Abstract
The phyllosphere is colonized by a wide variety of bacteria and fungi; it harbors epiphytes, as well as plant-pathogenic bacteria and even human pathogens. However, little is known about how the bacterial community composition on leafy greens develops over time. The bacterial community of the leafy-green phyllosphere obtained from two plantings of rocket salad (Diplotaxis tenuifolia) and three plantings of lettuce (Lactuca sativa) at two farms in Norway were profiled by an Illumina MiSeq-based approach. We found that the bacterial richness of the L. sativa samples was significantly greater shortly (3 weeks) after planting than at harvest (5 to 7 weeks after planting) for plantings 1 and 3 at both farms. For the second planting, the bacterial diversity remained consistent at the two sites. This suggests that the effect on bacterial colonization of leaves, at least in part must, be seasonally driven rather than driven solely by leaf maturity. The distribution of phyllosphere communities varied between D. tenuifolia and L. sativa at harvest. The variability between these species at the same location suggests that the leaf-dwelling bacteria are not only passive inhabitants but interact with the host, which shapes niches favoring the growth of particular taxa. This work contributes to our understanding of host plant-specific microbial community structures and shows how these communities change throughout plant development.
INTRODUCTION
The phyllosphere is a habitat on the surface of plant leaves colonized by a variety of bacteria, yeasts, and fungi (1). It harbors epiphytes, as well as plant-pathogenic bacteria and even human pathogens. Microbial populations on leaf surfaces are highly influenced by rapid fluctuations in UV radiation, temperature, and humidity and are restricted by limited access to nutrients (1, 2). Resident bacteria on leaves can have neutral, negative, or positive influences on their host plants by serving as pathogens, preventing leaf colonization by pathogens, or acting as growth promoters (3). Traditionally, phyllosphere bacteria have been characterized by using culture-based approaches and much of the work on produce-associated bacteria has focused on a small number of pathogenic species. Culture-based methods will not include bacteria that are not able to grow on standard artificial media or are slow growing. This limits our understanding of the phyllosphere microbial community's ecology, genetics, and physiology (4). Bacterial communities associated with leafy greens have already been described by several culture-independent studies. “First-generation” molecular techniques have been used to describe variation in community structure in the context of leaf surface properties and microbial interactions (5), seasonal variation in the community structure (6), and monitoring of bacterial communities in the food chain (7). The introduction of culture-independent methods, in particular, microbial profiling using high-throughput sequencing to study microorganisms, has revealed more complexity and diversity of the phyllosphere microbiota and has dramatically changed the landscape of microbial ecology (5, 7–10). The 454 pyrosequencing platform has been used in different studies of bacterial communities associated with leafy greens (11–15), but Illumina MiSeq has lately been the dominant platform for microbial profiling.
Previous studies have shown that the impact of leaf age and seasonal variations on the phyllosphere communities is not clear (14–18). It can be hypothesized that phyllosphere communities will change with time because of a decline in the nutrient supply when the leaf matures, selection of specific microbiota by different leafy greens, or weather effects. The objective of our study was to determine the succession patterns in microbial communities on the surface of leafy greens throughout the growing season (April to September) to see how the communities change over time and with leaf maturity. Because of the increased concern about vegetables as vehicles for transmission of human pathogens, we also wanted to investigate how frequently potential human pathogens were present in conventionally grown leafy greens at different maturity stages.
MATERIALS AND METHODS
Sample collection.
Conventionally grown lettuce (Lactuca sativa) and rocket salad (Diplotaxis tenuifolia) were collected from two farms in southeastern Norway during the growing season of 2013 (April to September), for microbial profiling of the phyllosphere bacteria. Farm Vestfold is located near the sea in Vestfold County, a region known for early-season vegetable production. Farm Buskerud is located at the head of the Drammensfjord, 55 km north of Farm Vestfold. Both farms use overhead irrigation; the water used at Farm Vestfold comes from the public drinking water supply and that used at Farm Buskerud comes from a nearby river. The soil type at both farms is sandy clay loam. L. sativa seedlings were locally produced in greenhouses and planted either on bare soil (Farm Vestfold) or on plastic-covered beds (Farm Buskerud). L. sativa heads were harvested by hand. D. tenuifolia was directly sown on raised beds, and the leaves were harvested by cropping machines. L. sativa samples were collected twice per planting (3 weeks after planting in the field and at harvest) and from three subsequent plantings. Four samples were collected per field, and each sample consisted of three L. sativa heads. Two leaves were picked from each L. sativa head (six leaves in total), one from the outer leaf circle and one from the inner leaf circle. Samples were also taken from L. sativa plants the day they were planted in the field (day zero). However, we were not able to amplify bacterial DNA from these samples; hence, they were not included in the sequencing experiment. D. tenuifolia was collected just after harvest with cropping machines from three subsequent plantings. Three samples were taken from two plastic bags freshly packed (not washed) for retail at the farm. The type of leafy green and replicates are described in detail in Table 1 (for some of the L. sativa samples, sequencing results were obtained from only three out of four subsamples because of sequencing errors). The leaves were briefly rinsed in running distilled water to remove traces of soil and pooled in an Erlenmeyer flask. A 50-ml salt-Tween solution (0.15 M NaCl, 0.1% Tween 20) (19) was added to each sample in the flask, and it was placed in a rotary shaker (100 rpm) at room temperature (RT) for 15 min. Aliquots of 100 μl of the solution were plated on nutrient glucose agar (NGA; 23 g nutrient agar [Difco], 5 g yeast extract, 10 g glucose, 1,000 ml distilled water) and incubated at RT. Bacteria in the solution were collected on 0.2-μm filters by vacuum filtration. The filters were stored at −20°C prior to molecular analysis.
TABLE 1.
