Briefly, L. delbrueckii subsp. lactis strain LdlS was grown in 50 ml of MRS-LCT to an OD600 of ∼0.15. Cells were then harvested at 5,580 × g and resuspended in 500 μl of MRS-LCT. Five hundred microliters of Ldl1 lysate was added to produce a multiplicity of infection (MOI) of ∼0.001, and cells were incubated at 42°C for 5 min, followed by centrifugation at 18,000 × g for 30 s, removing unadsorbed phage and thus ensuring a synchronous infection. The resultant pellet was resuspended in 50 ml of MRS-LCT and incubated at 42°C. This step represents time zero (T0), where phage population monitoring was initiated by the removal of 100 μl every 10 min and centrifugation at 18,000 × g for 30 s. Phages in the supernatant were then enumerated by employing the double-layer titration technique mentioned above (16). The burst size was calculated by using the following formula, where “titer after burst” is the phage titer after the initial burst (50 min postinfection here) and “phage added” is the phage titer added before adsorption (1.78 × 105 PFU/ml): burst size = (titer after burst − titer at T0)/(phage added − titer at T0).
Genome sequencing.
Five micrograms of DNA of Ldl1, as determined by Nanodrop quantification, was isolated from fresh CsCl-purified lysates according to a previously described method (20), prior to shipment to the contract sequencing facility (Macrogen Inc., South Korea). Sequencing of the genomes was conducted by using a GS-FLX Titanium sequencer, yielding a 109-fold coverage of the phage genome. The reads generated by the 454 FLX instrument were assembled with GSassembler (454 Lifesciences, Branford, CT, USA) to generate a consensus sequence. Quality improvement of the genome sequence involved sequencing of PCR products across the entire genome to ensure correct assembly and double stranding and the resolution of any remaining base conflicts occurring within homopolymer tracts.
In silico analysis.
Protein-encoding open reading frames (ORFs) were predicted by using GeneMark (21). ORFs with an AUG, UUG, or GUG start codon, encoding at least 30 amino acids (aa), and preceded by a sequence resembling the consensus Shine-Dalgarno sequence (AGGAGG) (22) were accepted. Initial functional annotation of the ORFs and percent amino acid identities between the deduced proteome of Ldl1 and its nearest relative were determined by using BLASTP (23), and functions were further confirmed by querying the PFAM (24) and NCBI Conserved Domain Database (25) protein domain databases and by performing homology prediction searches using HHPred (26). Phage genome maps were built with Clonemanager Suite 7 (Scientific & Educational Software, Morrisville, NC, USA), with identity scores being added between the ORFs in Adobe Illustrator v15.0.0 (Adobe, San Jose, CA, USA). The tRNAscan-SE Search server (27) was used to search for putative tRNAs.
Phage structural proteome and mass spectrometry.
Purified phage proteins were extracted from CsCl-purified Ldl1 phage particles by performing a single methanol-chloroform extraction (1:1:0.75, vol/vol/vol) and subsequently precipitated by the addition of an equal volume of methanol. Proteins were pelleted by centrifugation at 20,800 × g for 6 min and resuspended in 100 μl TBT buffer. The structural protein profile was generated by standard Tris-glycine sodium dodecyl sulfate (SDS)–12% polyacrylamide gel electrophoresis (PAGE). Gel slices were then excised, trypsinized, and analyzed by using electrospray ionization tandem mass spectrometry (ESI-MS/MS), as previously described (28).
Lactobacillus phage proteomic tree.
To gain an understanding of the evolutionary relationship between Ldl1 and phages of other Lactobacillus species, a phylogenetic tree was constructed. The genomes of all available Lactobacillus phages were downloaded from the NCBI database and arranged as previously described (29). Briefly, all encoded proteins were extracted and concatenated beginning with the terminase, with all proteins following the same gene order as that of the phage genome. The resultant concatemers were then aligned by using ClustalW (30) and a BLOSUM matrix using MEGA 5.10 (31). A phylogenetic tree was constructed by using the neighbor-joining method, using the number-of-differences method. The phylogeny was then tested by using bootstrap assessment based on 2,000 replicates.
Nucleotide sequence accession number.
The complete sequence data for Ldl1 are available in the GenBank database under accession number KM514685.
RESULTS AND DISCUSSION
Morphological analysis of Ldl1.
Ldl1 possesses a long noncontractile tail (399 ± 11 nm; n = 20) and a large head (73 ± 2 nm; n = 15) with a defined baseplate structure (Fig. 1). This phage morphology is unique among phages infecting L. delbrueckii, as the tail is significantly longer than those of other reported typical L. delbrueckii phages, which range from 116 to 290 nm (32). In fact, among phages that infect Lactobacillus species, the Ldl1 tail length is second only to that of L. plantarum phage B2, which was isolated in 1971 and which was originally reported to display a tail length of 500 nm (33). However, a recent study found the tail length of this phage to be 240 ± 3 nm, with no reasons being given for the discrepancy (34). Also evident is a minihead variant of Ldl1, exhibiting a much smaller head with the long tail still present. Such a variant could represent a phage particle where the head has not been filled with DNA, thus preventing the expansion of the prohead following the DNA packaging process, as previously observed for bacteriophage T4 (35). Alternatively, these minihead variants may represent naturally occurring mutant phages similar to those observed for T4, where mutations in the major capsid protein resulted in variants with both smaller and larger heads, which were found to be capable of infection (36).