Samples of leafy greens collected at two farms in southeastern Norway throughout the growing season of 2013 that were subjected to microbial profiling of the phyllosphere-inhabiting bacteria
| Farm and leafy green (variety) | Planting | Day/mo of sampling (no of samples sequenced) |
Plant age (wk) | |
|---|---|---|---|---|
| 3 wka | Harvest | |||
| Vestfold | ||||
| L. sativa (Little Gem) | 1 | 15/5 (3) | 12/6 (4) | 9 |
| L. sativa (Little Gem) | 2 | 03/7 (4) | 17/7 (4) | 7 |
| L. sativa (Little Gem) | 3 | 20/8 (4) | 13/9 (4) | 8.5 |
| Buskerudb | ||||
| L. sativa (Iceberg) | 1 | 23/5 (3) | 17/6 (4) | 9,5 |
| L. sativa (Frillice) | 2 | 29/7 (4) | 19/8 (4) | 8 |
| L. sativa (Frillice) | 3 | 19/8 (3) | 17/9 (4) | 10 |
| D. tenuifolia (Rocket) | 1 | 08/7 (3) | 5 | |
| D. tenuifolia (Rocket) | 2 | 29/7 (3) | 5 | |
The plants that were sampled 3 weeks after planting in the field were 5 weeks old upon sampling.
For the purpose of this work, communities developing on the different lettuce varieties at Farm Buskerud were assumed to be similar.
DNA isolation from filters and pure bacterial cultures.
The filters were ground in liquid nitrogen with a pestle and mortar, and added directly to the lysis buffer of the DNeasy Plant minikit (Qiagen, GmbH, Hilden, Germany). From each NGA plate, up to 10 phenotypically different single bacterial colonies were repeatedly streaked onto new NGA plates to obtain pure cultures. The isolates were grown on NGA agar plates at RT and stored in glycerol stocks at −80°C. One loop with bacterial growth from the pure culture was suspended in 1 ml sterile MilliQ water and incubated at 100°C on a heating block. The lysate was used directly as the template for PCR amplification.
Analysis of the 16S rRNA gene by Sanger sequencing.
A 500-bp fragment of the 16S rRNA gene (variable regions V6 to V8) was amplified for sequence analysis by PCR with the primer pair F985PTO/R1378 (F985PTO, 5′-AACGCGAAGAACCTTACSC-3′; R1378, 5′-CGGTGTGTACAAGGCCCGGGAACG-3′) (20). The PCR mixture (25 μl) contained 0.4 mM each primer, 0.2 mM deoxynucleoside triphosphates, 1.5 U AmpliTaq DNA polymerase (Applied Biosystems, Foster City, CA), 0.25 μl bovine serum albumin, and 2.5 μl template. The PCR cycle consisted of denaturation at 95°C for 3 min; 35 cycles of 95°C for 1 min, 55°C for 30 s, and 72°C for 30 s; and a final extension at 72°C for 5 min. PCR products were analyzed on a 1% agarose gel in TBE buffer by electrophoresis. Sequencing was performed at GATC Biotech, and the sequences were assembled and analyzed with CLC Main Workbench 6.
Microbial profiling with Illumina MiSeq.
Fifty-one L. sativa and D. tenuifolia samples (Table 1) were selected for sequencing with the Illumina MiSeq platform. The NEXTflex 16S V4 Amplicon-Seq kit (Bioo Scientific, Austin, TX), which amplifies the fourth hypervariable (V4) domain of the microbial 16S rRNA, was used to label all of the samples prior to sequencing according to the manufacturer's instructions. The samples were analyzed by agarose gel electrophoresis (1% agarose in TBE buffer), and the DNA fragments of the expected size were excised from the gel under UV light. The DNA amplicons were purified with the QIAquick gel extraction kit (Qiagen, GmbH, Hilden, Germany). Sequencing was performed at the Norwegian Sequencing Center, Oslo, Norway, on the Illumina MiSeq platform by the 300-bp paired-end protocol.
Sequence processing, analysis, and community comparisons.
Sequences were preprocessed and analyzed with Mothur v1.33 (21) by following the Mothur standard operating procedure (22, 23). In short, the sequences were trimmed and processed and aligned by using the Bacterial SILVA SEED database as the template (http://www.mothur.org/wiki/Silva_reference_files). Sequences that originated from chloroplasts and mitochondria were removed from the data set. Potentially chimeric sequences were detected and removed with chimera uchime (24), and the remaining sequences were called “filtered sequences.” The aligned filtered sequences were classified with Trainset9_032012.pds from the Ribosomal Database Project (25) and clustered into operational taxonomic units (OTUs) defined by 97% similarity.
The sequence libraries were further characterized to determine the degrees of diversity and similarity of the microbial communities present on the different samples. The Shannon (H0) and Simpson (1/D) indices were used to establish relative diversity levels, and Chao I was used to provide estimates of species richness that might be expected if more exhaustive sampling were done (see Table S1 in the supplemental material). Pairwise comparison of a subset (based on 9,549 random sequences from each sample) with weighted UniFrac (26) was performed to test if the bacterial communities in the fields were significantly different. Metastats, which is based on a nonparametric t test, Fisher's exact test, and the false-discovery rate (27), was used to find OTUs that were statistically significantly different when groups were compared (P < 0.05). The phylogenetic β-diversity tree was made with tools from Interactive Tree Of Life (28). In an effort to investigate if different L. sativa varieties hosted different bacterial families, we analyzed the abundances of selected bacterial families in the three L. sativa varieties Little Gem, Iceberg, and Frillice at 3 weeks after planting and at harvest, also including D. tenuifolia at harvest.
Nucleotide sequence accession number.
The sequences obtained in this study are available in the European Nucleotide Archive database under project identification number PRJEB6233.
RESULTS
Altogether, 51 samples of conventional field-grown leafy greens (D. tenuifolia and L. sativa) were collected from two farms in southeastern Norway during three subsequent plantings in the growing season of 2013 (Table 1).
Identification of culturable bacteria by Sanger sequencing.
Aliquots of the rinsing solution from L. sativa samples were plated onto NGA agar, and a total of 162 bacterial isolates were obtained. Sequencing of part of the 16S rRNA gene showed that the isolates represented 26 genera and four bacterial phyla, Proteobacteria, Actinobacteria, Firmicutes, and Bacteroidetes. Pseudomonas (30%) was the most abundant genus, followed by Arthrobacter (12%), Pantoea (10%), and Acinetobacter (8%).
Comparison of phyllosphere communities with Illumina MiSeq.
Out of 51 samples of leafy greens (Table 1), a total of 4,363,870 filtered bacterial 16S rRNA sequences were obtained by MiSeq sequencing (see Table S1 in the supplemental material). There was a significantly higher number of OTUs from samples collected at 3 weeks after planting (Fig. 1A) than from samples collected at harvest, according to a basic one-way analysis of variance with Tukey's range test (P = 0.002). These results were not consistent in all of the samples, and the second planting (V3-2 and VH-2, B3-2 and BH-2) was separated from the others at both locations with similar OTU abundances at 3 weeks and at harvest (Fig. 1B). A higher number of genera was also found at 3 weeks after planting (540 genera) than at harvest (472 genera).
FIG 1.
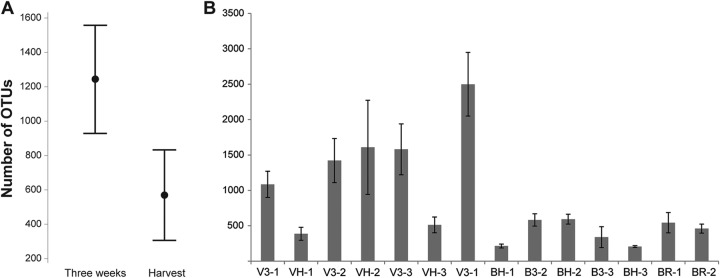
(A) Mean numbers of OTUs obtained from L. sativa samples collected 3 weeks after planting and at harvest at Farm Buskerud and Farm Vestfold. Also shown are the 95% confidence intervals. (B) OTU abundances on planting samples of L. sativa and D. tenuifolia (BR) from Farm Buskerud (B) and Farm Vestfold (V) at 3 weeks (3) and at harvest (H). Standard-error bars are shown. The number after the hyphen in each designation on the x axis is the planting number.
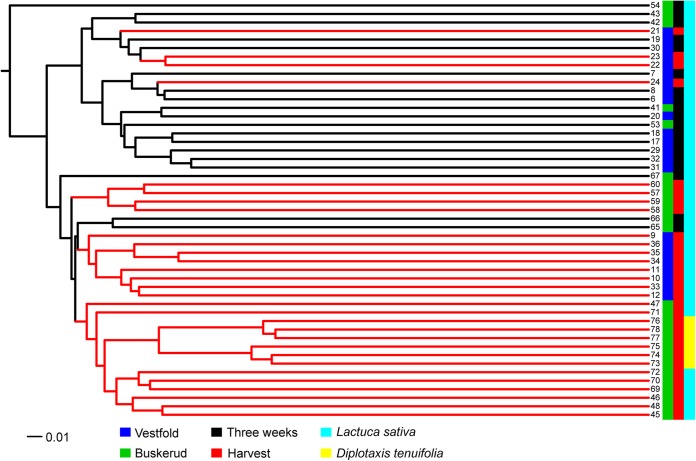
The weighted UniFrac method showed that the bacterial communities identified in the samples collected at harvest and 3 weeks after planting were significantly different (P < 0.001). The comparison of L. sativa sampled 3 weeks after planting and at harvest was illustrated by β-diversity phylogeny with jclass calculator (Jaccard index) on the basis of the weighted UniFrac distances (Fig. 2). The β-diversity tree shows that samples collected 3 weeks after planting mainly clustered separately from the samples collected at harvest.
FIG 2.
β-Diversity tree of all of the samples made with jclass calculator (Jaccard index) on the basis of a subset of 9,549 random sequences from each sample. Samples: 6 to 8, V3-1; 9 to 12, VH-1; 17 to 20, V3-2; 21 to 24, VH-2; 29 to 32, V3-3; 33 to 36, VH-3; 41 to 43, B3-1; 45 to 48, BH-1; 53 to 56, B3-2; 57 to 60, BH-2; 65 to 67, B3-3; 69 to 72, BH-3; 73 to 75, BR-1; 76 to 78, BR-2. Code: Farm Buskerud, B; Farm Vestfold, V; 3 weeks after planting, 3; harvest, H.
Identification of bacterial community compositions by Illumina MiSeq.
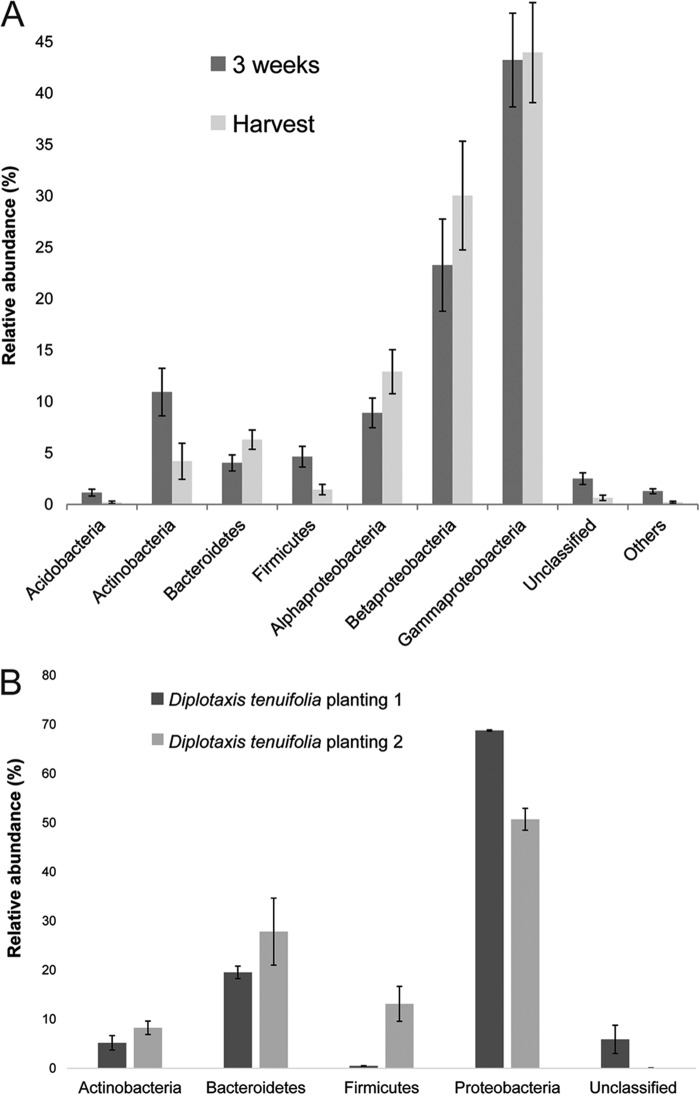
The distribution of sequences from the L. sativa samples collected 3 weeks after planting could be assigned to the phyla Proteobacteria (76%), Actinobacteria (11%), Firmicutes (5%), and Bacteroidetes (4%) (Table 2; Fig. 3 and 4A). The dominant phyla associated with L. sativa leaves at harvest could be assigned to Proteobacteria (87%), Bacteroidetes (6%), and Actinobacteria (4%) (Table 2; Fig. 3 and 4A). Members of the phylum Proteobacteria were also the most abundant bacteria (60%) found in the D. tenuifolia samples collected at harvest (Table 2; Fig. 3B and 4A).