FIG 1.
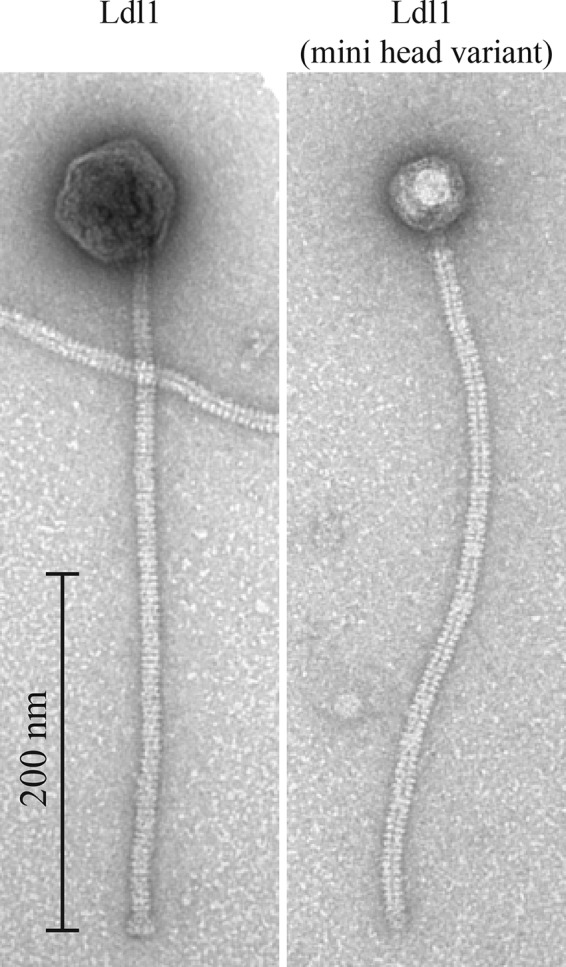
Transmission electron micrographs of L. delbrueckii subsp. lactis phage Ldl1 and an Ldl1 minihead variant, stained with uranyl acetate.
Phage population dynamics.
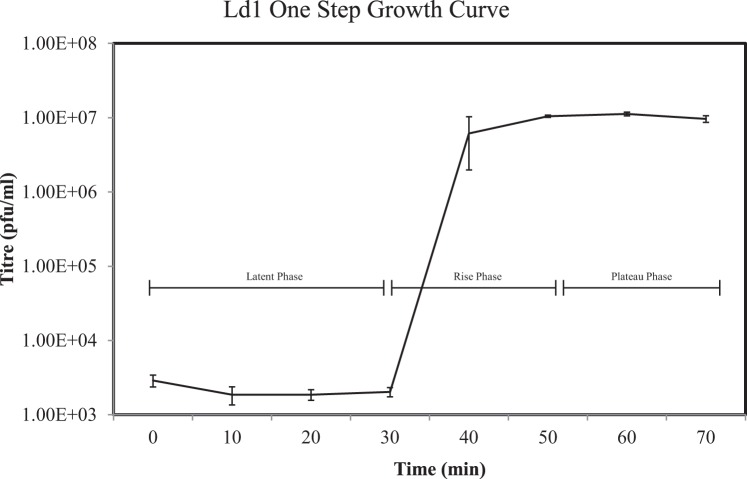
The population kinetics of Ldl1 was investigated by performing a one-step growth curve (see Materials and Methods). The latent period was found to be 30 to 40 min (Fig. 2). This is one of the shorter latent periods reported for a phage infecting L. delbrueckii subsp. lactis, being almost 40 min shorter than that observed for the group a phage LL-H (37). The burst size was determined to be 59.9 ± 1.9 (n = 3) phage particles, which is comparable to those of other L. delbrueckii subsp. lactis phages, such as YAB and lb3, with burst sizes of 48 and 27, respectively (38), but lower than the value observed for the group a phage LL-H, with a burst size ranging from 100 to 200 (36).
FIG 2.
One-step growth curve of the L. delbrueckii subsp. lactis phage performed with MRS-LCT at 42°C. Three replicates were performed to generate the presented data.
Genome analysis.
Ldl1 genomic DNA was isolated and sequenced, revealing a genome of 74,806 bp with 79 putative ORFs (see Table S1 in the supplemental material), overtaking the group c phage JCL1032 (49,433 nucleotides [nt]) in having the largest genome of an L. delbrueckii-infecting phage. The genomes of L. delbrueckii phages typically display a GC content of 49.6 to 49.7% (39–41), considerably higher than that of the Ldl1 genome (37.8%), suggesting that some elements of the phage genome may have been acquired from other phages infecting hosts with a lower GC content or could have evolved from an ancestor that infected a host with a lower percent GC content. Ldl1 bears little resemblance to any other phage infecting L. delbrueckii. The nearest relative of Ldl1 appears to be L. plantarum phage ATCC 8014-B2 (here called B2) (34), with limited levels of homology in the structural region (Fig. 3) and several other conserved regions, as discussed further below. The Ldl1 genome is organized into the following functional modules, which are discussed in further detail below: DNA packaging, morphogenesis, and DNA replication and lysis (Fig. 3).