TABLE 2.
Comparison of bacterial communities on leafy greens at different harvest time points at Farm Buskerud and Farm Vestfold
| Bacterial phylum and genus | Avg % representation ± SE |
||||||
|---|---|---|---|---|---|---|---|
|
L. sativa (Vestfold) at: |
L. sativa (Buskerud) at: |
L. sativa (Vestfold and Buskerud) at: |
D. tenuifolia at harvest | ||||
| 3 wk | Harvest | 3 wk | Harvest | 3 wk | Harvest | ||
| Actinobacteria | 13 ± 3.3 | 6 ± 3.1 | 9 ± 3.5 | 4 ± 1.5 | 11 ± 2.3 | 4 ± 1.8 | 7 ± 1.5 |
| Arthrobacter | 5 ± 1.3 | 3.5 ± 2.6 | 3.2 ± 1.0 | <1 ± 0.2 | 4.5 ± 0.9 | 2 ± 1.4 | 2.5 ± 0.7 |
| Nocardioides | 2 ± 0.6 | <1 ± 0.2 | 1 ± 0.7 | <1 ± 0.2 | 1.5 ± 0.4 | <1 ± 0.1 | 1 ± 0.8 |
| Rhodococcus | <1 ± 0.1 | <1 ± 0.03 | <1 ± 0.2 | <1 ± 0.3 | <1 ± 0.1 | <1 ± 0.1 | 1 ± 1.2 |
| Bacteroidetes | 6 ± 0.22 | 6 ± 0.1 | 2 ± 0.4 | 6.3 ± 1.2 | 4 ± 0.8 | 6 ± 0.9 | 24 ± 4.1 |
| Chryseobacterium | <1 ± 0.4 | 2 ± 0.8 | 0.4 ± 0.1 | 2 ± 0.8 | <1 ± 0.2 | 2 ± 0.5 | 9 ± 1.3 |
| Flavobacterium | 1.2 ± 0.4 | 1.6 ± 0.6 | 0.5 ± 0.2 | <1 ± 0.2 | <1 ± 0.3 | 1 ± 0.4 | 11 ± 1.2 |
| Hymenobacter | 1.5 ± 0.6 | <1 ± 0.2 | 0.7 ± 0.2 | 2 ± 1.5 | 1 ± 0.3 | 1.5 ± 0.7 | <1 ± 0.0 |
| Firmicutes | 3 ± 1.6 | 1.5 ± 0.7 | 6 ± 0.6 | 1 ± 0.4 | 5 ± 1.0 | 1 ± 0.5 | 7 ± 6.3 |
| Exiguobacterium | 1.5 ± 0.9 | <1 ± 0.04 | 4 ± 1.3 | 1 ± 0.9 | 2.5 ± 0.9 | <1 ± 0.4 | 6 ± 5.8 |
| Proteobacteria | 73 ± 6.2 | 86 ± 4.6 | 76 ± 10.5 | 87 ± 12 | 76 ± 3.8 | 87 ± 2.9 | 60 ± 9.0 |
| Acinetobacter | <1 ± 0.6 | <1 ± 0.02 | 1 ± 1.1 | 3 ± 1.0 | 1 ± 0.6 | 1.5 ± 0.8 | 1 ± 1.1 |
| Alkanindiges | 4 ± 1.8 | 12 ± 5.2 | 1 ± 0.4 | 3 ± 0.5 | 2.5 ± 1.0 | 7 ± 3.1 | <1 ± 0.0 |
| Brevundimonas | <1 ± 0.3 | 1.5 ± 0.9 | <1 ± 0.2 | <1 ± 0.1 | <1 ± 0.2 | <1 ± 0.5 | 2.5 ± 0.6 |
| Duganella | 8 ± 3.5 | 9 ± 3.9 | 5 ± 1 | 29 ± 0.8 | 7 ± 1.8 | 19 ± 4.8 | 5.5 ± 1.6 |
| Massilia | 12 ± 4 | 4 ± 0.8 | 13 ± 5.4 | 8 ± 3.2 | 12 ± 3.0 | 6 ± 1.6 | 8 ± 3.5 |
| Methylophilus | <1 ± 0.3 | 2 ± 0.5 | <1 ± 0.1 | <1 ± 0.7 | <1 ± 0.2 | 1.5 ± 0.5 | 1 ± 0.7 |
| Pantoea | 5 ± 3.6 | 14 ± 7.2 | 15 ± 10 | 5 ± 2.4 | 10 ± 5.6 | 10 ± 4.0 | 5 ± 4.6 |
| Pseudomonas | 26 ± 10 | 22 ± 7.0 | 22 ± 10 | 24 ± 5.1 | 24 ± 6.6 | 23 ± 3.9 | 25 ± 11.5 |
| Rhizobium | <1 ± 0.1 | 4 ± 1.9 | <1 ± 0.2 | 2 ± 0.9 | <1 ± 0.1 | 3 ± 1.1 | 1 ± 0.2 |
| Sphingomonas | 5 ± 0.4 | 5 ± 0.7 | 4 ± 1.3 | 8 ± 3.9 | 4.5 ± 0.6 | 6 ± 2.0 | 4.5 ± 3.0 |
| Xanthomonas | <1 ± 0.03 | 2 ± 0.6 | 3 ± 1.3 | <1 ± 0.03 | 1.5 ± 0.9 | 1 ± 0.5 | 2.5 ± 1.9 |
| Othera | 5 ± 0.14 | 1 ± 0.15 | 3 ± 0.11 | 1 ± 0.02 | 4 ± 0.1 | 2 ± 0.04 | 2 ± 0.16 |
| Unclassified/otherb | 25 ± 3.4 | 14 ± 3.6 | 22 ± 8.0 | 8 ± 0.7 | 23 ± 1.0 | 11 ± 3.0 | 13 ± 0.8 |
Acidobacteria, Armatimonadetes, Chlamydiae, Chlorobi, Chloroflexi, Deinococcus, Thermus, Fusobacteria, Gemmatimonadetes, Nitrospora, Planctomycetes, Spirochaetes, Tenericutes, Verrucomicrobia, “Candidatus BRC1,” “Candidatus OD1,” “Candidatus OP11,” “Candidatus TM7,” “Candidatus WS3,” and unclassified taxa.