FIG 3.
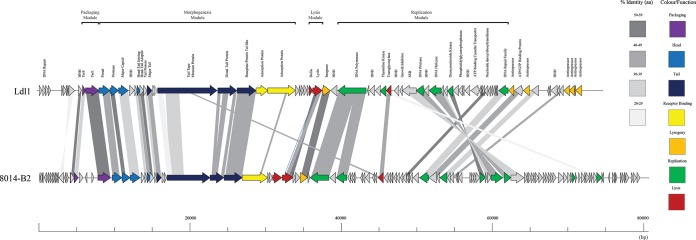
Genomic organization of Ldl1 compared with that of L. plantarum phage ATCC 8014-B2. The scale at the base of the genomes is in base pairs. Each arrow represents an ORF, with the color representing the putative function of the encoded protein indicated on the right. Percent amino acid identity between adjacent genomes is color coded as outlined on the right. HNH, HNH homing endonuclease; SSB, single-strand-binding protein.
Morphogenesis module.
The structural module of Ldl1 bears resemblance in its gene organization and deduced amino acid sequences of its encoded proteins to the corresponding properties of L. plantarum phage B2. This applies in particular to the genes that are predicted to encode the portal protein through to that specifying the major tail protein (Fig. 3), with percent identities outlined in Table S1 in the supplemental material.
Downstream of the gene encoding the major tail protein, the Ldl1 genome encompasses additional genes that specify putative structural proteins. The putative tail tape measure protein (TMP), which is specified by ORF23Ldl1 and which is known to determine the tail length of phages, is 2,627 aa long, with a predicted molecular mass of 288.2 kDa, corresponding to its unusually long tail (Fig. 1; see also Table S1 in the supplemental material). In its C terminus, this predicted TMP contains a number of notable domains, including a LysM domain, which is known to bind peptidoglycan (42), and a transglycosylase (lytic transglycosylase [LT]) domain, which catalyzes peptidoglycan cleavage. The presence of peptidoglycan-binding and -degrading domains indicates that it plays an important, probably multifunctional role (in addition to determining tail length) in the phage infection process, similar to those described for coliphage T5 and Staphylococcus aureus phage vb_SauS-philPLA35 (43), where, for example, the T5 protein Pb2 performs tail length determination functions as well as possessing confirmed in vitro muralytic activity (44).
HHPred analysis of the protein specified by ORF24Ldl1 (located downstream of the TMP-encoding gene) reveals significant similarity to the distal tail protein (Dit) of Bacillus phage SPP1 (E value, 5.3E−23) and lactococcal phage TP901-1 (E value, 1.8E−20), and therefore, it is proposed to encode the central hub upon which the baseplate apparatus is assembled, as has been established for the Dit proteins of both the Bacillus and lactococcal phage models mentioned above.
HHPred analysis of the deduced protein product of ORF25Ldl1 reveals two distinct domains, with residues 11 to 444 exhibiting significant similarity (E value, 6.5E−38) to various predicted phage tail/baseplate proteins known as “Gp27-like” proteins, which are exposed at the distal region of the baseplate (45). The second half of ORF25Ldl1 exhibits similarity to several beta propeller domains, which have previously been observed in bacteriophage endosialidases (46), suggesting that there is both a structural and an enzymatic function associated with this protein, being analogous to Tal (tail-associated lysin) in Tuc2009 (47, 48).
Receptor-binding proteins.
ORF26Ldl1 and ORF27Ldl1 encode two potential host recognition proteins, with the product of ORF26Ldl1 being similar to the N terminus of gp71 LL-H (76% across 112 aa), the confirmed receptor-binding protein of the latter phage. The protein encoded by ORF27Ldl1 is the most likely candidate for the receptor-binding protein, showing similarity to the identified adsorption proteins of LL-H (gp71) and ORF20JCL1032 (ORF20), with identities of 81% (across 330 aa) and 63% (across 424 aa), respectively, at the C terminus. The C terminus of gp71 LL-H was previously implicated in host recognition (10). Also, ORF27Ldl1 contains several repeat regions, most notably those from residues 161 to 255 and 412 to 526, where 11 copies of an 11-residue repeat are observed [consensus, NAEGN(V/I)S(S/Q)LQQ], and from residues 606 to 767, with 9 copies of a loosely repeated 10-residue sequence that shows homology to the repeated regions in the antireceptors of the group b phages (7). A role for these repeats has yet to be elucidated, but their presence in a putative adsorption protein of another distantly related L. delbrueckii phage lends further credence to a role in host recognition.