About 640 taxa are included in the “other” category.
FIG 3.
Relative abundances (percent) of phyla in L. sativa samples after 3 weeks and at harvest (A) and in D. tenuifolia plantings 1 and 2 (B). Standard error bars are shown.
FIG 4.
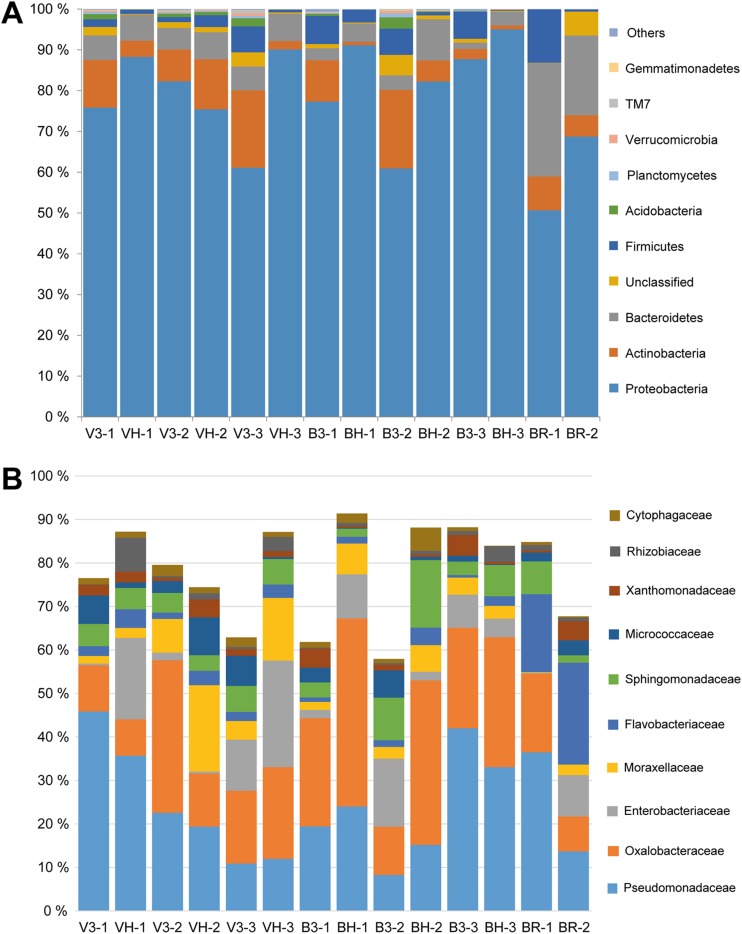
Relative abundances (percent) of bacterial phyla (A) and the most abundant families (B) on L. sativa and D. tenuifolia samples.
The most common genus overall on the L. sativa and D. tenuifolia samples was Pseudomonas (Table 2), which was present on all of the L. sativa samples and all but one of the D. tenuifolia samples. Pantoea and Sphingomonas were the only genera identified on all of the samples. The predominant families across all of the samples were Pseudomonadaceae (23%), Oxalobacteraceae (22%), and Enterobacteriaceae (10%) (Fig. 4B). The genera Salmonella and Escherichia, which include potentially harmful human pathogens, were not found in our study. The lactic acid bacterial genera Aerococcus, Enterococcus, Lactobacillus, Lactococcus, Leuconostoc, Streptococcus, and Weissella were present on the samples but at very low abundances.
The distribution of bacterial families among different L. sativa varieties was investigated (Fig. 5). Members of the family Enterobacteriaceae were more abundant on Frillice (12%) than on the other varieties at 3 weeks after planting. At harvest, members of the family Oxalobacteraceae were more abundant on Iceberg (43%) and Frillice (34%) than on Little Gem (14%). There were more Flavobacteriaceae bacteria (21%) on D. tenuifolia and more Sphingomonadaceae bacteria (11%) on Frillice than on the other varieties at harvest.
FIG 5.
Relative abundances of selected bacterial families on D. tenuifolia and three varieties of L. sativa (Frillice, Iceberg, and Little Gem).
Phyllosphere bacterial communities varied according to season.
To investigate more closely the main bacterial distribution at 3 weeks after planting and at harvest and the possible seasonal effect observed in both plantings 2 and 3, we used the Metastats method (27) to statistically evaluate the differential abundance of the OTUs in selected groups (see Table S2 in the supplemental material). This analysis showed that Actinobacteria, Acidobacteria, Planctomycetes, and Verrucomicrobia were the significantly different phyla 3 weeks after planting, while members of the phylum Proteobacteria (genera Duganella, Pedobacter, and Rhizobium) were most abundant at harvest. Samples collected at harvest at the second planting, from both Buskerud and Vestfold, showed similar microbial profiles regarding phyla and genera.
DISCUSSION
This comprehensive study used Illumina MiSeq to study bacterial communities associated with leafy greens across plant development and time. Illumina-based 16S rRNA gene sequencing has higher accuracy and greater throughput than previously used pyrosequencing (29), thus allowing, in principle, deeper insight into the microbial communities of the leafy-green phyllosphere.
The present study identified Proteobacteria, Bacteroidetes, Actinobacteria, and Firmicutes as the most prevalent phyla in the phyllosphere of leafy greens, regardless of culture or culture-independent analysis. This is consistent with previous studies of bacterial communities on leafy greens (11, 13–15, 30). Our results showed that Proteobacteria and Bacteroidetes were the most abundant phyla at harvest. This is consistent with the findings of Jackson et al. (11). However, Firmicutes replaced Bacteroidetes as one of the dominant phyla in other surveys of the leafy-green phyllosphere (13–15).
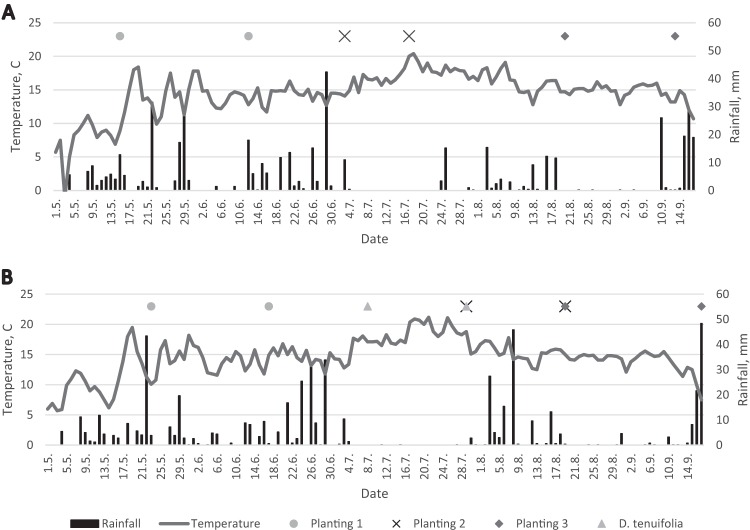
Comparison of the bacterial diversity on L. sativa leaves during the three plantings revealed some significant trends. We found that the bacterial richness of L. sativa samples was significantly greater shortly after planting (3 weeks) than at harvest (5 to 7 weeks after planting) for plantings 1 and 3 at both sites. This means that the bacterial communities in the phyllosphere of our leafy-green samples changed with the maturation of the plants, and it may indicate that plants exert a selective force on the colonizing bacteria. A few studies have already investigated the impact of leaf age and seasonal effects on phyllosphere communities (14–18). Our findings are consistent with previous studies. Thompson et al. (16) reported that the greatest number of bacterial species could be isolated from young plants. A study of the phyllosphere of spinach showed that the largest numbers of bacteria were present on newly emerged leaves (18). The richness and abundance of the spinach phyllosphere bacterial community decrease as the plant leaf matures (18). On the other hand, a study of leaf age and the seasonality of the tree species Quercus ilex in the Mediterranean forest revealed an increase in the richness of epiphytic bacteria with increasing time of colonization (17). With respect to planting 2 in our study, the bacterial richness remained consistent at both sites, and they also had similar microbial profiles that separated them from plantings 1 and 3 (Fig. 1B; see Table S2 in the supplemental material). Figure 6 shows that planting 2 had a higher temperature at harvest at both locations than plantings 1 and 3, which may partly explain the separation of planting 2 from plantings 1 and 3. The β-diversity tree (Fig. 2) showed that not all of the samples collected 3 weeks after planting and at harvest clustered. Two samples, VH-2 collected at harvest in Vestfold and B3-3 collected 3 weeks after planting in Buskerud, were in the “3 weeks after planting” and “harvest” clades, respectively (Fig. 2), suggesting that the bacterial community composition changes with season and not solely with leaf maturation. This confirms the findings of Williams et al. (15), where, for the first time, a seasonal effect was reported. Other studies have also observed seasonal variations in terms of the bacterial community composition of the phyllosphere of different plant species (8, 14, 31), but clearly, further in-depth studies on seasonal effects are needed.
FIG 6.
Daily mean temperatures and accumulated rainfall amounts at Farm Vestfold (A) and Farm Buskerud (B) from 1 May through 17 September 2013. Sampling dates are indicated for L. sativa plantings 1, 2, and 3 (3 weeks after planting and at harvest) and D. tenuifolia (two samplings at harvest).
Actinobacteria were more prevalent in the samples collected at harvest at Farm Vestfold (6%) than in those collected at harvest at Farm Buskerud (4%) (Table 2). This is likely due to the fact that the L. sativa plants at Farm Vestfold were exposed to open soil while the soil at Farm Buskerud was covered with plastic film. Consequently, the L. sativa plants at Farm Vestfold could easily be colonized by soil bacteria since the plastic barrier was lacking and Actinobacteria are abundant in soil (32). Our study may suggest that the growing environment (soil) also affects the composition of bacterial communities associated with leafy greens. D. tenuifolia and L. sativa at Farm Buskerud were irrigated with water from the same source (a river) and grown on the same type of soil. However, river water cannot be considered a homogeneous inoculum source for different crops at a single farm. Recently, airborne bacteria have been shown to be important in forming initial phyllosphere communities (33–35). Further studies of air, soil, and water samples should be done to identify possible inoculum sources. It should be mentioned that all of our L. sativa samples were sown in a greenhouse and grown for 2 weeks before planting in the field, but previous studies have shown that L. sativa plants grown indoors have lower total bacterial quantities than those grown in the field and that field microbiota can successfully colonize plants grown indoors (35). This suggests that the greenhouse period did not greatly influence the lettuce microbiota studied.
A notable variability in the distribution of phyllosphere communities in the D. tenuifolia samples compared to those in the L. sativa samples was observed at harvest, despite the fact that they were grown on the same type of soil at Farm Buskerud (sandy clay loam). This supports other studies showing a pronounced interspecies variability in phyllosphere communities (3, 36). The large variability of phyllosphere communities between D. tenuifolia and L. sativa at the same location and the observation that there was a shift in bacterial diversity during the planting suggest that the leaf-dwelling bacteria are not only passive and random inhabitants but are indeed influenced by the leafy-green species they colonize.