It is probable that the four ORFs downstream of the TMP-encoding gene specify the baseplate structure at the tail tip of Ldl1 (Fig. 1). In well-characterized bacteriophages infecting Gram-positive organisms, the baseplate-containing tail tip consists of a TMP, a distal tail protein (Dit), and a protein with enzymatic activity (Tal), which together form the so-called initiation complex, which is used as a scaffold for one or more receptor-binding proteins/adsorption proteins, which then form the baseplate (44, 49–52).
Ldl1 structural proteome.
Many of the proteins encoded by genes within the structural module of Ldl1 were confirmed to be part of the structural proteome by mass spectrometric analysis, including the predicted portal protein, the major capsid protein, the head-tail-joining protein, the major tail protein, the distal tail protein, and the two putative adsorption proteins (Table 1). The TMP was absent from the data, possibly due to its low abundance and/or size (predicted molecular mass of 288.2 kDa) preventing its migration through the SDS-PAGE gel. The deduced product of ORF25Ldl1, i.e., the putative Tal-like protein, was present in the mass spectrometric data, but with only one unique peptide and 1.6% coverage, it fell below our limits for inclusion (two unique peptides and/or 5% coverage). Also present in the data was the ATP-binding cassette transporter, the deduced product of ORF56Ldl1, although this protein is unlikely to be part of the structural proteome. It is possible that the gene encoding this function has been integrated into the Ldl1 genome from the host. Therefore, if this protein is expressed at a high level by the host, it is plausible that it could have been present in the lysate as contamination. The presence of the prohead protease in the structural proteome in group b phages Ld17 and Ld25A was observed previously (7) but may be explained by the protease remaining bound to the head after assembly.
TABLE 1.
Confirmed structural proteins encoded by genes in Ldl1a
| ORF | Function | No. of unique peptides | Coverage (%) |
|---|---|---|---|
| 11 | Terminase small subunit | 1 | 8.1 |
| 13 | Portal | 21 | 46.6 |
| 14 | Prohead protease | 9 | 14.8 |
| 15 | Capsid | 31 | 75.4 |
| 21 | Major tail | 7 | 38.1 |
| 24 | Distal tail protein | 4 | 5.4 |
| 26 | Adsorption protein | 2 | 5.3 |
| 27 | Adsorption protein | 4 | 4.6 |
| 56 | ATP-binding cassette transporter | 2 | 7.7 |
Structural proteins extracted from purified phage particles were identified by ESI-MS/MS after separation on a 12% SDS-PAGE gel. A minimum of two independent unique peptides or 5% coverage was used as the threshold value.
Lysogeny elements.
Ldl1 contains genetic elements associated with the lysogenic life-style, such as those encoding six putative nonidentical antirepressors (specified by ORFs 67, 69, 76, 77, 78, and 79) and a predicted recombinase/integrase (ORF35) showing 53% amino acid identity to the recombinase/integrase of B2. Despite the presence of these ORFs associated with the lysogenic life-style, Ldl1 does not appear to exhibit lysogenic characteristics, suggesting that it may have evolved from a lysogenic ancestor.
Replication module.
The replication module of Ldl1 shares limited homology with a similarly annotated region of the L. plantarum phage B2 genome. Like B2, Ldl1 appears to encode its own DNA polymerase, which is specified by ORF37Ldl1 and which is similar to the deduced products of ORF45ATCC 8014-B2 and ORF47ATCC 8014-B2. The latter two proteins in turn exhibit similarity to a DNA polymerase III protein (α subunit) from Bacillus phage SPBc2 (34). ORF37Ldl1 also shows 41% identity to a putative DNA polymerase of Bacillus subtilis subsp. natto across the entire protein. ORF45ATCC 8014-B2 and ORF47ATCC 8014-B2 of B2 appear to be interrupted by a group I intron, which harbors ORF46ATCC 8014-B2, thus indicating that the corresponding mRNA transcript will be processed to be translated as a single protein with very a high level of similarity to the product of ORF37Ldl1 across its full length. It is plausible to suggest that the phage-encoded polymerase is utilized for replication of the phage genome. The protein product of ORF66Ldl1 exhibits substantial similarity (37%) to that of ORF70ATCC 8014-B2, which is probably a double-stranded DNA repair enzyme due to the presence of the Mre11 nuclease and an N-terminal metallophosphatase domain (42).
ORF46Ldl1 and ORF48Ldl1 appear to specify a primase and a helicase, respectively, and thus are presumed to have crucial functions in replication, while ORF41Ldl1 and ORF60Ldl1, whose deduced products contain a thymidine kinase domain and a nucleoside 2-deoxyribosyltransferase domain, respectively, may play a role in nucleotide modification. The presence of a single tRNA gene in the Ldl1 genome at positions 66964 to 66893 is also noted, and this tRNA corresponds to the amino acid isoleucine (anticodon, UAU). Comparison of the usage of codon ATA in the genomes of Ldl1 and two L. delbrueckii strains (L. delbrueckii subsp. bulgaricus ATCC BAA-365 and L. delbrueckii subsp. bulgaricus ATCC 11842) shows a frequency of 45.3% in the Ldl1 genome, compared to frequencies of 19.1 and 19.7% in the genomes of the other L. delbrueckii strains, respectively, suggesting that this tRNA may compensate for the relatively low abundance of the TAT anticodon in the host.
Ldl1 necessitates the creation of a new group of L. delbrueckii-infecting phages.
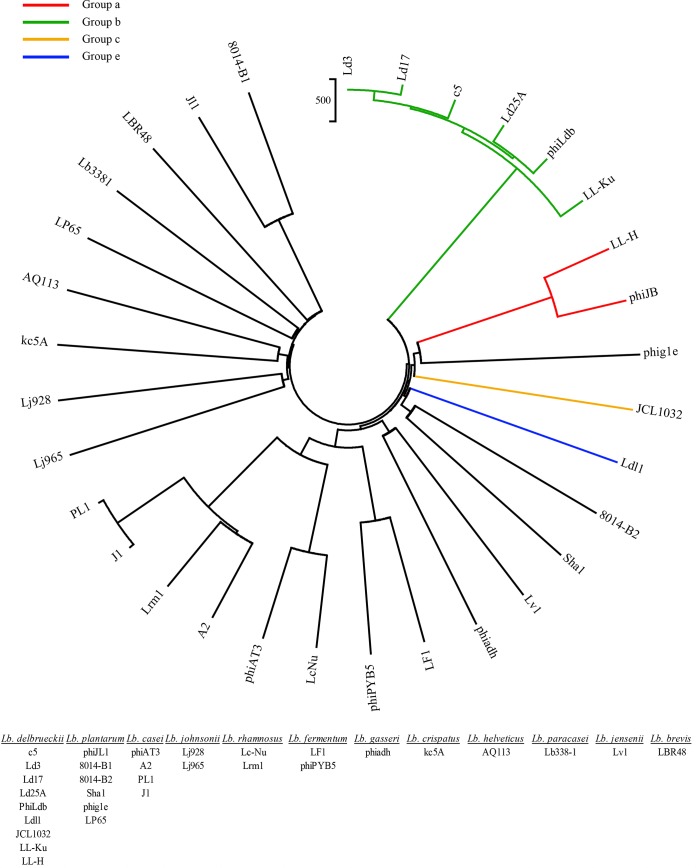
In order to classify Ldl1, a proteomic tree was constructed for comparison with all previously sequenced Lactobacillus phages. The tree also provides valuable insights into the diversity of L. delbrueckii phages and also provides some interesting clusters of phages that infect different species of Lactobacillus. Particularly evident in the tree is the relationship between Ldl1 and B2, where Ldl1 is shown to be more closely related to L. plantarum phages B2 and Sha1 than to any of the other previously sequenced L. delbrueckii phages. The previously established groups of phages infecting L. delbrueckii are evident in the tree (Fig. 4), with group a (indicated in red), group b (green), and group c (purple) occupying distinct branches on the tree, confirming their division into specific groups based on genetic distinction. This leads to the proposal of a new group (group e) of phages (Fig. 4, blue) infecting L. delbrueckii, of which Ldl1 is, as yet, the only member. Also evident are the close phylogenetic relationships between phages infecting different species of Lactobacillus, particularly the relationship between phages infecting Lactobacillus rhamnosus and those infecting Lactobacillus casei. This is highlighted by L. rhamnosus phages Lrm1 and Lc-Nu sharing a clade with L. casei phages PL-1, J1, A2, and phiAT3 (Fig. 4). This observed relationship between these phages may be explained by a common temperate evolutionary origin due to their ability to infect these closely related host species (53). Furthermore, this finding suggests that the related phages Ldl1 and B2 may also have shared a common evolutionary host albeit more distant than the L. casei and L. rhamnosus phages.
FIG 4.
Proteomic tree of all Lactobacillus phages constructed by using the neighbor-joining method according to the number-of-differences model. The species that each phage on the tree is known to infect are indicated at the bottom.
Conclusion.
In this study, we describe the isolation and characterization of a novel phage, Ldl1, infecting L. delbrueckii subsp. lactis.
The discovery of Ldl1 highlights the heterogeneity of phages infecting L. delbrueckii but also accentuates the crucial common elements that appear to be necessary for L. delbrueckii infection. It is clear that Ldl1 represents a new group of phages infecting L. delbrueckii, as the Ldl1 structural proteins and genome architecture exhibit remarkable similarity to those of a phage infecting another Lactobacillus species, L. plantarum phage B2. Interestingly, the baseplate region of Ldl1 is distinct from that of B2 and, as expected given its host preference, is more akin to the receptor-binding proteins of phages infecting L. delbrueckii. Two potential receptor-binding proteins show amino acid identity to those of L. delbrueckii phages LL-H and JCL1032, members of groups a and c, respectively, linking the three groups. The second putative receptor-binding protein shares homology with the previously noted (7, 13) repeat sequence in the antireceptors of group b phages, showing that there are common elements among all sequenced groups infecting L. delbrueckii in the receptor-binding regions, allowing them to infect strains of the same species.