Pantoea and Sphingomonas were the only genera identified in all of our samples. This is in contrast to other surveys of leafy greens, were Pseudomonas, Bacillus, Massilia, and Arthrobacter were among the genera present in all of the samples (11, 14). Bulgarelli et al. (30) included Pseudomonas, Pantoea, Massilia, and Sphingomonas as part of the core phyllosphere communities. Pseudomonas bacteria were also found in several other studies to dominate the phyllosphere (5–7, 13, 14). Sphingomonas bacteria were identified in all our samples, in contrast to previous investigations of the leafy-green phyllosphere (14). However, Sphingomonas bacteria have been reported to be prevalent in other studies of the leaf microbiota on other plants (31, 37) and are known to contribute to plant health (30). A comprehensive knowledge of the drivers of bacterial community structure in the phyllosphere is of the utmost importance in developing new strategies for plant protection.
The genera identified in this study also include important plant pathogens like Pantoea, Pseudomonas, Rhodococcus, and Xanthomonas, as well as plant symbiotic Rhizobium. The genus Xanthomonas was present in 44 of 51 samples in various abundances (0.1 to 14%). In contrast, Xanthomonas was present in only 33 of 88 samples in a survey in California (14). Pantoea, which is a member of the Enterobacteriaceae family, is also found to be one of the major genera in other studies of the leafy-green phyllosphere (5, 11, 14). The genus Pantoea includes several species that are generally associated with plants as either epiphytes or pathogens, and some species can cause disease in humans (38). One sample (no. 54, from B3-2, Fig. 2) was separated from the clades, and we found that most of the sequences (97%) belonged to genus Pantoea. This suggests a plant pathogen infection; however, no symptoms were visible on the L. sativa plants. Other genera in the Enterobacteriaceae family, such as Cronobacter, Proteus, Providencia, and Yersinia, were also prevalent across all of the samples. Other genera in the Enterobacteriaceae family that include potentially harmful human pathogens, such as Salmonella and Escherichia, were not found in our study. Our study did not confirm the findings of Williams et al. (15), which identified OTUs from the Enterobacteriaceae family that were not classified in any known genera and therefore suggested the presence of novel bacterial species unique to the phyllosphere.
This study provides novel ecological insight into the development of bacterial communities on leafy greens. A shift in bacterial community composition and richness could be observed over time throughout the growing season. We suggest that host-microbe interactions play a role over time in shaping niches favoring the growth of particular taxa. A notable degree of variability in the distribution of phyllosphere communities could be observed among leafy-green species.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Farm Vestfold and Farm Buskerud for providing samples of leafy greens; G. Johannessen for helpful advices regarding the sampling of leafy greens; and I.-L. Akselsen, P. Grønn, and E. S. Riiser for assistance in sample preparation and PCR.
This study was supported by a grant from the Norwegian Research Council (221663/F40).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03470-14.
REFERENCES
- 1.Lindow SE, Brandl MT. 2003. Microbiology of the phyllosphere. Appl Environ Microbiol 69:1875–1883. doi: 10.1128/AEM.69.4.1875-1883.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kinkel LL. 1997. Microbial population dynamics on leaves. Annu Rev Phytopathol 35:327–347. doi: 10.1146/annurev.phyto.35.1.327. [DOI] [PubMed] [Google Scholar]
- 3.Whipps JM, Hand P, Pink D, Bending GD. 2008. Phyllosphere microbiology with special reference to diversity and plant genotype. J Appl Microbiol 105:1744–1755. doi: 10.1111/j.1365-2672.2008.03906.x. [DOI] [PubMed] [Google Scholar]
- 4.Schloss PD, Handelsman J. 2005. Metagenomics for studying unculturable microorganisms: cutting the Gordian knot. Genome Biol 6:1. doi: 10.1186/gb-2005-6-8-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hunter PJ, Hand P, Pink D, Whipps JM, Bending GD. 2010. Both leaf properties and microbe-microbe interactions influence within-species variation in bacterial population diversity and structure in the lettuce (Lactuca species) phyllosphere. Appl Environ Microbiol 76:8117–8125. doi: 10.1128/AEM.01321-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zwielehner J, Handschur M, Michaelsen A, Irez S, Demel M, Denner EB, Haslberger AG. 2008. DGGE and real-time PCR analysis of lactic acid bacteria in bacterial communities of the phyllosphere of lettuce. Mol Nutr Food Res 52:614–623. doi: 10.1002/mnfr.200700158. [DOI] [PubMed] [Google Scholar]
- 7.Handschur M, Pinar G, Gallist B, Lubitz W, Haslberger AG. 2005. Culture free DGGE and cloning based monitoring of changes in bacterial communities of salad due to processing. Food Chem Toxicol 43:1595–1605. doi: 10.1016/j.fct.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 8.Redford AJ, Fierer N. 2009. Bacterial succession on the leaf surface: a novel system for studying successional dynamics. Microb Ecol 58:189–198. doi: 10.1007/s00248-009-9495-y. [DOI] [PubMed] [Google Scholar]
- 9.Yang CH, Crowley DE, Borneman J, Keen NT. 2001. Microbial phyllosphere populations are more complex than previously realized. Proc Natl Acad Sci U S A 98:3889–3894. doi: 10.1073/pnas.051633898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rastogi G, Coaker GL, Leveau JHJ. 2013. New insights into the structure and function of phyllosphere microbiota through high-throughput molecular approaches. FEMS Microbiol Lett 348:1–10. doi: 10.1111/1574-6968.12225. [DOI] [PubMed] [Google Scholar]
- 11.Jackson C, Randolph K, Osborn S, Tyler H. 2013. Culture dependent and independent analysis of bacterial communities associated with commercial salad leaf vegetables. BMC Microbiol 13:274. doi: 10.1186/1471-2180-13-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leff JW, Fierer N. 2013. Bacterial communities associated with the surfaces of fresh fruits and vegetables. PLoS One 8:e59310. doi: 10.1371/journal.pone.0059310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lopez-Velasco G, Welbaum GE, Boyer RR, Mane SP, Ponder MA. 2011. Changes in spinach phylloepiphytic bacteria communities following minimal processing and refrigerated storage described using pyrosequencing of 16S rRNA amplicons. J Appl Microbiol 110:1203–1214. doi: 10.1111/j.1365-2672.2011.04969.x. [DOI] [PubMed] [Google Scholar]
- 14.Rastogi G, Sbodio A, Tech JJ, Suslow TV, Coaker GL, Leveau JH. 2012. Leaf microbiota in an agroecosystem: spatiotemporal variation in bacterial community composition on field-grown lettuce. ISME J 6:1812–1822. doi: 10.1038/ismej.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Williams TR, Moyne AL, Harris LJ, Marco ML. 2013. Season, irrigation, leaf age, and Escherichia coli inoculation influence the bacterial diversity in the lettuce phyllosphere. PLoS One 8:e68642. doi: 10.1371/journal.pone.0068642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thompson IP, Bailey MJ, Fenlon JS, Fermor TR, Lilley AK, Lynch JM, McCormack PJ, McQuilken MP, Purdy KJ, Rainey PB, Whipps JM. 1993. Quantitative and qualitative seasonal changes in the microbial community from the phyllosphere of sugar beet (Beta vulgaris). Plant Soil 150:177–191. doi: 10.1007/BF00013015. [DOI] [Google Scholar]
- 17.Peñuelas J, Rico L, Ogaya R, Jump AS, Terradas J. 2012. Summer season and long-term drought increase the richness of bacteria and fungi in the foliar phyllosphere of Quercus ilex in a mixed Mediterranean forest. Plant Biol 14:565–575. doi: 10.1111/j.1438-8677.2011.00532.x. [DOI] [PubMed] [Google Scholar]
- 18.Tydings H, Lopez-Velasco G, Ponder M, Welbaum G. 2011. Alterations to the phylloepiphytic bacterial community of spinach with leaf age. Acta Hort 917:211–216. [Google Scholar]
- 19.Paulino LC, Tseng CH, Strober BE, Blaser MJ. 2006. Molecular analysis of fungal microbiota in samples from healthy human skin and psoriatic lesions. J Clin Microbiol 44:2933–2941. doi: 10.1128/JCM.00785-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heuer H, Smalla K. 1997. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) for studying soil microbial communities, p 353–373. In van Elsas J, Trevors J, Wellington E (ed), Modern soil microbiology. Marcel Dekker, New York, NY. [Google Scholar]
- 21.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. 2013. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schloss PD, Gevers D, Westcott SL. 2011. Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS One 6:e27310. doi: 10.1371/journal.pone.0027310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37(Database issue):D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lozupone C, Knight R. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.White JR, Nagarajan N, Pop M. 2009. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput Biol 5:10. doi: 10.1371/journal.pcbi.1000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Letunic I, Bork P. 2011. Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res 39:W475–W478. doi: 10.1093/nar/gkr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loman NJ, Constantinidou C, Chan JZM, Halachev M, Sergeant M, Penn CW, Robinson ER, Pallen MJ. 2012. High-throughput bacterial genome sequencing: an embarrassment of choice, a world of opportunity. Nat Rev Microbiol 10:599–606. doi: 10.1038/nrmicro2850. [DOI] [PubMed] [Google Scholar]
- 30.Bulgarelli D, Schlaeppi K, Spaepen S, Ver Loren van Themaat E, Schulze-Lefert P. 2013. Structure and functions of the bacterial microbiota of plants. Annu Rev Plant Biol 64:807–838. doi: 10.1146/annurev-arplant-050312-120106. [DOI] [PubMed] [Google Scholar]
- 31.Jackson CR, Denney WC. 2011. Annual and seasonal variation in the phyllosphere bacterial community associated with leaves of the southern magnolia (Magnolia grandiflora). Microb Ecol 61:113–122. doi: 10.1007/s00248-010-9742-2. [DOI] [PubMed] [Google Scholar]
- 32.Ventura M, Canchaya C, Tauch A, Chandra G, Fitzgerald GF, Chater KF, van Sinderen D. 2007. Genomics of Actinobacteria: tracing the evolutionary history of an ancient phylum. Microbiol Mol Biol Rev 71:495–548. doi: 10.1128/MMBR.00005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maignien L, DeForce EA, Chafee ME, Eren AM, Simmons SL. 2014. Ecological succession and stochastic variation in the assembly of Arabidopsis thaliana phyllosphere communities. mBio 5(1):e00682-13. doi: 10.1128/mBio.00682-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vokou D, Vareli K, Zarali E, Karamanoli K, Constantinidou HI, Monokrousos N, Halley JM, Sainis I. 2012. Exploring biodiversity in the bacterial community of the Mediterranean phyllosphere and its relationship with airborne bacteria. Microb Ecol 64:714–724. doi: 10.1007/s00248-012-0053-7. [DOI] [PubMed] [Google Scholar]
- 35.Williams TR, Marco ML. 2014. Phyllosphere microbiota composition and microbial community transplantation on lettuce plants grown indoors. mBio 5(4):e01564-14. doi: 10.1128/mBio.01564-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Redford AJ, Bowers RM, Knight R, Linhart Y, Fierer N. 2010. The ecology of the phyllosphere: geographic and phylogenetic variability in the distribution of bacteria on tree leaves. Environ Microbiol 12:2885–2893. doi: 10.1111/j.1462-2920.2010.02258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yashiro E, Spear RN, McManus PS. 2011. Culture-dependent and culture-independent assessment of bacteria in the apple phyllosphere. J Appl Microbiol 110:1284–1296. doi: 10.1111/j.1365-2672.2011.04975.x. [DOI] [PubMed] [Google Scholar]
- 38.Delétoile A, Decre D, Courant S, Passet V, Audo J, Grimont P, Arlet G, Brisse S. 2009. Phylogeny and identification of Pantoea species and typing of Pantoea agglomerans strains by multilocus gene sequencing. J Clin Microbiol 47:300–310. doi: 10.1128/JCM.01916-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.