The Ldl1 phage is one of the largest phages described for the genus Lactobacillus and is unlike any previously reported L. delbrueckii phage from both a morphological and a genomic perspective. This led us to propose the creation of a new group, group e, of which Ldl1 is currently the sole member. Thus, our discovery appears to be due to the recent emergence of a new phage group whose ancestor possibly originated from another (Lactobacillus) host yet has acquired characteristics to attack commercial L. delbrueckii starter strains.
Supplementary Material
ACKNOWLEDGMENTS
E.C. is the recipient of a scholarship from the Irish Research Council (IRC) Enterprise Partnership Scheme. D.V.S. is supported by a Principal Investigator award (reference no. 13/IA/1953) through Science Foundation Ireland (SFI). Orbitrap mass spectrometric services provided in this work were supported by Hercules Foundation (Belgium) project R-3986, with technical assistance from Erik Royackers.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03413-14.
REFERENCES
- 1.El Kafsi H, Binesse J, Loux V, Buratti J, Boudebbouze S, Dervyn R, Kennedy S, Galleron N, Quinquis B, Batto J-M, Moumen B, Maguin E, van de Guchte M. 2014. Lactobacillus delbrueckii ssp. lactis and ssp. bulgaricus: a chronicle of evolution in action. BMC Genomics 15:407. doi: 10.1186/1471-2164-15-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Kranenburg R, Kleerebezem M, van Hylckama Vlieg J, Ursing BM, Boekhorst J, Smit BA, Ayad EH, Smit G, Siezen RJ. 2002. Flavour formation from amino acids by lactic acid bacteria: predictions from genome sequence analysis. Int Dairy J 12:111–121. doi: 10.1016/S0958-6946(01)00132-7. [DOI] [Google Scholar]
- 3.Klaenhammer T, Fitzgerald G. 1994. Bacteriophages and bacteriophage resistance, p 106–168. In Glassen MJ, de Vos WM (ed), Genetics and biotechnology of lactic acid bacteria. Springer, New York, NY. [Google Scholar]
- 4.Lahbib-Mansais Y, Mata M, Ritzenthaler P. 1988. Molecular taxonomy of Lactobacillus phages. Biochimie 70:429–435. doi: 10.1016/0300-9084(88)90217-9. [DOI] [PubMed] [Google Scholar]
- 5.Forsman P. 1993. Characterization of a prolate-headed bacteriophage of Lactobacillus delbrueckii subsp. lactis, and its DNA homology with isometric-headed phages. Arch Virol 132:321–330. doi: 10.1007/BF01309542. [DOI] [PubMed] [Google Scholar]
- 6.Sechaud L, Cluzel P-J, Rousseau M, Baumgartner A, Accolas J-P. 1988. Bacteriophages of lactobacilli. Biochimie 70:401–410. doi: 10.1016/0300-9084(88)90214-3. [DOI] [PubMed] [Google Scholar]
- 7.Casey E, Mahony J, O'Connell-Motherway M, Bottacini F, Cornelissen A, Neve H, Heller KJ, Noben J-P, Dal Bello F, van Sinderen D. 2014. Molecular characterization of three Lactobacillus delbrueckii subsp. bulgaricus phages. Appl Environ Microbiol 80:5623–5635. doi: 10.1128/AEM.01268-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alatossava T, Pyhtila M. 1980. Characterization of a new Lactobacillus lactis bacteriophage. IRCS Med Sci Libr Compend 8:297–298. [Google Scholar]
- 9.Mikkonen M, Räisänen L, Alatossava T. 1996. The early gene region completes the nucleotide sequence of Lactobacillus delbrueckii subsp. lactis phage LL-H. Gene 175:49–57. [DOI] [PubMed] [Google Scholar]
- 10.Ravin V, Räisänen L, Alatossava T. 2002. A conserved C-terminal region in Gp71 of the small isometric-head phage LL-H and ORF474 of the prolate-head phage JCL1032 is implicated in specificity of adsorption of phage to its host, Lactobacillus delbrueckii. J Bacteriol 184:2455–2459. doi: 10.1128/JB.184.9.2455-2459.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Räisänen L, Draing C, Pfitzenmaier M, Schubert K, Jaakonsaari T, von Aulock S, Hartung T, Alatossava T. 2007. Molecular interaction between lipoteichoic acids and Lactobacillus delbrueckii phages depends on D-alanyl and α-glucose substitution of poly(glycerophosphate) backbones. J Bacteriol 189:4135–4140. doi: 10.1128/JB.00078-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Räisänen L, Schubert K, Jaakonsaari T, Alatossava T. 2004. Characterization of lipoteichoic acids as Lactobacillus delbrueckii phage receptor components. J Bacteriol 186:5529–5532. doi: 10.1128/JB.186.16.5529-5532.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Riipinen K-A, Forsman P, Alatossava T. 2011. The genomes and comparative genomics of Lactobacillus delbrueckii phages. Arch Virol 156:1217–1233. doi: 10.1007/s00705-011-0980-5. [DOI] [PubMed] [Google Scholar]
- 14.Riipinen KA, Räisänen L, Alatossava T. 2007. Integration of the group c phage JCL1032 of Lactobacillus delbrueckii subsp. lactis and complex phage resistance of the host. J Appl Microbiol 103:2465–2475. doi: 10.1111/j.1365-2672.2007.03479.x. [DOI] [PubMed] [Google Scholar]
- 15.Wang S, Kong J, Gao C, Guo T, Liu X. 2010. Isolation and characterization of a novel virulent phage (phiLdb) of Lactobacillus delbrueckii. Int J Food Microbiol 137:22–27. doi: 10.1016/j.ijfoodmicro.2009.10.024. [DOI] [PubMed] [Google Scholar]
- 16.Svensson U, Christiansson A. 1991. Methods for phage monitoring. Bulletin, vol 263 International Dairy Federation, Brussels, Belgium. [Google Scholar]
- 17.Lillehaug D. 1997. An improved plaque assay for poor plaque-producing temperate lactococcal bacteriophages. J Appl Microbiol 83:85–90. doi: 10.1046/j.1365-2672.1997.00193.x. [DOI] [PubMed] [Google Scholar]
- 18.Vegge CS, Brøndsted L, Neve H, Mc Grath S, van Sinderen D, Vogensen FK. 2005. Structural characterization and assembly of the distal tail structure of the temperate lactococcal bacteriophage TP901-1. J Bacteriol 187:4187–4197. doi: 10.1128/JB.187.12.4187-4197.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu Z, Breidt F Jr, Fleming H, Altermann E, Klaenhammer T. 2003. Isolation and characterization of a Lactobacillus plantarum bacteriophage, ΦJL-1, from a cucumber fermentation. Int J Food Microbiol 84:225–235. doi: 10.1016/S0168-1605(03)00111-9. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 21.Besemer J, Borodovsky M. 1999. Heuristic approach to deriving models for gene finding. Nucleic Acids Res 27:3911–3920. doi: 10.1093/nar/27.19.3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mikkonen M, Vuoristo J, Alatossava T. 1994. Ribosome binding site consensus sequence of Lactobacillus delbrueckii subsp. lactis bacteriophage LL-H. FEMS Microbiol Lett 116:315–320. [DOI] [PubMed] [Google Scholar]
- 23.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer EL. 2004. The Pfam protein families database. Nucleic Acids Res 32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR. 2011. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res 39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Söding J, Biegert A, Lupas AN. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res 33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS Web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res 33:W686–W689. doi: 10.1093/nar/gki366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ceyssens P-J, Mesyanzhinov V, Sykilinda N, Briers Y, Roucourt B, Lavigne R, Robben J, Domashin A, Miroshnikov K, Volckaert G. 2008. The genome and structural proteome of YuA, a new Pseudomonas aeruginosa phage resembling M6. J Bacteriol 190:1429–1435. doi: 10.1128/JB.01441-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Samson JE, Moineau S. 2010. Characterization of Lactococcus lactis phage 949 and comparison with other lactococcal phages. Appl Environ Microbiol 76:6843–6852. doi: 10.1128/AEM.00796-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar S, Nei M, Dudley J, Tamura K. 2008. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform 9:299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Villion M, Moineau S. 2008. Bacteriophages of Lactobacillus. Front Biosci 14:1661–1683. doi: 10.2741/3332. [DOI] [PubMed] [Google Scholar]
- 33.Nes IF, Brendehaug J, von Husby KO. 1988. Characterization of the bacteriophage B2 of Lactobacillus plantarum ATCC 8014. Biochimie 70:423–427. doi: 10.1016/0300-9084(88)90216-7. [DOI] [PubMed] [Google Scholar]
- 34.Briggiler Marcó M, Garneau JE, Tremblay D, Quiberoni A, Moineau S. 2012. Characterization of two virulent phages of Lactobacillus plantarum. Appl Environ Microbiol 78:8719–8734. doi: 10.1128/AEM.02565-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jardine PJ, Coombs DH. 1998. Capsid expansion follows the initiation of DNA packaging in bacteriophage T4. J Mol Biol 284:661–672. doi: 10.1006/jmbi.1998.2179. [DOI] [PubMed] [Google Scholar]
- 36.Doermann AH, Eiserling FA, Boehner L. 1973. Genetic control of capsid length in bacteriophage T4. J Virol 12:374–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alatossava T, Forsman P, Mikkonen M, Vasala A. 1991. Molecular biology of Lactobacillus phage LL-H. Finn J Dairy Sci 49:1–13. [Google Scholar]
- 38.Quiberoni A, Guglielmotti D, Binetti A, Reinheimer J. 2004. Characterization of three Lactobacillus delbrueckii subsp. bulgaricus phages and the physicochemical analysis of phage adsorption. J Appl Microbiol 96:340–351. doi: 10.1046/j.1365-2672.2003.02147.x. [DOI] [PubMed] [Google Scholar]
- 39.Makarova K, Slesarev A, Wolf Y, Sorokin A, Mirkin B, Koonin E, Pavlov A, Pavlova N, Karamychev V, Polouchine N. 2006. Comparative genomics of the lactic acid bacteria. Proc Natl Acad Sci U S A 103:15611–15616. doi: 10.1073/pnas.0607117103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van de Guchte M, Penaud S, Grimaldi C, Barbe V, Bryson K, Nicolas P, Robert C, Oztas S, Mangenot S, Couloux A. 2006. The complete genome sequence of Lactobacillus bulgaricus reveals extensive and ongoing reductive evolution. Proc Natl Acad Sci U S A 103:9274–9279. doi: 10.1073/pnas.0603024103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun Z, Chen X, Wang J, Zhao W, Shao Y, Guo Z, Zhang X, Zhou Z, Sun T, Wang L. 2011. Complete genome sequence of Lactobacillus delbrueckii subsp. bulgaricus strain ND02. J Bacteriol 193:3426–3427. doi: 10.1128/JB.05004-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buist G, Steen A, Kok J, Kuipers OP. 2008. LysM, a widely distributed protein motif for binding to (peptido)glycans. Mol Microbiol 68:838–847. doi: 10.1111/j.1365-2958.2008.06211.x. [DOI] [PubMed] [Google Scholar]
- 43.Rodríguez-Rubio L, Gutiérrez D, Martínez B, Rodríguez A, Götz F, García P. 2012. The tape measure protein of the Staphylococcus aureus bacteriophage vB_SauS-phiIPLA35 has an active muramidase domain. Appl Environ Microbiol 78:6369–6371. doi: 10.1128/AEM.01236-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boulanger P, Jacquot P, Plançon L, Chami M, Engel A, Parquet C, Herbeuval C, Letellier L. 2008. Phage T5 straight tail fiber is a multifunctional protein acting as a tape measure and carrying fusogenic and muralytic activities. J Biol Chem 283:13556–13564. doi: 10.1074/jbc.M800052200. [DOI] [PubMed] [Google Scholar]
- 45.Veesler D, Cambillau C. 2011. A common evolutionary origin for tailed-bacteriophage functional modules and bacterial machineries. Microbiol Mol Biol Rev 75:423–433. doi: 10.1128/MMBR.00014-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stummeyer K, Dickmanns A, Mühlenhoff M, Gerardy-Schahn R, Ficner R. 2005. Crystal structure of the polysialic acid-degrading endosialidase of bacteriophage K1F. Nat Struct Mol Biol 12:90–96. doi: 10.1038/nsmb874. [DOI] [PubMed] [Google Scholar]
- 47.Kenny JG, McGrath S, Fitzgerald GF, van Sinderen D. 2004. Bacteriophage Tuc2009 encodes a tail-associated cell wall-degrading activity. J Bacteriol 186:3480–3491. doi: 10.1128/JB.186.11.3480-3491.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stockdale SR, Mahony J, Courtin P, Chapot-Chartier M-P, van Pijkeren J-P, Britton RA, Neve H, Heller KJ, Aideh B, Vogensen FK, van Sinderen D. 2013. The lactococcal phages Tuc2009 and TP901-1 incorporate two alternate forms of their tail fiber into their virions for infection specialization. J Biol Chem 288:5581–5590. doi: 10.1074/jbc.M112.444901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Collins B, Bebeacua C, Mahony J, Blangy S, Douillard FP, Veesler D, Cambillau C, van Sinderen D. 2013. Structure and functional analysis of the host recognition device of lactococcal phage Tuc2009. J Virol 87:8429–8440. doi: 10.1128/JVI.00907-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mc Grath S, Neve H, Seegers JF, Eijlander R, Vegge CS, Brøndsted L, Heller KJ, Fitzgerald GF, Vogensen FK, van Sinderen D. 2006. Anatomy of a lactococcal phage tail. J Bacteriol 188:3972–3982. doi: 10.1128/JB.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sciara G, Bebeacua C, Bron P, Tremblay D, Ortiz-Lombardia M, Lichière J, van Heel M, Campanacci V, Moineau S, Cambillau C. 2010. Structure of lactococcal phage p2 baseplate and its mechanism of activation. Proc Natl Acad Sci U S A 107:6852–6857. doi: 10.1073/pnas.1000232107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Veesler D, Spinelli S, Mahony J, Lichière J, Blangy S, Bricogne G, Legrand P, Ortiz-Lombardia M, Campanacci V, van Sinderen D. 2012. Structure of the phage TP901-1 1.8 MDa baseplate suggests an alternative host adhesion mechanism. Proc Natl Acad Sci U S A 109:8954–8958. doi: 10.1073/pnas.1200966109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tuohimaa A, Riipinen K-A, Brandt K, Alatossava T. 2006. The genome of the virulent phage Lc-Nu of probiotic Lactobacillus rhamnosus, and comparative genomics with Lactobacillus casei phages. Arch Virol 151:947–965. doi: 10.1007/s00705-005-0672-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.